ABSTRACT
Human enterovirus 68 (EV-D68) is a member of the EV-D species, which belongs to the EV genus of the Picornaviridae family. Over the past several years, clusters of EV-D68 infections have occurred worldwide. A recent outbreak in the United States is the largest one associated with severe respiratory illness and neurological complication. Although clinical symptoms are recognized, the virus remains poorly understood. Here we report that EV-D68 inhibits innate antiviral immunity by downregulation of interferon regulatory factor 7 (IRF7), an immune factor with a pivotal role in viral pathogenesis. This process depends on 3Cpro, an EV-D68-encoded protease, to mediate IRF7 cleavage. When expressed in host cells, 3Cpro targets Q167 and Q189 within the constitutive activation domain, resulting in cleavage of IRF7. Accordingly, wild-type IRF7 is fully active. However, IRF7 cleavage abrogated its capacity to activate type I interferon expression and limit replication of EV-D68. Notably, IRF7 cleavage strictly requires the protease activity of 3Cpro. Together, these results suggest that a dynamic interplay between 3Cpro and IRF7 may determine the outcome of EV-D68 infection.
IMPORTANCE EV-D68 is a globally emerging pathogen, but the molecular basis of EV-D68 pathogenesis is unclear. Here we report that EV-D68 inhibits innate immune responses by targeting an immune factor, IRF7. This involves the 3C protease encoded by EV-D68, which mediates the cleavage of IRF7. These observations suggest that the 3Cpro-IRF7 interaction may represent an interface that dictates EV-D68 infection.
INTRODUCTION
Enterovirus D68 (EV-D68) was first isolated from children with lower respiratory tract infections in California, USA, in 1962 and belongs to the species Enterovirus D within the Enterovirus genus, Picornaviridae (1). A global upsurge of EV-D68 infections in patients with respiratory tract infections (RTIs) has been observed in recent years (2–21). In 2014, a large outbreak of EV-D68 infections occurred in the United States, which raised public health concern owing to severe respiratory illness and neurological complications (22–30).
Although linked to clinical disease, EV-D68 remains poorly characterized. EV-D68 is structurally similar to other enteroviruses (31). The virus possesses a genome approximately 7.4 kb in size, with the capacity to encode a large precursor that is processed into structural proteins (VP1, VP2, VP3, and VP4) and nonstructural proteins (2A, 2B, 2C, 3A, 3B, 3C, and 3D) (17). Viral infection initiates with sialic acids of the epithelial cells (32). In this process, the virus preferentially binds to α2,6-linked sialic acids rather than to α2,3-linked sialic acids (33). In addition, EV-D68 is able to infect leukocyte cells (34). As such, active replication of EV-D68 is thought to trigger cytokine responses (35).
It is well established that the pattern-recognition receptors (PRRs) initiate innate antiviral immunity through activation of interferon regulatory factor 3 (IRF3), interferon regulatory factor 7 (IRF7), and/or nuclear factor-κB (NF-κB) (36). This leads to the induction of type I interferons (IFNs) and inflammatory cytokines (37). IRF3 is a major player in the early phase of IFN induction, whereas IRF7 is critical in the late phase because its expression requires IFN derived from the initial infection (38, 39). Once activated, IRF7 cooperates with IRF3 to mediate antiviral responses. Recently, we reported that 3Cpro of EV-D68 perturbs the Toll-like receptor 3 (TLR3) pathway that controls cytokine expression (35). Nonetheless, whether EV-D68 targets other immune factors is unknown.
In this study, we found that EV-D68 suppresses expression of type I IFNs through cleavage of IRF7 in infected cells. This activity requires a functional viral 3Cpro. Furthermore, we show that IRF7 cleavage occurs at two sites located in the constitutive activation domain (CAD), resulting in inactive IRF7 fragments. Together, these results suggest that control of IRF7 by 3Cpro may be a viral mechanism that contributes to EV-D68 disease.
MATERIALS AND METHODS
Cell lines and viruses.
293T (CRL-11268; ATCC) cells and HeLa (CCL-2; ATCC) cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (HyClone, Logan, UT), 100 U/ml penicillin, and 100 U/ml streptomycin at 37°C in a 5% CO2 humidified atmosphere. Human monocytic THP1 (TIB-202; ATCC) cells were cultured in RPMI 1640 media supplemented with 10% FBS. EV-D68 infection was carried out as described previously (35). Peripheral blood mononuclear cells (PBMCs) isolated from healthy donors were cultured in RPMI 1640 media supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 U/ml), glutamine (2 mM), HEPES (5 mM), and sodium pyruvate (0.5 mM).
Plasmids.
Plasmids pEGFPC1, pEGFP-3C and pEGFP-3C variants, pCMV6-Flag-Myc-IRF7, pGL3-IFN-β–Luc, IFN-α4–Luc, and pRL-SV40 have been described elsewhere (35, 40). The IRF7 mutants were constructed by using Pfu DNA polymerase (Stratagene, La Jolla, CA). Plasmid pCDNA3.1-V5-IRES-EV71-2A was a gift from Zhendong Zhao (Institute of Pathogen Biology, Chinese Academy of Medical Sciences, China) and was described before (41). To construct pCDNA3.1-V5-IRES-EV-D68-2A, the sequences of EV71 were replaced with that of EV-D68. All variants were confirmed by subsequent sequencing.
Antibodies and reagents.
Antibodies against Flag, Myc, green fluorescent protein (GFP), V5, and β-actin were purchased from Sigma (St. Louis, MO). Rabbit antibodies against TBK1 and IRF7 (target amino acids near the N terminus of human IRF7) were purchased from Cell Signaling Technology (Danvers, MA). Rabbit antibody against IRF7 that recognizes the C terminus of human IRF7 was purchased from Novus Biologicals (Littleton, CO). Rabbit antibody against IRF3 was purchased from Epitomics (Burlingame, CA). Mouse anti-EV-D68 has been described before (35). IRDye 800-labeled IgG and IRDye 680-labeled IgG secondary antibodies were purchased from Li-Cor Biosciences (Lincoln, NE). Rupintrivir was purchased from Santa Cruz (Santa Cruz, CA).
Reporter assays.
293T cells were seeded in 24-well plates at a cell density of 3 × 105 cells per well. At 16 h after plating, cells were transfected with a control plasmid or a plasmid expressing IRF7, IRF7 mutants, and 3Cpro or its variants along with pGL3-IFN-β–Luc, IFN-α4–Luc, and pRL-SV40 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). At 24 h after transfection, cells were harvested and cell lysates were used to determine luciferase activities using a dual-luciferase reporter system (Promega, Madison, WI) according to the manufacturer's instructions. The firefly luciferase activities were normalized to the Renilla luciferase activities.
Quantitative real-time reverse transcription-PCR (RT-PCR).
Total RNA was extracted using TRIzol reagent (Invitrogen) and treated with DNase I (Pierce, Rockford, IL). Aliquots of RNA were reverse transcribed to cDNA using a Superscript cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions. Samples were then subjected to quantitative real-time PCR analysis using primers specific for detection of IFN-β, IFN-α1, interferon-stimulated gene 56 (ISG56), ISG54, interleukin-6 (IL-6), IL-8, RANTES, and IFN-γ-inducible protein 10 (IP10) using a SYBR green kit (TaKaRa Bio, Otsu, Japan), according to the manufacturer's instructions. Expression of IFN-β, IFN-α1, ISG56, ISG54, IL-6, IL-8, RANTES, and IP10 mRNA was normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA expression. The sequences of primers have been described elsewhere (42, 43).
Western blot analysis.
Cells were pelleted by centrifugation and lysed in buffer containing 150 mM NaCl, 25 mM Tris (pH 7.4), 1% NP-40, 0.25% sodium deoxycholate, and 1 mM EDTA with protease inhibitor cocktail (Roche, Indianapolis, IN). Aliquots of cell lysates were electrophoresed on 12% SDS-PAGE gels and transferred to a nitrocellulose membrane (Pall, Port Washington, NY). The membranes were blocked with 5% nonfat dry milk, and then proteins on the membrane were incubated with the indicated primary antibodies at 4°C overnight. This was followed by incubation with the corresponding IRD Fluor 800-labeled IgG or IRD Fluor 680-labeled IgG secondary antibody (Li-Cor Inc., Lincoln, NE). After washing, the membranes were scanned with an Odyssey infrared imaging system (Li-Cor, Lincoln, NE) at a wavelength of 700 or 800 nm, and the molecular sizes of the developed proteins were determined by comparison with prestained protein markers (Fermentas, Hanover, MD).
RNA interference (RNAi).
To generate control or IRF7-knockdown cell lines, THP1 and HeLa cells were seeded onto 24-well plates. The next day, cells were infected with lentiviruses expressing scrambled or IRF7-specific short hairpin RNA (shRNA) (GenePharma) at a multiplicity of infection (MOI) of 100. After 72 h, cells were selected by the use of 1 μg/ml puromycin for 1 week. The sequence of short hairpin RNA targeting IRF7 was 5′-CCAAGAGCTGGTGGAATTC-3′ as described previously (44).
Study approval.
This study was approved by the ethical review committee of the Institute of Pathogen Biology, Chinese Academy of Medical Sciences. The methods were carried out in accordance with the approved guidelines.
RESULTS
EV-D68 infection does not stimulate the type I IFN response in HeLa cells.
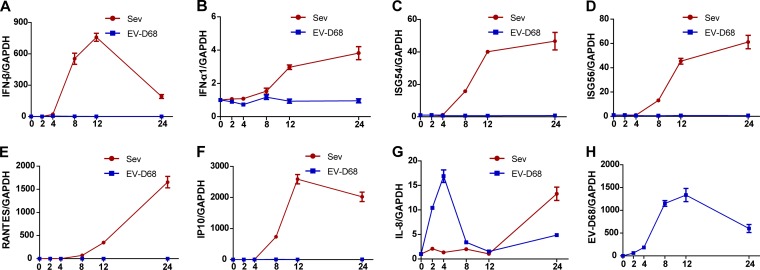
Innate antiviral immunity plays a pivotal role in controlling virus infection (36). To assess the impact of EV-D68, we investigated the cytokine responses in virus-infected cells. As such, HeLa cells were infected with EV-D68. At different time points postinfection, the cells were examined for the expression of cytokines or IFN-stimulated genes by quantitative real-time RT-PCR analysis. In parallel, Sendai virus was included as a control. As illustrated in Fig. 1A, Sendai virus significantly stimulated IFN-β expression, which diminished as virus infection progressed, and, with a different kinetics or magnitude, also stimulated expression of ISG56, ISG54, IL-8, RANTES, and IP10 (panels B to G). Under these experimental conditions, EV-D68 barely induced expression of IFN-β, IFN-α1, ISG56, ISG54, RANTES, and IP10 (panels A to F), although EV-D68 replicated in infected cells (panel H). However, EV-D68 triggered a rapid induction of IL-8 expression which peaked at 4 h postinfection and declined thereafter. These results suggest that EV-D68 infection negatively modulates the type I IFN response.
FIG 1.
Innate immune responses are not activated in EV-68-infected HeLa cells. HeLa cells were infected with EV-D68 at an MOI of 2. In parallel, Sendai virus (Sev) was included as a control. At different time points (hours) postinfection, total levels of RNA extracted from cells and expression of IFN-β (A), IFN-α1 (B), ISG54 (C), ISG56 (D), RANTES (E), IP10 (F), IL-8 (G), EV-D68, and GAPDH genes were evaluated by quantitative real-time PCR using SYBR green. Results are expressed as increases in mRNA levels relative to those seen in cells collected at 0 h and were normalized by using the GAPDH housekeeping gene.
EV-D68 promotes the cleavage of IRF7 in infected cells.
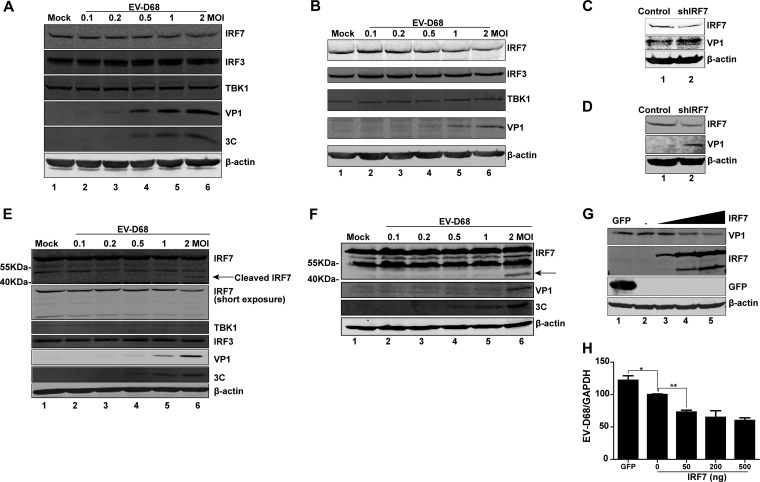
Previous studies suggest that IRF7 is essential for type I IFN induction (45). As IRF7 protects against picornavirus infection (45), we hypothesized that EV-D68 may inhibit the type I IFN response by targeting IRF7. To evaluate this, we analyzed protein levels in EV-D68-infected cells. Specifically, cells were mock infected or infected with increasing doses of EV-D68. At 24 h after infection, cell lysates were subjected to Western blot analysis. As illustrated in Fig. 2A, in HeLa cells infected with EV-D68, expression of IRF3, TBK1, and β-actin exhibited little change, but the level of IRF7 expression decreased as the infection dose of EV-D68 increased. This coincided with the appearance of VP1 expression. Similarly, this phenotype was observed in THP-1 cells (Fig. 2B) and PBMCs (Fig. 2E). Moreover, a cleaved fragment (about 40 kDa) of IRF7, representing the C terminus, was detectable in PBMCs (Fig. 2E, lane 6). When ectopically expressed Flag-Myc-tagged IRF7 (Flag-Myc was added to the C terminus of IRF7) in 293T cells, this cleaved IRF7 fragment was also seen in EV-D68-infected cells (Fig. 2F, lane 6). Therefore, EV-D68 induced IRF7 cleavage in infected cells. Notably, small interfering RNA (siRNA) knockdown of IRF7 increased viral replication (Fig. 2C and D). Moreover, ectopic expression of IRF7 decreased viral replication (Fig. 2G and H). We conclude that the interaction between IRF7 and EV-D68 determines the outcome of viral infection.
FIG 2.
Effects of EV-D68 infection on IRF7. (A) HeLa cells were mock infected or infected with an increasing dose of EV-D68. At 24 h after infection, cell lysates were subjected to Western blot analysis with antibodies against IRF7 (target amino acids near the C terminus of human IRF7), IRF3, TBK1, VP1, 3C, and β-actin. (B) THP1 cells were mock infected or infected with an increasing dose of EV-D68. At 24 h after infection, cell lysates were subjected to Western blot analysis with antibodies against IRF7, IRF3, TBK1, VP1, and β-actin. (C) Control or IRF7-knockdown HeLa cells were infected with EV-D68 (MOI = 1) for 24 h. Cell lysates were subjected to Western blot analysis with antibodies against IRF7, VP1, and β-actin. (D) Control or IRF7-knockdown THP1 cells were infected with EV-D68 (MOI = 2) for 24 h. Cell lysates were subjected to Western blot analysis with antibodies against IRF7, VP1, and β-actin. (E) Peripheral blood mononuclear cells isolated from healthy donors were mock infected or infected with an increasing dose of EV-D68. At 24 h after infection, cell lysates were subjected to Western blot analysis with antibodies against IRF7, IRF3, TBK1, VP1, 3C, and β-actin. (F) 293T cells were transfected with pCMV6-Flag-Myc-IRF7 for 24 h. Cells were then mock infected or infected with an increasing dose of EV-D68. At 24 h after infection, cell lysates were analyzed by Western blotting using antibodies against Myc (C terminal of IRF7), VP1, 3C, and β-actin. (G) 293T cells were transfected with a control plasmid or with increasing amounts (50 ng, 200 ng, and 500 ng; wedge) of pCMV6-Flag-Myc-IRF7. At 24 h after transfection, cells were infected with EV-D68 (MOI = 2) for 24 h. Cell lysates were subjected to Western blot analysis with antibodies against Flag (C terminal of IRF7), VP1, and β-actin. (H) 293T cells were treated as described for panel G. Total RNA was extracted, and the viral RNA levels of EV-D68 were evaluated by quantitative real-time PCR using SYBR green. Results are expressed as viral RNA levels relative to GAPDH RNA levels. *, P < 0.05; **, P < 0.01. In panels E and F, arrows denote EV-D68-induced cleavage fragments.
Expression of 3Cpro induces cleavage of IRF7 in 293T cells.
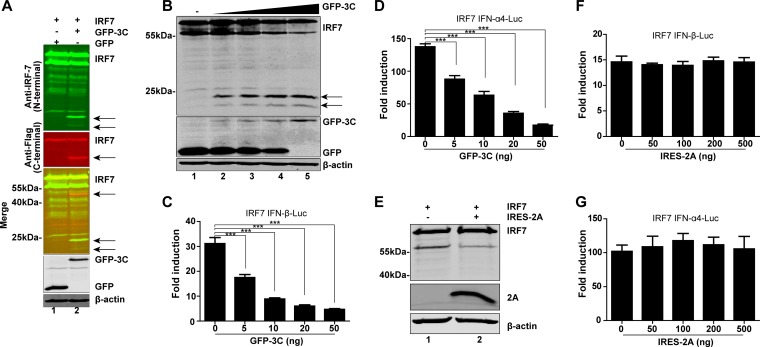
EV-D68 encodes two proteases, 2Apro and 3Cpro. To determine whether viral proteases are responsible for the cleavage of IRF7, we performed additional assays. 293T cells were transfected with 2Apro or 3Cpro along with IRF7 and subjected to Western blot analysis. As indicated in Fig. 3A, 3Cpro cleaved IRF7, producing a fragment of 40 kDa identical to that seen with EV-D68-infected cells. Furthermore, an antibody that recognizes the N terminus of human IRF7 detected two fragments of approximately 20 kDa. Such cleavage correlated with the extent of 3Cpro expression (Fig. 3B). In reporter assays, 3Cpro of EV-D68 also inhibited IRF7-stimulated IFN-β (Fig. 3C) and IFN-α (Fig. 3D) promoter activation in a dose-dependent manner. On the other hand, when expressed, 2Apro did not produce any visible cleavage product of IRF7 (Fig. 3E). Consistently, 2Apro had no inhibitory effect on promoter activation of IFN-β and IFN-α by IRF7 (Fig. 3F and G). These results suggest that 3Cpro of EV-D68 mediates IFR7 cleavage and subsequently inhibits its activity.
FIG 3.
The 3C protease of EV-D68 cleaves IRF7. (A) 293T cells were transfected with plasmids encoding Myc-Flag-IRF7 along with GFP or GFP-3C. At 24 h after transfection, cells were lysed and analyzed by Western blotting with antibodies against IRF7 (targeting the N terminus of human IRF7), Flag, GFP, and β-actin. (B) 293T cells were transfected with Flag-IRF7 and a control plasmid or increasing amounts (5 ng, 10 ng, 20 ng, and 50 ng; wedge) of plasmids expressing GFP-3C. (C and D) At 24 h after transfection, cells were analyzed by Western blotting. Effects of 3Cpro of EV-D68 on IFN-β (C) or IFN-α4 (D) promoter activation are indicated. 293T cells were transfected with IRF7 and GFP-3C along with IFN-β–Luc or IFN-α4–Luc; pRL-SV40 was used as an internal control. At 24 h after transfection, cell lysates were assayed for luciferase activities. (E) 293T cells were transfected with Myc-Flag-IRF7 along with a control plasmid or pCDNA3.1-V5-IRES-2A. At 24 h after transfection, cells were lysed and analyzed by Western blotting with antibodies specific for Flag, V5, and β-actin. (F and G) Effects of 2Apro of EV-D68 on IFN-β (F) or IFN-α4 (G) promoter activation. 293T cells were transfected with IRF7 and IRES-2A along with IFN-β–Luc or IFN-α4–Luc; pRL-SV40 was used as an internal control. At 24 h after transfection, cell lysates were assayed for luciferase activities. ***, P < 0.001. In panels A and B, arrows denote 3Cpro-induced cleavage fragments.
3C protease activity is crucial for cleavage and inhibition of IRF7.
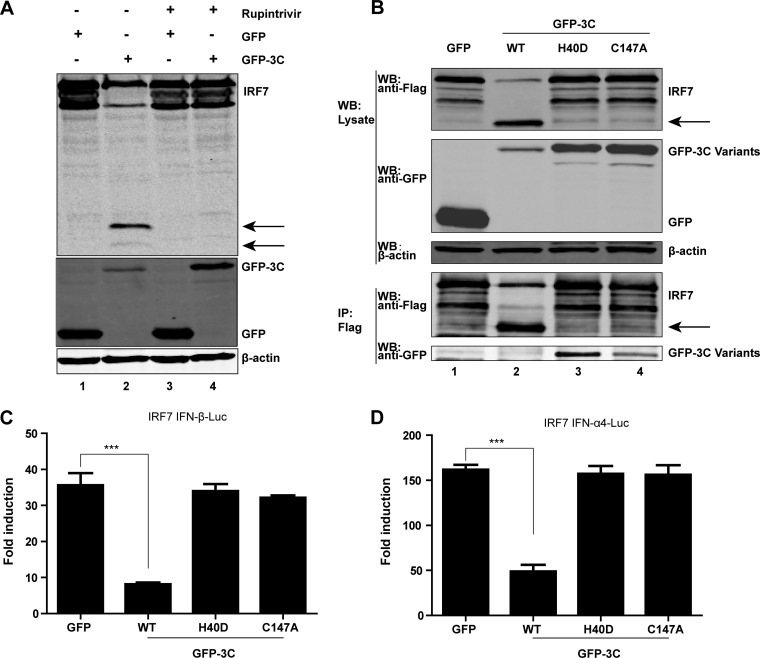
To probe the nature of 3Cpro-mediated IRF7 cleavage, we determined whether its proteolytic activity is involved. Rupintrivir is an inhibitor of 3Cpro with a broad spectrum of activity against picornaviruses (46). As such, we examined its effect on IRF7 cleavage in transfected cells. As indicated in Fig. 4A, expression of 3Cpro resulted in a cleaved IRF7 fragment in the absence of rupintrivir (lane 2). However, when cells were treated with rupintrivir, 3Cpro of EV-D68 failed to mediate the cleavage of IRF7 (lane 4), suggesting a role of protease activity. As 3Cpro of EV-D68 bears a catalytic triad consisting of Cys147, His40, and Glu71, we also carried out mutational analysis. As shown in Fig. 4B, wild-type 3Cproof EV-D68 effectively induced expression of a cleaved IRF7 product. However, neither the H40D nor the C147A variant exerted IRF7 cleavage as measured by Western blotting (top panel). Immunoprecipitation assays revealed that both the H40D and C147A variants associated with IRF7 (lower panel). These data indicate that 3Cpro forms a complex with IRF7. Notably, wild-type 3Cpro and IRF7 were not coimmunoprecipitated, presumably due to cleavage of full-length IRF7 (Fig. 4B). In reporter assays, wild-type 3Cpro inhibited IFN-β and IFN-α promoter activation mediated by IRF7 whereas the H40D and C147A variants had no inhibitory effect (Fig. 4C and D). Hence, the protease activity of 3Cpro of EV68 is essential to inhibit IRF7 function.
FIG 4.
(A) Effect of the protease inhibitor rupintrivir on IRF7 cleavage. 293T cells were transfected with Flag-IRF7 along with GFP or GFP-3C. Four hours after transfection, cells were incubated with the protease inhibitor rupintrivir (1 μM) for 24 h. Cell lysates were then processed for Western blot analysis. (B) Interaction of 3Cpro variants of EV-D68 with IRF7. 293T cells were transfected with Flag-IRF7 and GFP or GFP-3C variants as indicated. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody. Immunoprecipitates and aliquots of cell lysates were subjected to Western blot analysis (WB) with antibodies against Flag, GFP, and β-actin. (C and D) Effects of 3Cpro variants on IRF7-mediated IFN-β and IFN-α4 promoter activation. 293Tcells were transfected with plasmids encoding IRF7 and IFN-β–Luc (C) or IFN-α4–Luc (D), along with GFP or GFP-3C variants. pRL-SV40 was included as an internal control. Twenty-four hours after transfection, cells were harvested to determine luciferase activities. ***, P < 0.001. In panels A and B, arrows denote 3Cpro-induced cleavage fragments.
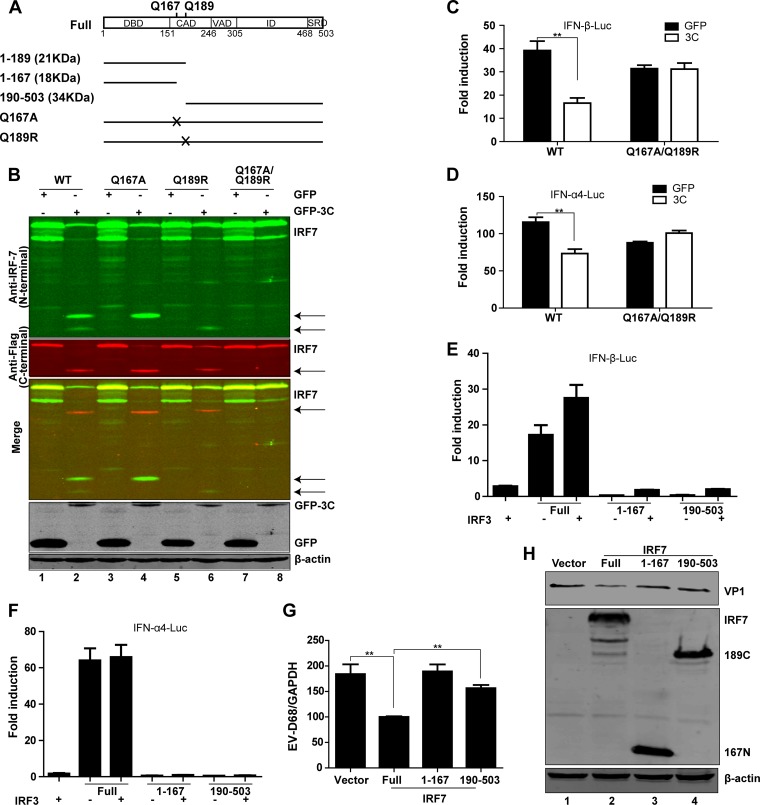
3Cpro cleaves IRF7 at Gln-167 and Gln-189.
Since IRF7 cleavage produced two 20-kDa fragments representing the N terminus and one 40-kDa fragment representing the C terminus (Fig. 5A), we inferred that the IRF7 cleavage sites might be located in the N-terminal domain. Therefore, we focused on the sites with a signature sequence (AXXQ↓G/S) of 3C protease. We constructed a series of mutants in which Q was replaced with A or R. As illustrated in IRF7 cleavage assays, the Q167A mutation blocked appearance of the lower 20-kDa fragment (Fig. 5B, lane 4), the Q189R mutation blocked the appearance of the upper 20-kDa fragment (Fig. 5B, lane 6), and the Q167A/Q189R combined mutation blocked the appearance of both (Fig. 5B, lane 8). These two cleavage sites were verified by the antibodies targeting the C terminus of human IRF7 plasmids (Fig. 5B). Q167 and Q189 lie close to each other within the protein, which might account for the fact that these two cleavage fragments could not be distinguished from each other by the antibodies targeting the C terminus in the gel. 3Cpro predominantly mediated IRF7 cleavage at Q189 because the cleavage fragment from Q167A was more abundant than that from Q189R (Fig. 5B, lanes 4 and 6). In reporter assays, the Q167A/Q189R mutant was able to activate the IFN-β and IFN-α promoters and reverse the inhibition of GFP-3C (Fig. 5C and D). Collectively, these data suggest Q167 and Q189 as 3Cpro cleavage sites within IRF7.
FIG 5.
3Cpro of EV-D68 cleaves IRF7 at two specific sites of the N-terminal domain. (A) Schematic diagrams of IRF7 denoting the cleavage sites and fragments for 3Cpro of EV-D68. (B) Cleavage analysis of wild-type IRF7 or 3Cpro-resistant mutants. 293T cells were transfected with wild-type IRF7 or IRF7 mutants with alanine or arginine substitutions for glutamine, along with GFP (lanes 1, 3, 5, and 7) or GFP-3C (lanes 2, 4, 6, and 8), as indicated. Twenty-four hours after transfection, cell lysates were subjected to Western blot analysis with antibodies against IRF7, Flag, GFP, and β-actin. (C and D) Effects of EV-D68 3Cpro on IFN-β or IFN-α4 promoter activation mediated by wild-type IRF7 or 3Cpro-resistant mutants. 293T cells were transfected with wild-type IRF7 or the Q167A/Q189R mutant of IRF7 along with IFN-β–Luc (C) or IFN-α4–Luc (D). pRL-SV40 was used as a control. Twenty-four hours after transfection, cell lysates were assayed for luciferase activities. **, P < 0.01. (E and F) Effects of putative IRF7 cleavage fragments on IFN-β or IFN-α4 promoter activation. 293T cells were transfected with IRF7 or Myc-Flag-IRF3 (full length and fragments consisting of amino acids 1 to 167 and 190 to 503) along with IFN-β–Luc (E) or IFN-α4–Luc (F). pRL-SV40 was used as a control. Twenty-four hours after transfection, cell lysates were assayed for luciferase activities. (G) 293T cells were treated as described for panel G. Total RNA was extracted, and the viral RNA levels of EV-D68 were evaluated by quantitative real-time PCR using SYBR green. Results are expressed as viral RNA levels relative to the GAPDH RNA level. (H) 293T cells were transfected with pCMV6 vector or with IRF7 (500 ng) (full length and fragments consisting of amino acids 1 to 167 and 190 to 503). Twenty-four hours after transfection, cells were infected with EV-D68 (MOI = 2) for 24 h. Cell lysates were subjected to Western blot analysis with antibodies against Myc, VP1, and β-actin. **, P < 0.01.
3Cpro-mediated cleavage inactivates IRF7.
To test whether IRF7 cleavage mediated by 3Cpro has a functional consequence, we generated IRF7 variants with site-specific mutations or deletions (Fig. 5A). In reporter assays, full-length IRF7 activated the IFN-β (Fig. 5E) and IFN-α (Fig. 5F) promoters. When cotransfected with IRF3, IRF7 had an enhanced activity (Fig. 5E). In contrast, the 1-to-167 and 190-to-503 fragments lost the ability to activate the IFN-β (Fig. 5E) and IFN-α (Fig. 5F) promoters. Intriguingly, the 1-to-167 fragment suppressed IRF3-mediated IFN-β promoter activation and the 190-to-503 fragment lost the synergist effect with IRF3 (Fig. 5E). Consistently, in 293T cells, ectopic expression of full-length IRF7 significantly inhibited EV-D68 replication, whereas the 1-to-167 and 190-to-503 fragments failed to inhibit viral replication (Fig. 5G and H).
DISCUSSION
EV-D68 is a human pathogen that primarily causes respiratory illness (17). Asthma or wheezing appears to be a risk factor for severe disease (30, 47). Although the underlying events are largely unknown, aberrant immunity is speculated to mediate viral pathogenesis (17). In this study, we provide evidence that EV-D68 inhibits the expression of type I IFN-, chemokine-, and interferon-stimulated genes in infected cells. In doing so, the 3C protease mediates IRF7 cleavage to facilitate viral replication. Intriguingly, EV-D68 stimulates the expression of inflammatory cytokines (e.g., IL-8). These results are consistent with the notion that EV-D68 differentially modulates cytokine responses, which may affect viral replication or infection.
Although actively replicated in infected cells, EV-D68 induced little expression of endogenous type I IFN, ISGs, and chemokines. This is quite different from Sendai virus, which stimulated robust antiviral immunity in infected cells. These results are in line with the model that EV-D68 potently suppresses the type I IFN response. MDA-5 is generally thought to detect picornaviruses (48–50). However, evidence suggests a pivotal role for TLR3 and RIG-I as well (51–55). We recently reported that EV-D68 perturbs TLR3 via proteolytic cleavage of the TIR-domain-containing adapter-inducing interferon-β (TRIF) adaptor (35). As multiple pathogen recognition sensors exist, this alone is insufficient to explain the effective silencing of IFN expression mediated by EV-D68. In support of this, we noted that EV-D68 infection reduced the expression of IRF7 in epithelial cells, cells of a human macrophage cell line, and PBMCs. Indeed, viral protease 3C, rather than 2A, mediated the proteolytic cleavage of IRF7. The precise relation of 3C to TRIF and IRF7 in EV-D68 infection has yet to be established. It is likely that dual targeting of these immune factors may render EV-D68 an advantage to negate antiviral immunity.
The 3C protease of EV-D68 adopts a chymotrypsin-like fold with two β-barrel domains that harbor the catalytic triad of H40, E71, and C147 (56). We demonstrated that H40D or C147A substitution in the catalytic triad eliminated its capacity to cleave IRF7. In correlation, the 3C mutants failed to block IFN induction. Interestingly, H40D or C147A substitution had no effect on the physical interaction between 3Cpro and IRF7. While the IRF7 binding site(s) is undefined, these data argue that the 3C protease may recruit IRF7 via a separate region or motif that directs IRF7 to the catalytic site. As the result of IRF7 cleavage, EV-D68 3Cpro interrupts the type I IFN response. This is reminiscent of the 3C protease encoded by EV71 that causes hand-foot-and-mouth disease and neurological complications (40). Although it does contain amino acid sequence variations, the 3C protease of EV-D68 is 53% identical to its counterpart from EV71 (56). The fact that both EV-D68 and EV71 target IRF7 likely implies a convergent evolution of viral evasion strategy. It also suggests the importance of IRF7 signaling in the control of enteroviruses.
IRF7 is a master regulator of the IFN immune response to virus infections (45). Our data suggest that replication of EV-D68 is functionally linked to IRF7. Knockdown of IRF7 enhanced viral replication. Conversely, overexpression of IRF7 reduced viral replication. Previous work suggests that, in addition to the N-terminal DNA-binding domain, IRF-7 contains a constitutive activation domain (CAD), a virus-activated domain, an inhibitory domain, and a signal response domain (39). Upon viral infection, these domains work coordinately to regulate the activity of IRF7. We observed that 3CPro of EV-D68 cleaved IRF7 at two sites (Gln167 and Gln189) within the CAD, resulting in the amino-terminal domain (fragment 1 to 167) and the carboxyl-terminal domain (fragment 190 to 503). Consequently, IRF7 cleavage may uncouple the DNA binding domain from the activation domain. Alternatively, it may simply destroy the constitutive activation domain, resulting in nonfunctional IRF7. It should be pointed out that IRF7 cleavage fragments also exerted a dominant negative effect on IRF3-mediated gene expression. As such, IRF7 cleavage by EV-D68 adds another layer of viral control, which may provide an additional benefit for EV-D68 infection.
Type I IFN is the first line of host defense against virus infection. It is, therefore, not surprising that enteroviruses have evolved various evasion strategies (57). For example, 3Cpro from coxsackievirus B3 (CVB3), enterovirus 71 (EV71), and poliovirus (PV) cleaves RIG-I (58). On the other hand, 2Apro from these viral pathogens targets mitochondrial antiviral-signaling protein (MAVS) and melanoma differentiation-associated protein 5 (MDA5). Moreover, 3Cpro of EV71 and CVB3 cleaves TRIF (43, 59). The present study further revealed that EV-D68 negatively regulates IRF7 through the viral 3C protease. It is notable that the 3Cpro proteases of enteroviruses are closely related phylogenetically (56). These observations support the view that enteroviruses likely share common mechanisms to inhibit IFN induction, although elucidation of the relative contribution of each one in viral replication awaits further investigation. Given that IRF7 deficiency predisposes humans to severe viral illness (60), we speculate that the interplay of EV-D68 3Cpro and IRF7 may represent an interface that determines EV-D68 replication or pathogenesis in vivo. Additional work is required to test this hypothesis.
ACKNOWLEDGMENTS
This work was supported by grants from the 973 Project (grant 2011CB504903) (Zichun Xiang, Lulu Liu, Xiaobo Lei, Zhuo Zhou, and Jianwei Wang), the National Science Foundation for Outstanding Young Scientists (grant 81225014) (Zichun Xiang, Lulu Liu, Xiaobo Lei, Zhuo Zhou, and Jianwei Wang), the Program for Changjiang Scholars and Innovative Research Team in University (IRT13007) (Zichun Xiang, Lulu Liu, Xiaobo Lei, Zhuo Zhou, and Jianwei Wang), and the National Institute of Allergy and Infectious Diseases of the United States (AI112755) (Bin He).
REFERENCES
- 1.Schieble JH, Fox VL, Lennette EH. 1967. A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol 85:297–310. [DOI] [PubMed] [Google Scholar]
- 2.Imamura T, Fuji N, Suzuki A, Tamaki R, Saito M, Aniceto R, Galang H, Sombrero L, Lupisan S, Oshitani H. 2011. Enterovirus 68 among children with severe acute respiratory infection, the Philippines. Emerg Infect Dis 17:1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaida A, Kubo H, Sekiguchi J, Kohdera U, Togawa M, Shiomi M, Nishigaki T, Iritani N. 2011. Enterovirus 68 in children with acute respiratory tract infections, Osaka, Japan. Emerg Infect Dis 17:1494–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC). 2011. Clusters of acute respiratory illness associated with human enterovirus 68—Asia, Europe, and United States, 2008–2010. MMWR Morb Mortal Wkly Rep 60:1301–1304. [PubMed] [Google Scholar]
- 5.Rahamat-Langendoen J, Riezebos-Brilman A, Borger R, van der Heide R, Brandenburg A, Scholvinck E, Niesters HG. 2011. Upsurge of human enterovirus 68 infections in patients with severe respiratory tract infections. J Clin Virol 52:103–106. doi: 10.1016/j.jcv.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda T, Mizuta K, Abiko C, Aoki Y, Itagaki T, Katsushima F, Katsushima Y, Matsuzaki Y, Fuji N, Imamura T, Oshitani H, Noda M, Kimura H, Ahiko T. 2012. Acute respiratory infections due to enterovirus 68 in Yamagata, Japan between 2005 and 2010. Microbiol Immunol 56:139–143. doi: 10.1111/j.1348-0421.2012.00411.x. [DOI] [PubMed] [Google Scholar]
- 7.Linsuwanon P, Puenpa J, Suwannakarn K, Auksornkitti V, Vichiwattana P, Korkong S, Theamboonlers A, Poovorawan Y. 2012. Molecular epidemiology and evolution of human enterovirus serotype 68 in Thailand, 2006–2011. PLoS One 7:e35190. doi: 10.1371/journal.pone.0035190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tokarz R, Firth C, Madhi SA, Howie SR, Wu W, Sall AA, Haq S, Briese T, Lipkin WI. 2012. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol 93:1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang Z, Gonzalez R, Wang Z, Ren L, Xiao Y, Li J, Li Y, Vernet G, Paranhos-Baccala G, Jin Q, Wang J. 2012. Coxsackievirus A21, enterovirus 68, and acute respiratory tract infection, China. Emerg Infect Dis 18:821–824. doi: 10.3201/eid1805.111376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meijer A, Benschop KS, Donker GA, van der Avoort HG. 2014. Continued seasonal circulation of enterovirus D68 in the Netherlands, 2011–2014. Euro Surveill 19:pii=20935. doi: 10.2807/1560-7917.ES2014.19.42.20935. [DOI] [PubMed] [Google Scholar]
- 11.Bal A, Schuffenecker I, Casalegno JS, Josset L, Valette M, Armand N, Dhondt PB, Escuret V, Lina B. 15 May 2015, posting date Enterovirus D68 nosocomial outbreak in elderly people, France, 2014. Clin Microbiol Infect doi: 10.1016/j.cmi.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bragstad K, Jakobsen K, Rojahn AE, Skram MK, Vainio K, Holberg-Petersen M, Hungnes O, Dudman SG, Kran AM. 2015. High frequency of enterovirus D68 in children hospitalised with respiratory illness in Norway, autumn 2014. Influenza Other Respir Viruses 9:59–63. doi: 10.1111/irv.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drews SJ, Simmonds K, Usman HR, Yee K, Fathima S, Tipples G, Tellier R, Pabbaraju K, Wong S, Talbot J. 2015. Characterization of enterovirus activity, including that of enterovirus D68, in pediatric patients in Alberta, Canada, in 2014. J Clin Microbiol 53:1042–1045. doi: 10.1128/JCM.02982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyrdak R, Rotzén-Östlund M, Samuelson A, Eriksson M, Albert J. 14 May 2015, posting date Coexistence of two clades of enterovirus D68 in pediatric Swedish patients in the summer and fall of 2014. Infect Dis (Lond) doi: 10.3109/23744235.2015.1047402. [DOI] [PubMed] [Google Scholar]
- 15.Esposito S, Zampiero A, Ruggiero L, Madini B, Niesters H, Principi N. 2015. Enterovirus D68-associated community-acquired pneumonia in children living in Milan, Italy. J Clin Virol 68:94–96. doi: 10.1016/j.jcv.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 16.Gimferrer L, Campins M, Codina MG, Esperalba J, Martin MD, Fuentes F, Pumarola T, Anton A. 28 February 2015, posting date First Enterovirus D68 (EV-D68) cases detected in hospitalised patients in a tertiary care university hospital in Spain, October 2014. Enferm Infecc Microbiol Clin doi: 10.1016/j.eimc.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imamura T, Oshitani H. 2015. Global reemergence of enterovirus D68 as an important pathogen for acute respiratory infections. Rev Med Virol 25:102–114. doi: 10.1002/rmv.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Midgley SE, Christiansen CB, Poulsen MW, Hansen CH, Fischer TK. 2015. Emergence of enterovirus D68 in Denmark, June 2014 to February 2015. Euro Surveill 20:pii=21105. doi: 10.2807/1560-7917.ES2015.20.17.21105. [DOI] [PubMed] [Google Scholar]
- 19.Pfeiffer HC, Bragstad K, Skram MK, Dahl H, Knudsen PK, Chawla MS, Holberg-Petersen M, Vainio K, Dudman SG, Kran AM, Rojahn AE. 2015. Two cases of acute severe flaccid myelitis associated with enterovirus D68 infection in children, Norway, autumn 2014. Euro Surveill 20:21062. doi: 10.2807/1560-7917.ES2015.20.10.21062. [DOI] [PubMed] [Google Scholar]
- 20.Reiche J, Bottcher S, Diedrich S, Buchholz U, Buda S, Haas W, Schweiger B, Wolff T. 2015. Low-level circulation of enterovirus D68-associated acute respiratory infections, Germany, 2014. Emerg Infect Dis 21:837–841. doi: 10.3201/eid2105.141900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres JP, Farfan MJ, Izquierdo G, Piemonte P, Henriquez J, O'Ryan ML. 2015. Enterovirus D68 infection, Chile, spring 2014. Emerg Infect Dis 21:728–729. doi: 10.3201/eid2104.141766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention. 2014. Enterovirus D68 in the United States. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/non-polio-enterovirus/outbreaks/EV-D68-outbreaks.html Accessed 22 December 2014. [Google Scholar]
- 23.The Lancet Infectious Diseases. 2014. Enterovirus D68: the unexpected guest. Lancet Infect Dis 14:1023. doi: 10.1016/S1473-3099(14)70968-5. [DOI] [PubMed] [Google Scholar]
- 24.Brown BA, Nix WA, Sheth M, Frace M, Oberste MS. 2014. Seven strains of enterovirus D68 detected in the United States during the 2014 severe respiratory disease outbreak. Genome Announc 2:e01201–14. doi: 10.1128/genomeA.01201-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Midgley CM, Jackson MA, Selvarangan R, Turabelidze G, Obringer E, Johnson D, Giles BL, Patel A, Echols F, Oberste MS, Nix WA, Watson JT, Gerber SI. 2014. Severe respiratory illness associated with enterovirus D68—Missouri and Illinois, 2014. MMWR Morb Mortal Wkly Rep 63:798–799. [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson R. 2014. Outbreaks of enterovirus D68 continue across the USA. Lancet Respir Med 2:791. doi: 10.1016/S2213-2600(14)70190-0. [DOI] [PubMed] [Google Scholar]
- 27.Stephenson J. 2014. CDC tracking enterovirus D-68 outbreak causing severe respiratory illness in children in the Midwest. JAMA 312:1290. doi: 10.1001/jama.2014.13256. [DOI] [PubMed] [Google Scholar]
- 28.Farrell JJ, Ikladios O, Wylie KM, O'Rourke LM, Lowery KS, Cromwell JS, Wylie TN, Melendez EL, Makhoul Y, Sampath R, Bonomo RA, Storch GA. 2015. Enterovirus D68-associated acute respiratory distress syndrome in adult, United States, 2014. Emerg Infect Dis 21:914–916. doi: 10.3201/eid2105.142033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messacar K, Schreiner TL, Maloney JA, Wallace A, Ludke J, Oberste MS, Nix WA, Robinson CC, Glode MP, Abzug MJ, Dominguez SR. 2015. A cluster of acute flaccid paralysis and cranial nerve dysfunction temporally associated with an outbreak of enterovirus D68 in children in Colorado, USA. Lancet 385:1662–1671. doi: 10.1016/S0140-6736(14)62457-0. [DOI] [PubMed] [Google Scholar]
- 30.Oermann CM, Schuster JE, Conners GP, Newland JG, Selvarangan R, Jackson MA. 2015. Enterovirus d68. A focused review and clinical highlights from the 2014 U.S. outbreak. Ann Am Thorac Soc 12:775–781. doi: 10.1513/AnnalsATS.201412-592FR. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Sheng J, Fokine A, Meng G, Shin WH, Long F, Kuhn RJ, Kihara D, Rossmann MG. 2015. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children. Science 347:71–74. doi: 10.1126/science.1261962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uncapher CR, DeWitt CM, Colonno RJ. 1991. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology 180:814–817. doi: 10.1016/0042-6822(91)90098-V. [DOI] [PubMed] [Google Scholar]
- 33.Imamura T, Okamoto M, Nakakita S, Suzuki A, Saito M, Tamaki R, Lupisan S, Roy CN, Hiramatsu H, Sugawara KE, Mizuta K, Matsuzaki Y, Suzuki Y, Oshitani H. 2014. Antigenic and receptor binding properties of enterovirus 68. J Virol 88:2374–2384. doi: 10.1128/JVI.03070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smura T, Ylipaasto P, Klemola P, Kaijalainen S, Kyllonen L, Sordi V, Piemonti L, Roivainen M. 2010. Cellular tropism of human enterovirus D species serotypes EV-94, EV-70, and EV-68 in vitro: implications for pathogenesis. J Med Virol 82:1940–1949. doi: 10.1002/jmv.21894. [DOI] [PubMed] [Google Scholar]
- 35.Xiang Z, Li L, Lei X, Zhou H, Zhou Z, He B, Wang J. 2014. Enterovirus 68 3C protease cleaves TRIF to attenuate antiviral responses mediated by Toll-like receptor 3. J Virol 88:6650–6659. doi: 10.1128/JVI.03138-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawai T, Akira S. 2006. Innate immune recognition of viral infection. Nat Immunol 7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 37.Bowie AG, Unterholzner L. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol 8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Pagano JS. 2002. Structure and function of IRF-7. J Interferon Cytokine Res 22:95–101. doi: 10.1089/107999002753452700. [DOI] [PubMed] [Google Scholar]
- 39.Ning S, Pagano JS, Barber GN. 2011. IRF7: activation, regulation, modification and function. Genes Immun 12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lei X, Xiao X, Xue Q, Jin Q, He B, Wang J. 2013. Cleavage of interferon regulatory factor 7 by enterovirus 71 3C suppresses cellular responses. J Virol 87:1690–1698. doi: 10.1128/JVI.01855-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang B, Xi X, Lei X, Zhang X, Cui S, Wang J, Jin Q, Zhao Z. 2013. Enterovirus 71 protease 2Apro targets MAVS to inhibit anti-viral type I interferon responses. PLoS Pathog 9:e1003231. doi: 10.1371/journal.ppat.1003231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lei X, Liu X, Ma Y, Sun Z, Yang Y, Jin Q, He B, Wang J. 2010. The 3C protein of enterovirus 71 inhibits retinoid acid-inducible gene I-mediated interferon regulatory factor 3 activation and type I interferon responses. J Virol 84:8051–8061. doi: 10.1128/JVI.02491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lei X, Sun Z, Liu X, Jin Q, He B, Wang J. 2011. Cleavage of the adaptor protein TRIF by enterovirus 71 3C inhibits antiviral responses mediated by Toll-like receptor 3. J Virol 85:8811–8818. doi: 10.1128/JVI.00447-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang Q, Deng H, Sun CW, Townes TM, Zhu F. 2011. Negative regulation of IRF7 activation by activating transcription factor 4 suggests a cross-regulation between the IFN responses and the cellular integrated stress responses. J Immunol 186:1001–1010. doi: 10.4049/jimmunol.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 46.De Palma AM, Vliegen I, De Clercq E, Neyts J. 2008. Selective inhibitors of picornavirus replication. Med Res Rev 28:823–884. doi: 10.1002/med.20125. [DOI] [PubMed] [Google Scholar]
- 47.Hasegawa S, Hirano R, Okamoto-Nakagawa R, Ichiyama T, Shirabe K. 2011. Enterovirus 68 infection in children with asthma attacks: virus-induced asthma in Japanese children. Allergy 66:1618–1620. doi: 10.1111/j.1398-9995.2011.02725.x. [DOI] [PubMed] [Google Scholar]
- 48.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 49.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A 103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, Hur S, Semler BL, van Rij RP, van Kuppeveld FJ. 2012. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep 2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slater L, Bartlett NW, Haas JJ, Zhu J, Message SD, Walton RP, Sykes A, Dahdaleh S, Clarke DL, Belvisi MG, Kon OM, Fujita T, Jeffery PK, Johnston SL, Edwards MR. 2010. Co-ordinated role of TLR3, RIG-I and MDA5 in the innate response to rhinovirus in bronchial epithelium. PLoS Pathog 6:e1001178. doi: 10.1371/journal.ppat.1001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng Q, Langereis MA, Olagnier D, Chiang C, van de Winkel R, van Essen P, Zoll J, Hiscott J, van Kuppeveld FJ. 2014. Coxsackievirus cloverleaf RNA containing a 5′ triphosphate triggers an antiviral response via RIG-I activation. PLoS One 9:e95927. doi: 10.1371/journal.pone.0095927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Papon L, Oteiza A, Imaizumi T, Kato H, Brocchi E, Lawson TG, Akira S, Mechti N. 2009. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology 393:311–318. doi: 10.1016/j.virol.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 54.Oshiumi H, Okamoto M, Fujii K, Kawanishi T, Matsumoto M, Koike S, Seya T. 2011. The TLR3/TICAM-1 pathway is mandatory for innate immune responses to poliovirus infection. J Immunol 187:5320–5327. doi: 10.4049/jimmunol.1101503. [DOI] [PubMed] [Google Scholar]
- 55.Tatematsu M, Nishikawa F, Seya T, Matsumoto M. 2013. Toll-like receptor 3 recognizes incomplete stem structures in single-stranded viral RNA. Nat Commun 4:1833. doi: 10.1038/ncomms2857. [DOI] [PubMed] [Google Scholar]
- 56.Tan J, George S, Kusov Y, Perbandt M, Anemuller S, Mesters JR, Norder H, Coutard B, Lacroix C, Leyssen P, Neyts J, Hilgenfeld R. 2013. 3C protease of enterovirus 68: structure-based design of Michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J Virol 87:4339–4351. doi: 10.1128/JVI.01123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Q, Langereis MA, van Kuppeveld FJ. 2014. Induction and suppression of innate antiviral responses by picornaviruses. Cytokine Growth Factor Rev 25:577–585. doi: 10.1016/j.cytogfr.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng Q, Langereis MA, Lork M, Nguyen M, Hato SV, Lanke K, Emdad L, Bhoopathi P, Fisher PB, Lloyd RE, van Kuppeveld FJ. 2014. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J Virol 88:3369–3378. doi: 10.1128/JVI.02712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, Coyne CB. 2011. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog 7:e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ciancanelli MJ, Huang SX, Luthra P, Garner H, Itan Y, Volpi S, Lafaille FG, Trouillet C, Schmolke M, Albrecht RA, Israelsson E, Lim HK, Casadio M, Hermesh T, Lorenzo L, Leung LW, Pedergnana V, Boisson B, Okada S, Picard C, Ringuier B, Troussier F, Chaussabel D, Abel L, Pellier I, Notarangelo LD, Garcia-Sastre A, Basler CF, Geissmann F, Zhang SY, Snoeck HW, Casanova JL. 2015. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 348:448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]