Abstract
Sialyltransferases (STs) catalyze the addition of sialic acids to the non-reducing ends of glycoproteins and glycolipids. In this work, we examined the acceptor specificity of five human α(2,3)sialyltransferases, namely ST3Gal-I, -II, -III, -IV and -VI. KM values for each of these enzymes is presented using radioactivity for acceptors containing Type-I (Galβ1,3GlcNAc), Type-II (Galβ1,4GlcNAc), Type-III (Galβ1,3GalNAc) and Core-2 (Galβ1,3(GlcNAcβ1,6)GalNAc) reactive groups. Several variants of acceptors inhibited ST3Gal activity emphasizing structural role of acceptor in enzyme-catalyzed reactions. In some cases, mass spectrometry was performed for structural verification. The results demonstrate human ST3Gal-I catalysis towards Type-III and Core-2 acceptors with KM = 5–50μM and high VMax values. The KM for ST3Gal-I and ST3Gal-II was 100 and 30-fold lower, respectively, for Type-III compared to Type-I acceptors. Variants of Type-I and Type-II structures characterized ST3Gal-III, -IV and -VI for their catalytic specificity. This manuscript also estimates KM for human ST3Gal-VI using Type-I and Type-II substrates. Together, these findings built a platform for designing inhibitors of STs having therapeutic potential.
Keywords: Sialyltransferase, Michaelis-Menten, Product Inhibition, Mass Spectrometry
INTRODUCTION
The glycosyltransferases (GTs) are a class of enzymes that participate in the post-translational glycosylation of proteins. GTs initializes multi-substrate reactions through nucleotide-sugar (donor) binding, thereby creating an enzyme-donor complex. This enzyme-donor complex then results in a conformational change in the enzyme and prepares it for subsequent acceptor interaction. These catalytic reactions follow an ordered sequential bi-bi mechanism [1]. However, a class of GTs belonging to GT29 family, termed sialyltransferases (STs), follows random sequential bi-bi mechanism. In other words, enzymatic reaction mechanism varies based on their structural differences. Furthermore, these eukaryotic STs are inverting enzymes such that the catalysis occurs via an SN2 reaction resulting in an oxocarbenium ion-like transition state [2]. This transition state determines the catalytic rate of the enzyme and is strongly influenced by the structure of the acceptor. Thus, detailed acceptor specificity and kinetic analysis is indispensible to understand the Structure Activity Relation (SAR) of an enzyme.
Amongst the known STs, human α2,3-sialyltransferases (ST3Gal) participates in a variety of inflammatory, immunological and cancerous conditions [3,4,5]. ST3Gals synthesize sialoglycans (sialic acid terminated glycans) by transferring sialic acid from its activated donor, CMP-Neu5Ac, onto the 3rd carbon of galactose (Gal) residue of the acceptor glycoconjugate (Fig. 1A, Table S1). Such Gal residue are commonly a part of three different lactosamine (LacNAc) structures termed Type-I (Galβ1,3GlcNAc), Type-II (Galβ1,4GlcNAc) and Type-III (Galβ1,3GalNAc), respectively. Heterogeneity in sialoglycans at specific site results from enzyme specificity, activity and competition with other Golgi-resident enzymes. For instance, Type-II (Neu5Acα2,3Galβ1,4GlcNAc) sialoglycans are likely synthesized by the competition between ST3Gal-I, -II, -III, -IV and -VI [6,7]. Thus, the specific sialoglycan-form depends not only on ST3Gal expression and abundance, but also acceptor-specific KM values.
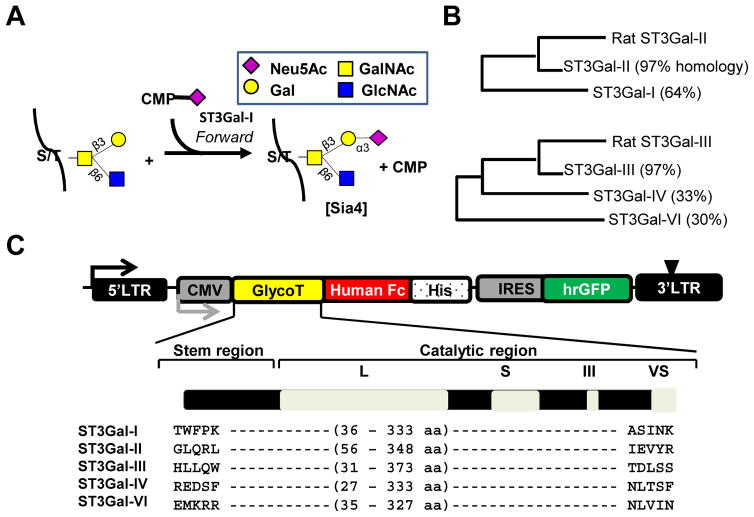
Figure 1. Sialyltransferase expression and activity.
A. Sialylation mediated by human ST3Gal-I results in the transfer of sialic acid from the donor, CMP-Neu5Ac, to the glycoprotein acceptor. Glycans are represented using the Consortium of Functional Glycomics nomenclature (http://www.functionalglycomics.org/static/consortium/Nomenclature.shtml). B. Similarity in the sequence of the catalytic domains of rat and human ST3Gals analyzed using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). C. pCSCG vector expresses human α2,3 sialyltransferases, ST3Gal-I, -II, -III, -IV and -VI, as fusion protein with C-terminal Fc and his-tag. Amino acids expressed are annotated. Expression of IRES-GFP reporter protein confirms stable enzyme synthesis in mammalian cells.
Previous studies by Kono et al. [8] and Williams et al. [9] report sialylation kinetics of mammalian enzymes using glycoprotein substrates. As glycoproteins contain micro and macro glycan heterogeneity, studies are required to determine substrate specificity using defined chemical substrate as addressed in the current work [10]. Additionally, quantitative comparison of kinetic parameters requires concurrent comparison of acceptor KM values. In this context, while some kinetic data are available for human ST3Gals, detailed catalytic data for a panel of acceptors is lacking [11]. Homologs of human ST3Gal-I and ST3Gal-II in rat exhibited unconventional pH-dependent catalytic activity termed ‘reverse’ sialylation [12]. Thus, the key contribution of the current manuscript lies in the systematic concurrent acceptor specificity and kinetic analysis for human ST3Gal-I, -II, -III, -IV and -VI. The results conclude that ST3Gal-I, ST3Gal-II and ST3Gal-IV show activity specific to Type-III glycans with ST3Gal-I also sialylating Core-2. ST3Gal-III, -IV and -VI acted, both, on Type-I and Type-II glycans [13]. Furthermore, inhibition studies using human ST3Gal-I dictate either iso- or random-ordered bi-bi mechanism. Taken together, biochemical characterization of five human ST3Gals emphasizes on structure specific inhibitor design.
MATERIALS AND METHODS
Materials
CMP (cytidine 5′-monophosphate) and CMP-Neu5Ac (cytidine 5′-monophosphate N-Acetylneuraminic acid) were from Sigma (St. Louis, MO). Synthetic oligosaccharides coupled to methyl (-Me), benzyl (-Bn), paranitrophenol (-pNp) and naphthalene (-Nap) aglycones at the reducing end were available from previous studies ([13,14], Table S1). Radioactive CMP-[14C]Neu5Ac was from American Radiolabeled Chemicals (55mCi/mmol specific activity, St. Louis, MO). CMP-Neu5Gc was kindly gifted by Dr. Willie F. Vann (FDA, New Hampshire, MD).
Recombinant soluble ST3Gal production
Soluble forms of the ST3Gals, ST3Gal-I, -II, -III, -IV and - VI were stably expressed in Chinese Hamster Ovary (CHO) and Human Embryonic Kidney (HEK293T) cells with a C-terminal Human Fc and his-tag. To this end, individual ST3Gal cDNA were obtained from the Mammalian Gene Collection (Thermo-Fisher, Waltham, MA). Coding regions corresponding to the soluble forms of each of these enzymes (listed in Fig. 1C) were PCR amplified with 5′-AgeI and 3′-HpaI restriction enzyme sites. The amplified products were inserted into a cloning vector containing a C-terminal human Fc and poly-histidine (6×His) sequence. This product was then excised and inserted downstream of the VWF signal peptide (SS) in the lentiviral plasmid pCSCG-KZK-SS-A2-FLAG-6×His [15]. A reporter gene, IRES-hrGFPII (Agilent, West Cedar Creek, TX), was PCR amplified with 5′- and 3′-XhoI sites. This was inserted into the pCSCG plasmid downstream of the ST3Gals. Lentivirus was generated using the above vectors and calcium phosphate method [16]. These were transduced into CHO-S and HEK293T cells to generate stable cell lines that secrete the soluble ST3Gals. For enzyme production, individual cell lines were scaled up in 10–20 T150 flasks. Upon reaching 60–70% confluence, the media was replaced with CHO/HEK-cell serum free media (Life Technologies, Grand Island, NY). 1L of cell culture supernatant was collected 3–4 days thereafter.
ST3Gals were purified using a HisTrap column (GE Healthcare, Piscataway, NJ). To this end, the column was equilibrated with 20mM HEPES containing 500mM NaCl, pH 7.4. Culture supernatant was then passed through the column to capture his-tagged protein. Following wash with 10 column volumes of HEPES buffer containing 10mM imidazole, the soluble ST3Gals were eluted using buffer containing 100–250mM imidazole. ST3Gal activity was measured in 1mL fractions using enzymatic assays (see next section) and relevant fractions were pooled. The buffer was exchanged to 10mM Tris, 150mM NaCl using a 7K Zeba spin desalting column (Thermo-Pierce, Rockford, IL), concentrated by a 10kD cut-off concentrator (Millipore, Billerica, MA), aliquot and stored at −80 °C for later use. Western-blot analysis of ST3Gals was performed by resolving the enzymes on 4–20% gradient gels (Thermo-Fisher) under reducing conditions. Following transfer onto 0.2μm nitrocellulose membranes, the enzymes were detected using horse-radish peroxidase (HRP) conjugated goat anti-human Fc antibody.
Enzymology using radioactivity
Miniaturized enzymatic assays were performed as described previously in 3μL volume [17]. Unless otherwise mentioned, the ‘standard’ reaction mixture contained 100mM sodium cacodylate (pH 6.0) buffer, 0.6mM acceptor, 0.0025μCi CMP-[14C]Neu5Ac and 200μM cold-CMP-Neu5Ac at room temperature. Following reaction for 2–16h, 1μl of the reaction mixture was spotted on a reverse phase TLC plate (silica gel 60 RP-TLC plates, EMD-Chemicals), and separation was carried out using water containing 0.2% w/w CH3COOH as the mobile phase. The hydrophobic radiolabeled glycoconjugates were retained at the initial spot while CMP-[14C]Neu5Ac moved with the mobile front. After separation, the TLC plates were dried, wrapped in thin plastic, exposed overnight to a phosphorscreen (Super Resolution (SR) Medium, Perkin Elmer, Akron, OH) and imaged using a cyclone storage phosphorimager (Perking Elmer). Radioactivity was quantified using NIH ImageJ software. Reaction velocity was then quantified after subtraction of background signal in runs without acceptor as described previously. In all studies, 1 unit (U) of ST3Gal enzyme activity is defined to transfer 1μmole of sialic acid from CMP-sialic acid to asialofetuin in 1 min. at 25°C.
Enzymatic Rate Expression
Multi-substrate enzymatic reactions follow either ping-pong or sequential bi-bi mechanisms. Among these, the sequential bi-bi mechanism requires that both substrates bind, either random or ordered manner, for product synthesis (Appendix, part A and B). The ping-pong mechanism results from the binding of a substrate A and its conversion into product P, prior to the binding of the second substrate B and its conversion into product Q. The detailed derivations for these reaction mechanisms are illustrated as: ordered sequential bi-bi (Eq. 1, from Eq. 2b Appendix), random sequential bi-bi (Eq. 2, from Eq. 5 Appendix) and ping-pong (Eq. 3, from Eq. 8 Appendix).
| Equation 1 |
| Equation 2 |
| Equation 3 |
Product inhibition studies data were fit to determine nature of sequential reaction. To this end, mixed inhibition scheme (Eq. 4, from Eq. 3c Appendix) is employed.
| Equation 4 |
| Equation 5 |
Glycomics using Mass Spectrometry
All enzymatic reactions were performed in 20μL volume. After fixed reaction time, 20μL acetonitrile was added to terminate the reaction and precipitate proteins. Following centrifugation at 20,000g for 30min at 4°C, the supernatant was collected and dried using an Eppendorf Vacufuge concentrator (Hauppauge, NY). The sample was dissolved in aqueous solution and subjected to LC-MS analysis using an API3000 Triple Quadrupole mass spectrometer (AB Sciex, San Diego). Chromatography was performed at 0.2mL/min using a C18 column (XTerra MS, 5μm, 2.1mm×250 mm, Waters, Milford, MA). For MS separation, gradient started with 100% buffer A (water+0.1% formic acid (FA)) and linearly reached 20% buffer B (acetonitrile+0.1%FA) at 10 min. and 100% B at 25min. Product detection was performed using Xtracted Ion Chromatogram (XIC) based on theoretical m/z values.
Statistics
Unless specified otherwise, all data are presented as mean ± standard error mean for ≥ 3 independent runs. P<0.05 considered as statistically significant.
RESULTS
Expression and purification of human ST3Gals
Soluble forms of ST3Gal-I, -II, -III, -IV and -VI were expressed as fusion proteins with the C-terminal human Fc tag. Homology assessment of the catalytic domains for ST3Gals was conducted using Rat ST3Gal-II and ST3Gal-III as the template sequence (Fig. 1B). As seen, human ST3Gal-I and -II possessed 64% and 97% sequence homology with rat ST3Gal-II/α2,3(O)ST [12]. The remaining human enzymes clustered with the rat ST3Gal-III/α2,3(N)ST [14]. These recombinant enzymes contain the stem and catalytic domain including the L (Large), S (Small), III and VS (Very Small), respectively (Fig. 1C). Theoretical molecular mass for all enzymes matched with bands detected on SDS-PAGE (Fig. S1).
Substrate specificity for ST3Gals
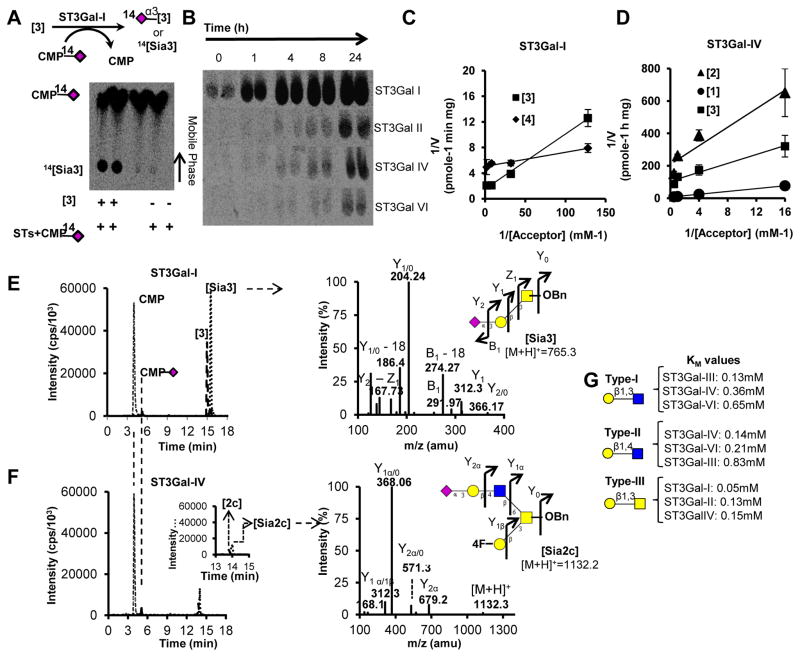
Specificity analysis was performed using acceptors listed in Table S1. Table 1 and Fig. 2 summarize the KM values for all enzymes. The overall radioactivity RP-TLC protocol is illustrated in Fig. 2A. Reaction time used for such analysis was 2h for ST3Gal-I, -II using acceptor [3] and 16h for ST3Gal-IV, -VI using acceptor [2] based on the linear range for initial velocities (Fig. 2B). ST3Gal-I, -II and -IV catalyzed sialic acid transfer to [3] resulting in Neu5Acα2,3Galβ1,3GalNAc [Sia3] (Fig. 2C, 2D, S2A). This transfer was specific towards the C-3 hydroxyl of Gal since MeO-3-Galβ1,3GalNAc-OBn [3a] was a poor acceptor for these enzymes (Table 2). The enzymatic activity towards [3] followed the sequence: ST3Gal-I (KM =0.05mM) > ST3Gal-II (KM =0.13mM) ~ST3Gal-IV (KM =0.14mM). ST3Gal-I, also, efficiently catalyzed core-2 trisaccharide [4] with a lower KM of 8μM (Fig. 2C). LC-MS/MS confirmed the synthesis of linear [Sia3] by ST3Gal-I. The MS2 fragmentation spectra of [Sia3] depicted sialic acid (m/z=292), Gal (m/z=168), GalNAc (m/z=204) and GalNAc-OBn (m/z=312) in the positive ion mode (Fig. 2E).
Table 1.
Michaelis-Menten constants for ST3Gals
| Enzyme | Species | KM (mM)~ | Acceptor |
|---|---|---|---|
| ST3Gal I | Type I | ||
| Human | 3.23# | [1] | |
| Mouse | 3.20$ | Galβ1,3GlcNAc | |
| Type II | |||
| Human | NR | [2] | |
| Type III | |||
| Human | 0.05# | [3] | |
| Rat | 0.05$ | [3] | |
|
| |||
| ST3Gal II | Type I | ||
| Human | 1.71# | [1] | |
| Mouse | 2.30$ | Galβ1,3GlcNAc | |
| Type II | |||
| Human | NR | [2] | |
| Type III | |||
| Human | 0.13# | [3] | |
| Rat | 0.16$ | [3] | |
| Mouse | 0.45@ | [3] | |
|
| |||
| ST3Gal III | Type I | ||
| Human | 0.13# | [1] | |
| Mouse | 0.32$ | Galβ1,3GlcNAc | |
| Type II | |||
| Human | 0.83# | [2] | |
| Mouse | 3.00$ | Galβ1,4GlcNAc | |
| Type III | |||
| Human | NP# | [3] | |
| Mouse | 2.2$ | [3] | |
|
| |||
| ST3Gal IV | Type I | ||
| Human | 0.65# | [1] | |
| Rat | 0.11$ | [1] | |
| Mouse | 0.75& | [1] | |
| Type II | |||
| Human | 0.14# | [2] | |
| Rat | 0.49* | [2] | |
| Mouse | 0.22& | [2] | |
| Type III | |||
| Human | 0.15# | [3] | |
| Rat | 3.00$ | [3] | |
|
| |||
| ST3Gal VI | Type I | ||
| Human | 0.36# | [1] | |
| Type II | |||
| Human | 0.21# | [2] | |
| Type III | |||
| Human | NR# | [3] | |
Figure 2. Sialylation enzymology.
Enzymatic reactions performed using recombinant ST3Gals and acceptors listed in Table 1 under ‘standard’ conditions described in Methods. Products resolved using RP-TLC or LC-MS. A. Synthesis of 14[Sia3] upon incubation of [3] with ST3Gal-I and CMP-[14C]Neu5Ac for 2h. CMP-[14C]Neu5Ac separated from radiolabeled product seen as an upper dark spot on RP-TLC. Right lanes show negative controls lacking acceptor. B. [14C] signal exhibiting product synthesis on varying reaction time from 0 – 24h. C. Lineweaver-Burk plot for reactions containing 0.008 – 0.5mM [3] or [4], 0.4mU/mL ST3Gal-I and 0.5mM CMP-Neu5Ac. D. Lineweaver-Burk plot for mixture containing 0.0625–2.0mM of either [1], [2], or [3] along with 1.1mU/mL ST3Gal-IV and 0.5mM CMP-Neu5Ac. E. 0.4mU/mL ST3Gal-I was incubated with 0.5mM CMP-Neu5Ac and 0.5mM [3] for 12h. MS/MS spectra for [Sia3] is shown to the right. F. Identical reaction as in panel D was performed only with 1.1mU/mL ST3Gal-IV, 0.5mM CMP-Neu5Ac and 0.5mM [2c]. MS/MS profile for the reaction product [Sia2c] is shown to the right. Data are from 3 independent runs. G. KM estimated for various ST3Gals that act on Type-I, -II and -III substrates.
Table 2.
Substrate specificity analysis
| ST3Gal-I | ST3Gal-II | ST3Gal-III | ST3Gal-IV | ST3Gal-VI | |
|---|---|---|---|---|---|
| Acceptor | Conversion (%dpm/μl) | ||||
| Type-I | |||||
| Galβ1,3GlcNAcβ1,3lac-β-pNp [1] | 0.47 | 8.75 | 59.9 | 19.2 | 7.62 |
| MeO-2-Galβ1,3GlcNAc-OBn [1a] | Nil | Nil | 32.88 | 5.91 | 4.53 |
|
| |||||
| Type-II | |||||
| Galβ1,4GlcNAcβ1,3Gal-β-pNp [2] | - | - | 16.78 | 79.5 | 5.4 |
| Galβ1,4GlcNAcβ1,3Galβ1,4GlcNAc-pNp [2a] | - | - | 15.21 | 20.87 | 13.14 |
| MeO-4-Galβ1,4GlcNAc-OBn [2b] | - | - | 15.56 | 66.97 | 100* |
| Galβ1,4GlcNAcβ1,6(4FGalβ1,3)GalNAc-OBn [2c] | - | - | 100* | 100* | 35.1 |
|
| |||||
| Type-III | |||||
| Galβ1,3GalNAc-OBn [3] | 100* | 100* | - | - | - |
| MeO-3-Galβ1,3GalNAc-OBn [3a] | Nil | Nil | - | - | - |
| Galβ1,3(GlcNAcβ1,6)GalNAc-OBn [4] | 48.53 | 100 | - | - | - |
Conversion quantified as % transfer of radioactivity from CMP-[14C]Neu5Ac to acceptors.
Data normalized to the acceptor exhibiting maximum activity for that enzyme, set as 100%. Nil: Conversion < 3×background.
ST3Gal-III, -IV and -VI exclusively sialylated Galβ1,4GlcNAcβ1,3Gal-pNp [2] and its variants. Based on KM estimates, the enzyme preference for [2] varies as: ST3Gal-IV (KM =0.14mM) > ST3Gal-VI (KM =0.21mM) > ST3Gal-III (KM =0.83mM) (Fig. 2G, S2B, C). The activity of these enzymes towards Galβ1,3GlcNAcβ1,3lac-pNp [1], on the contrary, follows as: ST3Gal-III (KM =0.13mM) > ST3Gal-VI (KM =0.36mM) > ST3Gal-IV (KM =0.65mM). Fig. 2F presents representative MS study shows Neu5Acα2,3Galβ1,4GlcNAcβ1,6(4F-Galβ1,3)GalNAc-OBn [Sia2c] formation upon addition of ST3Gal-IV and CMP-Neu5Ac to [2c]. MS2 fragmentation of this reaction product [Sia2c] results in the expected fragments: 4F-Galβ1,3GalNAc (m/z=368.4), GalNAc-OBn (m/z=312) along with other peaks. In general, [2c] was a better acceptor for both ST3Gal-IV and -VI compared to linear LacNAc based acceptors [2] and [2a] (Table 2). ST3Gal-IV and -VI also sialylated MeO-4-Galβ1,4GlcNAc-OBn [2b].
Product inhibition studies for ST3Gal-I
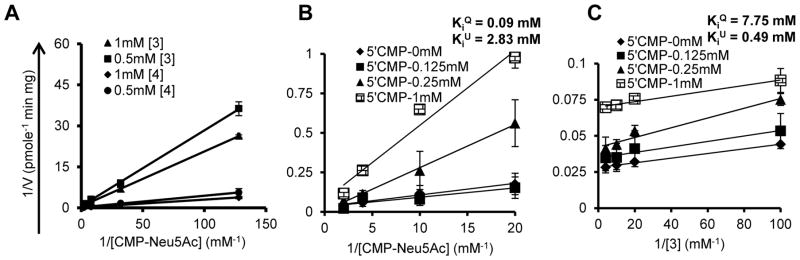
As STs catalyze multi-substrate reactions, identification of catalytic mechanism may facilitate enzyme-specific inhibitor design. Lineweaver-Burk plots resulted in a family of intersecting lines on varying CMP-Neu5Ac at fixed concentrations of [3] and [4] substrates (Eq. 1 or 2, Fig. 3A), suggesting sequential bi-bi mechanism. To further identify the nature of sequential bi-bi mechanism, mixed inhibition studies were conducted (Eq.4). Here, either CMP-Neu5Ac or [3] were varied at fixed CMP concentrations (0, 0.125mM, 0.25mM, 1mM) while leaving the other substrate in larger excess. On varying CMP-Neu5Ac, kinetic parameters arranged as suggesting partial competition with CMP (Fig. 3B). Also, as seen from the hyperbolic secondary plot, curve saturates for (Eq.5, Fig. S3A). Further, increases with CMP suggesting that product abrogates reaction velocity (Eq.5, Fig. S3B). Uncompetitive inhibition occurred between CMP and [3] as apparent from and secondary plots (Fig. 3C, S3C, D) [18].
Figure 3. ST3Gal-I reaction mechanism.
A. Michaelis-Menten plot obtained to study bi-bi mechanism by varying CMP-Neu5Ac from 0.008–0.5mM in the presence of 1mM and 0.5mM [3]/[4], respectively. B. ST3Gal-I reaction kinetics in the presence of 1mM [3] (Substrate B), and different fixed concentrations of CMP (Substrate Q). C. Experiments identical to panel B were performed only upon varying [3] in the presence of fixed 1mM CMP-Neu5Ac (Substrate A). All reactions were carried out in the presence of 1 mU/ml (panel A) and 0.1mU/ml (panel B, C) ST3Gal-I, respectively.
DISCUSSION
Structure Activity Relation (SAR) for ST3Gals
Homology assessment between human and rat ST3Gal translates to acceptor specific KM values. However, lack of crystallography data on mammalian STs limits our ability to link KM differences to their protein sequence. However, we note that the protein sequence for human ST3Gal-III, -IV and -VI compared to ST3Gal-I, -II has a prominent absence of Tyr230 and Tyr266 in the sialyl motif L and S. Thus, activity towards [1], [2] and its variants by these enzymes may be attributed to these residues. In particular, Tyr230 is replaced by aromatic residue Trp255 on ST3Gal-III. This disrupts interaction with Gal OH-6 thus justifying specificity of ST3Gal-III towards [2] substrate [19]. In addition, conservation of Glu residue on ST3Gal-IV that aligns with Glu196 on ST3Gal-I presumably participates in its activity towards [3] substrate.
In comparison to ST3Gal-II, ST3Gal-I catalyzes [3] with low KM of ~0.06mM. ST3Gal-II acts primarily on linear glycans belonging to globo series, as observed from its KM of 0.17mM towards [3]. Presence of conserved Tyr266 in the motif L on ST3Gal-I and -II induces favorable interaction with the axial hydroxyl on C-4 bound to Gal and GalNAc monosaccharides [20,21]. This agrees with low affinity of ST3Gal-I (KM = 3mM) towards [1] that has equatorial hydroxyl on C-4. In addition, ST3Gal-I forms [Sia4] with even higher efficiency as noted from its KM of 8μM. In this context, it’s often said that C2GnTI and ST3Gal-I compete each other for the common substrate core 1 i.e. [3] containing glycan (Fig. 4A). Our kinetic studies reveal that these two enzymes co-operate in an efficient assembly process of core2 sialylated glycan i.e. [Sia4] [22,23].
Figure 4. Reaction Schematic.
A. Graphical representation of the kinetic analysis performed using synthetic glycoconjugates found on cell-surface bound or circulatory glycoproteins. B. Illustrative schema for the random sequential bi-bi (upper panel) and iso sequential bi-bi (lower panel) reaction mechanism described for ST3Gal-I using production inhibition studies.
Enzymatic mechanism and inhibitor design
STs catalyze multi-substrate sialylation reaction, with the relative concentration of reactants dictating the catalytic rate progression. To facilitate specific inhibitor design, sequence in which substrate-binding results in transition state formation must be well understood. In this regard, product inhibition studies for ST3Gal-I postulate either an iso-sequential or random bi-bi mechanism (Fig. 4B) [24,25,26,27]. Overall, either of these mechanisms suggests identification of atomic interaction specific to catalytic amino acids and reacting substrates. As understood from the porcine ST3Gal-I crystal structure, its TS involve His302 and His319 interaction with phosphate group on 5′CMP and 3rd OH on Galactose moiety of [3] substrate [28]. Further, as Galβ1,3GalNAc forms primary substrate over Galβ1,3GlcNAc, designing sugar analog that disrupts interaction between 4-OH of GalNAc and N atom of Gln108 may inhibit sialylation. In a recent study, Brockhausen’s group reports that Galβ1,3(4-deoxy)GalNAcαO benzyl is an effective acceptor for ST3Gal-I. This suggests sialylation abrogation requires bulky group at 4th carbon [29]. In addition, transition state also proposes disruption of sialic acid interactions with the nearby residues. In this regard, CMP-Neu5Gc incubation with ST3Gal-I exhibited competitive binding with CMP-[14C]Neu5Ac with KM of 0.3 and 0.2 mM for substrates [3] and [4], respectively. Thus, sialic acid modification affects kinetic rate and abrogates sialylation (Fig. S4) [30]. Overall, comprehensive Structure-Activity-Relation (SAR) analysis dictates potential atomic perturbations leading to intelligent inhibitor design.
Supplementary Material
Highlights.
KM for five Human ST3Gals is reported towards Type-I, Type-II & Type-III acceptors.
LC-MS simultaneously quantifies CMP-Neu5Ac & Glycans in a sialylation reaction.
Efficient Core2 sialylation indicates co-operativitiy between ST3Gal-I & C2GnT1.
ST3Gal-I inhibition study proposes iso- or random-sequential bi-bi mechanism.
Acknowledgments
This study was funded by NIH HL63014.
ABBREVIATIONS
- Gal
Galactose
- GalNAc
N-acetylgalactosamine
- GlcNAc
N-acetylglucosamine
- CMP-[14C]Neu5Ac
C14-labeled cytidine 5′-monophosphate N-acetylneuraminic acid
- CMP-Neu5Gc
Cytidine 5′-monophosphate N-glycolylneuraminic acid
- LacNAc
N-acetyllactosamine
- LC-MS
Liquid Chromatography-Mass Spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Buschiazzo A, Alzari PM. Structural insights into sialic acid enzymology. Curr Opin Chem Biol. 2008;12:565–572. doi: 10.1016/j.cbpa.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 2.Audry M, Jeanneau C, Imberty A, et al. Current trends in the structure-activity relationships of sialyltransferases. Glycobiology. 2011;21:716–726. doi: 10.1093/glycob/cwq189. [DOI] [PubMed] [Google Scholar]
- 3.Mondal N, Buffone A, Jr, Stolfa G, et al. ST3Gal-4 is the primary sialyltransferase regulating the synthesis of E-, P-, and L-selectin ligands on human myeloid leukocytes. Blood. 2015;125:687–696. doi: 10.1182/blood-2014-07-588590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moody AM, North SJ, Reinhold B, et al. Sialic acid capping of CD8beta core 1-O-glycans controls thymocyte-major histocompatibility complex class I interaction. J Biol Chem. 2003;278:7240–7246. doi: 10.1074/jbc.M210468200. [DOI] [PubMed] [Google Scholar]
- 5.Ohnishi K, Komohara Y, Saito Y, et al. CD169-positive macrophages in regional lymph nodes are associated with a favorable prognosis in patients with colorectal carcinoma. Cancer Sci. 2013;104:1237–1244. doi: 10.1111/cas.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu G, Marathe DD, Matta KL, et al. Systems-level modeling of cellular glycosylation reaction networks: O-linked glycan formation on natural selectin ligands. Bioinformatics. 2008;24:2740–2747. doi: 10.1093/bioinformatics/btn515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang WH, Nussbaum C, Grewal PK, et al. Coordinated roles of ST3Gal-VI and ST3Gal-IV sialyltransferases in the synthesis of selectin ligands. Blood. 2012;120:1015–1026. doi: 10.1182/blood-2012-04-424366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kono M, Ohyama Y, Lee YC, et al. Mouse beta-galactoside alpha 2,3-sialyltransferases: comparison of in vitro substrate specificities and tissue specific expression. Glycobiology. 1997;7:469–479. doi: 10.1093/glycob/7.4.469. [DOI] [PubMed] [Google Scholar]
- 9.Williams MA, Kitagawa H, Datta AK, et al. Large-scale expression of recombinant sialyltransferases and comparison of their kinetic properties with native enzymes. Glycoconj J. 1995;12:755–761. doi: 10.1007/BF00731235. [DOI] [PubMed] [Google Scholar]
- 10.Stavenhagen K, Hinneburg H, Thaysen-Andersen M, et al. Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: an evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J Mass Spectrom. 2013;48:627–639. doi: 10.1002/jms.3210. [DOI] [PubMed] [Google Scholar]
- 11.Okajima T, Fukumoto S, Miyazaki H, et al. Molecular cloning of a novel alpha2,3-sialyltransferase (ST3Gal VI) that sialylates type II lactosamine structures on glycoproteins and glycolipids. J Biol Chem. 1999;274:11479–11486. doi: 10.1074/jbc.274.17.11479. [DOI] [PubMed] [Google Scholar]
- 12.Chandrasekaran EV, Xue J, Xia J, et al. Reversible sialylation: synthesis of cytidine 5′-monophospho-N-acetylneuraminic acid from cytidine 5′-monophosphate with alpha2,3-sialyl O-glycan-, glycolipid-, and macromolecule-based donors yields diverse sialylated products. Biochemistry. 2008;47:320–330. doi: 10.1021/bi701472g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marathe DD, Chandrasekaran EV, Lau JT, et al. Systems-level studies of glycosyltransferase gene expression and enzyme activity that are associated with the selectin binding function of human leukocytes. FASEB J. 2008;22:4154–4167. doi: 10.1096/fj.07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandrasekaran EV, Xue J, Xia J, et al. Analysis of the Specificity of Sialyltransferases toward Mucin Core 2, Globo, and Related Structures. Identification of the Sialylation Sequence and the Effects of Sulfate, Fucose, Methyl, and Fluoro Substituents of the Carbohydrate Chain in the Biosynthesis of Selectin and Siglec Ligands, and Novel Sialylation by Cloned α2,3(O)Sialyltransferase †. Biochemistry. 2005;44:15619–15635. doi: 10.1021/bi050246m. [DOI] [PubMed] [Google Scholar]
- 15.Madabhushi SR, Shang C, Dayananda KM, et al. von Willebrand factor (VWF) propeptide binding to VWF D′D3 domain attenuates platelet activation and adhesion. Blood. 2012;119:4769–4778. doi: 10.1182/blood-2011-10-387548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buffone A, Jr, Mondal N, Gupta R, et al. Silencing alpha1,3-fucosyltransferases in human leukocytes reveals a role for FUT9 enzyme during E-selectin-mediated cell adhesion. J Biol Chem. 2013;288:1620–1633. doi: 10.1074/jbc.M112.400929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patil SA, Chandrasekaran EV, Matta KL, et al. Scaling down the size and increasing the throughput of glycosyltransferase assays: activity changes on stem cell differentiation. Analytical biochemistry. 2012;425:135–144. doi: 10.1016/j.ab.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee HJ, Lairson LL, Rich JR, et al. Structural and kinetic analysis of substrate binding to the sialyltransferase Cst-II from Campylobacter jejuni. J Biol Chem. 2011;286:35922–35932. doi: 10.1074/jbc.M111.261172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeanneau C, Chazalet V, Auge C, et al. Structure-function analysis of the human sialyltransferase ST3Gal I: role of n-glycosylation and a novel conserved sialylmotif. The Journal of Biological Chemistry. 2004;279:13461–13468. doi: 10.1074/jbc.M311764200. [DOI] [PubMed] [Google Scholar]
- 20.Rao FV, Rich JR, Rakic B, et al. Structural insight into mammalian sialyltransferases. Nature structural & molecular biology. 2009;16:1186–1188. doi: 10.1038/nsmb.1685. [DOI] [PubMed] [Google Scholar]
- 21.Kim YJ, Kim KS, Kim SH, et al. Molecular cloning and expression of human Gal beta 1,3GalNAc alpha 2,3-sialytransferase (hST3Gal II) Biochem Biophys Res Commun. 1996;228:324–327. doi: 10.1006/bbrc.1996.1660. [DOI] [PubMed] [Google Scholar]
- 22.Dalziel M, Whitehouse C, McFarlane I, et al. The relative activities of the C2GnT1 and ST3Gal-I glycosyltransferases determine O-glycan structure and expression of a tumor-associated epitope on MUC1. J Biol Chem. 2001;276:11007–11015. doi: 10.1074/jbc.M006523200. [DOI] [PubMed] [Google Scholar]
- 23.Julien S, Grimshaw MJ, Sutton-Smith M, et al. Sialyl-Lewis(x) on P-selectin glycoprotein ligand-1 is regulated during differentiation and maturation of dendritic cells: a mechanism involving the glycosyltransferases C2GnT1 and ST3Gal I. J Immunol. 2007;179:5701–5710. doi: 10.4049/jimmunol.179.9.5701. [DOI] [PubMed] [Google Scholar]
- 24.Willemoes M, Hove-Jensen B, Larsen S. Steady state kinetic model for the binding of substrates and allosteric effectors to Escherichia coli phosphoribosyl-diphosphate synthase. J Biol Chem. 2000;275:35408–35412. doi: 10.1074/jbc.M006346200. [DOI] [PubMed] [Google Scholar]
- 25.Rearick JI, Sadler JE, Paulson JC, et al. Enzymatic characterization of beta D-galactoside alpha2 leads to 3 sialyltransferase from porcine submaxillary gland. J Biol Chem. 1979;254:4444–4451. [PubMed] [Google Scholar]
- 26.Cleland WW. The kinetics of enzyme-catalyzed reactions with two or more substrates or products: I. Nomenclature and rate equations. Biochimica et Biophysica Acta (BBA) - Specialized Section on Enzymological Subjects. 1963;67:104–137. doi: 10.1016/0006-3002(63)91800-6. [DOI] [PubMed] [Google Scholar]
- 27.Valenzuela-Soto EM, Munoz-Clares RA. Betaine-aldehyde dehydrogenase from leaves of Amaranthus hypochondriacus L. exhibits an Iso Ordered Bi Bi steady state mechanism. J Biol Chem. 1993;268:23818–23823. [PubMed] [Google Scholar]
- 28.Rao FV, Rich JR, Rakic B, et al. Structural insight into mammalian sialyltransferases. Nat Struct Mol Biol. 2009;16:1186–1188. doi: 10.1038/nsmb.1685. [DOI] [PubMed] [Google Scholar]
- 29.Czuchry D, Desormeaux P, Stuart M, et al. Identification and biochemical characterization of a novel alpha2,3-sialyltransferase WbwA from pathogenic Escherichia coli serotype O104. J Bacteriol. 2015 doi: 10.1128/JB.00521-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rillahan CD, Antonopoulos A, Lefort CT, et al. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat Chem Biol. 2012;8:661–668. doi: 10.1038/nchembio.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Dorst JA, Tikkanen JM, Krezdorn CH, et al. Exploring the substrate specificities of alpha-2,6-and alpha-2,3-sialyltransferases using synthetic acceptor analogues. Eur J Biochem. 1996;242:674–681. doi: 10.1111/j.1432-1033.1996.0674r.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.