Abstract
Immunotherapy exerts anticancer effects by activating competent immune effectors and inhibiting immunosuppressive cells, such as myeloid-derived suppressor cells (MDSCs). However, the mechanism underlying MDSCs-mediated immunosuppression in breast cancer is unclear. We have identified a poorly differentiated subset of MDSCs in breast cancer, which suppresses T-cell functions through STAT3-dependent IDO upregulation. Here, we aimed to investigate the mechanisms by which IDO expression is aberrant in MDSCs. We found increased STAT3 phosphorylation and NIK expression were correlated with upregulated IDO expression in MDSCs in human breast cancer. In mouse 4T1 mammary cancer model, blocking STAT3 signal significantly inhibited the activation of NF-κB and IDO expression in MDSCs, resulting in decrease of tumor growth and metastasis. We also induced MDSCs by co-culturing human CD33+ myeloid progenitors with MDA-MB-231 breast cancer cells. In these induced MDSCs, increased STAT3 activation was correlated with the activation of the noncanonical NF-κB pathway, including increased NIK protein level, phosphorylation of IKKα and p100 in cytoplasm, and RelB-p52 nuclear translocation. Blocking STAT3 activation significantly inhibited the accumulation of NIK and IDO expression in MDSCs. Knock-down of NIK by siRNA transfection in MDSCs suppressed IDO expression, but not STAT3 activation. Transcription factor assay and ChIP assay showed that RelB-p52 dimers directly bound to the IDO promoter, leading to IDO expression in MDSCs. These results suggest a STAT3-NF-κB-IDO pathway in breast cancer-derived MDSCs. Furthermore, IL-6 was found to stimulate STAT3-dependent NF-κB-mediated IDO upregulation in MDSCs. This study provides insights into understanding mechanisms through which MDSCs play an immunosuppressive role in breast cancer.
Keywords: Breast cancer, MDSCs, IDO, STAT3, Noncanonical NF-κB
Introduction
Myeloid-derived suppressor cells (MDSCs), a heterogeneous cell population, are composed of multiple myeloid cells arrested at various stages of lineage development. They serve as an important regulator for suppressive tumor microenvironment (1). MDSCs have been identified based on CD11b+ and GR-1+ expression in mice (1, 2). However, the markers for the phenotype of human MDSCs have not been defined well because different tumor types have different markers for MDSCs (1, 2). Human MDSCs are generally distinguished by their detectable immunosuppressive properties, definitive myeloid-lineage marker (CD33+), and poor antigen presentation capacity (HLA-DRlow/−) (1, 2). CD14 and CD15 have been reported as markers for two distinct subtypes of MDSCs in humans, monocyte MDSCs (Mo-MDSCs) and polymorphonuclear MDSCs (PMN-MDSCs) (1, 2).
In addition to the disparity in the phenotype of MDSCs, the molecular mechanisms underlying MDSCs-mediated immunosuppression vary among different tumor types, which are affected by the heterogeneity of tumors and complexity of tumor microenvironment. Several mechanisms have been reported to regulate the suppressive tumor milieu by MDSCs (1, 2). High expression levels of the enzymes, ARG1 and inducible nitric oxide synthase (iNOS) and production of reactive nitrogen species are involved in the identified mechanisms of MDSCs’ immunosuppressive effects (1). Other mechanisms include MDSCs-induced T cell immunosuppression, such as activation and expansion of regulatory T cell (TReg) population, cysteine sequestration, production of suppressive cytokines, such as IL-10 and TGF-β, and inhibition of CD62L-medated cell migration to lymph nodes (1). Recently, our and other groups have reported that upregulation of indoleamine 2,3-dioxygenase (IDO) might be a novel immunosuppressive mechanism of MDSCs in human breast cancer and hematological malignancies (3, 4).
IDO, an intracellular enzyme, activates kynurenine pathway of tryptophan to suppress T-cell proliferation and activate the cellular stress response pathway via the kinase GCN2 (5). Furthermore, tolerogenic kynurenine metabolites bind to aryl hydrocarbon receptor in T cells, resulting in inhibition of T cells-based anticancer immunity (5). In addition to macrophages, dendritic cells (DCs) and granulocytes (6, 7), IDO has been found in immature myeloid-derived cells (8). Mougiakakos and colleagues have reported a subset of MDSCs with high level of IDO protein in patients with hematologic malignancy after allogeneic hematopoietic stem cell transplantation therapy (3). IDO expression in MDSCs leads to a decreased IFN-γ release and CD3ζ-chain expression in T cells (3). Inhibition of IDO could reverse MDSCs-mediated suppression of T-cell proliferation (3). We have identified a subpopulation of MDSCs with the characteristic phenotype of CD45+ CD13+ CD33+ CD14− 15− in breast cancer. This subset of MDSCs suppresses T-cell functions in an IDO-dependent manner, and the number of this cell subpopulation correlated with increased infiltration of Foxp3+ Tregs in situ and lymph node metastasis in breast cancer patients (4).
The molecular mechanism regulating aberrant expression of IDO in MDSCs remains unclear. IFN-γ is the most potent inducer of IDO expression in macrophages and DCs (9). IFN-γ-dependent IDO expression is mainly through STAT1 pathway. STAT1 undergoes dimerisation and translocated to the nucleus where it directly binds to IDO gene or indirectly induces IDO transcription through IRF-1 protein (9). However, we and Mougiakakos’s groups did not observe significant increase of IFN-γ level in MDSCs from human breast cancer and hematological cancer (3, 4). Consistently, our previous study revealed low levels of phosphorylation of STAT1 protein and STAT1-dependent Arg-1 expression in MDSCs from human breast cancer (4). Therefore, STAT1 activation may not be the mechanism regulating aberrant upregulation of IDO in MDSCs in breast cancer. In our previous study, we demonstrated that STAT3, rather than STAT1, was a key potential upstream inducer of IDO expression in breast cancer-derived primary MDSCs. In this study, we investigated the mechanisms regulating IDO expression in MDSCs using induced MDSCs by co-culture of normal CD33+ progenitors and MDA-MB-231 breast cancer cells. We demonstrated that STAT3 activation stimulated the noncanonical NF-κB pathway, resulting in induction of IDO gene transcription via directly binding to the IDO promoter region. Furthermore, we found that IL-6 played a role in promoting STAT3-dependent NF-κB-mediated IDO upregulation in MDSCs. Our study suggests a novel regulatory mechanism of MDSCs-mediated immunosuppression in breast cancer and provides insights into developing more efficient immunotherapy for breast cancer treatment.
Materials and Methods Human
Samples
Umbilical cord blood (UCB) samples from 20 healthy donors (obtained from Tianjin Blood Center) were collected for isolation of CD33+ myeloid progenitors. This project was approved by the Ethics Committee of Tianjin Medical University. All experiments were performed in accordance with the principles of Declaration of Helsinki. Written consents were obtained from healthy donors.
Induction of MDSCs in vitro
CD33+ cells were isolated from UCB of healthy donors using human CD33 MicroBeads (130-045-501, Miltenyi Biotec, Germany) and cell viability analysis was performed using Trypan Blue staining method. MDSCs were induced by co-culturing CD33+ myeloid progenitors with breast cancer cell line MDA-MB-231 cells for 48 h in vitro and CD33+CD13+CD14−CD15− cells were sorted by flow cytometry as described before (4). MDSCs were treated with a specific STAT3 antagonist, JSI-124 (also called Cucurbitacin I, Sigma-Aldrich, St. Louis, MO, USA), at a concentration of 10 μM to block STAT3 phosphrylation. Untreated CD33+ myeloid cells from healthy UCB were used as the negative control.
siRNA Transfection
In order to define the regulatory effect of NF-κB pathway on STAT3-induced IDO expression, two siRNA candidates specifically targeting NIK mRNA sequence were synthesized (alias MAP3K14; siRNA1, sense 5′-CGCCAAAUCAAGCCAAUUATT -3′, anti-sense 5′-UAAUUGGCUUGAUUUGGCGTT-3′; siRNA2, sense 5′-GUGAGAAGAACCCAUCAAATT-3′, anti-sense 5′-UUUGAUGGGUUCUUCUCACTT-3′), and added into co-cultures system to inhibit NIK protein translation. A nontargeting control siRNA (sense 5′-UUCUCCGAACGUGUCACGUTT-3′, anti-sense 5′-ACGUGACACGUUCGGAGAATT-3′) was used to eliminate insignificant interference. Mixed siRNAs targeting C/EBPβ (Santa Cruz biotechnology, CA, USA) were also purchased for detecting whether the C/EBPβ pathway regulated IDO expression.
MDSCs were plated in 6-well plates without antibiotics for 24 h. Then cells were transfected with siRNA using Lipofectamine® RNAiMAX Reagent (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. After three days, cells were harvested for PCR and Western Blot analysis. The expression of C/EBPβ or NIK was evaluated to confirm full knock down of both genes.
Western Blot analysis
Western blot assay was performed to detect the expression levels of total protein and phosphorylated protein in MDSCs and CD33+ controls using anti-human IDO (Millipore Corporation, Billerica, MA), STAT1, STAT3, IKKα, IKKβ, NIK, phosphorylated (p) STAT1, pSTAT3, pC/EBPβ, p-p100, pIKKα/β, β-actin (Cell Signal Technology, Danvers, MA, USA) and pIKKα antibody (Signalway Antibody, College Park, Maryland, USA) antibodies as described previously (4). Horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit IgG antibody (Zhongshanjinqiao, Beijing, China) and SuperSignal West Pico Chemi- luminescent Substrate kit (Pierce Biotechnology, IL, USA) were used to visualize protein bands. The relative densities of protein were measured using the ChemiDoc XRS imaging system (Bio-Rad Laboratories, Hercules, CA, USA) and determined by normalizing to the β-actin, STAT1, STAT3, IKKα or IKKβ bands.
RT-PCR analysis
The mRNA levels of IDO, NIK, C/EBPβ and β-actin were analyzed using RT-PCR method as previously described (4). The primers for IDO, NIK, C/EBPβ and β-actin are shown in Table.1. PCR products were analyzed using agarose gel electrophoresis.
Table 1.
The primers for PCR
| Gene Name | Primer Sequences |
|---|---|
| IDO | UP: 5′-CATCTGCAAATCGTGACTAAG-3′ |
| DP: 5′-CAGTCGACACATTAACCTTCCTTC-3′ | |
| NIK | UP: 5′-AACGTCTATGTGTACAGATGAATGA-3′ |
| DP: 5′-GCAACAAGCCCGTAGAACAT-3′ | |
| C/EBPβ | UP: 5′-TTACAGGCCAGGCTCCTG-3′ |
| DP: 5′-GGAAGGTGGTCCTCAGCTC-3′ | |
| β-actin | UP: 5′-TGGCACCCAGCACAATGAA-3′ |
| DP: 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′ | |
| HIC2 | UP: 5′-GCTGACGGCAAAGGAACAG-3′ |
| DP: 5′-ACGCAGGCCCACGCTC-3′ | |
| AkAP12 | UP: 5′-CGGCTATTTATTTTTCCAGGTG-3′ |
| DP: 5′-GTCGCCCAAGTCCAAGAG-3′ | |
| Skp2 | UP: 5′-ACGAAGCGGGACGGAAACTA-3′ |
| DP: 5′-AGCTGCTCGCCTCCCAGAT-3′ |
Nuclear translocation assay
The nuclear extracts were prepared as described (10) to detect NF-κB nuclear translocation using Trans-AM NF-κB family kit (Active Motif Europe, Rixensart, Belgium) according to manufacturer’s instructions. A plate coated with immobilized oligonucleotides containing the consensus site (5′-GGGACTTTCC-3′) specific for NF-κB binding was used to enrich Rel/NF-κB dimers. To detect NF-κB subunits bound to its target DNA, mouse anti-human p50, p52, RelA and RelB monoclonal antibodies were incubated with the plate and followed by HRP-conjugated goat anti-mouse secondary antibody. The optical density was measured at 450 nm, and the relative level of NF-κB nuclear translocation was calculated by comparing the density treated group to that of the CD33+ control.
DNA binding sites Assay
A Trans AM Flexi kit was used to define specific binding sites on NF-κB (Active Motif Europe, Rixensart, Belgium). The biotinylated oligonucleotide (probe) containing the sites binding for NF-κB can form a complex with streptavidin coated on the plate. The presence of the DNA-bound transcription factor is then detected by an anti-NFκB antibody and measured by colorimetry (11). Four probes were generated using TaKaRa LA Taq® PCR kit (Takara Biotech, Kyoto, Japan). All probes were co-cultured with nuclear extracts isolated from MDSCs. Optical density was measured at 450 nm, and represented as relative binding activity of noncanonical NF-κB subunits (p52 and RelB) to certain probes. Identical oligonucleotide probes without biotinylation were used as competitive probes to eliminate the non-specific binding in this assay.
Chromatin immunoprecipitation (ChIP)-PCR assay
ChIP assay was performed to identify the specific binding sequences for the noncanonical NF-κB subunits, p52 and RelB, using an EZ-ChIP assay kit according to the manufacturer’s instructions (Millipore, Upstate Biotechnology). MiaPaCa2 cells were used as a positive control of p52 and RelB binding to skp2 gene (12, 13), while MDA-MB-231 cells served as a positive control of STAT3 binding to AKAP12 and HIC2 gene (14). The ChIP primers for detecting binding sites are listed in Table 1. Fragmented chromatin was immunoprecipitated with a ChIP-grade antibody against STAT3 (Cell Signal Technology, Danvers, MA, USA), RelB (Santa Cruz biotechnology, CA, USA) and p52 (Abcam, San Francisco, CA). After reversal of the cross-links and DNA precipitation, enriched DNA was analyzed by PCR amplification using primers specific for indicated regions. Quantification was performed by real-time qPCR, and enrichment was presented as the percentage of the test density to the total input, as described (15).
Cytokine array and ELISA
The levels of 42 cytokines in the supernatants of MDSCs and CD33+ controls were detected using RayBio® human cytokine antibody array (RayBiotech, Inc., Norcross, GA, USA) as described before (16). For validation, the levels of IL-1β, IL-6, IL-10, CM-CSF, IFN-γ and TGF-β1 in the cell culture supernatant were measured using ELISA kits (Genzyme, Cambridge, MA), as previously described (4).
Cytokine stimulation and blocking assay
MDSCs were treated with recombinant cytokines, IL-1β, GM-CSF, IL-6 and IL-10 (PeproTech, Rocky Hill, NJ) at a concentration of 1 μg/ml to examine the regulatory effects of these cytokines on IDO expression. MDSCs cultured with the supernatant of MDA-MB-231 cells were used as a positive control, and MDSCs cultured in FBS-supplemented RPMI 1640 medium were used as a negative control. In order to block the regulatory effects of cytokines on IDO expression, the specific neutralizing antibodies against IL-1β (30 μg/ml, Millipore, Upstate Biotechnology), IL-6 (50 μg/ml, Biolegend, San Diego, CA), GM-CSF (90 μg/ml, Biolegend, San Diego, CA) and IL-10 (30 μg/ml, Biolegend, San Diego, CA) were added to the co-culturing system of CD33+ myeloid progenitors and breast cancer cell line MDA-MB-231 cells. Expression of IDO, NIK, and STAT3 and STAT3 phosphorylation were detected using Western Blot analysis.
Immunofluorescence
Immunofluorescence was performed on 30 cases of formaldehyde-fixed, paraffin-embedded breast cancer tissues. Tissue sections were incubated with rabbit anti-human IDO (Millipore Corporation, Billerica, MA), rabbit anti-phospho-STAT3 (Cell Signaling Technology, Danvers, MA, USA), rabbit anti-human NIK (Santa Cruz biotechnology, CA, USA) and mouse anti-CD33 (Abcam, San Francisco, CA) antibodies overnight at 4°C, and Alexa Fluor® goat anti-mouse or anti-rabbit IgG secondary antibodies for 1 hour. 4′-6-diamidino-2-phenylindole (DAPI, Millipore Corporation, Billerica, MA) was used for nuclear counterstaining. Images were acquired using a Leica SP2 confocal microscope (Leica, Mannheim, Germany).
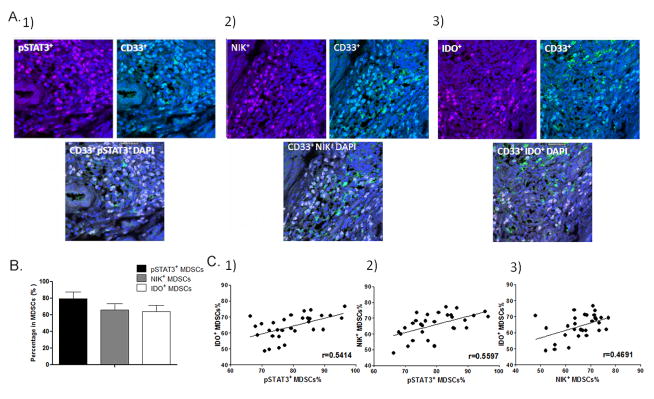
Breast cancer tissue sections were immunostained with phospho-STAT3 (pSTAT3) and CD33 for pSTAT3+ MDSCs, IDO and CD33 for IDO+ MDSCs, as well as NIK and CD33 for NIK+ MDSCs. Five representative high-power fields (×400 magnification) for each tissue section were selected for histology evaluation, and the percentages of pSTAT3+MDSCs, IDO+MDSCs and NIK+MDSCs in MDSCs were count and analyzed.
Mice and treatment
4T1 mouse mammary carcinoma cells (3 × 106/mouse) were injected into the mammary fat pads of BALB/c mice. After 5 days of tumor implantation, mice were randomly assigned to two groups (10 mice per group) Mice were then treated intraperitoneally with JSI-124 at dose of 1 mg/kg/d or vehicle control (PBS+DMSO) for 12 days. JSI-124 was initially dissolved in dimethyl sulphoxide (DMSO) at 1 mg/ml, and stored at −20°C. Before treatment, JSI-124 was diluted with PBS. On day16 after the treatment, mice were sacrificed. Tumor tissues were collected for preparation of paraffin-embedded sections. Spleens were prepared for CD11b+ MDSCs sorting.
Statistical analysis
All data were presented as mean ± SD. The statistical significance of the differences between mean values was determined using a SPSS 13.0 software package (SPSS, Chicago, IL). The one-way single-factor ANOVA and least significant difference method were used for comparison of the quantitative data. The level of statistical significance was set at p < 0.05.
Results
STAT3 activation-induced IDO expression is independent of direct binding of STAT3 to the promoter region of IDO gene
We tested whether STAT3 can regulate the IDO expression by directly binding to the IDO promoter. The whole DNA sequence of IDO promoter section was used to screen potential STAT3 binding sites using online prediction programs, including recommended eukaryotic transcription factors databases, TRANSFAC Database, and JASPAR Database. We found 5 potential binding sequences in IDO promoter for STATs binding (Table 2), 4 of them have been identified for STAT3-binding sequences of human genes (17–20).
Table 2.
Suspected binding sites specific for STAT3 in IDO promoter
| Sequence | Position 5′* | Position 3′* | Sequence Length | Consensus STAT3-Binding Sequences in Human genes |
|---|---|---|---|---|
| TTCCCAGAA | −3691 | −3683 | 9 | FCGR1A (17); PIM1 (18) |
| TTCCCAGAA | −3656 | −3648 | 9 | FCGR1A (17); PIM1 (18) |
| TCCCGGAAG | −3458 | −3450 | 9 | CDKN1A (19) |
| TTCCCCGAA | −3426 | −3418 | 9 | IRF1 (20) |
| TTCCTGTAA | −1119 | −1111 | 9 | Not Found |
The position of suspected binding sites from the first CDS.
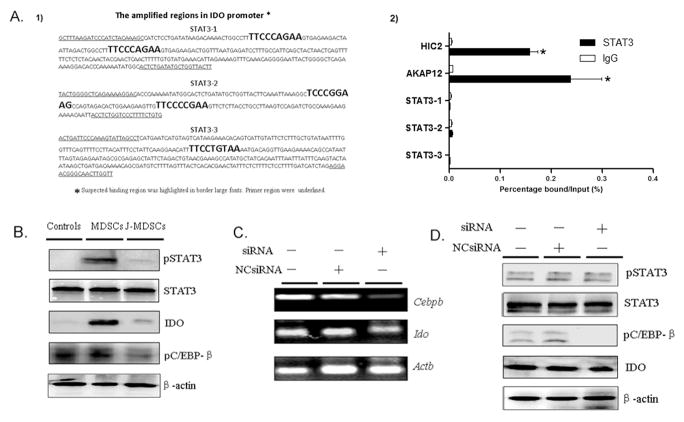
To test whether STAT3 directly binds the IDO promoter, ChIP analysis was conducted. Three pairs of primers were designed to amplify the regions containing 5 putative STAT3 binding sites (Figure 1A1). MDA-MB-231 cells served as the positive control of STAT3 binding to AKAP12 and HIC2 gene. However, none of these putative binding sequences in the IDO promoter region were amplified in the final PCR product (Figure 1A2), which indicated that there is no direct binding between STAT3 and the IDO promoter region. Thus STAT3 induced IDO expression is independent on direct binding of STAT3 to the promoter region of IDO gene.
Figure 1.
STAT3-induced IDO expression is not through a direct binding of STAT3 to the IDO promoter region and does not require C/EBPβ in MDSCs (A) Binding of STAT3 to three different regions in the IDO promoter containing 5 candidate STAT3-binding sites was measured by ChIP. (A1) Regions amplified are highlighted with bracketing the binding sites. (A2) ChIP-PCR assay was conducted to detect STAT3 binding to putative binding sequences. AKAP12 and HIC2 gene served as the positive. Quantitative values are calculated using the ratio of ChIP-PCR product to Input PCR product (relative ChIP value, enrichment ratio). * P<0.05. (B) Western blot was performed to detect the expression of phospho-(p)STAT3, pC/EBPβ and IDO in the whole cell extracts of CD33+ controls, MDSCs and JSI-124-treated MDSCs (J-MDSCs). STAT3 or β-actin blots were used as protein loading controls. (C) The mRNA of C/EBPβ and IDO was examined by RT-PCR in MDSCs transfected with C/EBPβ-specific siRNA or negative control. (D) Western blot was performed to examine phosphorylation of STAT3, C/EBPβ and IDO expression in MDSCs and C/EBPβ-specific siRNA transfected MDSCs.
STAT3 activation-induced IDO expression does not require C/EBPβ
It has been reported that CCAAT-enhancer-binding protein beta (C/EBPβ), also known as liver activating protein (LAP), is one of STAT3 downstream transcription factors which regulate myelopoiesis in healthy individuals and promote differentiation of myeloid progenitors to functional MDSCs (21). In addition, the IDO promoter region has binding sites for C/EBPβ (ATTCCCAAA, CTTCCTAAA and CATTCCAAAA) (22). Therefore we examined if the C/EBPβ signaling pathway regulates STAT3-mediated IDO upregulation. Since phosphorylation of C/EBPβ at specific sites is essential for its activation and transcriptional activity (23), we detected the phosphorylation level of C/EBPβ in MDSCs. There was no significant difference of the phosphorylation level of C/EBPβ in MDSCs, as compared to that in CD33+ controls (Figure 1B). JSI-124, a specific STAT3 antagonist, was used to block STAT3 activation. It is clear that JSI-124 provides a significant selective inhibition of JAK2/STAT3 pathway with scarcely any off-target effects, which has been used for examining the function of STAT3 pathway in MDSCs in vivo and in vitro (24, 25). Treatment of MDSCs with JSI-124, decreased STAT3 and C/EBPβ phosphorylation (Figure 1B). Then MDSCs were transfected with siRNA against C/EBPβ, or a nontargeting control siRNA (NCsiRNA). The level of C/EBPβ expression was examined by RT-PCR (Figure 1C). Transfection of C/EBPβ-specific siRNA dramatically decreased the phosphorylation level of C/EBPβ, but did not affect IDO expression in MDSCs significantly (Figure 1C–D). There is only a slight decrease for IDO expression after siRNA treatment, which implied that C/EBPβ activation could not completely account for the highly upregulated IDO expression, and other signal factors were bound to be involved in.
The noncanonical NF-κB pathway is activated in MDSCs
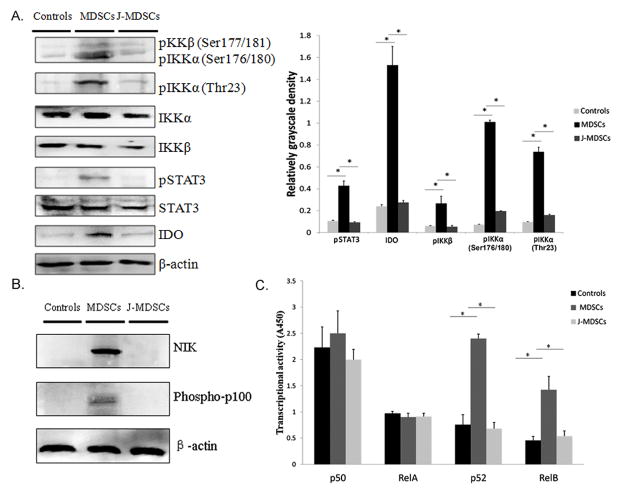
The interaction of STAT3 and NF-κB pathways in inflammatory or immune cells that infiltrate tumors serves as a key regulator in the suppressive environment of cancer by controlling the expression of anti-apoptotic, pro-proliferative and immune response genes (26, 27). NF-κB signaling also plays an important role in MDSCs’ suppressive activity and IDO expression in DCs (28–30). Therefore we examined the canonical and noncanonical NF-κB pathways in MDSCs. We detected the phosphorylation levels of inhibitor of nuclear factor kappa-B kinase subunit alpha (IKKα) and subunit beta (IKKβ), which represent the key signaling molecules in the noncanonical and canonical NF-κB pathway, respectively (31, 32). There was significant increase of phosphorylation levels of IKKα at Thr23 and Ser176/180 (P<0.05) and IKKβ at Ser177/181 (P<0.05), which is associated with increased STAT3 phosphorylation and expression of IDO in MDSCs, compared to those in CD33+ controls and JSI-124-treated MDSCs (Figure 2A).
Figure 2.
The noncanonical NF-κB pathway is activated in MDSCs. (A1) The phosphorylation levels of IKKβ (pIKKβ) and IKKα (pIKKα) in CD33+ controls, MDSCs and JSI-124-treated MDSCs (J-MDSCs). (A2) The levels of pIKKβ and pIKKα were compared using the density ratio of phosphorylated protein to total protein. * P<0.05. (B) The protein level of NIK and p100 phosphorylation in CD33+ controls, MDSCs and J-MDSCs. STAT3 or β-actin blots were used as protein loading controls. (C) Enzyme-linked immunosorbent assay was conducted to compare the levels of p50, p52, RelA and RelB subunits in nuclear extracts of CD33+ controls, MDSCs and J-MDSCs. The transcriptional activity was measured using absorbance at 450 nm (A450). * P<0.05.
Activation of noncanonical NF-κB pathway to induce the transcriptional activity is a complex process. High level of NF-κB-inducing kinase (NIK) leads to activation of IKKα, resulting in phosphorylation of p100 and nuclear translocation of RelB/p52 dimers (33). In untreated cells NIK is constitutively degraded to prevent unnecessary activation (33, 34). The level of NIK protein was low in CD33+ controls and increased in MDSCs. Treatment of MDSCs with JSI-124 to block STAT3 activation completely blocked NIK accumulation (Figure 2B). We also detected the phosphorylation level of p100, whose function is as important as its counterpart IκBα in the canonical NF-κB pathway (33). Consistently, the level of phosphorylated p100 notably increased in MDSCs, but not in CD33+ controls and JSI-124-treated MDSCs (Figure. 2B). Using an ELISA-based nuclear translocation assay, we compared the levels of p50, p52, RelA and RelB subunits in nuclear lysates of CD33+ controls and MDSCs. There was significant increase of the levels of noncanonical NF-κB subunits, p52 and RelB, but not canonical NF-κB subunits, p50 and RelA, in MDSCs, as compared to CD33+ controls (Figure 2C, P<0.05). Blocking STAT3 activation by JSI-124, dramatically decreased the levels of p52 and RelB in nuclei (Figure 2C, P<0.05). These data implied that noncanonical NF-κB pathway might be involved in STAT3-induced IDO expression.
STAT3 activation induced IDO expression through NF-κB activation
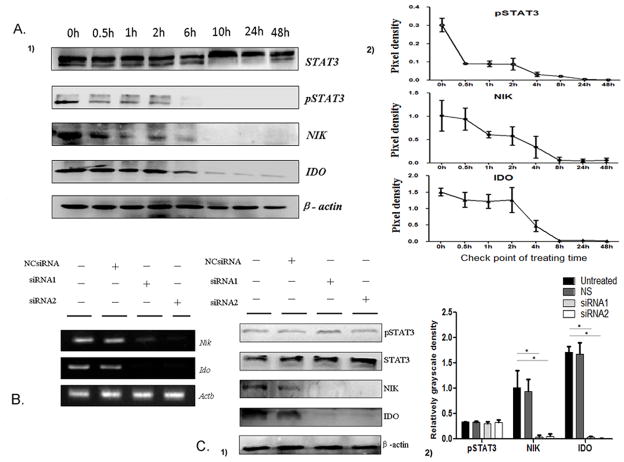
Next, we examined whether the activation of non-canonical NF-κB pathway is responsible for the upregulation of IDO in MDSCs. Firstly, we treated MDSCs with JSI-124 to block STAT3 pathway, and examined the changes of STAT3 phosphorylation, NIK and IDO proteins in MDSCs at different time points. The level of STAT3 phosphorylation in MDSCs was reduced at 30 min and completely suppressed at 4 h after treatment with JSI-124. Consistently, the level of NIK in MDSCs started to decline at 1 h and completely blocked at 8 h after treatment with JSI-124. The level of IDO was significantly decreased at 4 h after JSI-124 treatment (Figure 3A). These data suggest that the accumulation of NIK and upregulation of IDO is a consequence of STAT3 signaling activation.
Figure 3.
The noncanonical NF-κB pathway mediates STAT3-induced IDO expression in MDSCs. (A1) STAT3 phosphorylation and expression levels of NIK and IDO proteins in MDSCs at indicated time points. (A2) Quantification of density on immunoblots was performed by normalizing the density of each band to STAT3 or β-actin. (B) The mRNA of NIK and IDO in MDSCs transfected with NIK-specific siRNA or negative control was examined by RT-PCR. (C1) Western blot examined the expression of NIK and IDO and STAT3 phosphorylation in MDSCs and siRNA transfected MDSCs. (C2) Expression levels NIK and IDO and STAT3 phosphorylation were compared using the density ratio of indicated protein to β-actin respectively. * P<0.05.
In order to define if the noncanonical NF-κB activation is the upstream molecular event of IDO expression, MDSCs were transfected with NIK siRNA to knock down NIK. We found that two siRNA candidates efficiently knocked down NIK expression in MDSCs at RNA and protein levels, compared to NCsiRNA (nontargeting control siRNA, Figure 3B–C). In both NIK knock-down MDSCs, the levels of NIK and IDO proteins reduced significantly, but the level of STAT3 phosphorylation was not affected (Figure 3B–C). In contrast, both IDO and NIK expression were reduced by JSI-124 treatment in MDSCs (Figure 3A). Together, these data suggest that STAT3-induced IDO expression was through the noncanonical NF-κB activation in MDSCs.
RelB/p52 dimers directly bind to the IDO promoter region to regulate IDO expression in MDSCs
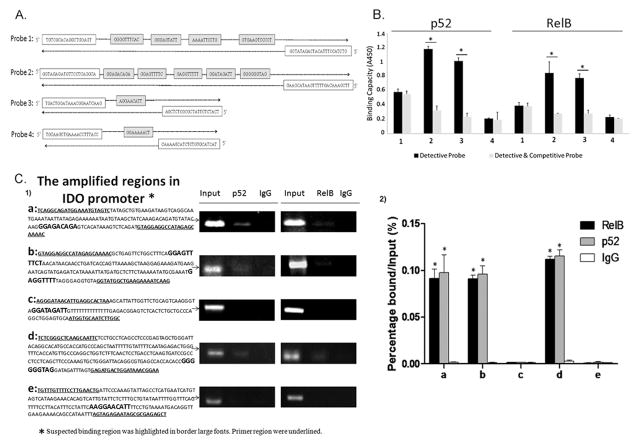
Recent study indicated that RelB/p52 dimers bind to IDO gene promoter and drive IDO protein expression in mouse DCs (28). There is a sequence diversity of the IDO promoter region between mouse and human and there is no report of similar mechanisms in human cells. Thus, we defined whether RelB/p52 dimers directly bind to the promoter region of IDO gene in human MDSCs. Bioinformatics-based screening method was used to find out potential NF-κB binding sites. Totally 11 putative p52-RelB binding sites were screened within 3000 bp of the IDO promoter region (Table. 3). To further determine the specificity of these binding sites to both noncanonical NF-κB subunits p52 and RelB, 4 biotinylated detective probes containing putative binding sequences were generated (Figure. 4A). Probe 2 and 3 displayed significantly stronger binding activity to RelB and p52 protein than the other 2 probes (Figure. 4B, p < 0.05).
Table 3.
Suspected binding sites specific for NF-κB factors within IDO promoter
| Sequence | Position 5′* | Position 3′* | Length | Methods |
|---|---|---|---|---|
| GGGGTTTCAC | −2984 | −2975 | 10 | prediction programs |
| GGGAGTATT | −2793 | −2785 | 9 | identified binding sequence |
| AAAATTCCTG | −2644 | −2635 | 10 | prediction programs |
| GTGAAGTCCCCT | −2307 | −2296 | 12 | prediction programs |
| AGGGAGACA | −2155 | −2147 | 9 | identified binding sequence |
| AGGAGTTTT | −2086 | −2078 | 9 | identified binding sequence |
| GAGGTTTTT | −1977 | −1969 | 9 | identified binding sequence |
| GGATAGATT | −1807 | −1799 | 9 | identified binding sequence & prediction programs |
| GGGGGGTAGGATAGATT | −1505 | −1489 | 9 | identified binding sequence & prediction programs |
| AGGAACATT | −1128 | −1120 | 9 | identified binding sequence & prediction programs |
The position of suspected binding sites from the first CDS.
Figure 4.
Noncanonical NF-κB subunits RelB and p52 regulate IDO expression via direct binding to the IDO promoter region in MDSCs. (A) Four detective biotinylated probes covering 11 putative sequences were synthesized. (B) Detective probes were incubated with MDSCs nuclear extracts with or without competitive probes to test the specificity of NF-κB-IDO binding. Forty fold dose of competitive probes was used to avoid NF-κB binding to the biotinylated detective probes. * P<0.05. (C1) Binding of p52 and RelB to five regions in the IDO promoter containing 6 putative noncanonical NF-κB binding sequences was measured by ChIP-PCR assay. Regions amplified are highlighted, bracketing putative noncanonical NF-κB binding sequences. MiaPaCa2 cells served as the positive control of p52 and RelB binding in skp2 gene. All products were analyzed on agarose gels. (C2) Quantitative values are calculated using the ratio of ChIP PCR product to Input PCR product (relative ChIP value, enrichment ratio). * P<0.05.
Then ChIP analysis was performed to test whether there is a direct binding of p52/RelB to the candidate binding sequences in the IDO promoter region. Five pairs of primers were designed to amplify the regions containing 6 putative noncanonical NF-κB binding sequnens (Figure. 4C1). The PCR results showed that positive amplification of 3 regions containing 4 putative RelB/p52-specific binding sequences in the IDO promoter after both RelB and p52 immunoprecipitation (Figure. 4C1–2). The results implied that STAT3 activation stimulates the noncanonical NF-κB subunits p52 and RelB translocation to nuclear and directly interact with the IDO promoter sequence to promote IDO transcription in MDSCs.
Increased STAT3 phosphorylation and NIK expression were correlated with upregulated IDO expression in situ in breast cancer
We investigated if the level of STAT3 phosphorylation and NIK expression were correlated with the IDO expression in MDSCs in situ in breast cancer using double immunostaining method (Figure 5A). Immunostaining results showed that 64. 2±7.4% of MDSCs were IDO+ MDSCs (defined as IDO and CD33 double positive karyocytes) in the stroma around cancer nests (CD33 positive karyocytes) (Figure 5A–B). 79.7±8.2% of MDSCs were with STAT3 phosphorylation (pSTAT3+MDSCs, defined as pSTAT3 and CD33 double positive cells) (Figure 5A–B). The percentage of NIK+MDSCs (defined as NIK and CD33 double positive cells) was 65.8±7.5% (Figure 5A–B). These data suggested that STAT3 and noncanonical NF-κB pathway were activated in most MDSCs in situ in breast cancer. We next compared the correlation among the numbers of pSTAT3+ MDSCs, NIK+ MDSCs and IDO+ MDSCs in primary cancer tissues. Significantly positive correlation was observed between the numbers of pSTAT3+ MDSCs and IDO+ MDSCs (p < 0.05, r=0.5414, Figure 5C1) which was consistent with our previous results (4). We also found a significant positive correlation between the numbers of pSTAT3+ MDSCs and NIK+ MDSCs, which implied activation noncanonical NF-κB pathway was correlated with STAT3 activation (p < 0.05, r=0.5597, Figure 5C2). Moreover there was a significant correlation between NIK+ MDSCs and IDO+MDSCs (p < 0.05, r=0.4691, Figure 5C3), which support our results that activation of noncanonical NF-κB pathway induces IDO expression in MDSCs. Together, these results suggest a STAT3-NF-κB-IDO pathway in breast cancer-derived MDSCs in situ in breast cancer.
Figure 5.
The correlation of STAT3 and NF-κB activation and IDO expression in MDSCs in human breast cancer. In this study, paraffin-embedded breast cancer tissues were collected from 30 patients who received radical mastectomy at the Department of Breast Oncology of Tianjin Medical University Cancer Institute and Hospital. (A) Breast cancer tissue sections were immunostained with phospho-STAT3 (pSTAT3) and CD33 for pSTAT3+ MDSCs, IDO and CD33 for IDO+ MDSCs, as well as NIK and CD33 for NIK+ MDSCs. (B) The percentages of pSTAT3+MDSCs, IDO+MDSCs and NIK+MDSCs in MDSCs are shown. (C) The correlations between the distribution of pSTAT3+ MDSCs, IDO+ MDSCs and NIK+ MDSCs are shown.
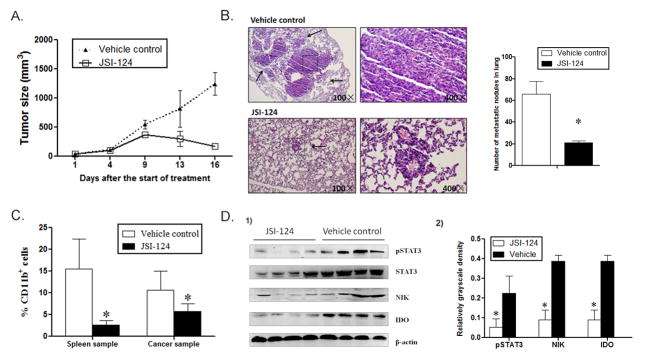
Blocking STAT3 signal inhibited tumor growth and metastasis in vivo
In order to investigate the regulatory effect of the STAT3-NF-κB-IDO pathway on MDSCs development in vivo, JSI-124 was used to block the STAT3-NF-κB-IDO pathway in MDSCs in mouse 4T1 mammary cancer model. The tumor-bearing mice were treated with JSI-124 or vehicle control (PBS+DMSO). Tumor growth was significantly inhibited after the JSI-124 treatment. On day16 after the treatment, the average tumor size in JSI-124-treated group shrunk to less than 200mm3, which decreased more than 7 folds compared to that in vehicle control group (Figure 6A). Metastasis was significantly reduced in JSI-124-treated mice compared to the control group, in which the number and size of metastatic nodules in lung dramatically decreased (Figure 6B). Simultaneously, significant decrease in the number of CD11b+ MDSCs was observed in spleen and cancer tissues of JSI-124-treated mice compared to that in the control group, which suggests that JSI-124 significantly inhibits MDSCs development (Figure 6C). We isolated CD11b+ MDSCs from the spleen of JSI-124-treated mice and found decreased expression of pSTAT3, NIK and IDO protein CD11b+ MDSCs (Figure 6D). Therefore, blocking STAT3 signal significantly inhibited the activation of NF-κB regulated IDO expression.
Figure 6.
Blocking STAT3 activation inhibits MDSCs and tumor metastasis in a mouse mammary cancer model. 4T1 mouse mammary carcinoma cells (3 × 106/mouse) were injected into the mammary fat pads of BALB/c mice. Mice were then treated intraperitoneally with JSI-124 at dose of 1 mg/kg/d or vehicle control (PBS+DMSO) for 12 days. (A) Tumor size in mice was monitored during JSI-124 treatment and measured using the following equation: Tumor volume= (longer diameter) × (shorter diameter) 2/2. (B) On day 16, mice were sacrificed and metastatic nodules in lungs were counted based on HE-stained slides. * P < 0.05. (C) Single cell suspensions of whole spleen and cancer tissues were prepared from tumor-bearing mice and the population of MDSCs were calculated by detecting CD11b+ subpopulation in CD45+ myeloid cells using flowcytometry. * P < 0.05. (D1) CD11b+ MDSCs were enriched using human CD11b MicroBeads and the expression of NIK and IDO, as well as STAT3 phosphorylation in MDSCs were detected by Western blot analysis. Four representative samples were displayed. (D2) Expression levels NIK and IDO and STAT3 phosphorylation were compared using the density ratio of indicated protein to β-actin or STAT3. * P<0.05.
IL-6 stimulated STAT3-dependent NF-κB-mediated IDO expression in MDSCs
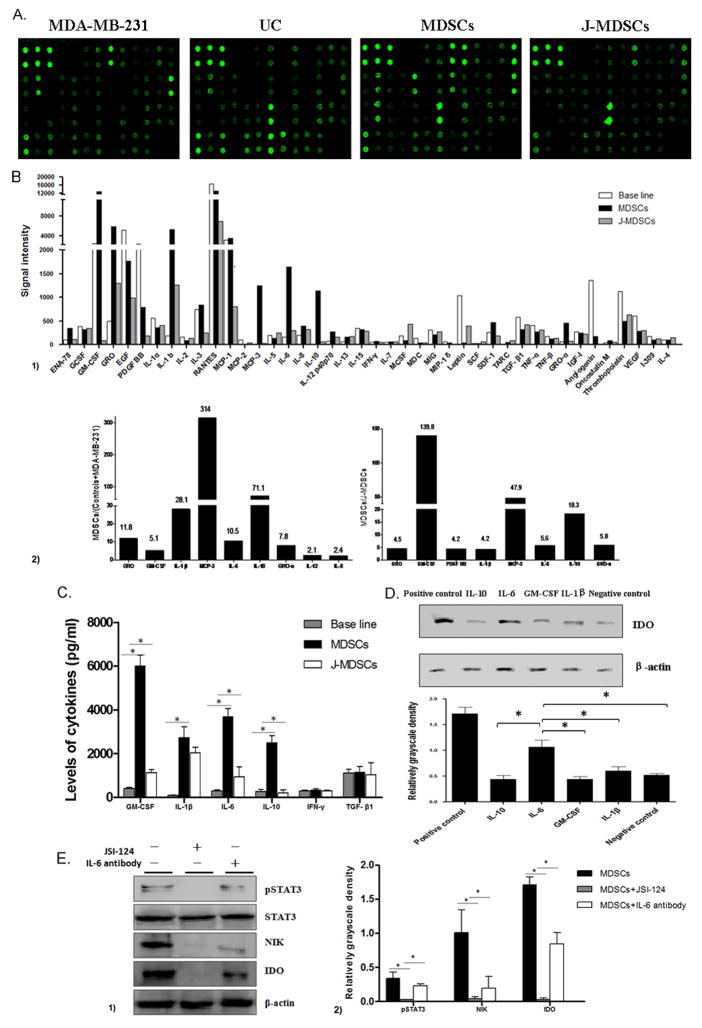
Several reports have indicated that cytokines participate in the expansion and accumulation of MDSCs (1, 35). Thus, we determined whether cytokines trigger STAT3 activation and promote IDO expression during MDSCs development. The culture supernatants were collected from CD33+ controls, MDA-MB-231 cells, MDSCs to detect the levels of 42 cytokines using human cytokine antibody array (Figure 7A). The cytokine signal intensities in MDA-MB-231 cell supernatant and CD33+ controls supernatant were combined to serve as the base line levels. The fold-changes were calculated by comparing the signal intensity of each cytokine in the supernatant of MDSCs to the respective base line level (Figure 7B1). Data shown the levels of nine cytokines increased more than 2 folds in the supernatants of MDSCs compared to the base line level (Figure 7B2). Interestingly after JSI-124 treatment, 8 cytokines decreased more than 2 folds compared to MDSCs (Figure. 7B2). Among them, GM-CSF, IL-6, IL-10 and IL-1β are well-defined inducers of the STAT3 pathway.
Figure 7.
IL-6 stimulates STAT3-dependent IDO expression in MDSCs. (A) Image of Human cytokine antibody array. The spot density represents the level of cytokine in the supernatants. UC represents CD33+ cells isolated from primary cord blood. (B1) The levels of 42 cytokines in the supernatant of MDSCs, J-MDSCs and the base line for the combination of MDA-MB-231 cell and CD33+ cells were displayed. (B2) Nine cytokines were increased at least 2 folds in MDSCs, compared to the base line, and 8 cytokines decreased at least 2 folds in J-MDSCs, compared to MDSCs. (C) The concentration of IL-1β, GM-CSF, IL-6 and IL-10 were detected by ELISA method. * P<0.05. (D) The expression level of IDO in the co-culture system for MDSCs development with or without co-treatment with IL-1β, GM-CSF, IL-6 and IL-10. Quantification of protein levels was evaluated using the density ratio of IDO protein to β-actin. * P<0.05. (E) Specific IL-6 neutralizing antibody was used in the co-culturing system of CD33+ progenitors and MDA-MB-231 cells to block IL-6 signal in the introduction of MDSCs. The IL-6 neutralizing antibody significantly decreased STAT3 phosphorylation and expression of NIK and IDO. * P<0.05.
Next, we detected the production of 6 cytokines using ELISA method, including GM-CSF, IL-6, IFN-γ, IL-1β, TGF-β and IL-10, which were reported to efficiently induce STAT3 activation and IDO expression (9, 36, 37). In consistent with results of cytokine array, the levels of GM-CSF, IL-6, and IL-10 dramatically increased in the supernatants of MDSCs compared to the controls (Figure 7C, P<0.05) which implied that these cytokines might be involved in activating of STAT3 signaling and IDO expression in MDSCs.
Next we treated MDSCs with recombinant cytokines, IL-1β, GM-CSF, IL-6 and IL-10, to detect their regulatory effects on IDO expression. Increased level of IDO was found in IL-6-treated MDSCs, compared to that in the other cytokines-treated MDSCs (Figure 7D, P<0.05). Furthermore, specific IL-6 neutralizing antibody significantly decreased the STAT3 phosphorylation, NIK protein level, and IDO expression in MDSCs (Figure 7E, P<0.05). These results indicate that IL-6 plays a role in STAT3-dependent IDO expression in MDSCs via inducing STAT3 and NF-κ B activation.
Discussion
MDSCs play vital roles in attenuating the therapeutic effects of immunotherapy (1, 35). Therefore, development of anti-MDSCs drugs has been a research focus. The progress in development innovative drugs has been made in recent years (1, 35). However any therapy does not display comparable anti-tumor efficiency in hematological cancer (1), as a result of the complicated phenotypes of tumor-derived MDSCs (3, 35).
Regarding to the MDSCs in breast cancer, most of efforts have been focused on mouse models rather than human beings because of uncertainty of cell phenotype and complicated regulatory mechanisms in human MDSCs (38–40). In previous study, our group has identified a poorly differentiated subpopulation of MDSCs with the characteristic phenotype of CD45+CD13+CD33+CD14−CD15− (4). This MDSCs subset significantly inhibited T cells immunity in breast cancer via a STAT3-induced IDO-dependent manner in breast cancer. Therefore, to elucidate concrete molecular mechanisms modulating the STAT3/IDO pathway in breast cancer-derived MDSCs may help us to develop novel therapies to suppress MDSCs-mediated immunosuppression in breast cancer patients.
In this study, we used a co-culture system of normal CD33+ progenitors and breast cancer cell line MDA-MB-231 cells to induce MDSCs with identical phenotype and immunosuppressive activity in vitro and define underlying molecular mechanisms regulating STAT3-dependent IDO expression in MDSCs (4). Because of limited information regarding molecular mechanisms for regulation of the STAT3/IDO pathway, we proposed three hypothesis: 1) activation of STAT3 directly regulates IDO expression by binding to the promoter region of IDO gene (9); 2) STAT3 indirectly regulates IDO via C/EBPβ pathway (37, 41); and 3) STAT3 indirectly regulates IDO gene via NF-κB pathway (28, 30). Our data indicated that the noncanonical NF-κB pathway is the main cause for IDO expression in MDSCs in response to STAT3 signaling.
It has been demonstrated that perturbation of STAT signalling plays a fundamental role in tumourigenesis (26). Among STAT members, STAT1 and STAT3 have shown important roles for IDO upregulation (9). STAT1 can directly bind to IDO promoter to induce its transcription in DCs (9). To define if STAT3 induced IDO expression in MDSCs directly, the binding activity of pSTAT3 to the IDO gene promoter was evaluated. 5 potential binding sequences for STATs binding in IDO promoter were found, and 4 of them are partly consistent with binding sites identified in other human genes (17–20). However no direct binding was observed using ChIP analysis which implied that other transcript factors rather than STAT3 regulated IDO expression in breast cancer-derived MDSCs.
We also detect the key downstream transcription factors of STAT3, C/EBPβ which has been suggested as an MDSC-determining transcription factor similar to FOXP3 in Tregs (37, 41). In fact, reports regarding C/EBPβ expression in human MDSCs have been inconsistent so far, and some study found no significant expression of this protein in MDSCs compared controls from healthy people (3, 41, 42). In the present study, we also found there is no significantly difference for the levels of 2 phosphorylated C/EBPβ subtypes which are essential for transcriptional activity (23), in controls and MDSCs. Then siRNA against C/EBPβ were used, and there is only a slight decrease for IDO expression after siRNA treatment. This indicated C/EBPβ activation could not completely account for the highly upregulated IDO expression, and other signal factors were bound to be involved in.
Then we detected NF-κB pathway. The NF-κB family has been considered the central mediator of the inflammatory process and a key participant in innate and adaptive immune responses (43). There are two independent signal transduction pathways of NF-κB family. The canonical NF-κB pathway induces IKKβ-dependent IκB phosphorylation and nuclear translocation of p65-containing heterodimers. The noncanonical NF-κB pathway triggers IKKα-mediated phosphorylation of p100 and generation of transcriptionally active p52-RelB complexes (33). Both NF-κB pathways are involved in the regulation of IDO expression in DCs (28–30). In the present study, we found that the noncanonical NF-κB pathway, but not the canonical NF-κB pathway accounts for STAT3-mediated IDO expression, which is consistent with the finding in DCs (9).
Although the biological roles of the noncanonical NF-κB pathway have been extensively studied, how this pathway functions in specific cell types is obscure (34). In cancer, the noncanonical NF-κB pathway is emerged as one of the most important drivers of the tumor promoting machinery (27, 44). Nadiminty and colleagues reported that the acetylation of STAT3 activates the noncanonical NF-κB pathway in breast cancer cell lines, but solid mechanism is lack(27). In present study we found it is phosphorylation of STAT3 leading to activation of the non-canonical NF-κB signal pathway in breast cancer-derived MDSCs, which was promoted by indirectly activated IKKα via non-canonical NF-κB-stimulating factors, since tremendous NIK accumulation was detected before IKKα activation and phosphorylation of p100 which is the core feature of factors-triggered activation of the noncanonical NF-κB pathway (34). As a result, IKKα was activated and promoted subsequent phosphorylation of p100, leading to nuclear translocation of RelB/p52 dimers.
Previous reports have shown that noncanonical NF-κB-dependent IDO expression was only observed in DCs, in which type I IFNs (IFN-α/β) can shift the balance of two NF-κB pathways and bias to the non-canonical pathway, thus lead to the transcription of IDO (9). In present study, we obtained consistent result that STAT3-induced activation of the noncanonical NF-κB pathway is responsible for aberrant IDO expression in MDSCs. With assistance of online prediction programs and eukaryotic transcription factors databases, 11 putative p52-RelB binding sites were screened in the IDO promoter region and 4 candidate sequences were demonstrated to display strong binding activity to p52-RelB dimmer by DNA-binding assay and ChIP analysis. We also found increased STAT3 phosphorylation and NIK expression were correlated with upregulated IDO expression in MDSCs in situ which implied that the STAT3-NF-κB-IDO pathway was activated in breast cancer-derived MDSCs both in vitro and vivo.
In order to further investigate the regulatory effects of the STAT3-NF-κB-IDO pathway in MDSCs development and function in breast cancer, a well-established 4T1 mouse mammary cancer model was used (34). Our pilot study demonstrated that 4T1 cells express very low level of IDO protein, however CD11b+ MDSCs isolated from 4T1 tumor-bearing mice expressed considerable level of IDO (data not shown). Moreover the proliferation rate of cells in Stat3 knockdown cells was similar to that of the normal 4T1 control; the cell cycle was also not affected (45). Here, we treated 4T1 tumor-bearing mice with JSI-124 to block STAT3 activation signal, and found a significant decrease of IDO expression coupled with consistent decrease of STAT3 phosphorylation and NIK expression in CD11b+ MDSCs, as well as dramatic decrease of tumor growth after JSI-124 treatment. These implied that blocking the activation of the STAT3-NF-κB-IDO pathway significantly affected the development and immune- suppressive activity of breast cancer-derived MDSCs in vivo, thus decreasing tumor growth via inhibiting MDSCs development and function.
But how the STAT3-NF-κB-IDO pathway was promoted in the microenvironment of breast cancer is not clear. Recently, Thomas and Dmitry reported that increased secretion of different soluble factors, such as IL-6, GM-CSF, IL-10 and IL-1β, in tumor sites provided a beneficial microenvironment for MDSCs development and functions (46). Signaling pathways triggered by most of above factors in MDSCs converge on STAT3 (35). Our study showed that IL-6 triggered STAT3 mediated-IDO expression in MDSCs in breast cancer. This result is consistent with Kim and colleagues’ study in which they demonstrated that IL-6 induced IDO upregulation in neuroblastoma cell line in vitro (47). In mouse models using 4T1-IL6 cells in vivo, a 4T1 cell population engineered to constitutively express IL-6, promoted IDO-driven MDSCs activity and pulmonary metastasis (48). Moreover, CD33+ cells with potent suppressive capacity were generated in vitro by cytokines contained IL-6 (42). Thus, we suggest that IL-6 might be a key factor to trigger the STAT3-NF-κB-IDO pathway in MDSCs in breast cancer.
In conclusion, we found that noncanonical NF-κB pathway is involved in STAT3-dependent IDO upregulation in breast cancer derived MDSCs, which displays potent immunosuppressive function on T cells immunity and efficiently promotes lymph node metastasis in patients. Therefore, specific blocking this signal pathway in MDSCs might improve the clinical efficiency of present immunotherapy.
Acknowledgments
This work was supported by National Basic Research Program of China (973 program, 2012CB9333004), National Natural Science Foundation of China (81072159), and Tianjin Municipal Education Commission (20090133). We thank Dr. Pengbo Zhou from Department of Pathology and Laboratory Medicine of Weill Cornell Medical College for providing constructive suggestions for this study.
Footnotes
Conflict of interest statement
The authors declare that they have no competing interests.
References
- 1.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peranzoni E, Zilio S, Marigo I, Dolcetti L, Zanovello P, Mandruzzato S, et al. Myeloid-derived suppressor cell heterogeneity and subset definition. Curr Opin Immunol. 2010;22:238–244. doi: 10.1016/j.coi.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 3.Mougiakakos D, Jitschin R, von Bahr L, Poschke I, Gary R, Sundberg B, et al. Immunosuppressive CD14+HLA-DRlow/neg IDO+ myeloid cells in patients following allogeneic hematopoietic stem cell transplantation. Leukemia. 2013;27:377–388. doi: 10.1038/leu.2012.215. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Du W, Yan F, Wang Y, Li H, Cao S, et al. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J Immunol. 2013;190:3783–3797. doi: 10.4049/jimmunol.1201449. [DOI] [PubMed] [Google Scholar]
- 5.Medzhitov R, Shevach EM, Trinchieri G, Mellor AL, Munn DH, Gordon S, et al. Highlights of 10 years of immunology in Nature Reviews Immunology. Nat Rev Immunol. 2011;11:693–702. doi: 10.1038/nri3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J Exp Med. 1999;189:1363–1372. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sim SH, Ahn YO, Yoon J, Kim TM, Lee SH, Kim DW, et al. Influence of chemotherapy on nitric oxide synthase, indole-amine-2,3-dioxygenase and CD124 expression in granulocytes and monocytes of non-small cell lung cancer. Cancer Sci. 2012;103:155–160. doi: 10.1111/j.1349-7006.2011.02158.x. [DOI] [PubMed] [Google Scholar]
- 8.Furset G, Floisand Y, Sioud M. Impaired expression of indoleamine 2, 3-dioxygenase in monocyte-derived dendritic cells in response to Toll-like receptor-7/8 ligands. Immunology. 2008;123:263–271. doi: 10.1111/j.1365-2567.2007.02695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Yang BH, Li H, Cao S, Ren XB, Yu JP. IDO+ DCs and Signalling Pathways. Curr Cancer Drug Targets. 2013;13:278–288. doi: 10.2174/15680096113139990073. [DOI] [PubMed] [Google Scholar]
- 10.Wen Z, Zhong Z, Darnell JJ. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 11.Renard P, Ernest I, Houbion A, Art M, Le Calvez H, Raes M, et al. Development of a sensitive multi-well colorimetric assay for active NFkappaB. Nucleic Acids Res. 2001;29:E21. doi: 10.1093/nar/29.4.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider G, Saur D, Siveke JT, Fritsch R, Greten FR, Schmid RM. IKKalpha controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression. Embo J. 2006;25:3801–3812. doi: 10.1038/sj.emboj.7601259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barre B, Perkins ND. A cell cycle regulatory network controlling NF-kappaB subunit activity and function. Embo J. 2007;26:4841–4855. doi: 10.1038/sj.emboj.7601899. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Oh YM, Kim JK, Choi Y, Choi S, Yoo JY. Prediction and experimental validation of novel STAT3 target genes in human cancer cells. PLoS One. 2009;4:e6911. doi: 10.1371/journal.pone.0006911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang BD, Yong-Gonzalez V, Strunnikov AV. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle. 2004;3:960–967. doi: 10.4161/cc.3.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Wang C, Yu J, Cao S, Wei F, Zhang W, et al. Dendritic cell-activated cytokine-induced killer cells enhance the anti-tumor effect of chemotherapy on non-small cell lung cancer in patients after surgery. Cytotherapy. 2009;11:1076–1083. doi: 10.3109/14653240903121252. [DOI] [PubMed] [Google Scholar]
- 17.Wehinger J, Gouilleux F, Groner B, Finke J, Mertelsmann R, Weber-Nordt RM. IL-10 induces DNA binding activity of three STAT proteins (Stat1, Stat3, and Stat5) and their distinct combinatorial assembly in the promoters of selected genes. Febs Lett. 1996;394:365–370. doi: 10.1016/0014-5793(96)00990-8. [DOI] [PubMed] [Google Scholar]
- 18.Matikainen S, Sareneva T, Ronni T, Lehtonen A, Koskinen PJ, Julkunen I. Interferon-alpha activates multiple STAT proteins and upregulates proliferation-associated IL-2Ralpha, c-myc, and pim-1 genes in human T cells. Blood. 1999;93:1980–1991. [PubMed] [Google Scholar]
- 19.Chen B, He L, Savell VH, Jenkins JJ, Parham DM. Inhibition of the interferon-gamma/signal transducers and activators of transcription (STAT) pathway by hypermethylation at a STAT-binding site in the p21WAF1 promoter region. Cancer Res. 2000;60:3290–3298. [PubMed] [Google Scholar]
- 20.Andersen P, Pedersen MW, Woetmann A, Villingshoj M, Stockhausen MT, Odum N, et al. EGFR induces expression of IRF-1 via STAT1 and STAT3 activation leading to growth arrest of human cancer cells. Int J Cancer. 2008;122:342–349. doi: 10.1002/ijc.23109. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Nguyen-Jackson H, Panopoulos AD, Li HS, Murray PJ, Watowich SS. STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood. 2010;116:2462–2471. doi: 10.1182/blood-2009-12-259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson CM, Hale PT, Carlin JM. The role of IFN-gamma and TNF-alpha-responsive regulatory elements in the synergistic induction of indoleamine dioxygenase. J Interferon Cytokine Res. 2005;25:20–30. doi: 10.1089/jir.2005.25.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, et al. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci U S A. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 2003;63:1270–1279. [PubMed] [Google Scholar]
- 25.Nefedova Y, Nagaraj S, Rosenbauer A, Muro-Cacho C, Sebti SM, Gabrilovich DI. Regulation of dendritic cell differentiation and antitumor immune response in cancer by pharmacologic-selective inhibition of the janus-activated kinase 2/signal transducers and activators of transcription 3 pathway. Cancer Res. 2005;65:9525–9535. doi: 10.1158/0008-5472.CAN-05-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21:11–19. doi: 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nadiminty N, Lou W, Lee SO, Lin X, Trump DL, Gao AC. Stat3 activation of NF-{kappa}B p100 processing involves CBP/p300-mediated acetylation. Proc Natl Acad Sci U S A. 2006;103:7264–7269. doi: 10.1073/pnas.0509808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pallotta MT, Orabona C, Volpi C, Vacca C, Belladonna ML, Bianchi R, et al. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat Immunol. 2011;12:870–878. doi: 10.1038/ni.2077. [DOI] [PubMed] [Google Scholar]
- 29.Puccetti P, Grohmann U. IDO and regulatory T cells: a role for reverse signalling and non-canonical NF-kappaB activation. Nat Rev Immunol. 2007;7:817–823. doi: 10.1038/nri2163. [DOI] [PubMed] [Google Scholar]
- 30.Pine R. Convergence of TNFalpha and IFNgamma signalling pathways through synergistic induction of IRF-1/ISGF-2 is mediated by a composite GAS/kappaB promoter element. Nucleic Acids Res. 1997;25:4346–4354. doi: 10.1093/nar/25.21.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozes ON, Mayo LD, Gustin JA, Pfeffer SR, Pfeffer LM, Donner DB. NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature. 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 32.Ling L, Cao Z, Goeddel DV. NF-kappaB-inducing kinase activates IKK-alpha by phosphorylation of Ser-176. Proc Natl Acad Sci U S A. 1998;95:3792–3797. doi: 10.1073/pnas.95.7.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun SC. The noncanonical NF-kappaB pathway. Immunol Rev. 2012;246:125–140. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun SC. Non-canonical NF-kappaB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 37.Babcock TA, Carlin JM. Transcriptional activation of indoleamine dioxygenase by interleukin 1 and tumor necrosis factor alpha in interferon-treated epithelial cells. Cytokine. 2000;12:588–594. doi: 10.1006/cyto.1999.0661. [DOI] [PubMed] [Google Scholar]
- 38.Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13:23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinha P, V, Clements K, Bunt SK, Albelda SM, Ostrand-Rosenberg S. Cross-talk between myeloid-derived suppressor cells and macrophages subverts tumor immunity toward a type 2 response. J Immunol. 2007;179:977–983. doi: 10.4049/jimmunol.179.2.977. [DOI] [PubMed] [Google Scholar]
- 40.Ozao-Choy J, Ma G, Kao J, Wang GX, Meseck M, Sung M, et al. The novel role of tyrosine kinase inhibitor in the reversal of immune suppression and modulation of tumor microenvironment for immune-based cancer therapies. Cancer Res. 2009;69:2514–2522. doi: 10.1158/0008-5472.CAN-08-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lechner MG, Megiel C, Russell SM, Bingham B, Arger N, Woo T, et al. Functional characterization of human Cd33+ and Cd11b+ myeloid-derived suppressor cell subsets induced from peripheral blood mononuclear cells co-cultured with a diverse set of human tumor cell lines. J Transl Med. 2011;9:90. doi: 10.1186/1479-5876-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lechner MG, Liebertz DJ, Epstein AL. Characterization of cytokine-induced myeloid-derived suppressor cells from normal human peripheral blood mononuclear cells. J Immunol. 2010;185:2273–2284. doi: 10.4049/jimmunol.1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DiDonato JA, Mercurio F, Karin M. NF-kappaB and the link between inflammation and cancer. Immunol Rev. 2012;246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi N, Ito T, Azuma S, Ito E, Honma R, Yanagisawa Y, et al. Constitutive activation of nuclear factor-kappaB is preferentially involved in the proliferation of basal-like subtype breast cancer cell lines. Cancer Sci. 2009;100:1668–1674. doi: 10.1111/j.1349-7006.2009.01228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ling X, Arlinghaus RB. Knockdown of STAT3 expression by RNA interference inhibits the induction of breast tumors in immunocompetent mice. Cancer Res. 2005;65:2532–2536. doi: 10.1158/0008-5472.CAN-04-2425. [DOI] [PubMed] [Google Scholar]
- 46.Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011;32:19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim H, Chen L, Lim G, Sung B, Wang S, McCabe MF, et al. Brain indoleamine 2,3-dioxygenase contributes to the comorbidity of pain and depression. J Clin Invest. 2012;122:2940–2954. doi: 10.1172/JCI61884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith C, Chang MY, Parker KH, Beury DW, DuHadaway JB, Flick HE, et al. IDO is a nodal pathogenic driver of lung cancer and metastasis development. Cancer Discov. 2012;2:722–735. doi: 10.1158/2159-8290.CD-12-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]