Abstract
Bacteria are known to consume some sugars over others, although recent work reported by Koirala and colleagues in this issue of the Journal of Bacteriology (S. Koirala, X. Wang, and C. V. Rao, J Bacteriol 198:386–393, 2016, http://dx.doi.org/10.1128/JB.00709-15) revealed that individual cells do not necessarily follow this hierarchy. By studying the preferential consumption of l-arabinose over d-xylose in Escherichia coli, those authors found that subpopulations consume one, the other, or both sugars through cross-repression between utilization pathways. Their findings challenge classic assertions about established hierarchies and can guide efforts to engineer the simultaneous utilization of multiple sugars.
TEXT
Bacteria possess remarkably accommodating palates when consuming nutrients present in their environment. They can consume dozens of different molecules as sole sources of carbon or energy and can take up other molecules that would otherwise be synthesized by the cells. Each nutrient is often consumed through a dedicated utilization pathway that imports the nutrient into the cytoplasm and then shunts the nutrient into cellular metabolism. Utilization pathways in bacteria have been extensively studied for over 60 years to understand the molecular basis of gene regulation (1), although nutrient utilization has received renewed interest because of its impact on how probiotics and pathogens colonize the human gut, how antibiotic resistance genes are spread through horizontal gene transfer, and how these properties can be used in engineering microbial chemical factories (2–5).
Despite their ability to accept a range of nutrients, bacteria can become extremely picky when presented with more than one option. As one classic example, Escherichia coli readily consumes d-glucose or l-lactose if either is present; however, when both sugars are combined, E. coli depletes virtually all of the d-glucose before transitioning to the l-lactose (6). This apparent preference has been traced to common molecular mechanisms, such as catabolite repression or inducer exclusion, that silence or block utilization pathways for nonpreferred nutrients. Although these mechanisms allow cells to consume preferred nutrients over nonpreferred nutrients, bacteria are known to exhibit preferences for some nonpreferred nutrients over others (7). For instance, E. coli consumes l-arabinose over d-xylose when both sugars are present, even though both are considered nonpreferred sugars (8, 9). These preferences can be viewed as a hierarchy in which cells follow a prescribed order when choosing which nutrients to consume. The hierarchy is thought to emerge as cells begin with nutrients that are more efficiently or quickly catabolized before proceeding to others. When only poor nutrients are present or the nutrients are at low concentrations, bacteria are thought to discard their pickiness and consume anything that is available.
The implication of the utilization hierarchy is that every bacterial cell is geared to consume the same sugar at any given time. However, work dating back to the 1950s revealed that an isogenic population of bacteria can exhibit remarkable phenotypic heterogeneity. In this seminal work in the 1950s, Novick and Weiner demonstrated that E. coli cells exposed to intermediate concentrations of the nonhydrolyzable l-lactose analog thiomethyl-β-d-galactoside (TMG) stably diverged into two subpopulations: one in which the l-lactose utilization pathway was fully induced and the other in which the pathway was uninduced (10). This “all-or-none” response was initially considered a biological artifact, because TMG circumvents catabolism; accordingly, l-lactose yields uniform, graded induction of the pathway across an isogenic population (11). However, single-cell analyses of other utilization pathways in E. coli (e.g., for l-arabinose, d-xylose, l-rhamnose, d-gluconate) revealed “all-or-none” behaviors and other distinct single-cell responses (12–14), suggesting more diverse and heterogeneous responses to individual nutrients in the environment.
Despite the critical role of single-cell analyses, our current perception of hierarchies in nutrient consumption has been overwhelmingly shaped via bulk characterization techniques. These techniques provide population averages and thus overlook individualistic behaviors. The ensuing question is whether the hierarchies observed through bulk characterization techniques extend to every cell in the population. And what about nutrients that are already known to drive heterogeneous behaviors? The most notable example in bacteria was previously reported by Ozbudak and coworkers, who measured the single-cell response of E. coli to d-glucose and TMG (11). They found that single cells retained the “all-or-none” response to TMG, where d-glucose desensitized cells to this l-lactose analog. What has remained poorly explored is the single-cell response to other combinations of sugars. The recent study from Koirala et al. published in this issue of the Journal of Bacteriology takes the first steps to test these combinations of sugars in individual cells (15).
In their work, Koirala and colleagues investigated how individual E. coli cells respond to a mixture of l-arabinose and d-xylose. These two sugars represent the major constituents of lignocellulosic biomass used for the production of biofuels and other chemical products (16). Both l-arabinose and d-xylose are also known to individually generate “all-or-none”-like responses in E. coli (12–14, 17). Finally, the Rao group had previously shown that l-arabinose is preferentially consumed over d-xylose through inhibition of the d-xylose catabolic operon xylAB by the l-arabinose regulator AraC. The questions being addressed in Koirala et al.'s most recent work (15) were focused on how the presence of both sugars influences pathway induction and on determining the underlying gene regulatory mechanisms.
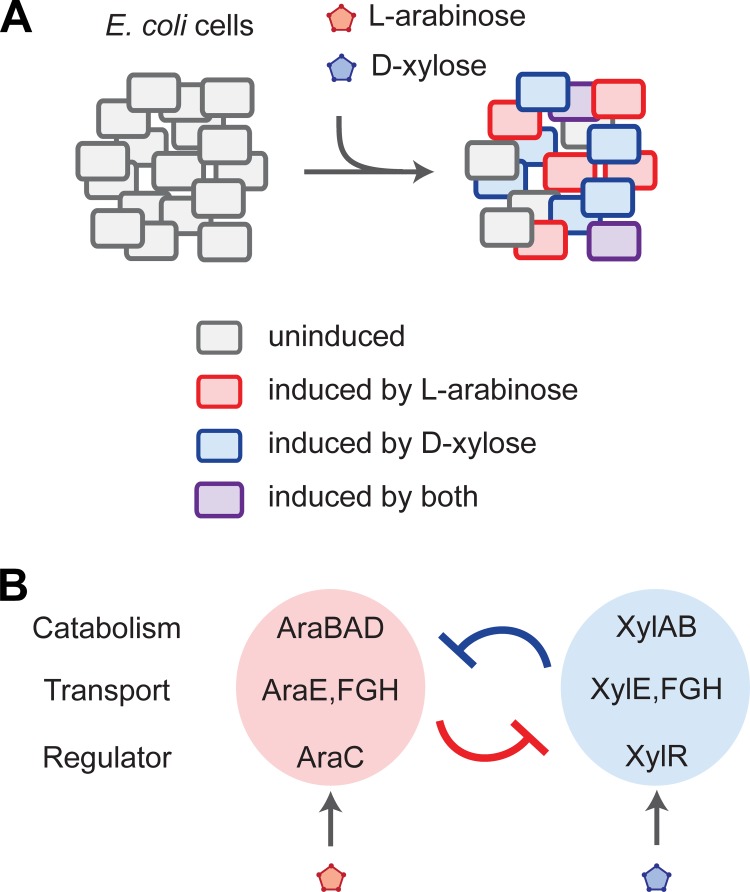
To evaluate how these sugars impact pathway induction in single cells, those authors performed flow cytometry analysis on E. coli cells with transcriptional fusions between fluorescent proteins and the promoters of l-arabinose and d-xylose utilization genes. The authors observed four different subpopulations in the presence of intermediate concentrations of l-arabinose and d-xylose: an uninduced subpopulation, a subpopulation with only the l-arabinose utilization pathway induced, a subpopulation with only the d-xylose utilization pathway induced, and finally a subpopulation with both pathways induced (Fig. 1A). By varying the concentration of each sugar, they further found that one sugar limited induction of the opposite pathway. l-Arabinose was much more inhibitory than d-xylose, in line with the preferred consumption of l-arabinose. However, the apparent cross-repression between the pathways and the ability of some cells to have both pathways induced suggested a more complex and nuanced hierarchy than previously envisioned.
FIG 1.
E. coli's single-cell response to the sugars l-arabinose and d-xylose. (A) A population of E. coli cells exposed to intermediate amounts of both sugars diverges into four subpopulations: uninduced cells, cells with the l-arabinose utilization pathway induced, cells with the d-xylose utilization pathway induced, and cells with both pathways induced. The fraction of each subpopulation depends on the concentrations of l-arabinose and d-xylose. (B) The two utilization pathways repress each other. In the presence of l-arabinose, AraC represses the expression of the d-xylose catabolic genes. In the presence of d-xylose, XylR represses the expression of araC.
To extend these insights, Koirala et al. evaluated the genetic basis of the observed cross-regulation (15). They found that d-xylose repressed induction of the l-arabinose utilization operons, where repression was dependent on the d-xylose-dependent regulator XylR. Further interrogation revealed a binding site within the promoter controlling expression of the l-arabinose-dependent regulator AraC. Factoring in their previous findings (9), the authors elucidated a cross-repression genetic circuit between XylR and AraC (Fig. 1B): XylR repressed expression from the promoter for araC, thereby limiting induction of the entire l-arabinose utilization pathway. Correspondingly, AraC repressed expression from the promoter controlling the xylAB catabolic operon, thereby limiting d-xylose consumption. This genetic circuit represents a relatively simple and direct mechanism in which these pathways could modulate each other's activity.
The insights from Koirala et al. beg a number of questions that could reshape our understanding of eating preferences in the bacterial world. The first is how frequently utilization pathways regulate each other. Extensive work has shown that catabolite repression represents a common mechanism through which bacteria consume preferred nutrients over nonpreferred nutrients (18). However, little work has been done to identify regulatory interactions between other utilization pathways, particularly in single cells. Exploring these pathways could yield not only equivalent examples to l-arabinose and d-xylose but also entirely new interactions, such as hierarchical activation, oscillations, excitatory behavior, or adaptive prediction (19–21). Elucidation of other types of behaviors will require more advanced techniques beyond flow cytometry, such as long-term, single-cell tracking using microfluidics and fluorescence microscopy (22). Through further work, we expect the cross-repression between the l-arabinose and d-xylose pathways to represent the tip of the iceberg for the myriad of regulatory interactions dictating what and when bacteria eat.
Beyond what types of regulatory interactions exist, an ensuing question is why cells would exhibit such complex responses to different sugars (23). Naively, one would expect every cell to consume nutrients that are most easily or efficiently catabolized in order to maximize growth. However, the nature of harsh, unpredictable, and fluctuating environments suggest that bacteria must adopt other strategies in order to best survive. For instance, the “all-or-none” response observed for l-arabinose and d-xylose could represent a division-of-labor strategy in which the population coordinately consumes all available nutrients even though individual cells consume only one nutrient. Another possibility is that the “all-or-none” response represents a bet-hedging strategy in which cells anticipate the sudden loss of one nutrient or the sudden appearance of another (24). These behaviors could also reflect a Pavlovian association with the environmental conditions in which the nutrients are found, such as E. coli being conditioned to associate high temperatures with oxygen deprivation (25). Finally, the behaviors could help limit attack by bacteriophages, toxins, or immune responses that recognize surface receptors associated with a given utilization pathway, such as the λ phage binding to the LamB receptor responsible for importing the sugar d-maltose. Understanding why a given strategy is adopted for a given environmental condition is expected to reveal the evolutionary principles by which microorganisms most effectively adapt to and exploit their surroundings.
A final set of questions involves the reciprocal repression of the l-arabinose and d-xylose pathways. The qualitative genetic circuitry mimics a synthetic toggle switch composed of two regulators that repress each other (26, 27). While the synthetic toggle switch yielded cells locked into expressing only one of the two regulators, the sugar-dependent circuit characterized by Koirala et al. yielded some cells expressing both pathways. Given this difference, can toggle switch behavior be achieved by rationally altering the circuitry (e.g., by strengthening the repression of AraC expression by XylR)? And will the resulting cells compete with wild-type cells better or worse when exposed to constant or fluctuating mixtures of l-arabinose and d-xylose? Finally, can cross-regulation be disrupted in order to promote the simultaneous consumption of these sugars? Answering these questions would further delve into cross-regulation between the l-arabinose and d-xylose utilization pathways—a convenient model to study utilization hierarchies—and work toward engineering the coutilization of multiple sugars in industrially relevant microbial strains.
Overall, the work from Koirala et al. hints at the sophistication of the bacterial palate. Only through further investigation will we learn how many bacteria share these eating preferences, the physiological, ecological, and evolutionary forces that shaped these single-cell choices, and how to optimize industrial strains for the efficient conversion of cheap carbon sources into valued-added chemicals and fuels.
ACKNOWLEDGMENTS
We thank Michelle Luo for critical feedback on this commentary.
This work is supported in part by a grant from the National Science Foundation (MCB-1452902 to C.L.B.).
We declare no conflicts of interest.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Jacob F, Monod J. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3:318–356. doi: 10.1016/S0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 2.Blaut M, Clavel T. 2007. Metabolic diversity of the intestinal microbiota: implications for health and disease. J Nutr 137(3 Suppl 2):751S–755S. [DOI] [PubMed] [Google Scholar]
- 3.Baharoglu Z, Krin E, Mazel D. 2012. Connecting environment and genome plasticity in the characterization of transformation-induced SOS regulation and carbon catabolite control of the Vibrio cholerae integron integrase. J Bacteriol 194:1659–1667. doi: 10.1128/JB.05982-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conrad RS, Wulf RG, Clay DL. 1979. Effects of carbon sources on antibiotic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 15:59–66. doi: 10.1128/AAC.15.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhamankar H, Prather KLJ. 2011. Microbial chemical factories: recent advances in pathway engineering for synthesis of value added chemicals. Curr Opin Struct Biol 21:488–494. doi: 10.1016/j.sbi.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Monod J. 1949. The growth of bacterial cultures. Annu Rev Microbiol 3:371–394. doi: 10.1146/annurev.mi.03.100149.002103. [DOI] [Google Scholar]
- 7.Aidelberg G, Towbin BD, Rothschild D, Dekel E, Bren A, Alon U. 2014. Hierarchy of non-glucose sugars in Escherichia coli. BMC Syst Biol 8:133. doi: 10.1186/s12918-014-0133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang HY, Song S, Park C. 1998. Priority of pentose utilization at the level of transcription: arabinose, xylose, and ribose operons. Mol Cells 8:318–323. [PubMed] [Google Scholar]
- 9.Desai TA, Rao CV. 2010. Regulation of arabinose and xylose metabolism in Escherichia coli. Appl Environ Microbiol 76:1524–1532. doi: 10.1128/AEM.01970-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novick A, Weiner M. 1957. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci U S A 43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozbudak EM, Thattai M, Lim HN, Shraiman BI, van Oudenaarden A. 2004. Multistability in the lactose utilization network of Escherichia coli. Nature 427:737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- 12.Afroz T, Biliouris K, Kaznessis Y, Beisel CL. 2014. Bacterial sugar utilization gives rise to distinct single-cell behaviours. Mol Microbiol 93:1093–1103. doi: 10.1111/mmi.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan-Kiss RM, Wadler C, Cronan JE Jr. 2002. Long-term and homogeneous regulation of the Escherichia coli araBAD promoter by use of a lactose transporter of relaxed specificity. Proc Natl Acad Sci U S A 99:7373–7377. doi: 10.1073/pnas.122227599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegele DA, Hu JC. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc Natl Acad Sci U S A 94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koirala S, Wang X, Rao CV. 2016. Reciprocal regulation of l-arabinose and d-xylose metabolism in Escherichia coli. J Bacteriol 198:386–393. doi: 10.1128/JB.00709-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Himmel ME, Ding S-Y, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 17.Afroz T, Biliouris K, Boykin KE, Kaznessis Y, Beisel CL. 2015. Trade-offs in engineering sugar utilization pathways for titratable control. ACS Synth Biol 4:141–149. doi: 10.1021/sb400162z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 19.Süel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. 2006. An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 20.Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. 2008. A fast, robust and tunable synthetic gene oscillator. Nature 456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, Dahan O, Pilpel Y. 2009. Adaptive prediction of environmental changes by microorganisms. Nature 460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- 22.Cookson S, Ostroff N, Pang WL, Volfson D, Hasty J. 2005. Monitoring dynamics of single-cell gene expression over multiple cell cycles. Mol Syst Biol 1:2005.0024. doi: 10.1038/msb4100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao CV, Koirala S. 2014. Black and white with some shades of grey: the diverse responses of inducible metabolic pathways in Escherichia coli. Mol Microbiol 93:1079–1083. doi: 10.1111/mmi.12734. [DOI] [PubMed] [Google Scholar]
- 24.Solopova A, van Gestel J, Weissing FJ, Bachmann H, Teusink B, Kok J, Kuipers OP. 2014. Bet-hedging during bacterial diauxic shift. Proc Natl Acad Sci U S A 111:7427–7432. doi: 10.1073/pnas.1320063111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tagkopoulos I, Liu Y-C, Tavazoie S. 2008. Predictive behavior within microbial genetic networks. Science 320:1313–1317. doi: 10.1126/science.1154456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardner TS, Cantor CR, Collins JJ. 2000. Construction of a genetic toggle switch in Escherichia coli. Nature 403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 27.Chen W, Bailey JE. 1994. Application of the cross-regulation system as a metabolic switch. Biotechnol Bioeng 43:1190–1193. doi: 10.1002/bit.260431124. [DOI] [PubMed] [Google Scholar]