Abstract
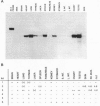
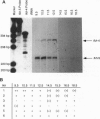
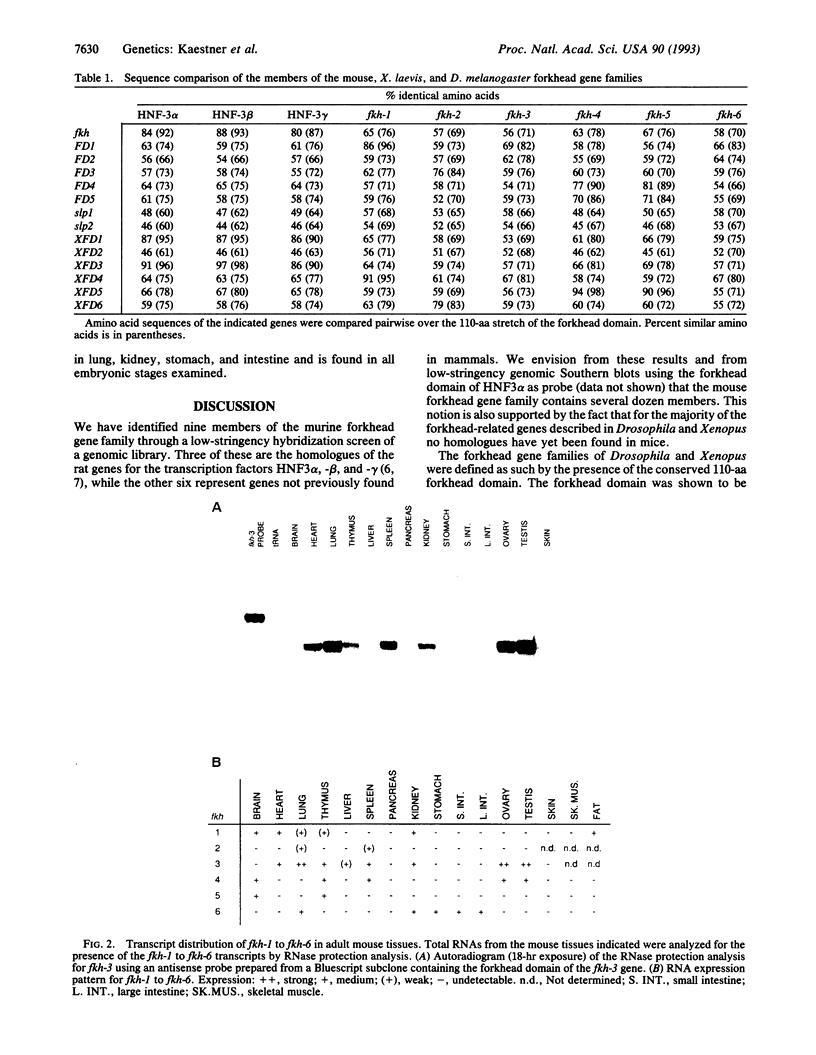
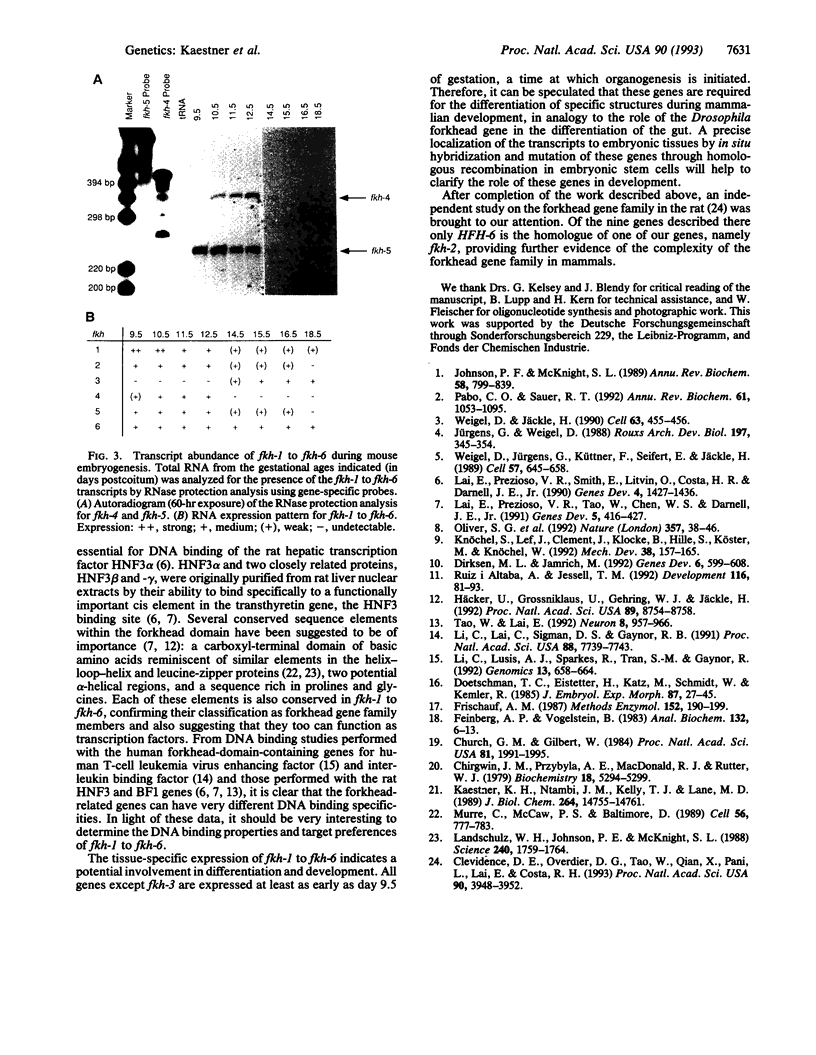
The 110-aa forkhead domain defines a class of transcription factors that have been shown to be developmentally regulated in Drosophila melanogaster and Xenopus laevis. The forkhead domain is necessary and sufficient for target DNA binding as shown for the rat hepatic nuclear factor 3 (HNF3) gene family. We have cloned six forkhead gene family members from a mouse genomic library in addition to the mouse equivalents of the genes for HNF3 alpha, -beta, and -gamma. The six genes, termed fkh-1 to fkh-6, share a high degree of similarity with the Drosophila forkhead gene, having 57-67% amino acid identity within the forkhead domain. fkh-1 seems to be the mammalian homologue of the Drosophila FD1 gene, as the sequences are 86% identical. fkh-1 to fkh-6 show distinct spatial patterns of expression in adult tissues and are expressed during embryogenesis.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevidence D. E., Overdier D. G., Tao W., Qian X., Pani L., Lai E., Costa R. H. Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc Natl Acad Sci U S A. 1993 May 1;90(9):3948–3952. doi: 10.1073/pnas.90.9.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen M. L., Jamrich M. A novel, activin-inducible, blastopore lip-specific gene of Xenopus laevis contains a fork head DNA-binding domain. Genes Dev. 1992 Apr;6(4):599–608. doi: 10.1101/gad.6.4.599. [DOI] [PubMed] [Google Scholar]
- Doetschman T. C., Eistetter H., Katz M., Schmidt W., Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. 1985 Jun;87:27–45. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M. Construction and characterization of a genomic library in lambda. Methods Enzymol. 1987;152:190–199. doi: 10.1016/0076-6879(87)52020-1. [DOI] [PubMed] [Google Scholar]
- Häcker U., Grossniklaus U., Gehring W. J., Jäckle H. Developmentally regulated Drosophila gene family encoding the fork head domain. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8754–8758. doi: 10.1073/pnas.89.18.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kaestner K. H., Ntambi J. M., Kelly T. J., Jr, Lane M. D. Differentiation-induced gene expression in 3T3-L1 preadipocytes. A second differentially expressed gene encoding stearoyl-CoA desaturase. J Biol Chem. 1989 Sep 5;264(25):14755–14761. [PubMed] [Google Scholar]
- Knöchel S., Lef J., Clement J., Klocke B., Hille S., Köster M., Knöchel W. Activin A induced expression of a fork head related gene in posterior chordamesoderm (notochord) of Xenopus laevis embryos. Mech Dev. 1992 Aug;38(2):157–165. doi: 10.1016/0925-4773(92)90007-7. [DOI] [PubMed] [Google Scholar]
- Lai E., Prezioso V. R., Smith E., Litvin O., Costa R. H., Darnell J. E., Jr HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990 Aug;4(8):1427–1436. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- Lai E., Prezioso V. R., Tao W. F., Chen W. S., Darnell J. E., Jr Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991 Mar;5(3):416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Li C., Lai C. F., Sigman D. S., Gaynor R. B. Cloning of a cellular factor, interleukin binding factor, that binds to NFAT-like motifs in the human immunodeficiency virus long terminal repeat. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7739–7743. doi: 10.1073/pnas.88.17.7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Lusis A. J., Sparkes R., Tran S. M., Gaynor R. Characterization and chromosomal mapping of the gene encoding the cellular DNA binding protein HTLF. Genomics. 1992 Jul;13(3):658–664. doi: 10.1016/0888-7543(92)90138-i. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Jessell T. M. Pintallavis, a gene expressed in the organizer and midline cells of frog embryos: involvement in the development of the neural axis. Development. 1992 Sep;116(1):81–93. doi: 10.1242/dev.116.Supplement.81. [DOI] [PubMed] [Google Scholar]
- Tao W., Lai E. Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron. 1992 May;8(5):957–966. doi: 10.1016/0896-6273(92)90210-5. [DOI] [PubMed] [Google Scholar]
- Weigel D., Jäckle H. The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell. 1990 Nov 2;63(3):455–456. doi: 10.1016/0092-8674(90)90439-l. [DOI] [PubMed] [Google Scholar]
- Weigel D., Jürgens G., Küttner F., Seifert E., Jäckle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989 May 19;57(4):645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]