Abstract
HY5 (Long Hypocotyles 5) is a key transcription factor in Arabidopsis thaliana that has a pivotal role in seedling development. Soil nitrogen is an essential macronutrient, and its uptake, assimilation and metabolism are influenced by nutrient availability and by lights. To understand the role of HY5 in nitrogen assimilation pathways, we examined the phenotype as well as the expression of selected nitrogen assimilation-related genes in hy5 mutant grown under various nitrogen limiting and nitrogen sufficient conditions, or different light conditions. We report that HY5 positively regulates nitrite reductase gene NIR1 and negatively regulates the ammonium transporter gene AMT1;2 under all nitrogen and light conditions tested, while it affects several other genes in a nitrogen supply-dependent manner. HY5 is not required for light induction of NIR1, AMT1;2 and NIA genes, but it is necessary for high level expression of NIR1 and NIA under optimal nutrient and light conditions. In addition, nitrogen deficiency exacerbates the abnormal root system of hy5. Together, our results suggest that HY5 exhibits the growth-promoting activity only when sufficient nutrients, including lights, are provided, and that HY5 has a complex involvement in nitrogen acquisition and metabolism in Arabidopsis seedlings.
Keywords: HY5, nitrite reductase NIR1, AMT1;2, light regulation, nitrogen metabolism, root development
1. Introduction
HY5 (Long Hypocotyles 5) is a nuclear bZIP type of transcription factor in Arabidopsis thaliana that has been extensively studied for its role in photomorphogenesis [1]. HY5 promotes photomorphogenesis in response to light signals of various wavelengths, and lack of HY5 weakens photomorphogenic responses [2-5]. In addition to light signaling, HY5 is critically involved in auxin mediated root growth [4, 6,7]. Analyses of genomic binding sits of HY5 have identified as many as 11797 target genes, which equates to approximately 44% of all genes in the Arabidopsis genome [8], highlighting the pivotal role that HY5 plays in Arabidopsis development.
Nitrogen is an essential macronutrient and a key factor limiting agricultural productivity [9,10]. Plants absorb inorganic nitrogen mainly in two forms, nitrate (NO3−) and ammonium (NH4+), and their nitrogen metabolism is dynamically regulated in response to ambient nitrogen sources and levels as well as other environmental factors [10,11]. Higher plants have both high- and low-affinity nitrate uptake systems (HATS and LATS, respectively), which operate under different nitrate concentrations and are thought to be genetically distinct [12]. In addition, nitrate and nitrite act both as nutrients as well as signals for the global regulation of gene expression in Arabidopsis roots [9,13]. Different nitrogen sources and varying nitrogen levels can also affect transcriptional profiles and various physiological processes of plants [11,14]. Light signals play a crucial role in regulating nitrogen uptake, translocation and assimilation into organic compounds [15]. The rates of photosynthesis and respiration are known to vary as a function of tissue nitrogen concentration in various species and growth conditions [16,17].
By analyzing null mutants of Arabidopsis for HY5 and HYH (an HY5 homologue) genes, Jonassen et al. showed that these genes are important for high nitrate reductase activity [18]. They further showed that HY5 and HYH are activators of NIA2, and are involved in light inhibition of NRT1.1 [19]. Based on these studies, a scheme of signal transduction pathway from light to nitrate translocation and assimilation has been proposed [15]. Notably, these studies used a hy5 hyh double mutant growing in a growth medium (half-strength Murashige and Skoog salts containing 3% sucrose) that contained nitrogen [18-20]. Moreover, hy5 hyh double mutant exhibits a root growth phenotype opposite to the hy5 single mutant [7]. It is therefore necessary to examine hy5 single mutant for nitrogen related morphological phenotype and gene expression alterations.
Improved understanding of the complex network of light, hormones, and nitrogen requires answers to further questions, including whether HY5 regulates nitrogen-related genes in a nitrogen concentration-dependent manner; how the nitrogen related genes respond to light; and what the role of HY5 is in their light regulation. To this end, here we characterized the phenotype of a hy5 mutant under different nitrogen conditions and examined the expression of representative genes involved in nitrogen assimilation using quantitative real-time polymerase chain reactions (qPCR) and biochemical methods. We found that HY5 regulates many nitrogen related genes in a nitrogen concentration dependent manner, and that it constitutively activates NIR1 (Nitrite reductase 1) while suppresses AMT1;2 (Ammonium transporter 1;2), two important genes in nitrogen metabolism.
2. Materials and methods
2.1. Plant materials and growth conditions
Wild-type (WT) A. thaliana used herein was the Col-0 ecotype. The hy5-ks50 mutant (Columbia background) has been described previously [4, 21-23]. All chemical reagents were purchased from Sigma. For root phenotype analysis, surface sterilized seeds were sown on 12×12 cm plates with 40 ml of solid medium (0.8% agar) containing 10 mM KH2PO4 2 mM MgSO4, 1 mM CaCl2, 0.1 mM Fe-EDTA, 50 μM H3BO4, 12 μM MnSO4, 1 μM ZnCl2, 1 μM CuSO4, 0.2 μM Na2MoO4, and 0.5% sucrose (pH 5.5) [24]. This basal medium was supplemented with (ammonium)2succinate, NH4NO3, or KNO3 as the nitrogen source as indicated in the text [25]. The final concentrations of nitrogen in the media were 0, 0.5, 2, 5 or 25 mM. After cold stratification for 3 days at 4°C in the dark, the plates were incubated at 22°C in continuous white light (WLc) at 180 μmol · m−2.s−1 or under a 16 h light/8 h dark regime. The experiments on nitrogen concentration series or when unspecified, were carried out using the 16 h light/8 h dark light period. The lengths of individual primary roots of seedlings were measured with Image J software (National Institutes of Health; http://rsb.info.nih.gov/ij) from images captured with a Canon G7 camera. For treatments requiring different light sources, seeds were surface-sterilized and sown on solid 1% Murashige and Skoog medium supplemented with 0.5% sucrose (GM). After 3 days of cold treatment, seeds were exposed to WLc (180 μmol · m−2.s−1), FRc(140 μmol · m−2.s−1), Rc (80 μmol · m−2.s−1), or dark (Dc) conditions at 22°C for 4 days as described previously [22]. All experiments were performed at least three times and the data represent one independent experiment.
2.2. RNA analyses
Total RNA was extracted from Arabidopsis seedlings using the RNeasy Plant Mini kit (Qiagen) [26]. Reverse transcription was performed using the SuperScript II First-strand cDNA Synthesis System (Invitrogen) according to manufacturer's instructions. qPCR analysis was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) with a Bio-Rad CFX96 real-time PCR detection system. Each experiment was repeated with three independent samples, and qPCR reactions were performed with three technical replicates for each sample. The primers used for qPCR are listed in Supplementary Table 1. Relative RNA expression was calculated with UBQ1 (AT3G52590) as the endogenous reference control. For Figure 3 showing N concentration series, the value of WT at zero N concentration was set at 1 for each gene. In Figure 4A, relative transcript levels of the genes were compared to NIA1 of WT Dc (set to 1). Representative gene profiles were repeated with ACTIN 2 (At3g18780) as endogenous reference. Data presented are means of three biological parallels, and error bars represent standard deviation of the sampling distribution of the mean. Analysis of variance were tested by least significant difference at P = 0.01(LSD0.01), based on SAS statistical analysis package (version 9.1.3, SAS Institute, Cary, NC, USA).
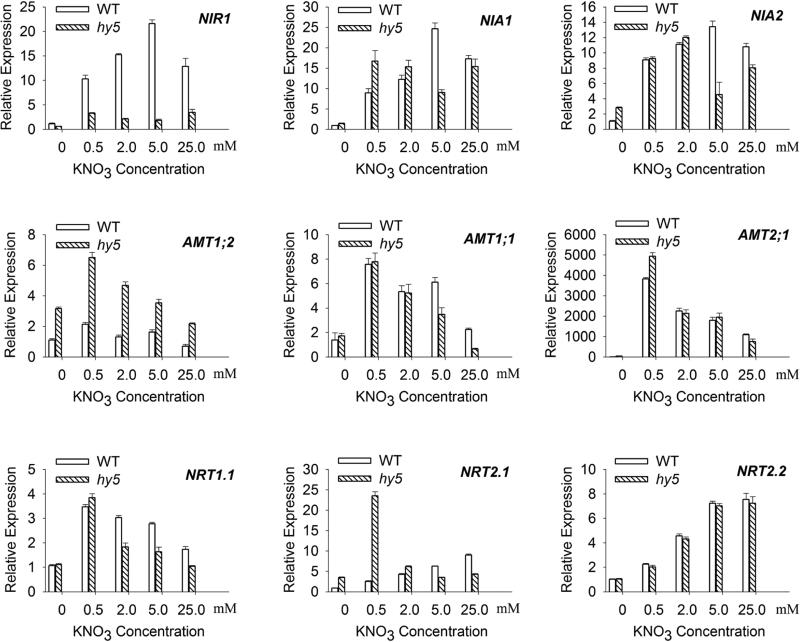
Figure 3.
Effect of the nitrogen supply on RNA expression of genes related to nitrogen metabolism. WT and hy5 mutant seedlings were grown for 10 days on plates with indicated concentration of potassium nitrate. The RNA samples were analyzed by real time RT-PCR against UBQ1 (At3g52590) on following genes: NIR1; NIA1; NIA2; AMT1;2; AMT1;1; AMT2;1; NRT1.1; NRT2.1; NRT2.2. The relative transcript level of WT under 0 mM KNO3 concentration is set to 1. Data presented are means of three repeats, and error bars are standard deviation.
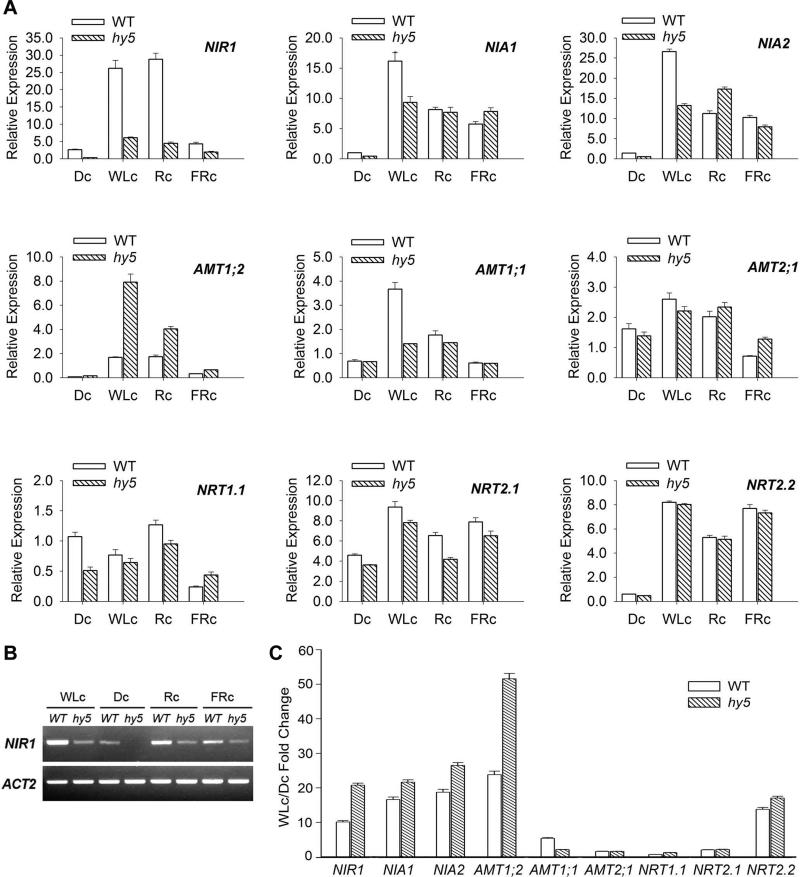
Figure 4.
Effect of different wavelengths of light on RNA transcript levels of genes related to nitrogen metabolism. A, WT and hy5 seedlings were grown on nitrogen sufficient media (GM) under indicated light source for 4 days, and RNA samples were analyzed by real time RT-PCR against UBQ1 (At3g52590) on following genes: NIR1; NIA1; NIA2; AMT1;2; AMT1;1; AMT2;1; NRT1.1; NRT2.1; NRT2.2. Dc, constant darkness; WLc, continuous white light; Rc, continuous red light; FRc, continuous far-red light. Relative transcript level of NIA1 in WT dark sample is set to 1. B, Decreased expression of NIR1 in hy5 under all light conditions was confirmed by RT-PCR DNA gel analysis, with ACTIN2 (ACT2 At3g18780) as the internal control. WLc, continuous white light; Dc, constant darkness; Rc, continuous red light; FRc, continuous far-red light. C, Fold of the light induction of tested genes WT or hy5. Light induction was calculated as the ratio (fold change) of WLc value over its corresponding Dc value (WLc/Dc) of each gene, according to the data in panel A.
2.3. Nitrite reduction activity assay
Tissue samples were extracted for nitrite reduction activity (NiRA) assays as described [27]. In brief, the assay mixture (pH 8.0) consisted of 75 mM Tris-HC1, 5 mM methyl-viologen and 50 mM sodium dithionite was prepared fresh in a solution of 30 mM sodium bicarbonate and 1 mM sodium nitrite. The reaction was initiated by adding freshly prepared sodium dithionite solution and followed by incubating at 25°C for 20 min. Then, the mixture was vortexed vigorously to oxidize the remaining dithionite and 0.1 ml of the mixture was diluted to 1 ml with water. To this mixture, 1 ml of 1% (w/v) sulfanilamide prepared in 1 N HCl and 1 ml of 0.01% (w/v) N-1-naphthylethylene-diamine-dihydrochloride prepared in water were added. After 30 min, the resultant color was measured with a spectrophotometer at 540 nm. NiRA is expressed as μM of nitrite consumed per hour per gram fresh weight (g−1.FW).
2.4. Determination of ammonium contents
Fresh seedlings of 500 mg of were added to 1 ml deionized water and shaken for 1 h at 45°C. Samples were centrifuged at 15000 g for 20 min. Ammonium content was determined for 50 μl of supernatant using 1 ml Nessler reagent (Merck). Optical density was measured at 404 nm and NH4+ content was determined using a standard curve and expressed as μmol NH4+ g−1.FW (Fresh Weight) [28].
2.5. Chlorophyll Measurement
Seedlings were collected and weighed for their fresh weight. Chlorophyll was extracted with ethanol and its content was assayed based on the absorbance of the extract at 645 and 663 nm [29].
3. Results
3.1. Phenotype of hy5 in response to different forms and concentrations of nitrogen nutrients
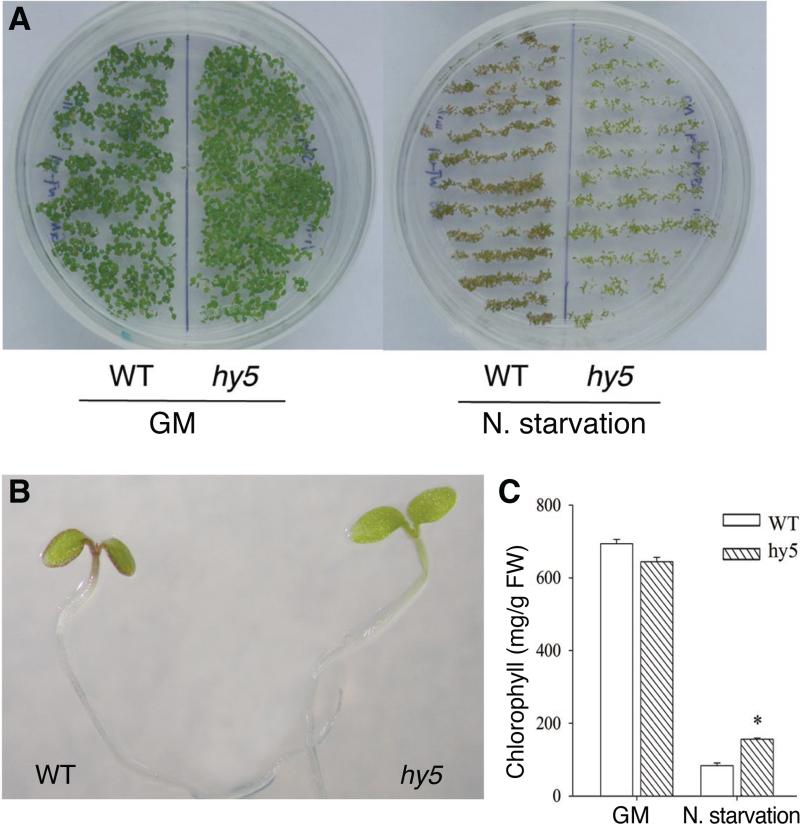
We examined the seedling phenotype of hy5 mutant growing under nitrogen deficient conditions. As a normal condition control, we used standard growth media (GM) that provided 60 mM of nitrogen, which is sufficient for plant growth. When grown on plates that had no nitrogen supplement (nitrogen starvation), wild type seedlings were purple, small, and about to die, confirming that nitrogen was necessary for plant survival. In comparison, the aerial part of hy5 mutants appeared greener than wild type under the same nitrogen starvation condition (Fig. 1A and 1B). While growing in the nitrogen sufficiency condition (GM), hy5 mutant was paler due to reduced chlorophyll content (92.7%) compared to that of wild type. Under nitrogen starvation however, the chlorophyll level of the hy5 mutant was 1.89-fold greater than wild type, though still substantially below normal chlorophyll levels (Fig. 1C). Thus, the reduction of chlorophyll content resulted from nitrogen deprivation was less severe in hy5 mutant, suggesting that hy5 was slightly more tolerant to nitrogen deficiency at least in the aerial part of the seedlings.
Figure 1.
Phenotype of the hy5 mutant growing under the nitrogen starvation condition. A. Wild type (WT) and hy5 seedlings of 4-day old growing on GM plate or on medium that lacked nitrogen nutrient (N. starvation). B. Enlarged image of WT and hy5 seedlings growing on nitrogen starvation plate for 4 days. C. Chlorophyll contents of 4-day-old WT and the hy5 mutant seedlings grown on GM plate and nitrogen starvation conditions.
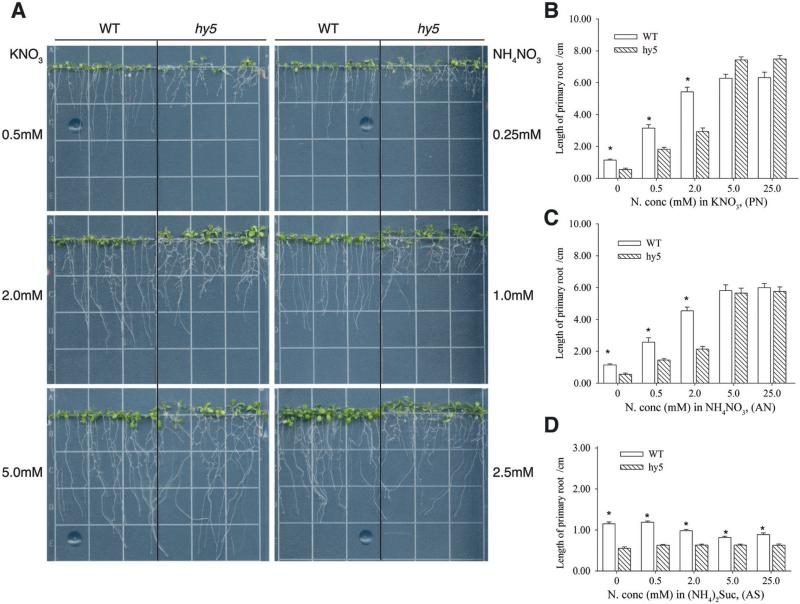
We examined the root growth of hy5 in a gradient concentration of nitrogen (0, 0.5 2, 5 and 25 mM) that were provided in three different forms (Fig. 2): Potassium nitrate (KNO3) was used as a nitrate salt; ammonium succinate [(NH4)2Suc] was used as ammonium salt; and ammonium nitrate (NH4NO3) was used as combination of nitrate and ammonium salt. The hy5 mutant displayed a drastically different root system architecture when grown at nitrogen-limiting conditions, such as in 0.5 mM of nitrogen supplied as potassium nitrate or as ammonium nitrate (equivalent to 0.25 mM NH4NO3) (Fig. 2A). Under those low nitrogen conditions, hy5 mutants had shorter primary roots but exaggerated lateral roots compared to wild type. We measured primary root length and found that hy5 had shorter primary roots than wild type at the nitrogen concentration range of 0-2 mM in both potassium nitrate (PN) or ammonium nitrate (AN) (Fig. 2B, 2C). Consistent with a previous report [4], wild type and hy5 mutant showed no significant differences in primary root length at nitrate concentrations higher than 5.0 mM (Fig. 2B, 2C).
Figure 2.
The root phenotype of hy5 seedlings growing in different nitrogen nutrient environments. A. The root system of WT and hy5 seedlings grown for 10 days on vertical plates that contained indicated concentrations of nitrogen in potassium nitrate (left) or in ammonium nitrate (right). On the same plate, left and right of the black line are WT and hy5 mutant seedlings, respectively. B-D. Primary root lengths of 10-day old WT and hy5 seedlings grown on B, potassium nitrate (PN); C, ammonium nitrate (AN); D, ammonium succinate (AS). Asterisks denote significant differences (* P<0.05) between WT and hy5 mutant.
Supplying ammonium salt alone did not promote root growth, and high concentration of (NH4)2Suc (higher than 0.25 mM) repressed root growth to less than 0.9 cm length in both wild type and hy5 mutant (Fig. 2D). This result is consistent with the reports that high concentrations of ammonium are toxic to plants [11, 28, 30, 31]. Under ammonium salt condition, the hy5 mutant showed 1.85−2.08-fold shorter primary root length than wild type. Considering its toxicity, (NH4)2Suc salt was not used as nitrogen nutrient in further experiments.
The different phenotypes of hy5 mutant in different nitrogen nutrition environments suggest that HY5 may have different roles under high-affinity nitrate uptake system (HATS) and low-affinity nitrate uptake system (LATS), which involve different nitrogen-related genes. It seems that HY5 participates in nitrogen management in a complex manner that depends on ambient nitrogen levels.
3.2. HY5 regulates genes involved in nitrogen metabolism in different ways
We next determined the transcript levels of nitrogen-related genes in seedlings growing in different concentrations of nitrogen nutrients, NH4NO3 (AN) or KNO3 (PN). Representative genes were examined, including those encoding enzymes for primary nitrogen metabolism such as NRT (Nitrate transporter), NIA (Nitrate reductase), NIR (Nitrite reductase), and AMT (Ammonium transporter). The expression of each gene in hy5 mutant was compared relative to that of the wild type under nitrogen starvation (0 mM) (set to 1 as reference value) (Fig. 3).
Amongst the genes examined, NIR1 and AMT1;2 were most strongly affected by hy5 mutation. NIR1 expression was strongly induced by nitrate, particularly potassium nitrate in wild type seedlings, but not in hy5 mutation (Fig.3 panel NIR1), indicating that HY5 is required for nitrate induced high level expression of NIR1. The ammonium transporter gene AMT1;2, which unlike AMT1;1 or AMT2;1, did not show significant nitrate induction in wild type seedlings. This gene was expressed in higher levels in hy5 than WT at all nitrogen conditions tested, indicating that HY5 has a negative role in AMT1;2 expression (Fig.3 panel AMT1;2). We also found that NRT2.2 expression was progressively induced by nitrate, but it was not affected by hy5 mutation under all nitrogen concentrations tested (Fig.3 panel NRT2.2).
The effect of hy5 mutation on the expression of NRT2.1, NIA1, NIA2 and AMT1;1 varied depending on the nitrogen growth conditions. AMT1;1 was only moderately decreased in hy5, and it occurred only at high nitrogen concentrations (greater than 5 mM) (Fig. 3). This observation also applied to NRT1.1, NIA1, NIA2; all of which showed moderately decreased expression only when grown in higher nitrogen concentrations, suggesting that HY5 positively influenced these genes expression under nitrogen sufficient conditions. However, under nitrogen starvation and low nitrogen condition (0.5 mM), NRT2.1 and NIA1 expression appeared higher in hy5 than in WT (Fig. 3). This result might suggest that HY5 could be involved in suppressing these genes during low nitrogen stress. Together we found that HY5 affects many nitrogen assimilation genes in a nitrogen-dependent manner, but HY5 constitutively activates NIR1 and inhibits AMT1;2 regardless of the nitrogen nutrient status.
3.3. Light induction of nitrogen related genes was largely preserved in hy5 seedlings
To determine the effect of HY5 on light regulation of nitrogen related genes, we examined RNA expression of these genes in wild type and hy5 seedlings grown in nitrogen sufficient conditions (GM) under continuous white light (WLc), far red light (FRc), red light (Rc), or darkness (Dc) (Fig. 4). Several genes examined were induced by light, including NIR1, AMT1;2, NRT2.2, NIA1, NIA2, and weakly, AMT1;1 and NRT2.1 (Fig. 4A, 4C). However the effects of hy5 mutation on those genes were different. In all light environments, NIR1 transcript levels were markedly reduced in hy5, whereas AMT1;2 transcript levels were markedly elevated in hy5 compared to wild type. Gel analysis of RT-PCR products further confirmed marked reduction of NIR1 in hy5 across all light conditions, while ACT2 (At3g18780) expression stayed largely constant (Fig. 4B). Together with data from nitrogen concentration series (Fig.3), our results indicate that HY5 constitutively promotes NIR1 and inhibits AMT1;2 regardless of the nitrogen nutrient concentration or light environments.
Nitrate transport gene NRT2.2 was induced by all wavelengths of lights in both wild type and hy5 mutant equally (Fig.4), indicating that lack of HY5 had little influence on NRT2.2 expression. This is also consistent with the NRT2.2 result in Figure 3, confirming that hy5 mutation has no detectable effect on NRT2.2 expression over different nitrogen concentrations or lights. NRT1.1, a dual transporter in both LATS and HATS, and NRT2.1, the major transporter in HATS, were moderately decreased in hy5 mutants compared to WT in the dark as well as in white light and red light (Fig.4). Again, these results are in agreement with the nitrogen concentration experiments, in which NRT1.1 and NRT2.1 expression moderately declined in hy5 in high concentration of nitrogen (Figure 3).
There are two functional genes in the nitrate reductase gene family, namely NIA1 (At1g77760) and NIA2 (At1g37130), with the latter reported to account for 90% of nitrate reductase activities in higher plants [32]. Expression of these two genes was very low under darkness, and was significantly induced by light. The expression of NIA1 and NIA2 in white light was notably decreased in the hy5 mutant compared with WT (by 62% and 68%, respectively). No significant reductions were observed in red light or far-red light. Similarly, AMT1;1 was under-expressed in hy5 mutant only under constant white light, suggesting that HY5 promoted these genes in white light.
To evaluate the effect of hy5 on light responsiveness of these genes, we calculated the light induction of each gene, or fold of change in gene expression increase (or decrease) in white light (WLc) over darkness (Dc) (Fig.4C). With exception of AMT1;1, light induction of most genes did not weaken due to lack of HY5. This surprising results suggest that, in contrast to most photosynthetic genes, HY5 is not essential for light-stimulated expression of many nitrogen related genes, such as NIR1, AMT1;2, NRT2.2, NIA1, and NIA2. Nonetheless, HY5 is important for overall high level expression of genes such as NIR1.
3.4. Changes in the nitrite reductase activity and the ammonium content in the hy5 mutant
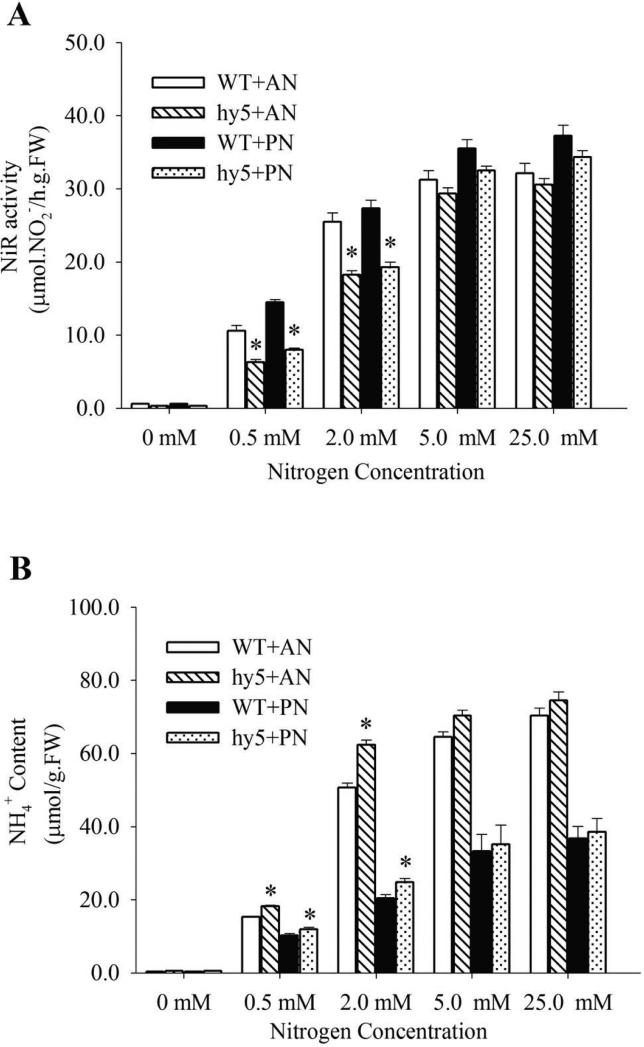
Prompted by nitrogen-dependent involvement of HY5 on genes involved in nitrogen uptake and assimilation, we examined the nitrite reductase activity (NiRA) and ammonium contents in hy5 seedlings growing in different nitrogen concentrations (Fig. 5). Under nitrogen depletion (0mM), NiRA and ammonium contents were almost undetectable in the hy5 mutant or wild type. With increasing nitrogen supply, the NiRA of seedlings rose steadily until the concentration of 5.0 mM, and then plateaued thereafter. hy5 showed lower NiRA compared to wild type, and the reduction was most significant under limited nitrogen supply of less than 2.0 mM ammonium nitrate or potassium nitrate (Fig.6A). This result suggests that HY5 has a greater role for seedling's NiRA when growing at limited nitrogen environment.
Figure 5.
Determination of nitrite reductase activity (NiRA) (A) and NH4+ content (B) in WT and hy5 mutants. Seedlings of 10-day old were grown in different nitrogen environments as indicated were used for the tests. Values are expressed relative to fresh weight of seedlings (FW). AN, ammonium nitrate; PN, potassium nitrate.
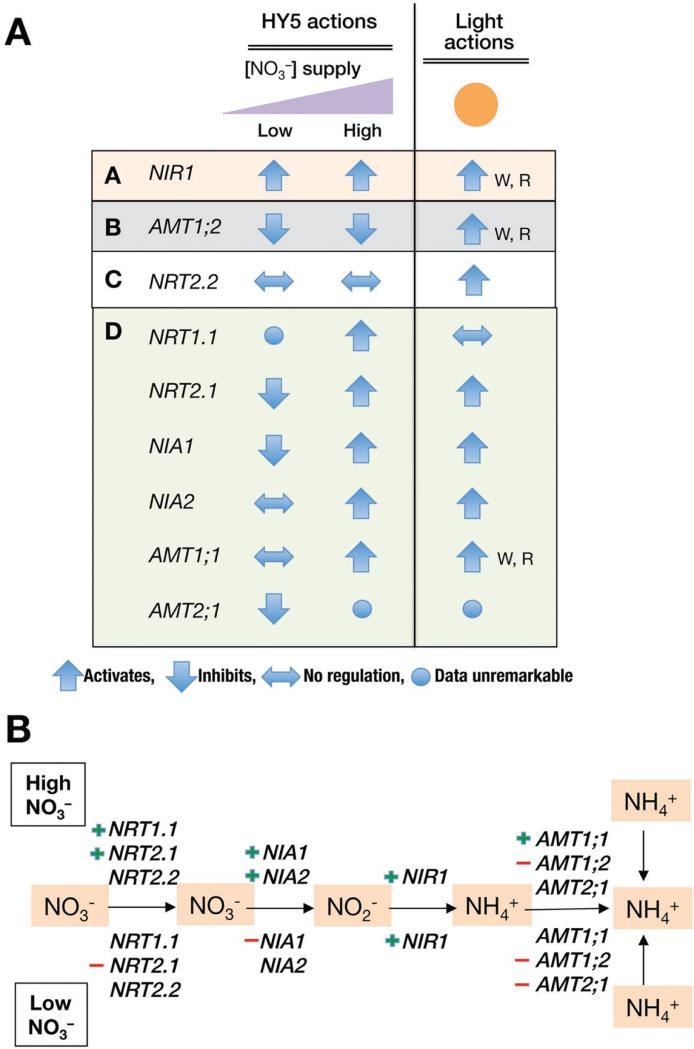
Figure 6.
Summaries on regulations of nitrogen transport and assimilation genes by HY5. A. Summary of the effect of HY5 on indicated nitrogen-related genes at low nitrogen (0, 0.5 2.0 mM KNO3) or high nitrogen (5.0 and 25 mM KNO3) conditions, according to the data shown in figure 3. Light actions of the genes are based on the data in Figure 4, by comparing the values of WLc to Dc in wild type seedlings. The label “W.R.” denotes that the induction only applies to WLc and Rc, but not FRc. B. A diagram showing tested nitrogen-related genes along a simplified the pathway of nitrogen uptake and assimilation. “+” signs indicate positive regulation by HY5, and “–“ signs indicate negative regulation by HY5, either in high NO3− conditions (upper side) or low NO3− (lower side) conditions.
At all concentrations of external nitrogen, the NH4+ contents of wild type and hy5 mutant seedlings were all significantly greater on ammonium nitrate medium compared with potassium nitrate medium, reflecting greater uptake and assimilation of NH4+ in the former medium by both genotypes. At 0.5 mM and 2 mM of ammonium nitrate, the NH4+ contents were 19% and 23% greater in the hy5 mutant than in WT, and when the nitrogen source was potassium nitrate, the NH4+ contents were 16% and 22% greater in the mutant than wild type respectively, thus showing significant differences between the two genotypes. At higher nitrogen level (5 mM and 25 mM), NH4+ contents did not differ significantly between wild type and hy5 on either the ammonium nitrate or potassium nitrate medium. It appeared that HY5 affects steps of NiRA and transportation of ammonium, resulting in abnormal nitrogen metabolism. The altered nitrite reductase activity and ammonium contents, particularly in the nitrogen-limiting environments, may contribute to the altered root system architecture of hy5 under those conditions.
4. Discussion
HY5 transcription factor is widely involved in growth and developmental processes of Arabidopsis. Here we studied the role of HY5 in nitrogen assimilation pathways by characterizing the hy5 mutant seedlings with respect to its nitrogen-dependent phenotype and the expression of selected genes related to nitrogen assimilation. Among the nine representative nitrogen-related genes selected for this study, five of which are known as sentinels for Primary Nitrate Response [33]: NRT1.1, NRT2.1, NIA1, NIA2, and NIR1. We presented evidence that HY5 positively regulates NIR1 expression, negatively regulates AMT1;2, and affects expression of several other genes in a manner dependent on the nitrogen nutrient supply. Surprisingly, HY5 does not appear to affect light responses of most of these genes, but it is necessary to support high level gene expression under optimal nutrient and light conditions. Under sustained low nitrogen conditions, lack of HY5 resulted in a more dramatic root phenotype, reduced NiRA activity, and increased ammonium content. The hy5 single mutant still expressed the HY5 homolog HYH, which might partially compensate the loss of HY5 functions in the mutant [34]. Despite of the partial redundancy, our data nonetheless showed that HY5 is an important player in the nitrogen assimilation pathway in Arabidopsis.
4.1. The hy5 mutant displays a complex effect on expression of nitrogen related genes
Among the nine representative nitrogen assimilation genes, we found that the expression of NIR1 and AMT1;2 changed most drastically in hy5, and the changes were consistent regardless of nitrogen supplement and light conditions tested (Fig. 3 and 4). Other genes examined were either not affected (NRT2.2) or weakly and conditionally affected by the lack of HY5, as summarized in Figure 6. We should mention that the experiment of nitrogen concentration series was not conducted by transient reduction or increase of the nutrient, but was conducted with seedlings that grew on specified conditions. Therefore our data cannot distinguish whether HY5 has a direct or indirect role in their gene expression. However, given the robust effect on NIR1 (stimulation) and AMT1;2 (inhibition), we think these two genes are likely directly regulated by HY5. We have searched the HY5 chromatin-IP genomic binding data [8]. Based on the binding criteria described in the report, all of the selected genes including NIR1 and AMT1;2 have been categorized as in vivo HY5 binding loci in light-grown plants (NIH Gene Expression Omnibus database accession number GSE24974).
Even on this small group of selected nitrogen-related genes, HY5 exhibited a complex pattern of regulation. According to how the genes are affected in hy5 mutant, they can be classified into 4 different types (Fig. 6A). Type A, as represented by NIR1, is activated by HY5 constitutively; Type B, as represented by AMT1;2, is inhibited by HY5 constitutively; Type C, as represented by NRT2.2, is not regulated by HY5; and Type D, which are affected by HY5 in a nitrogen concentration dependent manner. The functions of the tested genes in the nitrogen assimilation pathway are depicted in a simplified diagram in figure 6B. In general, it appears that HY5 tends to act negatively at low nitrate conditions (except for NIR1), while predominantly promotes nitrogen assimilation genes when nitrate are abundantly supplied (Fig. 6B). This is consistent with a general growth-stimulating role of HY5, either directly or indirectly [35,36].
We also found that different genes of the multigene family are distinctively regulated by HY5. For example, HY5 constitutively represses AMT1;2, but not AMT1;1 or AMT2;1. As a member of six isoforms in the AMT family, AtAMT1;2 is a low-affinity NH4+ transporter uniquely located in the plasma membrane of the root endodermis rather than in the rhizodermis and cortex as in high-affinity NH4+ transporter AMT1;1 and AMT1;3 [37,38]. AMT1;2 has been shown to be up-regulated in roots when internal ammonium concentration increases [39]. However, increased internal ammonium concentration in hy5 (Fig. 5) is unlikely the direct cause of the abnormal elevation of AMT1;2 in hy5. This is because hy5 seedlings exhibited increased internal ammonium level only at low nitrogen conditions, but exhibited increased AMT1;2 expression at all nitrogen conditions tested (Fig. 3 and 4). Further study is required to understand the mechanism of cell-specific regulation of different gene isoforms.
By investigating transient light responsiveness of nitrogen related genes in hy5 hyh double mutant, Lillo's team showed that HY5 and HYH positively regulate nitrate reductase activity as well as the main nitrate reductase gene NIA2 in Arabidopsis leaves [18-20], which is consistent with our observations. Interestingly, Jonassen et al., (2009b) did not find strong effect of hy5 hyh double mutant in transient (6 hours) induction of NIR1 by nitrate [19]. The discrepancy of the two studies on NIR1 maybe explained by one of the following reasons. 1) It is possible that the strong positive regulation of HY5 on NIR1 expression (Figs. 3 and 4) represent a persistent function of HY5, as oppose to a transient response. 2) Given that opposite root growth phenotypes have been observed in hy5 versus hy5 hyh double mutant [7], similar discrepancy in the single and double mutant might also occur in this case. 3) The internal reference genes in the RT-PCR analyses of RNA expression were different. Jonassen et al., (2009b) [19] used At3g02540, whereas we used UBQ1 (At3g52590) as the reference gene (Fig. 3 and Fig. 4A), and we further validated NIR1 result with ACT2 (At3g18780) (Fig. 4B). According to a microarray study of nitrate response submitted by Wolf Scheible lab to the database (https://www.arabidopsis.org/servlets/TairObject?type=hyb_descr_collection&id=1005823564) and a study of light response in hy5 (http://www.ncbi.nlm.nih.gov/geoprofiles) [8], UBQ1 and ACT2 show no significant responses to nitrate or light, and they are not regulated by HY5. However At3g02540 used in [19] showed weak nitrate response, with KNO3/KCl fold change as high as 2.378, while it is not subject to regulation by light or HY5 [8]. Detailed examination of the behaviors of internal control genes has lent further support to our result that HY5 is unequivocally required for high-level expression of NIR1.
The gene expression defects for certain genes in hy5 were found unremarkable and inconclusive (indicated by dots in Fig. 6A). This might be related to our protocol of using whole seedlings for RNA analysis, which would not allow us to clearly distinguish genes that are tissue-specifically regulated in roots or shoots. In the future, organ-specific sampling might provide more precise information on the function of HY5.
4.2. HY5 versus lights regulation of nitrogen related genes
HY5 and light signals both regulate the expression of nitrogen assimilation genes, but our results indicate that HY5 actions are not always in accord with light actions (Fig. 6A). For examples, AMT1;2 gene was induced by lights (WLc and Rc), but it was repressed by HY5; NRT2.2 gene was strongly induced by all wavelength of lights, but was not regulated by HY5. (Figs. 3, 4, and 6). In the case of NIR1, which was activated by both light and HY5, even though the hy5 mutation dramatically lowed the overall expression level of NIR1 (Fig. 4A), it did not weaken the light inducibility, the ratio of WLc over Dc, of the gene (Figs 4C). Similarly, the light induction of AMT1;2 was not weakened by hy5 mutation. In fact the folds of light induction for most genes examined, except for AMT1;2, were greater in hy5 than in wild type (Fig.4C). These results indicate that HY5 is not required for light activation of NIR1 and AMT1;2. This finding is in agreement with Jonassen et al. (2009b) [19] that HY5 and HYH are not required for transient light induction of several nitrogen assimilation genes, and that HY5 and HYH were involved in light repression of NRT1.1. Similarly, we found that light repression of NRT1.1 (Fig. 4A) did not occur in hy5, mainly due to its higher expression in the dark. Those results raise questions whether HY5 may have a distinct function in mediating gene expression of nitrogen assimilation pathway, besides its well-established role in light signaling.
It should be emphasized that our data support a pro-growth role of HY5 with regard to nitrogen nutrient and light nutrient. Under sufficient nutrient conditions, HY5 predominantly acts to promote expression of nitrogen-related genes (except for AMT1;2), as summarized in figure 6.
4.3. Enhanced root phenotype in hy5 when grown under chronic nitrogen stress
The hy5 mutant exhibited a drastically different root system that was exacerbated at low nitrogen conditions, as characterized by a significantly shorter primary root length and greater development of lateral roots (Fig. 2). The primary root phenotype of hy5 diminished when nitrogen was provided in sufficient amount (above 5 mM nitrate concentrations). Roots are known to sense and respond to local and global nutrient supply [30, 40-42] as well as other factors [43]. Nitrate itself is an important signal that modulates root growth [44]. Normally, low nitrogen limits, while high nitrate stimulates, the generation of lateral roots [41,45]. Extensive crosstalk between nitrate and auxin signaling has been reported to explain nitrate-dependent root morphogenesis [9,46], as auxin is a key modulator of root architecture: high auxin concentration inhibits primary root elongation and stimulates lateral root development. The hy5 and hy5 hyh double mutants, are known to have a root phenotype that is caused by increased auxin signaling in the mutant [4,6,7]. HY5 regulates a remarkable numbers of auxin responsive genes, and is critical in auxin-dependent regulation of root architecture and growth responses [6,7]. It is plausible that low nitrogen environment somehow enhanced the altered auxin signaling of hy5 roots, resulting in the drastic root system phenotype under sustained nitrogen deficiency. In addition, since excess ammonium in plants can cause ammonium poisoning and inhibit the growth of primary roots [11], the greater amounts of internal ammonium in hy5 at low nitrogen medium (less than 2 mM, Fig. 5B) likely contribute to the root phenotype of the mutant in the same medium.
Interestingly, the aerial part of hy5 appeared more tolerant to nitrogen starvation than wild type seedlings, as evidence by the alleviated reduction of chlorophyll under nitrogen starvation (Fig. 1). We suggest that the overall phenotypes of hy5 may imply that HY5 has important functions in the interplay of nitrogen signaling and auxin pathways in modulation of plant root growth.
Supplementary Material
Highlights.
The hy5 mutant displayed shorter primary root only at limiting nitrogen conditions.
The hy5 seedlings showed drastically reduced expression of NIR1 at all conditions.
The hy5 seedlings had higher expressions of AMT1;2 at all tested conditions.
Most genes tested displayed higher or unchanged light induction in hy5 mutant.
Higher NH4+ content and lower NiR activity occur in hy5 only at low N conditions
Acknowledgements
We thank Dr. Jigang Li (currently at China Agricultural University), Dr. Zhaohua Peng (Mississippi State University), and Dr. Christian Hardtke (University of Lausanne) for technical advice and helpful discussion. This work was supported jointly by the National Natural Science Foundation of China (31201154), the Natural Science Foundation of Jiangsu Province (BK2009187), the Priority Academic Program Development of Jiangsu Higher Education Institutions, National Institute of Health (US) grant 2R01GM047850-23A1, and the Agriculture and Food Research Initiative competitive grant (2011-67013-30125) of the USDA National Institute of Food and Agriculture (US).
Abbreviations
- NRT
Nitrate transporter
- NIA
Nitrate reductase
- NIR
Nitrite reductase
- AMT
Ammonium transporter
- HY5
Long Hypocotyles 5
- HYH
HY5 homologue
- HATS
high-affinity nitrate uptake systems
- LATS
low-affinity nitrate uptake systems
- WT
Wild-type
- Dc
constant darkness
- WLc
continuous white light
- Rc
continuous red light
- FRc
continuous far-red light
- PN
potassium nitrate
- AN
ammonium nitrate
- AS
ammonium succinate
- NiRA
nitrite reduction activity
- qPCR
quantitative PCR
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 2.Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Zeitschrift FÜR Pflanzenphysiologie. 1980;100:147–160. [Google Scholar]
- 3.Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- 4.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Gene Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbas N, Maurya JP, Senapati D, Gangappa SN, Chattopadhyay S. Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell. 2014;26:1036–1052. doi: 10.1105/tpc.113.122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cluis CP, Mouchel CF, Hardtke CS. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 2004;38:332–347. doi: 10.1111/j.1365-313X.2004.02052.x. [DOI] [PubMed] [Google Scholar]
- 7.Sibout R, Sukumar P, Hettiarachchi C, Holm M, Muday GK, Hardtke CS. Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. Plos Genet. 2006;2:e202. doi: 10.1371/journal.pgen.0020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011;65:346–358. doi: 10.1111/j.1365-313X.2010.04426.x. [DOI] [PubMed] [Google Scholar]
- 9.Vidal EA, Gutiérrez RA. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol. 2008;11:521–529. doi: 10.1016/j.pbi.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Xu G, Fan X, Miller AJ. Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol. 2012;63:153–182. doi: 10.1146/annurev-arplant-042811-105532. [DOI] [PubMed] [Google Scholar]
- 11.Britto DT, Kronzucker HJ. NH4+ toxicity in higher plants: a critical review. J Plant Physiol. 2002;159:567–584. [Google Scholar]
- 12.Liu KH, Huang CY, Tsay YF. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell. 1999;11:865–874. doi: 10.1105/tpc.11.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R, Xing X, Crawford N. Nitrite acts as a transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiol. 2007;145:1735–1745. doi: 10.1104/pp.107.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi YM, Wang RL, Zhu T, Rothstein SJ. Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genomics. 2007;8:281. doi: 10.1186/1471-2164-8-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lillo C. Signalling cascades integrating light-enhanced nitrate metabolism. Biochem J. 2008;415:11–19. doi: 10.1042/BJ20081115. [DOI] [PubMed] [Google Scholar]
- 16.Hachiya T, Noguchi K. Integrative response of plant mitochondrial electron transport chain to nitrogen source. Plant Cell Rep. 2011;30:195–204. doi: 10.1007/s00299-010-0955-0. [DOI] [PubMed] [Google Scholar]
- 17.Reich PB, Tjoelker MG, Machado JL, Oleksyn J. Universal scaling of respiratory metabolism, size and nitrogen in plants. Nature. 2006;439:457–461. doi: 10.1038/nature04282. [DOI] [PubMed] [Google Scholar]
- 18.Jonassen EM, Lea US, Lillo C. HY5 and HYH are positive regulators of nitrate reductase in seedlings and rosette stage plants. Planta. 2008;227:559–564. doi: 10.1007/s00425-007-0638-4. [DOI] [PubMed] [Google Scholar]
- 19.Jonassen EM, Sevin DC, Lillo C. The bZIP transcription factors HY5 and HYH are positive regulators of the main nitrate reductase gene in Arabidopsis leaves, NIA2, but negative regulators of the nitrate uptake gene NRT1.1. J Plant Physiol. 2009;166:2071–2076. doi: 10.1016/j.jplph.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Jonassen EM, Sandsmark BA, Lillo C. Unique status of NIA2 in nitrate assimilation: NIA2 expression is promoted by HY5/HYH and inhibited by PIF4. Plant Sign Behav. 2009;4:1084–1086. doi: 10.4161/psb.4.11.9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardtke C, Gohda K, Osterlund M, Oyama T, Okada K, Deng XW. HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 2000;19:4997–5006. doi: 10.1093/emboj/19.18.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sims L, Pastor J, Lee T, Dewey B. Nitrogen, phosphorus and light effects on growth and allocation of biomass and nutrients in wild rice. Oecologia. 2012;170:65–76. doi: 10.1007/s00442-012-2296-x. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Ribot C, Rezzonico E, Poirier Y. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 2004;135:400–411. doi: 10.1104/pp.103.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto M, Kumar A, Li W, Wang Y, Siddiqi MY, Crawford NM, Glass ADM. High-Affinity Nitrate Transport in Roots of Arabidopsis Depends on Expression of the NAR2-Like Gene AtNRT3.1. Plant Physiol. 2006;140:1036–1046. doi: 10.1104/pp.105.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, Ouyang X, Yang P, Lau OS, Li G, Li J, Chen H, Deng XW. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell. 2012;24:4590–4606. doi: 10.1105/tpc.112.103994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sivasankar S, Rothstein S, Oaks A. Regulation of the accumulation and reduction of nitrate by nitrogen and carbon metabolites in maize seedlings. Plant Physiol. 1997;114:583–589. doi: 10.1104/pp.114.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin C, Qian W, Wang W, Wu Y, Yu C, Jiang X, Wang D, Wu P. GDP-mannose pyrophosphorylase is a genetic determinant of ammonium sensitivity in Arabidopsis thaliana. P Natl Acad Sci USA. 2008;105:18308–18313. doi: 10.1073/pnas.0806168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wintermans JF, De MA. Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim Biophys Acta. 1965;109:448–453. doi: 10.1016/0926-6585(65)90170-6. [DOI] [PubMed] [Google Scholar]
- 30.Cerutti T, Delatorre CA. Nitrogen and phosphorus interaction and cytokinin: responses of the primary root of Arabidopsis thaliana and the pdr1 mutant. plant Sci. 2013;198:91–97. doi: 10.1016/j.plantsci.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 31.Sarasketa A, González-Moro MB, González-Murua C, Marino D. Exploring ammonium tolerance in a large panel of Arabidopsis thaliana natural accessions. J Exp Bot. 2014;65:6023–6033. doi: 10.1093/jxb/eru342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X, Sukumaran S, Márton L. Differential expression of the Arabidopsis Nia1 and Nia2 genes cytokinin-induced nitrate reductase activity is correlated with increased Nia1 transcription and mRNA levels. Plant Physiol. 1998;116:1091–1096. doi: 10.1104/pp.116.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medici A, Krouk G. The primary nitrate response: a multifaceted signalling pathway. J Exp Bot. 2014;65:5567–5576. doi: 10.1093/jxb/eru245. [DOI] [PubMed] [Google Scholar]
- 34.Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light dependent gene expression in Arabidopsis. Gene Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osterlund M, Hardtke C, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 37.Loqué D, Yuan L, Kojima S, Gojon A, Wirth J, Gazzarrini S, Ishiyama K, Takahashi H, Wirén NV. Additive contribution of AMT1; 1 and AMT1; 3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 2006;48:522–534. doi: 10.1111/j.1365-313X.2006.02887.x. [DOI] [PubMed] [Google Scholar]
- 38.Neuhäuser B, Dynowski M, Mayer M, Ludewig U. Regulation of NH4+ transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol. 2007;143:1651–1659. doi: 10.1104/pp.106.094243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan L, Loqué D, Ye F, Frommer WB, Wirén NV. Nitrogen-dependent posttranscriptional regulation of the ammonium transporter AtAMT1; 1. Plant Physiol. 2007;143:732–744. doi: 10.1104/pp.106.093237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desnos T. Root branching responses to phosphate and nitrate. Curr Opin Plant Biol. 2008;11:82–87. doi: 10.1016/j.pbi.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Forde BG, Walch-Liu P. Nitrate and glutamate as environmental cues for behavioural responses in plant roots. Plant Cell Environ. 2009;32:682–693. doi: 10.1111/j.1365-3040.2008.01927.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang R, Guegler K, LaBrie ST, Crawford NM. Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell. 2000;12:1491–1509. doi: 10.1105/tpc.12.8.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thum KE, Shin MJ, Gutiérrez RA, Mukherjee I, Katari MS, Nero D, Shasha D, Coruzzi GM. An integrated genetic, genomic and systems approach defines gene networks regulated by the interaction of light and carbon signaling pathways in Arabidopsis. BMC Syst Biol. 2008;2:31. doi: 10.1186/1752-0509-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.HO CH, Lin SH, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Mounier E, Pervent M, Ljung K, Gojon A, Nacry P. Auxin-mediated nitrate signalling by NRT1. 1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ. 2014;37:162–174. doi: 10.1111/pce.12143. [DOI] [PubMed] [Google Scholar]
- 46.Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, Zazimalova E, Benkova E, Nacry P, Gojon A. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell. 2010;18:927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 47.Vidal EA, Moyano TC, Riveras E, Contreras-López O, Gutiérrez RA. Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. P Natl Acad Sci USA. 2013;110:12840–12845. doi: 10.1073/pnas.1310937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canales J, Moyano TC, Villarroel E, A Gutiérrez R. Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Front Plant Sci. 2014;5:22. doi: 10.3389/fpls.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.