ABSTRACT
Streptococcus mutans, a causative agent of dental caries, relies on multiple quorum-sensing (QS) pathways that coordinate the expression of factors needed for colonization in the oral cavity. S. mutans uses small peptides as QS signaling molecules that typically are secreted into the outside milieu. Competence-stimulating peptide (CSP) is one such QS signaling molecule that functions through the ComDE two-component signal transduction pathway. CSP is secreted through NlmTE, a dedicated ABC transporter that cleaves off the N-terminal leader peptide to generate a mature peptide that is 21 residues long (CSP-21). We recently identified a surface-localized protease, SepM, which further cleaves the CSP-21 peptide at the C-terminal end and removes the last 3 residues to generate CSP-18. CSP-18 is the active QS molecule that interacts with the ComD sensor kinase to activate the QS pathway. In this study, we show that SepM specifically cleaves CSP-21 between the Ala18 and Leu19 residues. We also show that SepM recognizes only Ala at position 18 and Leu at position 19, although some CSP-18 variants with a substitution at position 18 can function equally as well as the QS peptide. Furthermore, we demonstrate that SepM homologs from other streptococci are capable of processing CSP-21 to generate functional CSP-18.
IMPORTANCE SepM is a membrane-associated streptococcal protease that processes competence-stimulating peptide (CSP) to generate an active quorum-sensing molecule in S. mutans. SepM belongs to the S16 family of serine proteases, and in this study, we found that SepM behaves as an endopeptidase. SepM displays strict substrate specificity and cleaves the peptide bond between the Ala and Leu residues. This is the first report of an endopeptidase that specifically cleaves these two residues.
INTRODUCTION
Streptococcus mutans is considered to be one of the primary etiological agents of dental caries. S. mutans forms robust biofilms on tooth surfaces and heart valves, a prerequisite for disease formation. Biofilm formation on the tooth surface requires a highly coordinated signaling pathway known as quorum sensing (QS). QS is a primary bacterial communication system that often uses secreted peptide pheromones to regulate the expression of various genes when the bacterial cell density reaches a certain threshold concentration (1). In addition to biofilm formation, numerous cellular processes, such as virulence factor expression, extracellular enzyme production, antibiotic production, and genetic exchanges, are coordinated by QS (2, 3). S. mutans employs a well-conserved QS system called ComDEC, which is required for the regulation of biofilm formation, stress responses, the expression of bacteriocin-encoding genes, and the development of genetic competence (4). S. mutans and other Gram-positive bacteria generally use peptides as QS molecules (5). These peptides typically are translated as prepeptides that undergo processing during export to the extracellular environment. In S. mutans, the peptide pheromone competence-stimulating peptide (CSP) is encoded by the comC gene as a prepeptide with a leader sequence containing a conserved double-glycine (GG) motif (4, 6). During secretion through NlmTE, a dedicated ABC transporter complex, the N-terminal leader peptide, is cleaved off by the proteolytic activity of the transporter to generate a mature peptide that is 21 residues long (CSP-21) (7, 8). When the extracellular CSP concentration reaches a certain threshold, ComD, a membrane-associated histidine kinase, senses the signal. ComD is activated by autophosphorylation and subsequently transfers the phosphate group to ComE, a cytoplasmic response regulator. The activated ComE then stimulates the expression of various mutacin-like genes by directly recognizing a conserved direct repeat sequence present in the promoter regions; ComE also indirectly activates about 20 early competence-related genes, genes related to biofilm formation, and virulence-associated genes (9, 10).
Interestingly, a derivative of CSP-21 lacking the last 3 residues (CSP-18) has been isolated from the S. mutans culture supernatant (11–13). Early reports indicated that CSP-18 is more potent and works at a much lower concentration (10-fold) than CSP-21 (13). We recently identified a membrane-associated protease called SepM, which is responsible for the processing of CSP-21 to generate the active QS peptide CSP-18 (8). Surprisingly, genome analysis suggests that S. mutans clinical isolates secrete various CSP subtypes, although the variation among them is not as diverse as that of the pneumococcus CSP (14–16). Furthermore, several isolates encode a subtype that is identical to the active CSP-18 peptide (16). This indicates that some S. mutans strains evolved to bypass the need for SepM processing, although they still contain the sepM gene. The other CSP subtypes include a CSP-19 peptide missing the first 2 N-terminal residues and a CSP-23 peptide with 2 additional residues at the C terminus (16). The significance of the presence of these various subtypes in S. mutans biology is currently unknown.
SepM is a membrane-associated 346-residue-long polypeptide. SepM contains at least three domains: a transmembrane domain spanning from residues 10 to 26, a eukaryotic-domain-like PDZ domain spanning residues 131 to 195, and a Lon-like proteolytic domain at the C-terminal end (8). As with the Lon protease, this S16 type protease SepM contains a Ser-Lys dyad (S235 and K280) in its active site, in which serine is the nucleophile. However, SepM does not share any similarity to the classical catalytic Ser-His-Asp triad of serine proteases (17). Unlike the Lon protease, SepM does not encode any canonical ATPase domain, such as Walker A and B motifs, and it can process CSP-21 in the absence of ATP. In Escherichia coli, the Lon protease recognizes the N-terminal end of a polypeptide that is rich in nonpolar residues or the C-terminal end that is marked with the SsrA tag (18, 19). Lon degrades the substrates in a progressive manner from one end to another, producing small oligopeptides (20). In contrast, SepM does not display processive protease activity; rather, it acts as a site-specific endopeptidase.
The gene that encodes SepM is the last gene within a three-gene operon. The first gene of the operon putatively encodes S-adenosylmethionine (SAM)-dependent methyltransferase, and the middle gene putatively encodes phosphopantetheine adenylyltransferase. SepM homologs are present in many Gram-positive organisms, including all streptococci, bacilli, lactobacilli, and others (8). Multiple-sequence alignment indicates that the primary sequence is highly conserved across the species. However, whether SepM from heterologous species can process CSP-21 is currently unknown. Furthermore, the substrate specificity for SepM and its homologs has not been studied.
In this study, using mutational analysis, we found that SepM absolutely requires alanine and leucine residues at the 18th and 19th positions, respectively, for the processing of the CSP-21 peptide. We also found that SepM can cleave CSP to CSP-18, irrespective of the length, since a synthetic CSP with 28 residues was successfully processed. Finally, we showed that SepM homologs from other streptococci are also capable of cleaving CSP-21, indicating a broader role in interspecies communication.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. E. coli DH5α was grown in Luria-Bertani medium supplemented with 100 μg/ml ampicillin (Ap), 300 μg/ml erythromycin (Em), or 100 μg/ml kanamycin (Km) when necessary. Streptococcal strains were routinely grown in Todd-Hewitt medium (BBL; BD) supplemented with 0.2% yeast extract (THY medium). When necessary, 10 μg/ml Em or 300 μg/ml Km was added to THY medium. For transformation, S. mutans strains were grown up to an A595 of 0.2, and 400 nM CSP-21 was added if needed, followed by 10 min of incubation at 37°C. A total of 1.0 μg of linear or circular DNA was added to 1 ml of culture, followed by incubation at 37°C for 2 h and plating onto THY agar plates with or without antibiotics. Synthetic CSPs were synthesized by GenScript (NJ) and were desalted. The purity of the peptides was ≥80%.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| S. mutans UA159 | Wild type | 37 |
| S. mutans IBS-D29 | Markerless deletion of SMU.518 (ΔsepM) | 8 |
| S. mutans IBS-N42 | Markerless deletion of comC (ΔcomC) | This study |
| S. mutans IBS-N43 | Markerless deletion of comC and sepM (ΔcomC ΔsepM) | This study |
| S. agalactiae NEM316 | Wild type | 38 |
| S. gordonii DL-1 | Wild type | 39 |
| S. pneumoniae ATCC 49619 | Wild type | ATCC |
| S. pyogenes AM3 | Wild type | 40 |
| S. ratti FA-1 | Wild type | ATCC |
| S. uberis UT387 | Wild type | 41 |
| Plasmids | ||
| pIB184-Km | E. coli-streptococcal shuttle plasmid | 21 |
| pIB-N1 | pIB184-Km with S. gordonii SepM | This study |
| pIB-N2 | pIB184-Km with S. pyogenes SepM | This study |
| pIB-N3 | pIB184-Km with S. agalactiae SepM | This study |
| pIB-N4 | pIB184-Km with S. ratti SepM | This study |
| pIB-N5 | pIB184-Km with SepM | This study |
| pIB-N6 | pIB184-Km with S. pneumoniae SepM | This study |
| pIB-N7 | pIB184-Km with S. uberis SepM | This study |
| pIB-N40 | pIB184-Km::SepM with an Asn141Asp substitution | This study |
| pIB-N41 | pIB184-Km::SepM with a Gln315Asp substitution | This study |
| pIB-N42 | pIB184-Km::SepM with a Gln315Glu substitution | This study |
| pIB-N43 | pIB184-Km::SGO with an Asp141Asn substitution | This study |
| pIB-N44 | pIB184-Km::SGO with a Glu315Gln substitution | This study |
| pIB-N35 | pGhost9 TR::PnlmA-gusA reporter | This study |
| pIB-D57 | pIB184Km::comC (18 residues) | 8 |
| pIB-D58 | pIB184Km::comC (21 residues) | 8 |
| pIB-D55 | pGEM-T-Easy::comC region | 8 |
| pIB-N36 | pIB-D55::ΔcomC with the loxP-Km cassette | This study |
| pIB-N11 | pIB184Km::comC with an Ala18Ala substitution | This study |
| pIB-N12 | pIB184Km::comC with an Ala18Asp substitution | This study |
| pIB-N13 | pIB184Km::comC with an Ala18Leu substitution | This study |
| pIB-N14 | pIB184Km::comC with an Ala18Gly substitution | This study |
| pIB-N15 | pIB184Km::comC with an Ala18Pro substitution | This study |
| pIB-N16 | pIB184Km::comC with an Ala18Phe substitution | This study |
| pIB-N17 | pIB184Km::comC with an Ala18Thr substitution | This study |
| pIB-N18 | pIB184Km::comC with an Ala18Val substitution | This study |
| pIB-N19 | pIB184Km::comC with an Ala18Cys substitution | This study |
| pIB-N20 | pIB184Km::comC with an Ala18Arg substitution | This study |
| pIB-N21 | pIB184Km::comC with an Ala18His substitution | This study |
| pIB-N22 | pIB184Km::comC with an Ala18Tyr substitution | This study |
| pIB-N23 | pIB184Km::comC with an Ala18Ser substitution | This study |
| pIB-N24 | pIB184Km::comC with an Ala18Glu substitution | This study |
| pIB-N25 | pIB184Km::comC with an Ala18Ile substitution | This study |
| pIB-N26 | pIB184Km::comC with an Ala18Trp substitution | This study |
| pIB-N27 | pIB184Km::comC with an Ala18Gln substitution | This study |
| pIB-N28 | pIB184Km::comC with an Ala18Lys substitution | This study |
| pIB-N29 | pIB184Km::comC with an Ala18Asn substitution | This study |
| pIB-N30 | pIB184Km::comC with an Ala18Met substitution | This study |
| pIB-N37 | pIB184Km::comC with a Leu18Ile substitution | This study |
| pIB-N38 | pIB184Km::comC with a Leu18Ala substitution | This study |
| pIB-N39 | pIB184Km::comC with a Leu18Val substitution | This study |
SGO, S. gordonii SepM.
Cloning of SepM and its homologs.
SepM and its homologs were amplified from the genomic DNA of the target strains with high-fidelity AccuTaq DNA polymerase (Sigma) using the primers indicated in Table 2. After restriction with BamHI and XhoI, the PCR fragments were cloned into the BamHI-XhoI-digested pIB184-Km plasmid (21). Positive clones were verified by restriction digestion and sequencing. The clones were named pIB-N1 through pIB-N7. SepM was also cloned into the pASK43+ (IBA) vector for protein purification as a His-tagged protein. For this, the sepM open reading frame was amplified using the BamH-SepM-F and Pst-SepM-R primers (Table 2). The PCR fragment was restricted with BamHI and PstI and cloned into the BamHI-PstI-digested pASK43+ vector to generate pIBN8. SepM was purified from E. coli, according to the manufacturer's instructions (IBA).
TABLE 2.
List of oligonucleotides used in this study
| Name | Sequence (5′ to 3′) | Purposea |
|---|---|---|
| BamH-SepM-F | CGGGATCCATGCCCGGCGGAGCCTAT | Purification of SepM |
| Pst-SepM-R | AAAACTGCAGAGTGTTTTCTTAGGTAA | Purification of SepM |
| BamHI SMU.518-F | GCGGGATCCTTGGGGCAGACATCAGTCCTT | Cloning of SepM in pIB184 |
| XhoI SMU.518-R | GCGCTCGAGTTAGTGTTTTCTTAGGTAATC | Cloning of SepM in pIB184 |
| BamHI GBS0502-F | GCGGGATCCGTTCCAAAGAGTGTTGTTAGA | Cloning of GBS homolog in pIB184 |
| XhoI GBS0502-R | GCGCTCGAGCCGAACTCTTTGAGTTTATTT | Cloning of GBS homolog in pIB184 |
| BamHI SGO_0652-F | GCGGGATCCTATGTGCCCCAAAGTGTAATC | Cloning of SGO homolog in pIB184 |
| XhoI SGO_0652-R | GCGCTCGAGGCCTCAGCTTTATTATTTATT | Cloning of SGO homolog in pIB184 |
| BamHI Spy1536-F | GCGGGATCCTAGAAGGTTTGGTTCCTCAGT | Cloning of GAS homolog in pIB184 |
| XhoI Spy1536-R | GCGCTCGAGTTATTTGCGAAGATAAACCAG | Cloning of GAS homolog in pIB184 |
| SMU.518 a421g-F | TTTATGTTTTGCAGGTTAGTAGGGATTCAACATTCAAGGGTGTTTTA | Mutagenesis of SepM (N141D) |
| SMU.518 a421g-R | TAAAACACCCTTGAATGTTGAATCCCTACTAACCTGCAAAACATAAA | Mutagenesis of SepM (N141D) |
| SMU.518 c943g-F | CCCAAAGCTCTCACTAATTATGAAGAAGCTAAGCAGGC | Mutagenesis of SepM (Q315E) |
| SMU.518 c943g-R | GCCTGCTTAGCTTCTTCATAATTAGTGAGAGCTTTGGG | Mutagenesis of SepM (Q315E) |
| SMU.518 c943g_a945c-F | AAAAAAATCCCAAAGCTCTCACTAATTATGACGAAGCTAAGCAGGCTG | Mutagenesis of SepM (Q315D) |
| SMU.518 c943g_a945c-R | CAGCCTGCTTAGCTTCGTCATAATTAGTGAGAGCTTTGGGATTTTTTT | Mutagenesis of SepM (Q315D) |
| SGO g421a-F | GTCTATGTTTTGCAAGTCGCAAAAAATTCGACTTTTAAGGGAATTTT | Mutagenesis of SGO (D141N) |
| SGO g421a-R | AAAATTCCCTTAAAAGTCGAATTTTTTGCGACTTGCAAAACATAGAC | Mutagenesis of SGO (D141N) |
| SGO g943c-F | CGATCCAAAAGCAAAGACAAACTATCAAACTGCAGTTGAG | Mutagenesis of SGO (E315Q) |
| SGO g943c-R | CTCAACTGCAGTTTGATAGTTTGTCTTTGCTTTTGGATCG | Mutagenesis of SGO (E315Q) |
| comC g127 c128-F | GGCTGTTTAACAGAAGTTTTACACAANNTTTGGGAAAATAACTCGAGATCTA | Mutagenesis of CSP-21 (A18) |
| comC g127 c128-R | TAGATCTCGAGTTATTTTCCCAAANNTTGTGTAAAACTTCTGTTAAACAGCC | Mutagenesis of CSP-21 (A18) |
| comC c128 t129-F | CTGTTTAACAGAAGTTTTACACAAGNNTTGGGAAAATAACTCGAGATCTATC | Mutagenesis of CSP-21 (A18) |
| comC c128 t129-R | GATAGATCTCGAGTTATTTTCCCAANNCTTGTGTAAAACTTCTGTTAAACAG | Mutagenesis of CSP-21 (A18) |
| comC g127a_c128t-F | GGCTGTTTAACAGAAGTTTTACACAAATTTTGGGAAAATAACTCGAGATCTA | Mutagenesis of CSP-21 (A18) |
| comC g127a_c128t-R | TAGATCTCGAGTTATTTTCCCAAAATTTGTGTAAAACTTCTGTTAAACAGCC | Mutagenesis of CSP-21 (A18) |
| comC g127t_c128g_t129g-F | TTTTTCCGGCTGTTTAACAGAAGTTTTACACAATGGTTGGGAAAATAACTCGAGATC | Mutagenesis of CSP-21 (A18) |
| comC g127t_c128g_t129g-R | GATCTCGAGTTATTTTCCCAACCATTGTGTAAAACTTCTGTTAAACAGCCGGAAAAA | Mutagenesis of CSP-21 (A18) |
| comC g127c_c128a_t129g-F | TTTTTCCGGCTGTTTAACAGAAGTTTTACACAACAGTTGGGAAAATAACTCGAGATC | Mutagenesis of CSP-21 (A18) |
| comC g127c_c128a_t129g-R | GATCTCGAGTTATTTTCCCAACTGTTGTGTAAAACTTCTGTTAAACAGCCGGAAAAA | Mutagenesis of CSP-21 (A18) |
| comC g127a_c128a_t129s-F | TTTTTCCGGCTGTTTAACAGAAGTTTTACACAAAASTTGGGAAAATAACTCGAGATC | Mutagenesis of CSP-21 (A18) |
| comC g127a_c128a_t129s-R | GATCTCGAGTTATTTTCCCAASTTTTGTGTAAAACTTCTGTTAAACAGCCGGAAAAA | Mutagenesis of CSP-21 (A18) |
| comC g127a_c128t_t129g-F | TTTTTCCGGCTGTTTAACAGAAGTTTTACACAAATGTTGGGAAAATAACTCGAGATC | Mutagenesis of CSP-21 (A18) |
| comC g127a_c128t_t129g-R | GATCTCGAGTTATTTTCCCAACATTTGTGTAAAACTTCTGTTAAACAGCCGGAAAAA | Mutagenesis of CSP-21 (A18) |
| comC t211g_t212c-F | GGCTGTTTAACAGAAGTTTTACACAAGCTGCGGGAAAATAACTCGAG | Mutagenesis of CSP-21 L19A |
| comC t211g_t212c-R | CTCGAGTTATTTTCCCGCAGCTTGTGTAAAACTTCTGTTAAACAGCC | Mutagenesis of CSP-21 L19A |
| comC t211g-F | GGCTGTTTAACAGAAGTTTTACACAAGCTGTGGGAAAATAACTCG | Mutagenesis of CSP-21 L19V |
| comC t211g-R | CGAGTTATTTTCCCACAGCTTGTGTAAAACTTCTGTTAAACAGCC | Mutagenesis of CSP-21 L19V |
| comC t211a_g213c-F | CGGCTGTTTAACAGAAGTTTTACACAAGCTATCGGAAAATAACTCGAGAT | Mutagenesis of CSP-21 L19I |
| comC t211a_g213c-R | ATCTCGAGTTATTTTCCGATAGCTTGTGTAAAACTTCTGTTAAACAGCCG | Mutagenesis of CSP-21 L19I |
| comC 5′-F | GTCTTTTTTACATATTTTATTCCCC | Deletion of comC |
| comC 3′-R | TTGATTCCAATGAATATTACTATGC | Deletion of comC |
| KpnI pnlmA-gusA-F | GGGGTACCTACAAATATGGCAATCGAAGTTTTGG | Amplification of PnlmA-gusA |
| PstI pnlmA-gusA-R | AACTGCAGTTGTTTGCCTCCCTGCTGCGGTTTT | Amplification of PnlmA-gusA |
| lox77-Kan-F | CGTACCGTTCGTATAGCATACATTATACGAAGTTATGAGGATGAAGAGGATGAGGAGGCAG | Amplification of lox-Km cassette |
| lox66-Kan-R | CGTACCGTTCGTATAATGTATGCTATACGAAGTTATGCTTTTTAGACATCTAAATCTAGG | Amplification of lox-Km cassette |
SGO, S. gordonii SepM.
Site-directed mutagenesis of SepM and Streptococcus gordonii SepM.
pIB-N1 and pIB-N5 were used as the template to mutate various residues. Site-directed mutagenesis was performed using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) with the mutagenic primers that encode the following replacements: Asp141Asn and Glu315Gln in pIB-N1 and Asn141Asp, Gln315Glu, and Gln315Asp in pIB-N5 (Table 2). The mutations were confirmed by DNA sequencing.
Site-directed mutagenesis of CSP-21.
Plasmid pIB-D58, which contains the entire comC open reading frame (8), was used as the template to mutate the alanine that corresponds to position 18 of the matured peptide (A18). Site-directed mutagenesis was performed using the QuikChange II XL site-directed kit with the various mutagenic primers to cover all 19 substitutions. Similarly, plasmid pIB-D58 was used as the template to mutate leucine at position 19 (Leu19). Site-directed mutagenesis was performed with the mutagenic primers that encode the Leu19Ala, Leu19Val, and Leu19Ile substitutions. All the mutations were confirmed by sequencing.
Construction of PnlmA-gus reporter plasmid.
Plasmid pGhost9-TR is a thermoresistant derivative of the pGhost-9 plasmid containing pWV01 (a broad-host-range replicon) (22) and was used to subclone the PnlmA-gusA reporter construct. A fragment carrying the PnlmA-gusA construct was amplified from plasmid pIB-D21 using the primers KpnI-PnlmA-gusA-F and PstI-PnlmA-gusA-R. The amplified fragment was digested with KpnI and PstI and cloned into KpnI–PstI-digested pGhost9-TR to generate pIB-N35. PCR amplification and restriction digestion were used to verify the reporter construct.
Construction of clean deletion mutants.
A clean markerless deletion of comC was achieved by using the Cre-loxP methodology (23). Briefly, a kanamycin resistance cassette with modified loxP sites (lox71-Km-lox66) was amplified from pIB184-Km with the primers listed in Table 2. A DNA fragment corresponding to sequences upstream and downstream of the comC gene was also amplified with the primers listed in Table 2 and inserted into pGEM-T-EZ. An inverse PCR was performed to remove an ∼180-bp comC coding region, and the product was ligated with the lox71-Km-lox66 cassette. The resultant construct was named pIBN36 and verified by sequencing. Plasmid pIBN36 was linearized with NotI restriction digestion and transformed into S. mutans UA159, and Km-resistant colonies were selected. A few representative clones were selected. To remove the lox71-Km-lox66 cassette, pCrePA, a thermosensitive plasmid that expresses Cre recombinase, was transformed into the selected clones. The chromosomally integrated lox71-Km-lox66 cassette was excised by Cre at 30°C. The pCrePA plasmid was cured from the resultant clones at 37°C. One such clone was renamed S. mutans IBS-N42. Similarly, the comC gene was also deleted from S. mutans IBS-D29, a ΔsepM derivative of S. mutans UA159 (8), to generate S. mutans IBS-N43, which lacks both comC and sepM (ΔcomC ΔsepM).
β-Glucuronidase assay.
Two different assays were performed. In the first assay, overnight cultures were diluted 1:20 and were incubated at 37°C until the optical density at 595 nm (OD595) reached 0.8. Three milliliters of culture was harvested, washed in saline, and resuspended in 500 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 20 mM dithiothreitol). Cells were homogenized by bead beating, 200 μl of the cell lysate was transferred to a new tube, and 100 μl of p-nitrophenyl-β-d-glucoside (PNPG) (at 4 mg/ml in Z buffer) was added. The mixture was incubated at 37°C, and when yellow color developed, the reaction was stopped by the addition of 200 μl of 1 M Na2CO3. The absorbance at 420 nm and the time period of the reaction were noted. β-Glucuronidase (Gus) activity was expressed as (1,000 × A420)/(time in minutes × OD595) in Miller units.
In the second assay, reporter strains were grown overnight at 37°C, diluted 1:20 in THY broth, and incubated at 37°C until the growth reached an OD595 of 0.2. Synthetic CSPs were then added (various concentrations), and the cultures were incubated until the OD595 reached 0.8. One milliliter of culture was harvested, washed in saline, resuspended in 760 μl Z buffer plus 40 μl of freshly prepared lysozyme (10 mg/ml), and incubated at 37°C for 10 min. Eight microliters of Triton X (10%) was added, and the reaction mixture was incubated for an additional 5 min. Two hundred microliters of PNPG (4 mg/ml) was added, and the mixture was incubated at 37°C until it turned yellow. The reaction was stopped with 400 μl of 1 M Na2CO3, and the mixture was spun down. The absorbance of the supernatant at 420 nm and the time period of the reaction were noted. Gus activity was defined as described above.
Mass spectrometry analysis.
Treated CSPs were desalted on a reverse-phase C4 column with a buffer containing acetonitrile and formic acid, and electrospray ionization (ESI) spectra were acquired on a Synapt G2 hybrid quadrupole-ion mobility-time of flight (TOF) mass spectrometer (Waters Corp., USA). Data were acquired at a 9,091-Hz pusher frequency covering the range from 100 to 3,000 mass units and accumulating data for 1.5 s per cycle. The time to mass calibration was made with NaI cluster ions acquired under the same conditions. Mass spectra of [Glu1]-fibrinopeptide B were acquired in parallel scans, and doubly charged ions at m/z 785.8426 were used as a lock mass reference. The MassLynx (4.1) software was used to acquire and process data.
RESULTS
SepM can efficiently process CSP.
In our previous study, we demonstrated that the cleavage of CSP-21 by SepM is absolutely necessary for the activation of the ComDE pathway, since CSP-18 is the active signaling peptide (8). We wanted to know whether SepM is able to process CSPs of various lengths in a comparable manner for the signaling to initiate. We also wanted to find the minimum concentration of CSP to generate maximum activation, regardless of the length. For this, we used IBS-N42 (ΔcomC), which contains the reporter plasmid pIB-N35 (IBS-N42/pIBN35). We added various concentrations of synthetic CSP-21 or CSP-18 and measured the activation of the PnlmA promoter, which is under direct regulation by ComE, by assaying Gus activity. As shown in Fig. 1, we observed that a peptide concentration of as little as 10 nM could induce the PnlmA promoter; however, the maximum induction is achieved at around 400 nM. The addition of a concentration of CSPs of ≥800 nM did not increase the expression from the PnlmA promoter (Fig. 1 and data not shown). Interestingly, we did not observe any significant (not >2-fold) difference in promoter activation between the CSP-21 and CSP-18 peptides. Since some naturally occurring S. mutans strains encode a mature CSP that is 23 residues long with additional Ile-Arg amino acids, we also included this CSP-23 variant in our assay. This CSP-23 peptide behaved similarly to the CSP-21 and CSP-18 peptides, indicating that SepM can successfully process CSP-23 to CSP-18, and there is no significant difference in promoter activation among the three peptides at a concentration of 400 nM. However, we did not check whether there is a difference in the rates of activation by these CSPs.
FIG 1.
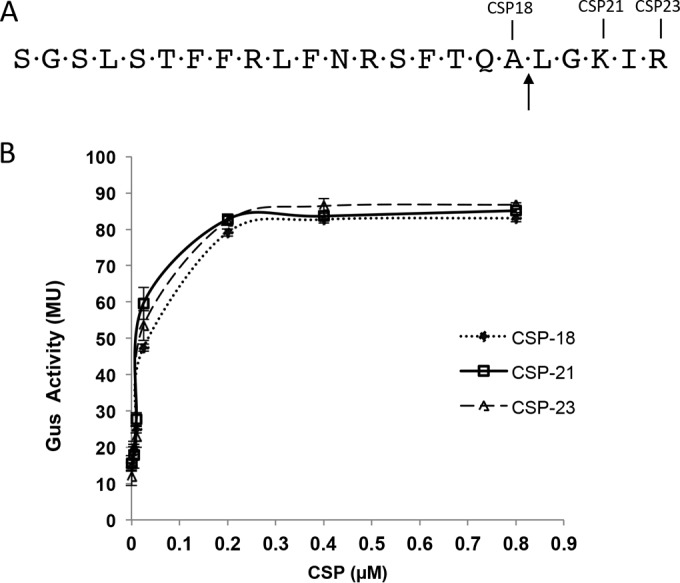
Activation of the nlmA promoter by various synthetic CSPs. (A) Primary amino acid sequence of naturally occurring matured CSP molecules of various lengths. The arrow marks the SepM processing site. (B) Induction of the PnlmA-gusA reporter fusion. An assay of the β-glucuronidase activity of the ΔcomC/pIBN35 reporter strain in the presence of various concentrations of CSPs was conducted. The assays were done at mid-exponential phase, as described in the text. The experiments were performed at least three times, and the means with standard deviations are plotted.
Substrate specificity for SepM.
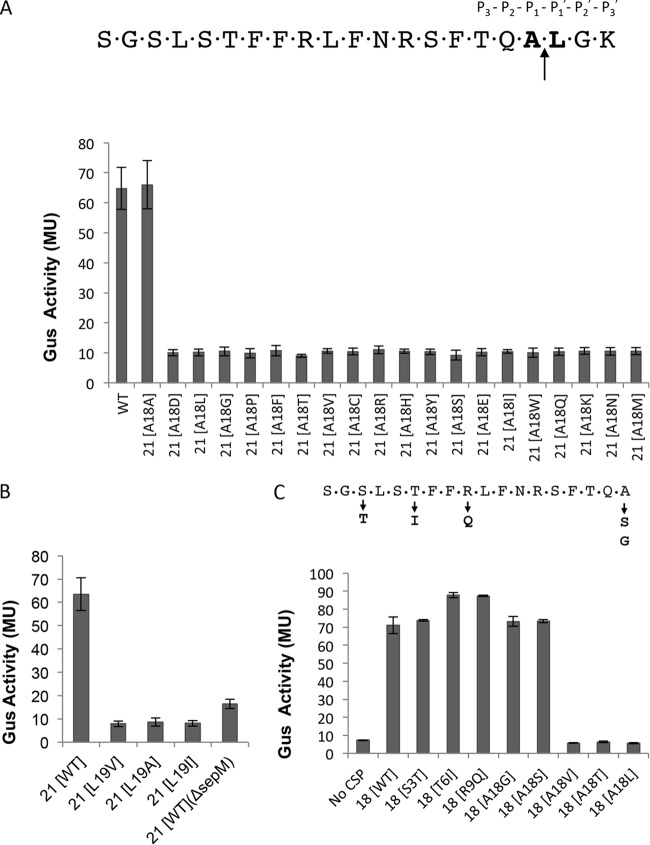
Since SepM appears to function as an endopeptidase and cleaves the matured CSP after position 18, we wanted to study the requirement for amino acid residues at position 18 to be cleaved by SepM. For this, we used the pIBD58 plasmid, which contains the comC gene under the control of the constitutive P23 promoter (8). We first targeted the Ala residue at position 18 (Ala18), which was replaced with all possible 20 amino acids, including Ala, by site-directed mutagenesis. These plasmids were then introduced into the IBS-N42/pIBN35 strain, and Gus activity was measured to assay the transcription from the PnlmA promoter. As shown in Fig. 2A, the replacement of the Ala residue at position 18 with any other residue completely abolished the capacity of CSP to function as a signaling peptide, suggesting that SepM is unable either to recognize and bind the mutated peptides or to cleave the mutated peptides. In either case, SepM appears to display a strict requirement for the Ala residue at position 18.
FIG 2.
SepM-mediated CSP-21 cleavage is highly site specific. The CSP sequence is shown with the conserved residues required for the generation of CSP-18. (A) Requirement for alanine at the 18th position. The 18th residue corresponding to the matured CSP was mutated to all possible 19 amino acids (the 18th and 19th residues are in bold). The constructs containing mutations were introduced into the ΔcomC strain (IBS-N42) carrying the reporter plasmid pIBN35 to produce endogenous CSP. β-Glucuronidase activity was measured at mid-exponential phase. (B) Requirement for leucine at the 19th position. The 19th residue corresponding to the matured CSP was mutated to alanine (L19A), valine (L19V), or isoleucine (L19I). The mutated constructs were introduced into the ΔcomC/pIBN35 strain to produce endogenous CSP, and β-glucuronidase activity was measured at mid-exponential phase. Wild-type (WT) CSP-21 served as a positive control, whereas the ΔsepM strain served as a negative control. (C) Verification of ComD activation by synthetic CSP-18 variants. Various synthetic CSP-18 peptides with substitutions at the 18th position were added to exponentially growing cultures of ΔsepM ΔcomC (IBS-N43)/pIBN35 cells, and β-glucuronidase activity was measured. Single amino acid substitutions shown below the CSP-18 sequence did not interfere with the activity. All experiments were repeated three times, and the means and standard deviations (error bars) are shown.
After knowing the absolute requirement for Ala at position 18, we then focused on position 19, which is occupied by a Leu residue. We replaced the Leu residue with Ala, Ile, or Val by site-directed mutagenesis. As shown in Fig. 2B, conservative substitutions of Leu at position 19 also prevented the processing of CSP by SepM.
Some earlier reports indicate that the Ala18 residue is also necessary for the peptide to be recognized by the ComD sensor kinase for signaling purposes. To confirm that SepM did not generate a CSP-18 peptide that is unable to function as a signaling molecule, we synthesized various CSP-18 derivatives. We also included two naturally occurring CSP-18 derivatives in which the Ser at position 3 was replaced with Thr (but retains Ala18; CSP-18 [S3T]) and Thr at position 6 is replaced with Ile but retains Ala18 (T6I). We added the synthetic peptides to the IBS-N42/pIBN35 strain and measured Gus activity. As shown in Fig. 2C, CSP-18 peptides in which the Ala residue was replaced with a Gly or Ser residue were active as signaling peptides, whereas those with a substitution of Leu, Val, or Thr did not function as signaling molecules. The CSP-18 S3T and T6I peptides were also able to induce ComDE pathway activity.
To confirm our in vivo results, we used mass spectrometry analysis of the synthetic peptides that correspond to Ala18 to Leu (CSP-21 LL), Asn (CSP-21 A18N), Thr (CSP-21 A18T), or Gly (CSP-21 A18G), as well as the Leu19-to-Ala (CSP-21 AA) substitutions. The synthetic peptides were treated with purified SepM protein (or wild-type SepM-containing bacteria) and then subjected to mass spectrometry analysis. We found that while SepM was able to cleave the original CSP-21, SepM could not process any of the CSP-21 variants that contain substitutions at position 18 or 19 (Fig. 3 and data not shown). Taken together, our results suggest that SepM displays strict substrate specificity and requires specific residues at both the 18th and the 19th position.
FIG 3.
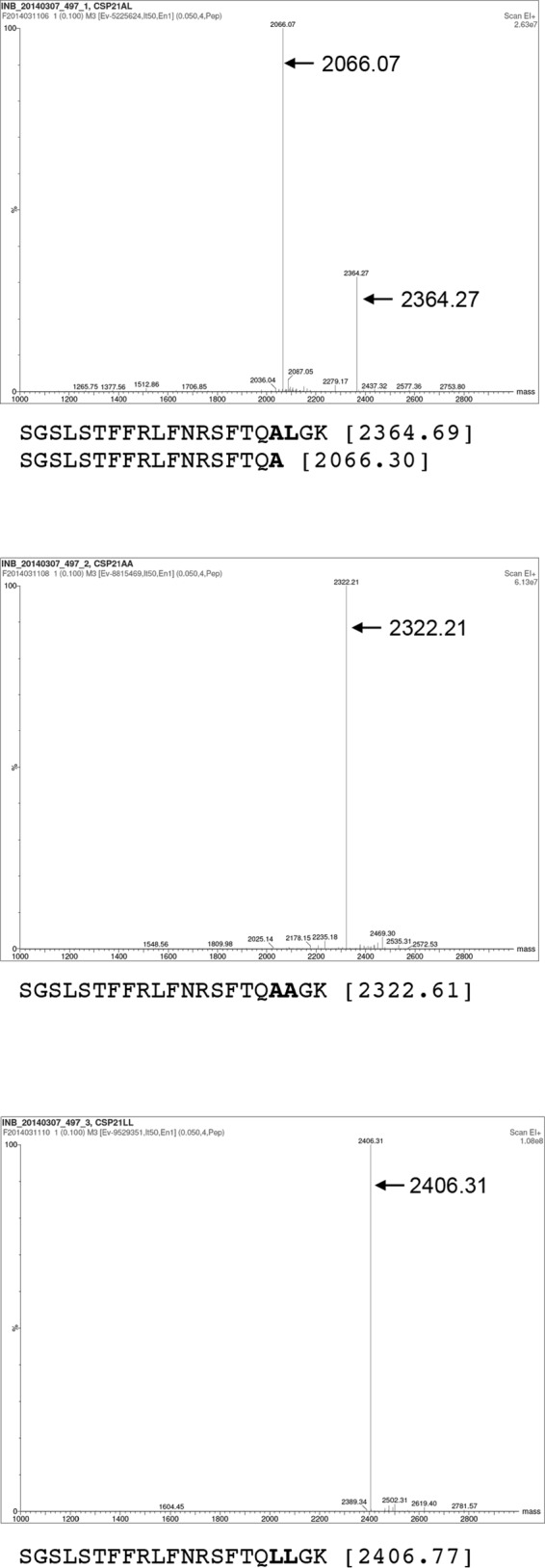
Mass spectrometric analysis of processed CSP. CSP samples were treated with purified SepM. After removal of the protein by centrifugation, the samples were desalted on a reverse-phase C4 column and then subjected to matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) ESI mass spectrometry analysis. Cleavage occurs between the residues shown in bold (top panel), which were mutated as indicated (middle and bottom panels). The theoretical peptide masses are shown next to the sequences in parentheses.
SepM can efficiently process most of the CSP variants that retain the cleavage site.
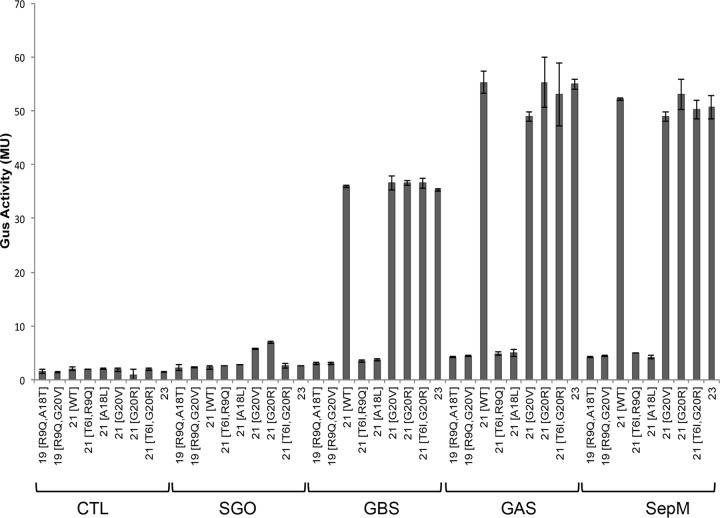
More than 200 S. mutans genome sequences are available in the public database, and an analysis of the comC sequence suggests that the sequences of the matured CSP-21 peptides vary in as many as 15% of the isolates (14–16). Primarily, the variations occur at positions 3, 6, 9, and 20 (corresponding to positions in the matured CSP-21 peptide). A search of the GenBank database returned a few variants that also lack the first 2 residues of the matured peptide (CSP-19*; strains SM24 and TCI-280). To study whether these naturally occurring CSP variants could be processed by SepM and recognized by ComD, we synthesized various CSP variants (Fig. 4). We added these peptides to the IBS-N42/pIB-N35 strain and measured Gus to determine the degree of PnlmA activation. We observed that the replacement of the Gly residue at the 20th position with Arg or Val did not interfere with SepM processing (G20R and G20V) (Fig. 4). SepM also processed CSP-21 containing the following substitutions: Ser to Thr at the 3rd position (S3T), Thr to Ile at the 6th position (T6I), and Arg to Gln at the 9th position (R9Q). Since these CSP variants induced PnlmA expression, the results also suggest that these peptides were recognized by the ComD sensor kinase.
FIG 4.
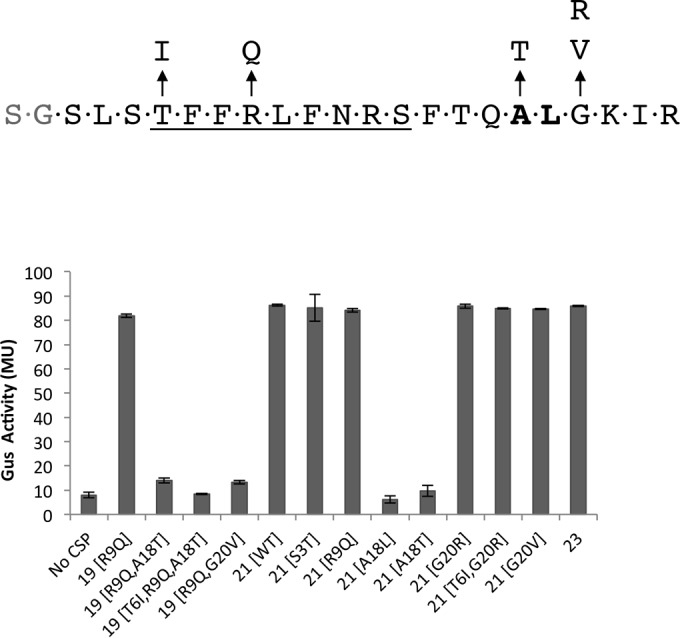
Processing of various synthetic CSPs by SepM. Naturally occurring full-length CSPs were synthesized and tested as SepM substrates. CSP-21 (G20V) and CSP-21 (A19L) served as positive and negative controls, respectively. The assays were done using the ΔcomC/pIBN35 reporter strain, as described in the legend to Fig. 1. A concentration of 400 nM CSP was used in each assay. The underlined sequence is known for specific binding to S. mutans.
As shown above, SepM can successfully process the naturally occurring CSP-23 that contains two additional residues at the C terminus. We wondered whether SepM could process a CSP that is even longer than CSP-23. For this, we synthesized a 28-residue-long CSP that carries 5 additional amino acids. The amino acid sequence was selected from an internal part of the matured CSP-21 sequence and does not end with a positively charged residue (Fig. 5). This CSP-28 peptide was added to the IBS-N42/pIB-N35 strain, and Gus activity was measured. As shown in Fig. 5, SepM also successfully processed this CSP-28 to generate a functional CSP-18 molecule. We further exposed the CSP-28 peptide to SepM, followed by mass spectrometry analysis, which confirmed the peptide cleavage after Ala18 (data not shown). Taken together, our results indicate that SepM can accommodate even longer peptides for processing.
FIG 5.
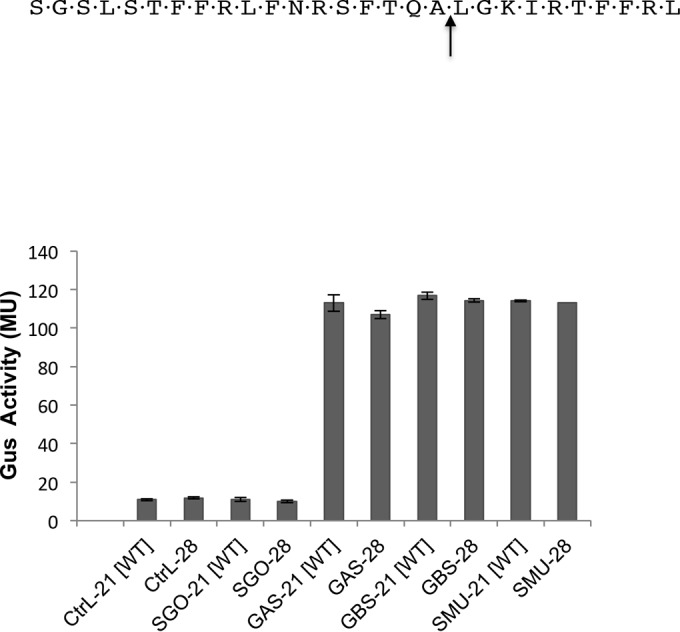
Processing of the CSP-28 peptide by SepM and its homologs. The CSP-28 peptide was synthesized and subjected to SepM-mediated processing. The assays were done using in vivo complementation of the ΔsepM ΔcomC/pIBN35 strain with SepM homologs from various streptococci. A concentration of 400 nM CSP was used in each assay. All experiments were repeated three times; the means and standard deviations (error bars) are shown. CtrL, control (vector only); GAS, S. pyogenes SepM; GBS, S. agalactiae SepM; SGO, S. gordonii SepM.
Substrate specificity for SepM homologs.
As mentioned earlier, SepM is a highly conserved protein, and its homologs are universally present in all streptococci. However, many of these streptococci neither produce bacteriocin nor are naturally competent. Furthermore, CSP-21 is uniquely produced by S. mutans. Since the sequence conservation among the various SepM homologs is very high (Fig. 6), we were interested to know whether SepM homologs from other streptococci could cleave CSP. For this, we selected Streptococcus pyogenes (group A streptococcus [GAS]), Streptococcus agalactiae (group B streptococcus [GBS]), and Streptococcus gordonii, an oral streptococcus. The sepM genes from these three organisms were cloned into a complementing vector, pIB184, and the resultant plasmids were introduced into the IBS-N43 (ΔcomC ΔsepM) strain carrying the pIB-N35 reporter plasmid. Various CSP variants were added to these strains, and the activity of Gus was measured. As shown in Fig. 5, we found that the SepM homologs from S. pyogenes and S. agalactiae displayed similar patterns of recognition and cleavage of CSP-21 (Fig. 7). Furthermore, we found that these SepM homologs could also process CSP-28 to generate CSP-18 (Fig. 5). We then tested several naturally occurring CSP variants that were successfully processed by S. mutans SepM. We found that the S. agalactiae and S. pyogenes SepM homologs display similar cleavage activities (Fig. 7). However, the activity of the S. agalactiae homolog was slightly lower than those of the S. pyogenes and S. mutans SepM homologs (Fig. 7). On the other hand, despite high sequence conservation, the SepM homolog from S. gordonii failed to process CSP-21 and its derivatives. We also found that the SepM homolog from Streptococcus uberis failed to process CSP-21 (data not shown). Taken together, our results indicate that SepM homologs from some streptococci are able to process CSP-21, despite the fact that these organisms do not encode CSP and lack the ComDE pathway.
FIG 6.
Multiple-sequence alignment of SepM and its homologs. The alignment was performed using Clustal W. The degrees of shading were done using BoxShade, in which black and gray blocks indicate identical and similar amino acids, respectively. Sequences were obtained from GenBank. The organisms are S. mutans (SMU [SepM]; GenBank accession no. NP_720956), S. pyogenes (GAS; GenBank accession no. WP_010922499.1), S. agalactiae (GBS; GenBank accession no. WP_000790649.1), S. gordonii (SGO; GenBank accession no. ABV09441.1), and S. uberis (SUB; GenBank accession no. WP_015911639.1). Asterisks indicates the residues that were targeted for mutagenesis.
FIG 7.
SepM homologs are capable of processing various CSP derivatives. In vivo complementation of the ΔsepM ΔcomC strain carrying pIBN35 with SepM homologs from various streptococci is shown. β-Glucuronidase activity was measured at mid-exponential phase after the addition of 200 nM CSPs. The data shown are the means and standard deviations derived from at least three independent experiments. CTL, control (vector only); GAS, S. pyogenes SepM; GBS, S. agalactiae SepM; SGO, S. gordonii SepM.
Although all the SepM homologs carry the conserved S16 peptidase domain with the conserved Ser-Lys dyad, the homologs from S. gordonii and S. uberis failed to cleave CSP-21. In an attempt to identify the residues that are critical for recognition of the peptide, we analyzed the sequences and identified a few conserved residues that are present in S. agalactiae, S. mutans, and S. pyogenes but are absent in S. gordonii and S. uberis, and vice versa. Specifically, we targeted two residues, Asn141 and Gln315, which are present in S. mutans SepM. We mutated Asn141 to Asp141 and Gln315 to Asp315 and Glu315. Similarly, we also introduced two substitutions in S. gordonii SepM, Asp141Asn and Glu315Gln. We tested the four mutants for their ability to process CSP-21. We found that both SepM mutants of S. mutans retained the cleavage activity, while both SepM mutants of S. gordonii did not cleave the CSP-21 (data not shown). Thus, these two residues alone do not contribute to substrate specificity.
DISCUSSION
Both Gram-negative and Gram-positive bacteria express a variety of endopeptidases, many of which are essential for cell viability. Some of these endopeptidases display strict substrate specificity. For example, SPase I, a signal peptidase responsible for the generation of matured polypeptides that are secreted through the general secretion (Sec) pathway, recognizes a conserved Ala-Xaa-Ala motif (where Xaa indicates any amino acid) present at the N-terminal region and cleaves the substrates after the second Ala residue (24). Similarly, C39 domain-containing peptidases that are responsible for the secretion of small peptides by ABC transporters generally recognize a Gly-Gly motif present at the N-terminal signal sequence (25, 26). Often, C39 peptidase domains are borne within the same ABC transporter peptide. NlmT/E of S. mutans is one such ABC transporter that recognizes and cleaves after the Gly-Gly motif of the CSP prepeptide to generate the matured CSP-21. NlmT/E also processes several other bacteriocin prepeptides containing the Gly-Gly motif (7). In this study, we characterized yet another endopeptidase, SepM, which displays strict sequence specificity. However, unlike the majority of the endopeptidases, which are cytoplasmic, SepM is associated with the surface, exposing the catalytic domain outward and cleaving CSP-21 once it is secreted outside.
SepM belongs to the S16 protease family that encodes a conserved catalytic Ser-Lys dyad, and the members of this family display diverse substrate specificities (27). We found that SepM has much stricter substrate specificity than the other members of the family. Based on the standard notation of peptide cleavage activity, we found that at the P1 position, SepM can accommodate an Ala residue only. The replacement of Ala with any other residues, including Gly or Val, abolished the cleavage activity. Similarly, at the P′1 position, SepM requires a Leu residue; the replacement of this residue with Ala, Ile, or Val also prevented CSP-21 processing. It was surprising to see that even the constitutional isomer, the Ile residue, could not serve as a target for SepM cleavage. Although Leu and Ile are isomers and have the same residue volume (166.7 Å3), there are some subtle differences between these two residues. The accessible surface areas of these residues when present in a peptide are slightly smaller for Leu than for Ile (175 versus 170 Å2) (28), and although both residues are hydrophobic, the hydropathy index is also lower in Leu than in Ile (3.8 versus 4.5) (29). As far as the P′2 position is concerned, we found that the Gly (small nonpolar) residue could be successfully replaced with a Val (large nonpolar) or Arg (large charged polar) residue. Although we have not tested other replacements, since SepM recognized two drastic changes at the P′2 position, we believe that SepM will be able to recognize and cleave other peptides with a different P′2 residue. At the P2 position, which is occupied by a Gln residue, we did not test any replacements, since all the naturally occurring CSPs contain Gln at this position. However, we speculate that this P2 position is also dispensable, and as long as the P1-P′1 residues are conserved, SepM will be able to cleave the peptide that it recognizes. As far as the length of the substrate peptide is concerned, it seems that SepM is flexible. For example, we found that SepM successfully cleaved CSPs that lacked 2 residues at the N terminus (CSP-19* [data not shown]). Similarly, SepM recognized at least 10 residues after the cleavage site at the C-terminal end (Fig. 5). Although we have not tested fewer than 3 residues at the C-terminal end, a recent study by Syvitski and colleagues (30) demonstrated that synthetic CSP molecules lacking the last two C-terminal residues (Gly-Lys) are functional. These authors also showed that synthetic CSPs lacking as many as 4 residues at the N terminus are also functional. Therefore, we speculate that SepM will be able to process peptide substrates with various lengths. At present, it is not clear how SepM recognizes its substrates. Since there are other peptides in the extracellular environment in which S. mutans colonizes, there must be a mechanism by which SepM can efficiently and quickly process CSP. One possibility is the physical proximity of SepM to the NlmT/E transporter, which secretes the matured CSP-21 into the external milieu. NlmT/E also secretes various bacteriocins, and the production of these bacteriocins is positively regulated by the CSP-mediated ComD two-component pathway. By coordinating secretion with the processing, S. mutans can control bacteriocin production. When bacteriocins are produced in excess, these peptides can nonspecifically bind to SepM and inhibit its CSP-21 processing activity, thus preventing further bacteriocin production.
The three-dimensional structures of the mature CSP-21 and a CSP-18 variant (called TPC3) have been determined by nuclear magnetic resonance (NMR) and circular dichroism (CD) spectroscopies (30, 31). Both CSP-21 and CSP-18 (S3T) adopt amphipathic α-helices with a well-defined hydrophobic face with four hydrophobic Phe residues (Phe-7, -8, -11, and -15) and hydrophilic residues on the other side. About 40% of the helix surface is a hydrophobic face, and the rest is composed of hydrophilic and charged residues. The structures also suggest that CSP-21 displays more rotational freedom at the C terminus than CSP-18 (TPC3). We believe that the relatively unstructured C-terminal region of CSP-21 is necessary for proper recognition and cleavage by SepM. A recent study by Eckert and colleagues (32) showed that when an antimicrobial peptide called G2 is conjugated with an 8-residue sequence (M8, TFFRLFNR) encoded within primary CSP-21, the conjugated peptide (M8G2) is specifically targeted to S. mutans. The M8 peptide can successfully distinguish S. mutans from other oral streptococci and selectively bind to S. mutans. This species-specific binding is not dependent on the ComD sensor kinase, since the M8 peptide also binds to comD mutant strains. We speculate that SepM is the primary target molecule with which the M8G2 peptide interacts. Further experiments are required to determine the minimum sequence required for the interaction between SepM and CSP.
Another interesting finding that emerged from our study is the sequence for ComD activation. We found that several CSP derivatives successfully induced PnlmA expression, suggesting that these peptides can act as QS molecules and are recognized by ComD. For example, we found that replacement of Ser3 with Thr (S3T) does not interfere with the QS signaling activity. This S3T variant is a naturally occurring mutation found in many clinical isolates (14, 16), and Allan and colleagues (14) have also shown that this S3T variant, called CSP-3, does not impair the peptide's function (16). In contrast, Syvitski and colleagues (30) have shown that the TCP3, which is similar to S3T, was inactive. The reason for the apparent discrepancy is currently unknown. We also found that several other peptide variants functioned equally well in our assay. Two such CSP-18 variants, in which the Ala18 residues were replaced with Gly or Ser, were active in our assay (Fig. 2). This observation was somewhat surprising, since SepM did not tolerate any substitutions at the Ala18 position. The other notable CSP variants that induced PnlmA expression are the Thr6-to-Ile (T6I) and Arg9-to-Gln (R9Q) substitutions. Interestingly, while the single-substitution peptides were functional as signaling molecules, the double-substitution (T6I R9Q) peptide was nonfunctional. Since both of these residues map within the M8 peptide, which is responsible for species-specific binding, we speculate that SepM did not recognize the peptide properly and therefore failed to cleave. Alternatively, SepM processed the T6I R9Q peptide well; however, ComD did not recognize the peptide, probably due to structural constraints.
It was surprising to find that several SepM homologs from other streptococci could process the CSP-21. Notably, SepM homologs from S. agalactiae, Streptococcus pneumoniae, S. pyogenes, and Streptococcus ratti all cleaved CSP-21 to produce an active CSP-18 molecule (Fig. 5 and data not shown). On the other hand, two SepM homologs from S. gordonii and S. uberis failed to process CSP-21. This was also surprising since these two homologs share >80% identity with SepM residues. Furthermore, we found that homologs from S. agalactiae and S. pyogenes retain the same substrate specificity as that of SepM (Fig. 3). Currently, it is not clear why some SepM homologs display CSP-21 cleavage activity while others do not. We speculate that the reason homologs failed to cleave CSP-21 might be either a lack of recognition of the peptide or a lack of protease activity, despite the fact that the homologs contain the conserved Ser-Lys catalytic dyad. In an attempt to identify the residues that might confer substrate specificity, we removed two residues that are well conserved among the homologs that cleave CSP-21 and replaced them with the corresponding residues from the noncleaving homologs. This approach, i.e., targeting an individual residue, was not successful, since we neither abolished SepM activity nor made the SepM homolog from S. gordonii active. We are now constructing various chimeric SepM proteins by combining the sequences from S. mutans and S. gordonii to locate the substrate recognition domain.
S. pyogenes and S. agalactiae lack the CSP-21 peptide but encode several small peptides, some of which are involved in quorum sensing. For example, some, but not all, strains of S. pyogenes encode SilC, a 39-residue-long peptide. However, SilC does not contain the Ala-Leu sequence and therefore might not be a substrate for the SepM homolog. The S. pyogenes SepM homolog is encoded by the Spy1536 gene, and a recent study suggests that Spy1536 is required for surface expression of two important virulence factors, the M protein and Spy0269 (33). Thus, it appears that SepM homologs might have different functions in different streptococci. It might be that in S. mutans, the processing of CSP-21 may not be the primary function of SepM; rather, it might be required for proper surface localization of some unknown proteins. It is noteworthy to mention that SepM also contains a PDZ domain, which is involved in protein-protein interaction. Further studies are necessary to unravel the true function of SepM and its homolog in other streptococci.
It is important to mention that SepM is not the only member of the S16 protease family that is encoded by S. mutans. A quick search in the MEROPS database (release 9.13 [34]) returned another protein, SMU.327, which also contains a Lon-like domain and an ATPase domain. This protein is annotated as a RadA-like protein, a protein involved in DNA repair in bacteria and archaea (35, 36). Nothing is known about the function of SMU.327 or its homolog in streptococcal biology. Surprisingly, our search also reveals that the numbers of S16 family proteins encoded by a given Streptococcus strain vary significantly. While most of the streptococci encode only 1 or 2 S16 family members, S. pneumoniae and Streptococcus sanguinis contain as many as 12 to 15 members. Currently, the significance and the function of these proteases are largely unknown.
ACKNOWLEDGMENTS
We thank Nadya Galeva (Mass Spectrometry Laboratory, University of Kansas) for the mass spectrometry analysis of the peptide samples.
This work was supported in part by an NIDCR grant awarded to I.B. (DE021664).
REFERENCES
- 1.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 2.Rutherford ST, Bassler BL. 2012. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2:a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suntharalingam P, Cvitkovitch DG. 2005. Quorum sensing in streptococcal biofilm formation. Trends Microbiol 13:3–6. doi: 10.1016/j.tim.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Senadheera D, Cvitkovitch DG. 2008. Quorum sensing and biofilm formation by Streptococcus mutans. Adv Exp Med Biol 631:178–188. doi: 10.1007/978-0-387-78885-2_12. [DOI] [PubMed] [Google Scholar]
- 5.Cook LC, Federle MJ. 2014. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev 38:473–492. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li YH, Lau PC, Tang N, Svensäter G, Ellen RP, Cvitkovitch DG. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J Bacteriol 184:6333–6342. doi: 10.1128/JB.184.22.6333-6342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hale JD, Heng NC, Jack RW, Tagg JR. 2005. Identification of nlmTE, the locus encoding the ABC transport system required for export of nonlantibiotic mutacins in Streptococcus mutans. J Bacteriol 187:5036–5039. doi: 10.1128/JB.187.14.5036-5039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hossain MS, Biswas I. 2012. An extracellular protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J Bacteriol 194:5886–5896. doi: 10.1128/JB.01381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemme A, Gröbe L, Reck M, Tomasch J, Wagner-Döbler I. 2011. Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J Bacteriol 193:1863–1877. doi: 10.1128/JB.01363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung DC, Downey JS, Ayala EA, Kreth J, Mair R, Senadheera DB, Qi F, Cvitkovitch DG, Shi W, Goodman SD. 2011. Characterization of DNA binding sites of the ComE response regulator from Streptococcus mutans. J Bacteriol 193:3642–3652. doi: 10.1128/JB.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kreth J, Merritt J, Shi W, Qi F. 2005. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol Microbiol 57:392–404. doi: 10.1111/j.1365-2958.2005.04695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Ploeg JR. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J Bacteriol 187:3980–3989. doi: 10.1128/JB.187.12.3980-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen FC, Fimland G, Scheie AA. 2006. Purification and functional studies of a potent modified quorum-sensing peptide and a two-peptide bacteriocin in Streptococcus mutans. Mol Microbiol 61:1322–1334. doi: 10.1111/j.1365-2958.2006.05312.x. [DOI] [PubMed] [Google Scholar]
- 14.Allan E, Hussain HA, Crawford KR, Miah S, Ascott ZK, Khwaja MH, Hosie AH. 2007. Genetic variation in comC, the gene encoding competence-stimulating peptide (CSP) in Streptococcus mutans. FEMS Microbiol Lett 268:47–51. doi: 10.1111/j.1574-6968.2006.00593.x. [DOI] [PubMed] [Google Scholar]
- 15.Cornejo OE, Lefebure T, Bitar PD, Lang P, Richards VP, Eilertson K, Do T, Beighton D, Zeng L, Ahn SJ, Burne RA, Siepel A, Bustamante CD, Stanhope MJ. 2013. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol Biol Evol 30:881–893. doi: 10.1093/molbev/mss278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song L, Wang W, Conrads G, Rheinberg A, Sztajer H, Reck M, Wagner-Döbler I, Zeng AP. 2013. Genetic variability of mutans streptococci revealed by wide whole-genome sequencing. BMC Genomics 14:430. doi: 10.1186/1471-2164-14-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botos I, Melnikov EE, Cherry S, Tropea JE, Khalatova AG, Rasulova F, Dauter Z, Maurizi MR, Rotanova TV, Wlodawer A, Gustchina A. 2004. The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site. J Biol Chem 279:8140–8148. doi: 10.1074/jbc.M312243200. [DOI] [PubMed] [Google Scholar]
- 18.Smith CK, Baker TA, Sauer RT. 1999. Lon and Clp family proteases and chaperones share homologous substrate-recognition domains. Proc Natl Acad Sci U S A 96:6678–6682. doi: 10.1073/pnas.96.12.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gur E, Sauer RT. 2008. Recognition of misfolded proteins by Lon, a AAA(+) protease. Genes Dev 22:2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gur E. 2013. The Lon AAA+ protease. Subcell Biochem 66:35–51. doi: 10.1007/978-94-007-5940-4_2. [DOI] [PubMed] [Google Scholar]
- 21.Hossain MS, Biswas I. 2011. Mutacins from Streptococcus mutans UA159 are active against multiple streptococcal species. Appl Environ Microbiol 77:2428–2434. doi: 10.1128/AEM.02320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. 1992. New thermosensitive plasmid for Gram-positive bacteria. J Bacteriol 174:5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banerjee A, Biswas I. 2008. Markerless multiple-gene-deletion system for Streptococcus mutans. Appl Environ Microbiol 74:2037–2042. doi: 10.1128/AEM.02346-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paetzel M, Karla A, Strynadka NC, Dalbey RE. 2002. Signal peptidases. Chem Rev 102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 25.Gebhard S. 2012. ABC transporters of antimicrobial peptides in Firmicutes bacteria—phylogeny, function and regulation. Mol Microbiol 86:1295–1317. doi: 10.1111/mmi.12078. [DOI] [PubMed] [Google Scholar]
- 26.Havarstein LS, Diep DB, Nes IF. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol 16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 27.Tripathi LP, Sowdhamini R. 2008. Genome-wide survey of prokaryotic serine proteases: analysis of distribution and domain architectures of five serine protease families in prokaryotes. BMC Genomics 9:549. doi: 10.1186/1471-2164-9-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chothia C. 1976. The nature of the accessible and buried surfaces in proteins. J Mol Biol 105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 29.Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Syvitski RT, Tian XL, Sampara K, Salman A, Lee SF, Jakeman DL, Li YH. 2007. Structure-activity analysis of quorum-sensing signaling peptides from Streptococcus mutans. J Bacteriol 189:1441–1450. doi: 10.1128/JB.00832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian X, Syvitski RT, Liu T, Livingstone N, Jakeman DL, Li YH. 2009. A method for structure-activity analysis of quorum-sensing signaling peptides from naturally transformable streptococci. Biol Proced Online 11:207–226. doi: 10.1007/s12575-009-9009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckert R, He J, Yarbrough DK, Qi F, Anderson MH, Shi W. 2006. Targeted killing of Streptococcus mutans by a pheromone-guided “smart” antimicrobial peptide. Antimicrob Agents Chemother 50:3651–3657. doi: 10.1128/AAC.00622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fritzer A, Senn BM, Minh DB, Hanner M, Gelbmann D, Noiges B, Henics T, Schulze K, Guzman CA, Goodacre J, von Gabain A, Nagy E, Meinke AL. 2010. Novel conserved group A streptococcal proteins identified by the antigenome technology as vaccine candidates for a non-M protein-based vaccine. Infect Immun 78:4051–4067. doi: 10.1128/IAI.00295-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawlings ND, Waller M, Barrett AJ, Bateman A. 2014. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seitz EM, Brockman JP, Sandler SJ, Clark AJ, Kowalczykowski SC. 1998. RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes Dev 12:1248–1253. doi: 10.1101/gad.12.9.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burghout P, Bootsma HJ, Kloosterman TG, Bijlsma JJ, de Jongh CE, Kuipers OP, Hermans PW. 2007. Search for genes essential for pneumococcal transformation: the RADA DNA repair protein plays a role in genomic recombination of donor DNA. J Bacteriol 189:6540–6550. doi: 10.1128/JB.00573-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ajdic D, McShan WM, McLaughlin RE, Savić G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glaser P, Rusniok C, Buchrieser C, Chevalier F, Frangeul L, Msadek T, Zouine M, Couvé E, Lalioui L, Poyart C, Trieu-Cuot P, Kunst F. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol Microbiol 45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- 39.Clemans DL, Kolenbrander PE, Debabov DV, Zhang Q, Lunsford RD, Sakone H, Whittaker CJ, Heaton MP, Neuhaus FC. 1999. Insertional inactivation of genes responsible for the d-alanylation of lipoteichoic acid in Streptococcus gordonii DL1 (Challis) affects intrageneric coaggregations. Infect Immun 67:2464–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whatmore AM, Kapur V, Sullivan DJ, Musser JM, Kehoe MA. 1994. Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol Microbiol 14:619–631. doi: 10.1111/j.1365-2958.1994.tb01301.x. [DOI] [PubMed] [Google Scholar]
- 41.Oliver SP, Almeida RA, Gillespie BE, Headrick SJ, Dowlen HH, Johnson DL, Lamar KC, Chester ST, Moseley WM. 2004. Extended ceftiofur therapy for treatment of experimentally-induced Streptococcus uberis mastitis in lactating dairy cattle. J Dairy Sci 87:3322–3329. doi: 10.3168/jds.S0022-0302(04)73468-2. [DOI] [PubMed] [Google Scholar]