ABSTRACT
Cyclic diadenylate monophosphate (c-di-AMP) is a second messenger utilized by diverse bacteria. In many species, including the Gram-positive human pathogen Listeria monocytogenes, c-di-AMP is essential for growth. Here we show that the single diadenylate cyclase of L. monocytogenes, CdaA, is an integral membrane protein that interacts with its potential regulatory protein, CdaR, via the transmembrane protein domain. The presence of the CdaR protein is not required for the membrane localization and abundance of CdaA. We have also found that CdaR negatively influences CdaA activity in L. monocytogenes and that the role of CdaR is most evident at a high growth temperature. Interestingly, a cdaR mutant strain is less susceptible to lysozyme. Moreover, CdaA contributes to cell division, and cells depleted of CdaA are prone to lysis. The observation that the growth defect of a CdaA depletion strain can be partially restored by increasing the osmolarity of the growth medium suggests that c-di-AMP is important for maintaining the integrity of the protective cell envelope. Overall, this work provides new insights into the relationship between CdaA and CdaR.
IMPORTANCE Cyclic diadenylate monophosphate (c-di-AMP) is a recently identified second messenger that is utilized by the Gram-positive human pathogen Listeria monocytogenes. Here we show that the single diadenylate cyclase of L. monocytogenes, CdaA, is an integral membrane protein that interacts with CdaR, its potential regulatory protein. We show that CdaR is not required for membrane localization or abundance of the diadenylate cyclase, but modulates its activity. Moreover, CdaA seems to contribute to cell division. Overall, this work provides new insights into the relationship between CdaA and CdaR and their involvement in cell growth.
INTRODUCTION
Bacteria from diverse phyla produce the cyclic dinucleotide cyclic diadenylate monophosphate (c-di-AMP) that is synthesized and degraded by specific diadenylate cyclases and phosphodiesterases, respectively (1). The DNA integrity scanning protein DisA from Thermotoga maritima was the first diadenylate cyclase structurally and biochemically characterized (2), and its characterization led to the discovery of c-di-AMP. Many bacteria possess only a single, DisA-type, diadenylate cyclase (1), which is involved in the maintenance of DNA integrity (3, 4, 5). The cyclase activity of DisA is modulated by unusual DNA recombination intermediates (2), but it is presently unclear how c-di-AMP signals the cell that the chromosome integrity is affected.
In addition to DisA, two diadenylate cyclases, CdaA and CdaS, are synthesized in the Gram-positive model bacterium Bacillus subtilis (6). While cdaA is expressed during vegetative growth, the cdaS gene is expressed during sporulation or germination of spores (7). The cdaS inactivation decreases the germination efficiency of spores, indicating a germination-specific function for this enzyme (8). Recently, it has been shown that c-di-AMP is essential for the growth of B. subtilis (6, 9, 10). c-di-AMP production by a single diadenylate cyclase is sufficient for viability of B. subtilis, but the existence of three diadenylate cyclases in this organism suggests that c-di-AMP levels need to be fine-tuned during different lifestyle conditions (6, 9). In contrast to DisA and CdaS, which have relatively limited phylogenetic distributions, CdaA is more widespread in bacteria (1). Some bacteria, such as the human pathogen Listeria monocytogenes, possess only a single, CdaA-type, diadenylate cyclase (11), which was shown to be essential for growth on rich medium (12), and CdaA depletion leads to cessation of bacterial growth (13).
Several c-di-AMP-binding proteins were recently identified (for recent reviews, see references 1, 14). Among these is the transcription factor DarR from Mycobacterium smegmatis, the DNA-binding activity of which is stimulated by c-di-AMP (15). c-di-AMP was also shown to bind to and to control the activities of the potassium transporters from Staphylococcus aureus and Streptococcus pneumoniae and to allosterically regulate the L. monocytogenes pyruvate carboxylase (16, 17, 18). Recently, members of a novel class of PII-like signal transduction proteins from B. subtilis, S. aureus, and L. monocytogenes were also shown to bind c-di-AMP (19, 20, 21, 22). However, the impact of c-di-AMP on the activity of the PII-like proteins remains to be uncovered.
CdaA from L. monocytogenes is a 273-amino acid (aa) protein consisting of an N-terminal transmembrane domain (aa 17 to 70) followed by a diadenylate cyclase (DAC) domain (aa 103 to 237), which is surrounded by two coiled-coil (CC) motifs (aa 82 to 103 and aa 240 to 273) (Fig. 1B) (1). Recently, an N-terminally truncated variant of the L. monocytogenes diadenylate cyclase CdaA lacking the membrane-binding domain and the adjacent CC motif was structurally and biochemically characterized (23). The functionality of the CdaA variant was demonstrated by expression of the truncated cdaA gene in Escherichia coli, an organism that does not produce c-di-AMP (6, 23), and c-di-AMP production was dependent on the presence of divalent ions in vitro (23).
FIG 1.
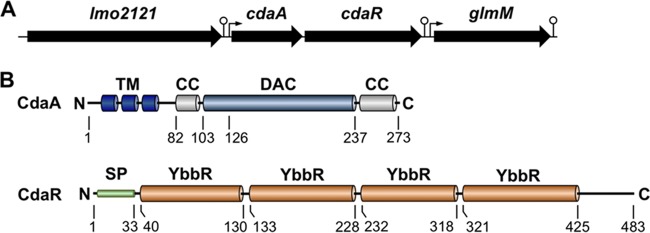
Genomic context of the cdaAR genes and the domains of CdaA and CdaR. (A) The diadenylate cyclase CdaA and its potential regulator CdaR are encoded by the bicistronic cdaAR operon (38), which is surrounded by the putative lmo2121 maltose phosphorylase gene and the glmM gene encoding the essential phosphoglucosamine mutase. Arrows and circles illustrate transcription start sites and terminators, respectively (38). (B) CdaA contains three N-terminally located α-helices that form a transmembrane (TM) domain. The TM domain is attached to the diadenylate cyclase (DAC) domain, which is surrounded by two coiled-coil (CC) motifs. CdaR is predicted to contain a signal peptide (SP) and four YbbR domains of unknown function (24, 25, 26).
The cdaA diadenylate cyclase gene is located in the conserved cdaR-cdaA-glmM module (Fig. 1A) that also encodes the phosphoglucosamine mutase GlmM and CdaR. The phosphoglucosamine mutase GlmM converts glucosamine 6-phosphate to the cell wall precursor glucosamine 1-phosphate. CdaR is a regulator controlling the CdaA activity in B. subtilis (6). It is predicted to contain an N-terminal signal peptide (aa 1 to 33) and four YbbR domains of unknown function (Fig. 1B) (24, 25, 26). This particular genetic context of the cdaA gene suggests that CdaA may be involved in cell envelope biosynthesis. Indeed, both depletion and overexpression of cdaA were shown to severely affect the integrity of the cell wall in several Gram-positive bacteria (6, 27). Moreover, alterations in the intracellular c-di-AMP pool affect the resistance to cell wall-targeting antibiotics (9, 27, 28, 29, 30). The underlying mechanism, explaining how c-di-AMP synthesis is linked to the cell wall integrity, remains to be elucidated.
In this study, we show that CdaA of L. monocytogenes is an integral membrane protein and that it directly interacts with CdaR. We also show that CdaR negatively controls the diadenylate cyclase activity of CdaA, but it is not needed for membrane localization of CdaA. We describe the phenotypes of the ΔcdaR mutant and cells depleted in CdaA with regard to growth, cell division, susceptibility to antibiotics, and osmotic pressure. This work expands our understanding of the complex relationship between the interacting partners, CdaA and CdaR, and provides new insights into their involvement in cell wall homeostasis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains used in this study are listed in Table 1. L. monocytogenes cells were grown in brain heart infusion (BHI) broth or on BHI agar plates at 37°C. When necessary, antibiotics and supplements were added to the growth medium at the following concentrations: erythromycin, 5 μg/ml; kanamycin, 50 μg/ml; IPTG (isopropyl-β-d-thiogalactopyranoside), 1 mM; NaCl, 20 g/liter; and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), 100 μg/ml. The Escherichia coli strains TOP10 and XL1-Blue (Invitrogen) were used for all cloning procedures (31).
TABLE 1.
Strains and plasmids used in this study
| Plasmid or strain | Construction/relevant characteristicsa | Reference or sourceb |
|---|---|---|
| Plasmids | ||
| pBP33 | bla PT7-Δ300cdaA | 23 |
| pBP224 | pUT18-cdaR | This work |
| pBP225 | pUT18C-cdaR | This work |
| pBP226 | p25-N-cdaR | This work |
| pBP227 | pKT25-cdaR | This work |
| pBP228 | pUT18-Δ84cdaR | This work |
| pBP229 | pUT18C-Δ84cdaR | This work |
| pBP230 | p25-N-Δ84cdaR | This work |
| pBP231 | pKT25-Δ84cdaR | This work |
| pBP232 | pUT18-cdaA | This work |
| pBP233 | pUT18C-cdaA | This work |
| pBP234 | p25-N-cdaA | This work |
| pBP235 | pKT25-cdaA | This work |
| pBP236 | pUT18-Δ240cdaA | This work |
| pBP237 | pUT18C-Δ240cdaA | This work |
| pBP238 | p25-N-Δ240cdaA | This work |
| pBP239 | pKT25-Δ240cdaA | This work |
| pBP353 | Phelp-lacO-cdaR lacI neo | This work |
| pMAD | bla erm bgaB | 34 |
| pIMK3 | Phelp-lacO lacI neo | 32 |
| pJR26 | Phelp-lacO-cdaA lacI neo | This work |
| pJR27 | bla erm bgaB cdaR | This work |
| pJR28 | bla erm bgaB cdaA | This work |
| pJR29 | bla erm bgaB ΔcdaR | This work |
| pJR31 | bla erm bgaB ΔcdaA | This work |
| pJR40 | bla erm bgaB ΔcdaA ΔcdaR | This work |
| pKT25 | Plac-cyaA-MCS kan | 36 |
| pKT25::zip | Plac- cyaA-zip kan | 36 |
| pUT18 | Plac-MCS-cyaA bla | 36 |
| pUT18C | Plac-cyaA-MCS bla | 36 |
| pUT18C::zip | Plac- cyaA-zip bla | 36 |
| p25-N | Plac-MCS-cyaA kan | 37 |
| L. monocytogenes strains | ||
| EGD-e | Wild type, serovar 1/2a strain | 49 |
| BPL16 | ΔcdaR attB::Phelp-lacO-cdaR lacI neo | pBP353 → LMJR45 |
| LMJR43 | attB::Phelp-lacO-cdaA lacI neo | pJR26 → EGD-e |
| LMJR45 | ΔcdaR | pJR29 ↔ EGD-e |
| LMJR46 | ΔcdaA attB::Phelp-lacO-cdaA lacI neo | pJR31 ↔ LMJR43 |
| LMJR65 | ΔcdaA ΔcdaR attB::Phelp-lacO-cdaA lacI neo | pJR40 ↔ LMJR43 |
Designations Δ300cdaA, Δ84cdaR, and Δ240cdaA represent truncated genes lacking the indicated number of nucleotides. MCS, multiple-cloning site.
A right-pointing arrow represents a transformation event. A double-headed arrow indicates gene deletions obtained by chromosomal insertion and subsequent excision of pMAD plasmid derivatives (see Materials and Methods for details).
General methods, manipulations of DNA, and oligonucleotide primers.
Transformation of E. coli and isolation of plasmid DNA were carried out using standard protocols (31). Generation of electro-competent L. monocytogenes cells and transformation of plasmid DNA into L. monocytogenes were performed as described earlier (32). Restriction and ligation of DNA were done as described by the manufacturer's instructions. QuikChange mutagenesis was used for restriction-free modifications of plasmids (33). The DNA sequences of the oligonucleotides are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence (5′ → 3′) |
|---|---|
| FC146 | CGATGCGTTCGCGATCCAGGC |
| FC147 | CCAGCCTGATGCGATTGCTGCAT |
| FC148 | GTCACCCGGATTGCGGCGG |
| FC150 | GATTCGGTGACCGATTACCTGGC |
| JH21 | AAACCATGGATCGAATTTTAAATAATAAATGGTCGATTC |
| JH22 | TTTGTCGACTTATGTGCTTTTGGAAGGTACTTCAATGG |
| JR50 | AAATCTAGAGATGGATTTTTCCAATATGTCGATATTGCA |
| JR51 | TTTGGTACCCGTTCGCTTTTGCCTCCTTTCCAT |
| JR52 | AAATCTAGAGATGGATCGAATTTTAAATAATAAATGGTCGA |
| JR53 | TTTGGTACCCGTGTGCTTTTGGAAGGTACT |
| JR54 | AAATCTAGAGTTCCAACCGGAATTACGCCG |
| JR55 | AAATCTAGAGAATAATAATAACGCCACGACTTTTTCTACG |
| JR83 | GCGCGGAATTCGGGAAGATATCGAGCATCGCATG |
| JR86 | CGCGGGATCCCTTTGACGGTATCTGCTTTGATTTTAG |
| JR87 | CGCGCCATGGATGGATTTTTCCAATATGTCGATATTGC |
| JR88 | CGCGGTCGACTCATTCGCTTTTGCCTCCTTTCC |
| JR90 | GCGCGGAATTCGAGGCACGGAGGTGAAGTGATGG |
| JR93 | CGCGGGATCCGAAGTTGCACCATTCGCACAATCAAG |
| JR97 | CATCATTCGCCATCACTTCACCTCCGTGC |
| JR98 | GTGATGGCGAATGATGGATCGAATTTTAAA |
| JR99 | CCTTATTTTACCATCATTCGCTTTTGCCTCCTTTCC |
| JR100 | GAATGATGGTAAAATAAGGAGTGTGGGGCGCTAG |
| JR125 | GTGATGGATTTTAGCTCTGTTGATGTAGAATTGAGC |
| JR126 | CATCAACAGAGCTAAAATCCATCACTTCACCTCC |
Construction of plasmids and strains.
The plasmid pJR31 was constructed for the deletion of cdaA from the L. monocytogenes chromosome. The primers JR83/JR86 were used for the amplification of the cdaA genomic region. The resulting PCR fragment was BamHI/EcoRI digested and ligated with pMAD cut with the same enzymes to give plasmid pJR28. Finally, the cdaA gene was removed from this plasmid in a PCR using the primer pair JR97/JR98. Plasmid pJR29 for the removal of the cdaR gene was constructed in a similar way. A fragment comprising the cdaR gene and up- and downstream regions was amplified by PCR using primers JR90/JR93, and the fragment produced was inserted into the plasmid pMAD via BamHI/EcoRI, resulting in plasmid pJR27. The cdaR gene was removed from this plasmid in a PCR using the primers JR99/JR100. In order to generate cdaA cdaR double deletions, the cdaR gene was removed from pJR31 by PCR with the primers JR125/JR126, resulting in plasmid pJR40. To allow for the IPTG-inducible expression of cdaA, the cdaA open reading frame was amplified from the chromosomal DNA using the primers JR87/JR88 and cloned into pIMK3 using NcoI/SalI restriction digestion. The resulting plasmid was designated pJR26. Plasmid pBP353 was constructed for the IPTG-dependent expression of the native cdaR gene using the primer pairs JH21/JH22. The PCR products were digested with NcoI and SalI and ligated with the plasmid pIMK3 that was cut with the same enzymes. The derivatives of pIMK3 were introduced into L. monocytogenes by electroporation, and kanamycin-resistant clones were isolated. Plasmid insertion at the attB site of the tRNAArg locus was confirmed by PCR. For removal of cdaA, cdaR, or cdaAR, the plasmid pJR31, pJR29, or pJR40, respectively, was used to transform the respective L. monocytogenes recipient strains, and the genes were removed as described elsewhere (34). Gene deletions were verified by PCR.
Microscopy.
Samples (0.4 μl) of logarithmically growing cultures were transferred onto microscope slides, which had been covered with a thin agarose film (1.5% in distilled water). After air drying, the samples were covered with a cover lid and examined by phase-contrast or fluorescence microscopy. For membrane staining, 100 μl of the cultures was mixed with 1 μl of Nile red (100 μg/ml in dimethyl sulfoxide [DMSO]) and shaken for 20 min at 37°C before the cells were prepared for microscopy. Images were taken with a Nikon Eclipse Ti microscope coupled to a Nikon DSMBWc charge-coupled device (CCD) camera and processed using the NIS-Elements AR software package (Nikon).
Autolysis assays.
Bacterial strains were cultivated in BHI broth containing 1 mM IPTG, if necessary, and incubated until the culture reached an optical density of around 0.8. The cells were collected by centrifugation (5 min, 6,000 × g) and resuspended in 50 mM Tris-HCl (pH 8.0) to an optical density at 600 nm (OD600) of 0.6. Penicillin (25 μg/ml final concentration) or lysozyme (2.5 μg/ml final concentration) was added, and the cells were shaken at 37°C. Autolysis was followed by measurement of the decline in the OD600 every 30 min (penicillin) or every 15 min (lysozyme).
Determination of MICs.
Test strips with the following concentration gradients were used to determine the MICs against selected antibiotics: amoxicillin, 0.016 to 256 μg/ml; ampicillin, 0.016 to 256 μg/ml; gentamicin, 0.016 to 256 μg/ml; meropenem, 0.002 to 32 μg/ml; penicillin G, 0.016 to 256 μg/ml; and vancomycin, (0.016 to 256 μg/ml) (all from bestbion dx, Germany). L. monocytogenes strains were grown on BHI plates (containing 1 mM IPTG, if necessary), and 4 to 5 morphologically identical colonies were then used to inoculate 5 ml BHI broth. Cell suspensions were used to swab-inoculate BHI agar plates, supplemented with 1 mM IPTG where indicated. MIC test strips were placed on top of the agar surface, and the plates were incubated at 37°C for 1 day.
Protein purification and generation of an anti-CdaA antiserum.
Plasmid pBP33 was used to overexpress the N-terminally Strep-tagged diadenylate cyclase CdaA from L. monocytogenes as described previously (23). The protein was purified with the Strep-tag II–Strep-Tactin purification system (IBA, Göttingen, Germany) and used for the generation of polyclonal antibodies in rabbits (Seqlab, Göttingen, Germany). The rabbit anti-CdaA antibodies were diluted 1:5,000 for Western blot analyses.
Isolation of protein fractions and Western blotting.
Cells were harvested by centrifugation and washed once with ZAP buffer (10 mM Tris-HCl [pH 7.5] and 200 mM NaCl). Cells were disrupted by sonication in ZAP buffer also containing 1 mM phenylmethylsulfonyl fluoride (PMSF). Cell debris was removed by centrifugation, and the resulting supernatant was considered the total cellular protein fraction. Membrane proteins were then separated from the soluble cytosolic proteins by ultracentrifugation (100,000 × g for 30 min at 4°C). The supernatant contained the soluble cytoplasmic proteins, and the pellet corresponded to the membrane fraction. The purified N-terminally Strep-tagged diadenylate cyclase CdaA lacking the N-terminal transmembrane domain (Fig. 1) (23) was used to generate rabbit polyclonal antibodies. For Western blot analysis, proteins were separated by 12% SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) or nylon membranes (Bio-Rad) by electroblotting. Rabbit anti-CdaA served as the primary antibody with visualization using anti-rabbit immunoglobulin–horseradish peroxidase-conjugated secondary antibodies and the ECL detection system (Thermo Scientific). Protein concentrations were determined as described previously (35).
Bacterial two-hybrid assay.
The primary protein-protein interactions were analyzed using the bacterial two-hybrid (B2H) system (36, 37). Plasmid pairs pUT18C/pKT25 and pUT18/p25N were used for the expression of proteins fused to the C and N termini, respectively, of the T18 and T25 fragments of CyaA (36). The plasmids constructed for the B2H analysis are listed in Table 2. The genes were amplified using the oligonucleotides listed in Table 1 and cloned between the XbaI and KpnI sites of the plasmids pUT18, pUT18C, p25-N, and pKT25. The full-length and truncated cdaR variants were amplified using the oligonucleotide pairs JR52/JR53 and JR55/JR53, respectively. The full-length and truncated cdaA variants were amplified using the oligonucleotide pairs JR50/JR51 and JR54/JR51, respectively. The DNA sequences were verified using the oligonucleotides FC146, FC147, FC148, and FC150. Plasmids were used for the cotransformation of E. coli BTH101, and the protein-protein interactions were then analyzed by plating the cells on LB plates containing 100 μg/ml ampicillin, 50 μg/ml kanamycin, 40 μg/ml X-Gal, and 0.5 mM IPTG. The plates were incubated for a maximum of 36 h at 30°C.
Analysis of the cyclic dinucleotide pools.
The concentration of c-di-AMP in L. monocytogenes cells was determined by a liquid chromatography-tandem mass spectrometry (LC-MS/MS) method. Briefly, a single colony was used to inoculate a reaction tube containing 4 ml BHI broth. The wild type (wt) and strains BPL16 (IcdaR, where “I” indicates IPTG-dependent expression of the gene), LMJR45 (ΔcdaR), LMJR46 (IcdaA), and LMJR65 (ΔcdaR IcdaA) were grown in BHI broth (including 1 mM IPTG, where necessary). The precultures were incubated overnight at 37°C and with agitation (220 rpm). On the next day, 2-ml aliquots of each culture were harvested by centrifugation, and the cell pellets were washed 4 times in 2 ml BHI broth. Then 100-ml shake flasks containing 25 ml BHI broth were inoculated with the washed cell suspensions to an optical density of 0.02. The main cultures were incubated for 4 to 5 h at 37°C with agitation (200 rpm) until they reached an OD600 of 1.0, and 10-ml aliquots of the cultures were transferred into precooled (−20°C) reaction tubes and swirled in liquid nitrogen for a maximum of 10 s to prevent freezing of the cultures. The cells were collected by centrifugation for 5 min at 4,000 rpm and 4°C, and the pellets were immediately frozen in liquid nitrogen and stored at −80°C. Two additional aliquots (1 ml each) were harvested for total protein determination (see below). The pellets were stored at −20°C. The cell pellets for c-di-AMP quantification were resuspended in 800 μl of extraction mixture I (acetonitrile-methanol [1:1]) and transferred to 2-ml screw-cap microtubes that contained 0.5 g of 0.1-mm-diameter glass beads. The tubes were vortexed for a short time, frozen in liquid nitrogen, and subsequently incubated for 10 min at 95°C and stored on ice. The tubes were transferred into a precooled (−20°C) TissueLyser II adapter, and the cells were disrupted by three pulses of 30 s each with an oscillation frequency of 30 Hz in a TissueLyser II (Qiagen, Hilden, Germany). The adapter was cooled on ice between each pulse. The reaction tubes were centrifuged for 10 min at 14,800 rpm and 4°C, and the supernatants were transferred into fresh tubes. Next, 200 μl of extraction mixture II (acetonitrile-methanol-H2O [2:2:1]) was added to the pellets, and the cell disruption procedure using the TissueLyser II was repeated as described above. The supernatants were collected by centrifugation and added to the supernatants obtained by the first extraction step, and the process was repeated a second time with 200 μl of extraction mixture II. The pooled supernatants were stored overnight at −20°C. The mixtures were centrifuged for 20 min at 14,800 rpm and 4°C, and the resulting supernatants were dried in a vacuum concentrator (SpeedVac; Thermo Scientific, Germany) for 2 to 4 h at 40°C. The nucleotides extracted were dissolved with 200 μl of H2O. After repeated centrifugation and addition of the internal standard [13C,15N]c-di-AMP, part of the extracts was analyzed by LC-MS/MS as described previously (10). The cell pellets for protein quantification were dissolved in 800 μl 0.1 M NaOH and heated for 10 min at 98°C. After a short centrifugation (5 min, 20,800 × g), the supernatant was transferred to a new tube. The pellet was processed a second time as described before. The resulting supernatant was pooled with the previous one. The cell pellets harvested for protein quantification were treated as described previously (10), and the protein concentration was determined using the Bradford assay (35).
RESULTS
CdaR does not affect synthesis and membrane localization of CdaA.
The L. monocytogenes cdaAR operon (38) encodes the diadenylate cyclase CdaA (Lmo2120) and a putative regulator of CdaA activity, CdaR (Lmo2119) (Fig. 1A and B) (6, 11). Lmo2120 was previously designated DacA (diadenylate cyclase A) (11). We suggest an alternative name, CdaA (cyclic di-AMP synthase A), for two reasons: DacA is commonly used to designate d-alanyl-d-alanine carboxypeptidases; and Lmo2120 is homologous to the B. subtilis diadenylate cyclase CdaA (65% identity).
To explore the mutual relationship between CdaA and CdaR, we first assessed whether CdaR affects the levels of CdaA by Western blot analyses using anti-CdaA antibodies. As shown in Fig. 2A, the wild-type strain EGD-e and the ΔcdaR mutant LMR45 produced similar amounts of CdaA, indicating that CdaR does not control the CdaA levels. Because cdaA is essential for growth in rich medium, we constructed a CdaA depletion strain, LMJR46 (IcdaA), in which the native cdaA gene is deleted but an ectopic cdaA copy is present at the tRNAArg attB site and expressed from an IPTG-inducible promoter. We observed that this strain indeed synthesized CdaA in an IPTG-dependent manner (Fig. 2A). Next, we assessed whether the inactivation of the cdaR gene affects membrane localization of CdaA. For this purpose, cell-free crude extracts from the strains EGD-e (wt) and LMJR45 (ΔcdaR) were separated into cytoplasmic and membrane fractions. As shown in Fig. 2B, the absence of CdaR did not affect the integration of CdaA into the membrane. Moreover, this analysis indicates that like CdaA from B. subtilis (10), the protein from L. monocytogenes is an integral membrane protein. To conclude, CdaR affects neither the amount nor the membrane localization of CdaA.
FIG 2.
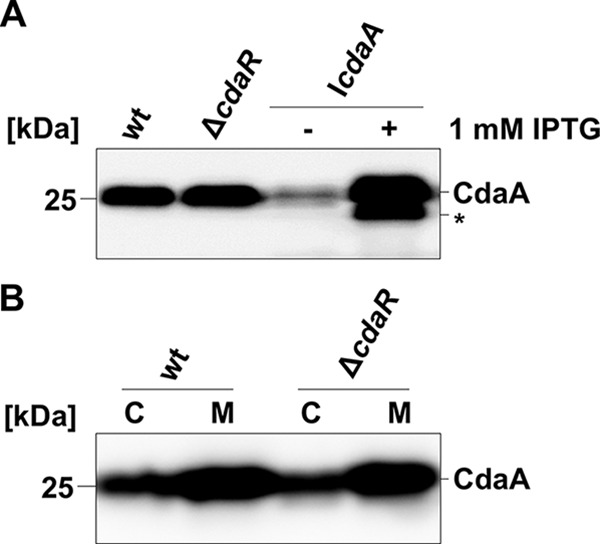
Synthesis and localization of the diadenylate cyclase CdaA. (A) Expression of cdaA in cdaR and cdaA mutant strains of L. monocytogenes was analyzed by Western blotting. The protein extracts of strains EGD-e (wild type), LMJR45 (ΔcdaR), and LMJR46 (IcdaA) grown with or without IPTG were separated by SDS-PAGE, and the CdaA levels were visualized using a polyclonal antiserum specific for L. monocytogenes CdaA. (B) Effect of cdaR inactivation on CdaA membrane localization. Strains EGD-e (wild type) and LMJR45 (ΔcdaR) were grown in BHI medium at 37°C, and the bacteria were harvested at an OD600 of 1.0. Cytosolic (C, 15 μg) and membrane (M) fractions of each strain were separated by SDS-PAGE (12%) and analyzed by Western blotting using CdaA antiserum. The protein labeled by an asterisk might be CdaA that is processed due to overproduction of the protein.
CdaR-CdaA complex formation.
Recently, it has been suggested that CdaR and CdaA interact with each other (1). Indeed, the orthologs from B. subtilis were shown to form a complex in vivo (10). To test whether CdaR and CdaA from L. monocytogenes are interaction partners, we performed a bacterial two-hybrid (B2H) experiment, which is based on the interaction-mediated reconstitution of the Bordetella pertussis adenylate cyclase in E. coli (36). The B2H experiment revealed that all proteins showed self-interaction (Fig. 3A). The analysis also revealed that CdaA interacts directly with CdaR when the diadenylate cyclase and the potential regulator harbor the adenylate cyclase domain at their C terminus (C-CdaA, pUT18 derivative) and N terminus (N-CdaR, pKT25 derivative), respectively, which are both likely cytoplasmic (Fig. 3B). Moreover, truncated CdaR variants lacking the signal peptide (24, 25, 26) showed interactions with the full-length N-CdaR (pKT25 derivative) and C-CdaR (p25-N derivative) proteins. This observation might indicate that the CdaR variants can interact with each other via their YbbR domains even though they are not properly inserted into the membrane and thus are cytoplasmic. We further tested whether the signal peptide of CdaR and the transmembrane domain of CdaA contribute to the formation of the CdaA-CdaR complex. For this purpose, we constructed B2H constructs encoding the Δ28CdaR and Δ80CdaA variants lacking the signal peptide of CdaR and the transmembrane domains of CdaA, respectively. We observed that the truncated CdaR and CdaA variants do not interact. To conclude, our findings indicate that the signal peptide of CdaR and the transmembrane domain of CdaA might be involved in the direct interaction between the two proteins (Fig. 3B).
FIG 3.
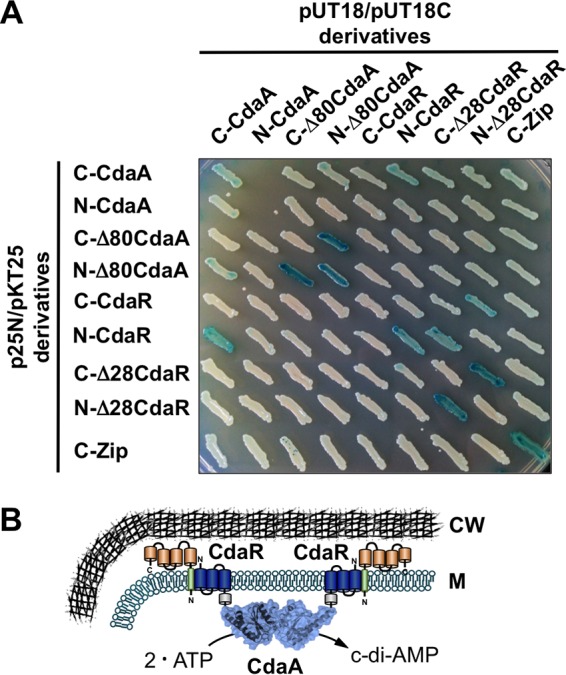
CdaA-CdaR complex formation. (A) B2H analysis for studying the interactions between full-length and truncated CdaA and CdaR variants. N-CdaA and C-CdaA represent domains of the Bordetella pertussis adenylate cyclase that are fused to the N and C termini of the proteins, respectively. Δ80CdaA and Δ28CdaR are truncated proteins lacking the N-terminal 80 and 28 amino acids, respectively. Probably due to the toxic effects of the diadenylate cyclase, we have failed to combine the N-CdaA fusion proteins in E. coli (23). C-CdaR and N-CdaA fusions are most likely inactive because many of the adenylate cyclase domains are not cytosolic. (B) Model illustrating the localization of CdaR and CdaA and the interaction of the proteins in L. monocytogenes. CW, cell wall; M, membrane.
CdaR regulates the activity of CdaA in L. monocytogenes.
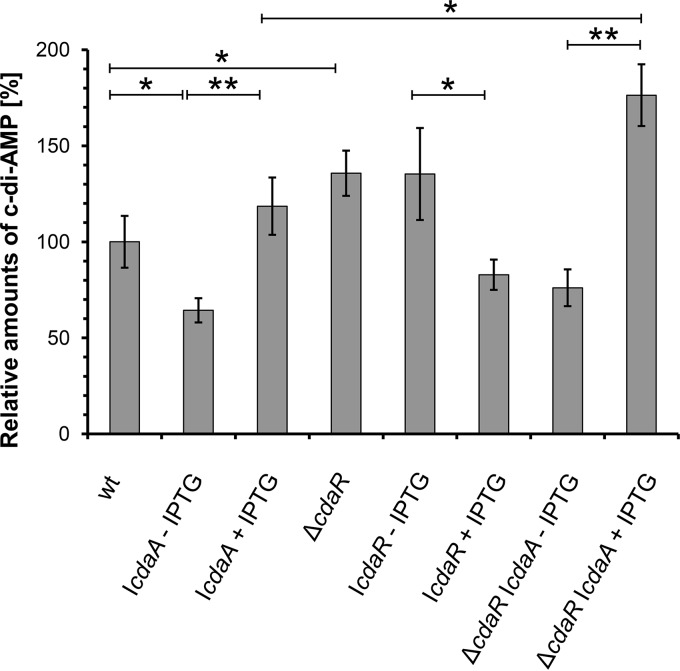
A CdaR protein homolog from B. subtilis has been reported to act as a potential regulator of CdaA activity (6); however, the role of CdaR in L. monocytogenes has not been studied so far. To address the question of how CdaR affects the activity of CdaA in its native environment, we determined the intracellular levels of c-di-AMP in the ΔcdaR mutant LMJR45 and in the strain BPL16 (IcdaR), which has the ΔcdaR mutation and the cdaR gene under the control of an IPTG-inducible promoter. The wild-type strain EGD-e, the cdaA depletion strain LMJR46 (IcdaA), and strain LMJR65 (ΔcdaR IcdaA) served as controls. The cells were grown to a point where the effects of CdaA depletion were detectable (OD600 of 1.0) (see Fig. 6A). As expected, the CdaA depletion strain LMJR46 (IcdaA) produced significantly less c-di-AMP than the wild type in the absence of IPTG (64% ± 6% of the wild-type level) (Fig. 4). In contrast, strain LMJR46 produced more c-di-AMP than the wild type (119% ± 15%) in the presence of the inducer, a finding which is in good agreement with the observed overexpression of CdaA during growth in the presence of IPTG (Fig. 2A). Both the inactivation of cdaR in strain LMJR45 (136% ± 12%) and the depletion of CdaR in strain BPL16 (136% ± 24%) resulted in elevated intracellular c-di-AMP levels. Accordingly, c-di-AMP levels were 1.5-fold higher in strain LMJR65 (ΔcdaR IcdaA) cells during growth in medium with IPTG compared to those in LMJR45 (IcdaA) cells cultivated in the presence of the inducer. Both comparisons consistently indicate that the presence of CdaR has a negative influence on cellular c-di-AMP levels and suggest that CdaR inhibits the activity of the diadenylate cyclase in L. monocytogenes.
FIG 6.
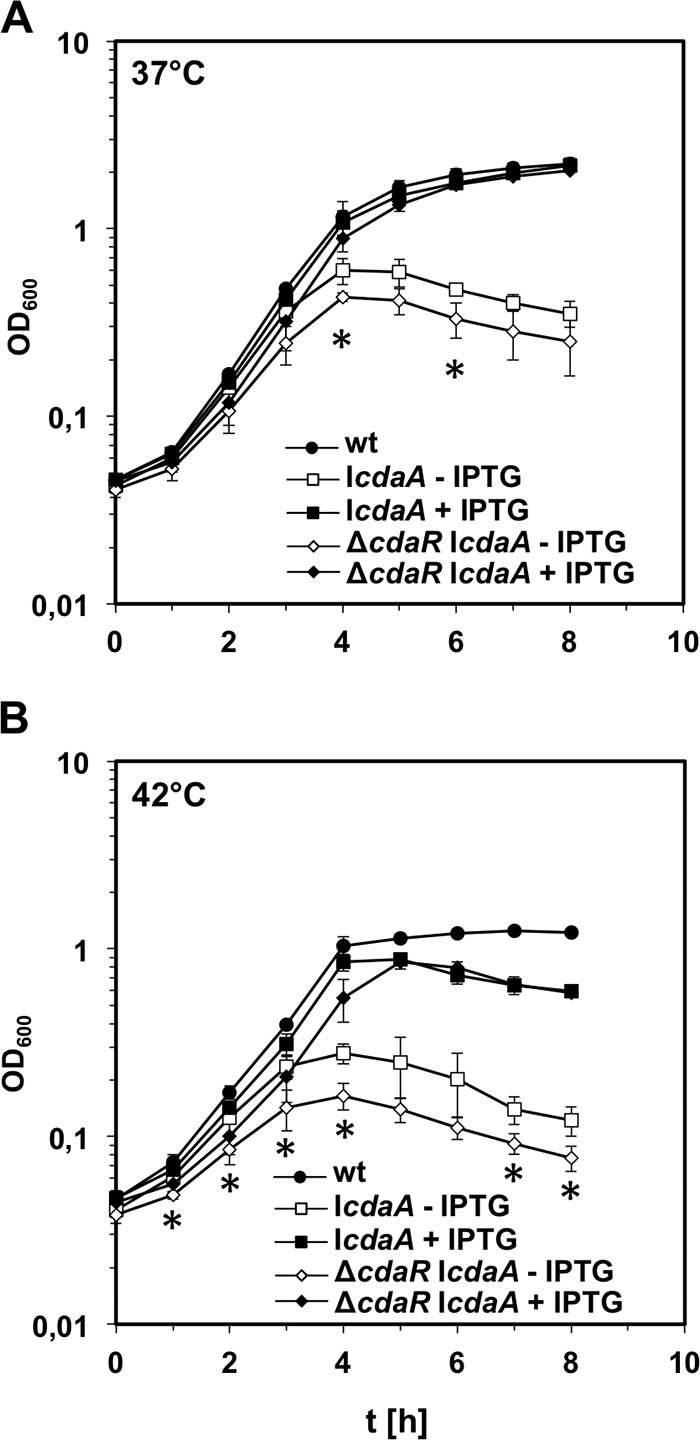
Growth of a cdaA cdaR double mutant. (A) Effect of a ΔcdaR deletion on growth of CdaA-depleted L. monocytogenes cells. Strain LMJR65 (ΔcdaR IcdaA) was cultivated in BHI broth, containing 1 mM IPTG where indicated, at 37°C, and optical density was recorded. Strains LMJR45 (ΔcdaR) and LMJR46 (IcdaA) were included for comparison. (B) Growth of the same set of strains under identical conditions at 42°C. The average values and standard deviations were calculated from three independent repetitions. *, P < 0.05, statistically significant difference between LMJR45 (ΔcdaR) and LMJR65 (ΔcdaR IcdaA without IPTG).
FIG 4.
Roles of CdaA and CdaR in c-di-AMP biosynthesis, as demonstrated by intracellular c-di-AMP levels in the wild-type (wt) strain EGD-e and in strains LMJR46 (IcdaA), LMJR45 (ΔcdaR), BPL16 (IcdaR), and LMJR65 (ΔcdaR IcdaA). All strains were grown in BHI broth at 37°C to an optical density of 1.0. For depletion of CdaA and CdaR, the strains LMJR46 (IcdaA), LMJR65 (ΔcdaR IcdaA), and BPL16 (IcdaR) were pregrown overnight in the presence of 1 mM IPTG. The precultures were washed and used to start a depletion culture without IPTG, and the cultures were supplemented with 1 mM inducer. Data from three biological replicates are shown; standard deviations are indicated as error bars. *, P < 0.05, and **, P < 0.005, significant changes detected using unpaired t tests.
Effect of cdaR deletion on L. monocytogenes growth.
Next, we explored the effect of cdaR deletion on L. monocytogenes growth. The cdaR mutant LMJR45 (ΔcdaR) grew normally at 37°C (Fig. 5A) but showed growth retardation and reduced optical densities in all growth phases at 42°C (Fig. 5B). IPTG-induced expression of CdaR in the ΔcdaR mutant strain BPL16 (IcdaR) restored normal growth at 42°C (Fig. 5C). These findings suggest that CdaR is required for optimal growth at elevated temperatures.
FIG 5.
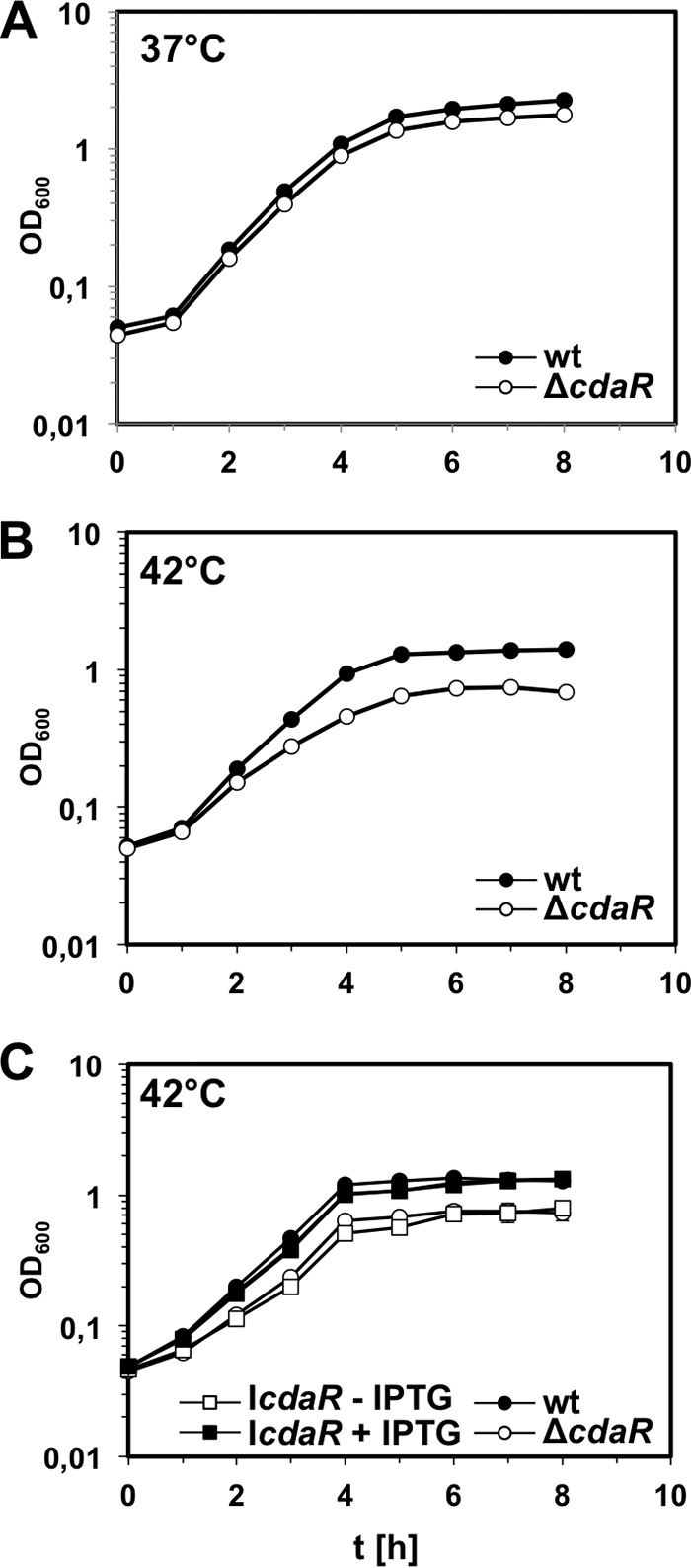
Growth defect of L. monocytogenes cdaR mutant strains. (A) Growth of an L. monocytogenes cdaR mutant strain in BHI broth at 37°C. Strains EGD-e (wild type) and LMJR45 (ΔcdaR) were grown in BHI broth at 37°C and the optical densities (OD600) were measured in hourly intervals. (B) Growth of the same set of strains in BHI broth at 42°C. (C) Complementation of the ΔcdaR growth defect at 42°C. Strains EGD-e (wild type), LMJR45 (ΔcdaR), and BPL16 (IcdaR) were cultivated in BHI broth with or without 1 mM IPTG at 42°C, and the OD600 values were recorded over time. Average values were calculated from three independent experiments, and standard deviations are shown.
To explore the effect of cdaR deletion in a CdaA depletion strain, we analyzed the growth of strain LMJR65 (ΔcdaR IcdaA). This strain grew as fast as the wild type and the ΔcdaR single mutant in the presence of IPTG. However, when no IPTG was added, its growth significantly lagged behind that of strain LMJR46 (IcdaA) at 37°C (Fig. 6A), and this effect was even more apparent at 42°C (Fig. 6B). This finding might indicate that in addition to the control of CdaA activity, CdaR might have additional functions. Interestingly, the growth defects associated with depletion of CdaA can be detected already after approximately three generations in culture medium without IPTG. This is in contrast to what we routinely observe with other strains allowing depletion of essential genes (such as secA), which usually need up to six generations to cause retardation of growth (39) and confirms that cdaA is an essential gene in L. monocytogenes during growth in rich medium (11, 12). To conclude, inactivation of cdaR and cdaA has additive negative effects on L. monocytogenes growth.
CdaA depletion affects cell division.
Several studies revealed that c-di-AMP is involved in cell wall homeostasis (9, 13, 30), which may affect the cell shape. Hence, we investigated the morphology of cdaA and cdaR mutant strains by microscopy. For this purpose, strains LMJR45 (ΔcdaR) and LMJR46 (IcdaA) were cultivated in BHI medium (containing 1 mM IPTG where indicated) at 37°C to an OD600 of 1.0, and the cell membranes were stained with Nile red. The depletion of CdaA caused cell elongation, a phenotype that was absent in the LMJR46 (IcdaA) cells grown in the presence of 1 mM IPTG (Fig. 7A). The cdaR deletion in strain LMJR45 did not affect the cell morphology (Fig. 7A). For quantification of these effects, we measured lengths of 310 cells per strain. A graphical illustration of the cell length distribution in these strains confirmed our qualitative observations (Fig. 7B), revealing that CdaA depletion in strain LMJR46 (IcdaA) caused cell elongation (average length, 1.59 ± 0.45 μm) compared to that of cells grown in the presence of IPTG (1.25 ± 0.34 μm) or to that of the wild-type cells (1.05 ± 0.2 μm). That induction of cdaA expression did not fully restore the cell lengths of LMJR46 (IcdaA) cells to the wild-type levels (Fig. 2A) might be explained by artificial CdaA overexpression (Fig. 2A) under this condition. This in turn indicates that even increased levels of CdaA affect cell division efficiency. In contrast, the average cell length of the ΔcdaR deletion mutant LMJR45 cells (1.03 ± 0.19 μm) was similar to that of the wild type.
FIG 7.

Phenotypes of L. monocytogenes cdaA and cdaR mutant strains. (A) Fluorescence micrographs of L. monocytogenes cdaA and cdaR mutant strains stained with Nile red. Strains EGD-e (wild type), LMJR45 (ΔcdaR), and LMJR46 (IcdaA) were grown in BHI broth at 37°C until the midlogarithmic growth phase. For depletion of CdaA, strain LMJR46 (IcdaA) was pregrown overnight in the presence of IPTG. The preculture was washed and used to start a depletion culture without IPTG. Bar, 5 μm. (B) The same strains used in panel A were stained as described above, and cell lengths of 310 cells per strain were measured. The distribution of cell lengths is illustrated as a frequency plot.
Effect of cdaA and cdaR mutations on antibiotic susceptibility.
It was shown earlier that depletion of CdaA affects the susceptibility of L. monocytogenes to cell wall-targeting antibiotics, such as penicillin, ampicillin, and cefuroxime (13). However, it remains unclear how deletion of cdaR affects antibiotic susceptibilities. To investigate this issue, MICs for different antibiotics were determined using E-tests. The depletion of the cdaA gene in strain LMJR46 (IcdaA) had a strong effect, causing 3- to 4-fold reductions in the resistance to β-lactam antibiotics, including ampicillin, penicillin, amoxicillin, and meropenem, and mild reductions for vancomycin and gentamicin (Table 3). The complete restoration of the wild-type susceptibilities to β-lactams was observed when strain LMJR46 (IcdaA) was grown in the presence of IPTG, while resistance to gentamicin was also slightly increased. The cdaR deletion had a minor effect on penicillin resistance and did not change the susceptibilities to any of the other antibiotics tested (Table 3).
TABLE 3.
Antibiotic susceptibilities of L. monocytogenes cdaA and cdaR mutants
| Strain | Genotype | MICa (μg/ml) of: |
|||||
|---|---|---|---|---|---|---|---|
| Amoxicillin | Ampicillin | Penicillin | Meropenem | Vancomycin | Gentamicin | ||
| EGD-e | Wild type | 0.058 ± 0.01 | 0.074 ± 0.017 | 0.074 ± 0.017 | 0.064 ± 0.00 | 1.5 ± 0.00 | 0.21 ± 0.035 |
| LMJR45 | ΔcdaR | 0.058 ± 0.01 | 0.074 ± 0.017 | 0.064 ± 0.00 | 0.064 ± 0.00 | 1.5 ± 0.00 | 0.21 ± 0.035 |
| LMJR46 without IPTG | IcdaA | <0.016 | <0.016 | 0.012 ± 0.004 | 0.01 ± 0.002 | 1 ± 0.00 | 0.17 ± 0.038 |
| LMJR46 with IPTG | IcdaA | 0.058 ± 0.01 | 0.074 ± 0.017 | 0.074 ± 0.017 | 0.074 ± 0.017 | 1.67 ± 0.29 | 0.46 ± 0.07 |
Antibiotic susceptibilities were calculated from three independent repetitions. Values are averages ± standard deviations.
To explain the increased penicillin susceptibilities of cdaA and cdaR mutant strains, we tested whether penicillin induces autolysis of cdaA and cdaR mutants. For this purpose, cells of strains EGD-e (wild type), LMJR45 (ΔcdaR), LMJR46 (IcdaA), and LMJR65 (ΔcdaR IcdaA) were resuspended in a Tris buffer, and the decline in optical density after penicillin addition was recorded. The deletion of cdaR had no effect on penicillin-induced autolysis (Fig. 8A). In contrast, depletion of CdaA from cells of strains LMJR46 (IcdaA) and LMJR65 (ΔcdaR IcdaA) stimulated autolysis in the presence of penicillin to the same degree (Fig. 8A). However, repetition of the same experiment in the absence of penicillin showed that this effect is due to intrinsic spontaneous autolysis and is penicillin independent. Since autolysis is clearly penicillin dependent in the mutants depleted for cell wall biosynthetic proteins such as PBP B1 (40), this result indicates that autolysis provoked by CdaA depletion might be due to the malfunction of other processes. Lysozyme treatment leads to a rapid decline in the optical density of L. monocytogenes wild-type cells, as it promotes massive lysis. However, the absence of CdaR intriguingly increased the resistance of the LMJR45 (ΔcdaR) cells to lysozyme (Fig. 8B). This indicates that the peptidoglycan of the ΔcdaR mutant is modified in a way that renders it more resistant against hydrolysis by lysozyme. The intrinsic autolysis of CdaA-depleted cells masks any possible effect on lysozyme resistance (data not shown). To conclude, a strain lacking cdaR is significantly less susceptible to lysozyme.
FIG 8.
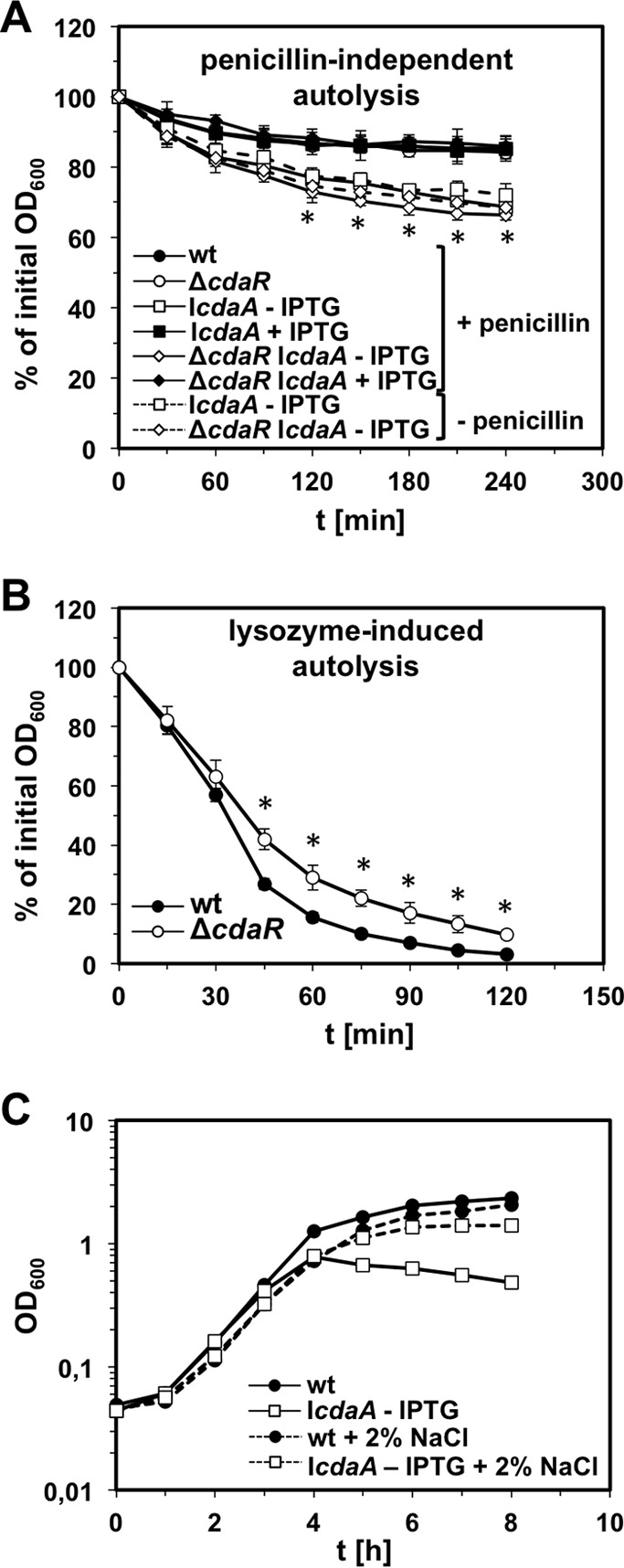
Effect of CdaAR on autolysis. (A) Autolysis assay of L. monocytogenes cdaAR mutant strains in the presence and absence of penicillin. Strains EGD-e (wild type), LMJR45 (ΔcdaR), LMJR46 (IcdaA), and LMJR65 (ΔcdaR IcdaA) were grown in BHI broth with or without 1 mM IPTG at 37°C until an OD600 of 0.8. Cells were harvested and resuspended in 50 mM Tris-HCl (pH 8.0) with or without 25 μg/ml penicillin. The decline in optical densities was recorded photometrically and is expressed as relative average values. Standard deviations are calculated from an experiment performed in triplicate. *, P < 0.05, data points at which differences in autolysis rates in the presence of penicillin between LMJR46 without IPTG and LMJR46 with IPTG and between LMJR65 without IPTG and LMJR65 with IPTG were statistically different. (B) Autolysis of L. monocytogenes strains EGD-e (wt) and LMJR45 (ΔcdaR) in the presence of 2.5 μg/ml lysozyme. Average values and standard deviations were calculated from three independent experiments. *, P < 0.05, statistically significant differences. (C) Suppression of the cdaA-dependent growth defect by osmoprotection of growth conditions. Strains EGD-e (wild type) and LMJR46 (IcdaA) were pregrown in BHI broth (containing 1 mM IPTG for LMJR46) at 37°C and used to inoculate fresh BHI broth (without IPTG) containing or not containing 2% NaCl. OD600 values were measured in hourly intervals.
Suppression of the cdaA growth defect by sodium chloride.
Others have shown that autolysis of L. monocytogenes and B. subtilis due to reduced intracellular amounts of c-di-AMP can be suppressed by increasing the osmolarity of the growth medium (9, 13, 27). We wondered whether increased autolysis of CdaA-depleted cells is the sole reason for their retarded growth. In order to address this point, we reanalyzed the growth of strain LMJR46 (IcdaA) in BHI broth in the presence and absence of 2% NaCl. During growth in BHI broth containing neither IPTG nor NaCl, strain LMJR46 (IcdaA) showed the same growth defect as described above. In contrast, the addition of 2% NaCl almost completely restored its growth to the wild-type levels (Fig. 8C). This result shows that osmotic instability of the CdaA-depleted LMJR46 (IcdaA) cells accounts for most of the growth defect in BHI broth and suggests that depletion of CdaA per se should not be detrimental to viability. If true, it should be possible to delete cdaA in the presence of 2% NaCl. We tested this hypothesis; however, we did not obtain cdaA deletion mutants. This indicates that osmoprotection is only part of the reason for CdaA essentiality.
DISCUSSION
In the present study, we have demonstrated that like in B. subtilis and Chlamydia trachomatis (10, 41), the diadenylate cyclase CdaA from L. monocytogenes is an integral membrane protein. Moreover, similar to what was reported for B. subtilis, the CdaR protein directly interacts with CdaA in L. monocytogenes (10), and membrane localization of CdaA is not perturbed by the absence of CdaR (see Fig. 2).
The fact that CdaA interacts with CdaR suggests that the regulator also comes in close proximity to the cell wall. The inactivation of both proteins affects the cellular integrity or the modification of the cell surface. This might imply the possibility that c-di-AMP homeostasis is linked to the metabolism of the cell envelope (the cell wall and cell membrane). In fact, (i) depletion of CdaA enhances the sensitivity of L. monocytogenes to cell wall-active antibiotics, and (ii) the lack of CdaR increases the resistance of the bacteria to lysozyme. Furthermore, for other bacteria, such as S. aureus, B. subtilis, and Streptococcus pyogenes, it has been reported that alterations of cellular c-di-AMP levels affect the integrity of the cell wall (6, 27). Moreover, elevated c-di-AMP levels resulted in the accumulation of extramembranous material, such as vesicles in S. aureus (28). However, it remains to be elucidated how c-di-AMP controls membrane biosynthesis in this organism. In addition to CdaR, yet another protein, the glucosamine-6-phosphate mutase GlmM, which synthesizes an essential precursor for the cell wall, directly interacts with CdaA, as revealed by a recent study (10). We furthermore showed that alterations in the intracellular c-di-AMP levels affect cell length, which also is an effect that might be caused by aberrations in peptidoglycan homeostasis (27). It is attractive to hypothesize that CdaR acts as a sensory protein that monitors the integrity of the cell envelope either by interacting with cell wall proteins or with peptidoglycan. Depending on the signal perceived by CdaR, the protein in turn might regulate the activity of the diadenylate cyclase CdaA to maintain cell wall integrity. Here, we showed that CdaR indeed can negatively control the diadenylate cyclase CdaA (see Fig. 4). Moreover, the complex between CdaA and CdaR may be important for adjustment of the synthesis of cell wall precursors, depending on the state of the cell envelope (10). It will be interesting to identify the signals perceived by CdaR and decipher how c-di-AMP produced by CdaA is linked to the metabolism of the cell envelope. Interestingly, it has been observed that the quality of the nitrogen source also affects c-di-AMP production (10). However, how the nitrogen source controls the activity of CdaA and whether this is linked to peptidoglycan metabolism remain to be elucidated.
Another recent study uncovered a link between c-di-AMP metabolism and integrity of the cell envelope in actinobacteria (42). The protein family of so-called resuscitation-promoting factors includes a variety of muralytic enzymes that are involved in the resuscitation of dormant cells (43, 44, 45). The expression and activities of the resuscitation-promoting factors are tightly regulated to prevent lysis due to degradation of the protective cell wall. In Streptomyces coelicolor, expression of the rpfA gene encoding the resuscitation-promoting factor RpfA is controlled by the upstream ydaO-like riboswitch in response to c-di-AMP (42, 46). The riboswitch specifically binds c-di-AMP, which leads to transcription attenuation and reduced synthesis of the muralytic enzyme RpfA. It has been suggested that the reduced RpfA levels prevent germination of S. coelicolor. Interestingly, resuscitation-promoting factors that are related to the actinobacterial proteins also exist in Firmicutes, the bacterial phylum to which L. monocytogenes belongs (43, 44, 47). In L. monocytogenes, it has been observed that accumulation of c-di-AMP results in the increased expression of the resuscitation factors Lmo2522 and Lmo0186 that are homologs of the B. subtilis YocH and YabE proteins, respectively (13, 43, 44). However, it remains to be elucidated how the synthesis of YocH and YabE is controlled by c-di-AMP.
As mentioned above, several studies suggest a link between c-di-AMP metabolism and the integrity of the cell envelope. However, it is as yet unclear why a decrease in the intracellular concentration of the nucleotide causes bacterial lysis. This work and other studies revealed that lysis of L. monocytogenes and B. subtilis cells that were depleted for CdaA can be rescued by increased medium osmolarity (Fig. 8) (9, 27). Since it has been reported that c-di-AMP negatively controls the influx of potassium ions into S. pneumoniae and S. aureus cells (16, 17, 48), one can imagine that the deregulation of potassium homeostasis increases the turgor pressure to lethal levels, which would result in cell lysis. This hypothesis is supported by the observation that the osmosensitive phenotype of an S. aureus ltaS mutant strain unable to synthesize lipoteichoic acid might be rescued by an increased intracellular c-di-AMP concentration (27). Thus, the c-di-AMP-dependent regulation of potassium uptake may indeed be essential for maintaining cell integrity. That processes other than peptidoglycan biosynthesis are affected by depletion of CdaA is in good agreement with our observation that penicillin had no effect on autolysis of CdaA-depleted cells. A recent study revealed that c-di-AMP is essential only for the growth of L. monocytogenes on rich medium (12). It has been shown that the lack of c-di-AMP led to a decrease in intracellular GTP levels and consequently to the activation of several genes that are usually repressed by the pleiotropic transcription factor CodY (50). Some derepressed genes code for an opp oligopeptide uptake system, suggesting that the unbalanced import of oligopeptides is lethal to the cell. Indeed, simultaneous inactivation of the cdaA and the opp genes enabled the bacteria to grow on rich medium (12). However, whether c-di-AMP is essential because it controls influx of compounds that either might lead to a change in the internal osmotic pressure or are simply toxic has to be investigated. Moreover, it is unclear how the activity of the uptake system is linked to other phenotypes associated with the cell envelope.
ACKNOWLEDGMENTS
We acknowledge Mark Gomelsky for helpful discussions and are grateful to Sabine Lentes and Annette Garbe for excellent technical assistance.
Funding Statement
This work was also supported by the Göttingen Centre for Molecular Biology (GZMB). Johannes Gibhardt and Jonathan Rosenberg were supported by the Göttingen Graduate School for Neuroscience, Biophysics and Molecular Biosciences (DFG grant GSC 226/2).
REFERENCES
- 1.Corrigan RM, Gründling A. 2013. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol 11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 2.Witte G, Hartung S, Büttner K, Hopfner KP. 2008. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell 30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 3.Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky I, Rouvinski A, Meyerovich M, Ben-Yehuda S. 2006. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell 125:679–690. doi: 10.1016/j.cell.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 4.Gándara C, Alonso J. 2015. DisA and c-di-AMP act at the intersection between DNA-damage response and stress homeostasis in exponentially growing Bacillus subtilis cells. DNA Repair (Amst) 27:1–8. doi: 10.1016/j.dnarep.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. 2011. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep 12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stülke J. 2013. Cyclic-di-AMP homeostasis in Bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem 288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessières P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 8.Mehne FM, Schröder-Tittmann K, Eijlander RT, Herzberg C, Hewitt L, Kaever V, Lewis RJ, Kuipers OP, Tittmann K, Stülke J. 2014. Control of the diadenylate cyclase CdaS in Bacillus subtilis: an autoinhibitory domain limits cyclic di-AMP production. J Biol Chem 289:21098–21107. doi: 10.1074/jbc.M114.562066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo Y, Helmann JD. 2012. Analysis of the role of Bacillus subtilis σ(M) in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol 83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gundlach J, Mehne FM, Herzberg C, Kampf J, Valerius O, Kaever V, Stülke J. 2015. An essential poison: synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J Bacteriol 197:3265–3274. doi: 10.1128/JB.00564-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodward JJ, Iavarone AT, Portnoy DA. 2010. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whiteley AT, Pollock AJ, Portnoy DA. 2015. The PAMP c-di-AMP is essential for listerial growth in macrophages and rich but not minimal medium due to a toxic increase in (p)ppGpp. Cell Host Microbe 17:788–798. doi: 10.1016/j.chom.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witte CE, Whiteley AT, Burke TP, Sauer JD, Portnoy DA, Woodward JJ. 2013. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. mBio 4:e00282–00213. doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Commichau FM, Dickmanns A, Gundlach J, Ficner R, Stülke J. 2015. A jack of all trades: the multiple roles of the unique essential second messenger cyclic di-AMP. Mol Microbiol 97:189–204. doi: 10.1111/mmi.13026. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Li W, He ZG. 2013. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem 188:3085–3095. doi: 10.1074/jbc.M112.428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai Y, Yang J, Zarrella TM, Zhang Y, Metzger DW, Bai G. 2014. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J Bacteriol 196:614–623. doi: 10.1128/JB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H, Youn SJ, Kim SO, Ko J, Lee JO, Choi BS. 2015. Structural studies of potassium transport protein KtrA regulator of conductance of K+ (RCK) C domain in complex with cyclic diadenosine monophosphate (c-di-AMP). J Biol Chem 290:16393–16402. doi: 10.1074/jbc.M115.641340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sureka K, Choi PH, Precit M, Delince M, Pensinger DA, Huynh TN, Jurado AR, Goo YA, Sadilek M, Iavarone AT, Sauer JD, Tong L, Woodward JJ. 2014. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell 158:1389–1401. doi: 10.1016/j.cell.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campeotto I, Zhang Y, Mladenov MG, Freemont PS, Gründling A. 2015. Complex structure and biochemical characterization of the Staphylococcus aureus cyclic diadenylate monophosphate (c-di-AMP)-binding protein PstA, the founding member of a new signal transduction protein family. J Biol Chem 290:2888–2901. doi: 10.1074/jbc.M114.621789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi PH, Sureka K, Woodward JJ, Tong L. 2015. Molecular basis for the recognition of cyclic-di-AMP by PstA, a PII-like signal transduction protein. MicrobiologyOpen 4:361–374. doi: 10.1002/mbo3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gundlach J, Dickmanns A, Schröder-Tittmann K, Neumann P, Kaesler J, Kampf J, Herzberg C, Hammer E, Schwede F, Kaever V, Tittmann K, Stülke J, Ficner R. 2015. Identification, characterization, and structure analysis of the cyclic di-AMP-binding PII-like signal transduction protein DarA. J Biol Chem 290:3069–3080. doi: 10.1074/jbc.M114.619619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller M, Hopfner KP, Witte G. 2015. c-di-AMP recognition by Staphylococcus aureus PstA. FEBS Lett 589:45–51. doi: 10.1016/j.febslet.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg J, Dickmanns A, Neumann P, Gunka K, Arens J, Kaever V, et al. 2015. Structural and biochemical analysis of the essential diadenylate cyclase CdaA from Listeria monocytogenes. J Biol Chem 290:6596–6606. doi: 10.1074/jbc.M114.630418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barb AW, Cort JR, Seetharaman J, Lew S, Lee HW, Acton T, Xiao R, Kennedy MA, Tong L, Montelione GT, Prestegard JH. 2011. Structures of domains I and IV from YbbR are representative of a widely distributed protein family. Protein Sci 20:396–405. doi: 10.1002/pro.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renier S, Micheau P, Talon R, Hébraud M, Desvaux M. 2012. Subcellular localization of extracytoplasmic proteins in monoderm bacteria: rational secretomics-based strategy for genomic and proteomic analyses. PLoS One 7:e42982. doi: 10.1371/journal.pone.0042982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 27.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. 2011. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog 7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corrigan RM, Bowman L, Willis AR, Kaever V, Gründling A. 2015. Cross-talk between two nucleotide pathways in Staphylococcus aureus. J Biol Chem 290:5826–5839. doi: 10.1074/jbc.M114.598300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho KH, Kang SK. 2013. Streptococcus pyogenes c-di-AMP phosphodiesterase, GdpP, influences SpeB processing and virulence. PLoS One 8:e69425. doi: 10.1371/journal.pone.0069425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dengler V, McCallum N, Kiefer P, Christen P, Patrignani A, Vorholt JA, et al. 2013. Mutation in the c-di-AMP cyclase dacA affects fitness and resistance of methicillin resistant Staphylococcus aureus. PLoS One 8:e73512. doi: 10.1371/journal.pone.0073512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 32.Monk IR, Gahan CG, Hill C. 2008. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl Environ Microbiol 74:3921–3934. doi: 10.1128/AEM.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng L, Baumann U, Reymond JL. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradford MM. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Claessen D, Emmins D, Hamoen LW, Daniel RA, Errington J, Edwards DH. 2008. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol Microbiol 68:1029–1046. doi: 10.1111/j.1365-2958.2008.06210.x. [DOI] [PubMed] [Google Scholar]
- 38.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Régnault B, Coppée JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 39.Halbedel S, Hahn B, Daniel RA, Flieger A. 2012. DivIVA affects secretion of virulence-related autolysins in Listeria monocytogenes. Mol Microbiol 83:821–839. doi: 10.1111/j.1365-2958.2012.07969.x. [DOI] [PubMed] [Google Scholar]
- 40.Rismondo J, Möller L, Aldridge C, Gray J, Vollmer W, Halbedel S. 2015. Discrete and overlapping functions of peptidoglycan synthases in growth, cell division and virulence of Listeria monocytogenes. Mol Microbiol 95:332–351. doi: 10.1111/mmi.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, Valdivia RH. 2013. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. mBio 4:e00018–13. doi: 10.1128/mBio.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.St-Onge RJ, Haiser HJ, Yousef MR, Sherwood E, Tschowri N, Al-Bassam M, Elliot MA. 2015. Nucleotide second messenger-mediated regulation of a muralytic enzyme in Streptomyces. Mol Microbiol 96:779–795. doi: 10.1111/mmi.12971. [DOI] [PubMed] [Google Scholar]
- 43.Commichau FM, Halbedel S. 2013. The resuscitation promotion concept extends to firmicutes. Microbiology 159:1298–1300. doi: 10.1099/mic.0.069484-0. [DOI] [PubMed] [Google Scholar]
- 44.Pinto D, Sao-José C, Santos MA, Chambel L. 2013. Characterization of two resuscitation promoting factors of Listeria monocytogenes. Microbiology 159:1390–1401. doi: 10.1099/mic.0.067850-0. [DOI] [PubMed] [Google Scholar]
- 45.Mukamolova GV, Kaprelyants AS, Young DI, Young M, Kell DB. 1998. A bacterial cytokine. Proc Natl Acad Sci U S A 95:8916–8921. doi: 10.1073/pnas.95.15.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson JW, Sundarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. 2013. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol 9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravagnani A, Finan CL, Young M. 2005. A novel firmicute protein family related to the actinobacterial resuscitation-promoting factors by non-orthologous domain displacement. BMC Genomics 6:39. doi: 10.1186/1471-2164-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Gründling A. 2013. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci U S A 110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett HJ, Pearce DM, Glenn S, Taylor CM, Kuhn M, Sonenshein AL, Andrew PW, Roberts IS. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol Microbiol 63:1453–1467. doi: 10.1111/j.1365-2958.2007.05597.x. [DOI] [PubMed] [Google Scholar]
- 50.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, et al. 2001. Comparative genomics of Listeria species. Science 294:849–852. [DOI] [PubMed] [Google Scholar]