ABSTRACT
Regulation of gene transcription in varicella-zoster virus (VZV), a ubiquitous human neurotropic alphaherpesvirus, requires coordinated binding of multiple host and virus proteins onto specific regions of the virus genome. Chromatin immunoprecipitation (ChIP) is widely used to determine the location of specific proteins along a genomic region. Since the size range of sheared virus DNA fragments governs the limit of accurate protein localization, particularly for compact herpesvirus genomes, we used a quantitative PCR (qPCR)-based assay to determine the efficiency of VZV DNA shearing before ChIP, after which the assay was used to determine the relationship between transcript abundance and the occupancy of phosphorylated RNA polymerase II (RNAP) on the gene promoter, body, and terminus of VZV genes 9, 51, and 66. The abundance of VZV gene 9, 51, and 66 transcripts in VZV-infected human fetal lung fibroblasts was determined by reverse transcription-linked quantitative PCR. Our results showed that the C-terminal domain of RNAP is hyperphosphorylated at serine 5 (S5P) on VZV genes 9, 51, and 66 independently of transcript abundance and the location within the virus gene at both 1 and 3 days postinfection (dpi). In contrast, phosphorylated serine 2 (S2P)-modified RNAP was not detected at any virus gene location at 3 dpi and was detected at levels only slightly above background levels at 1 dpi.
IMPORTANCE Regulation of herpesvirus gene transcription is an elaborate choreography between proteins and DNA that is revealed by chromatin immunoprecipitation (ChIP). We used a quantitative PCR-based assay to determine fragment size after DNA shearing, a critical parameter in ChIP assays, and exposed a basic difference in the mechanism of transcription between mammalian cells and VZV. We found that hyperphosphorylation at serine 5 of the C-terminal domain of RNAP along the lengths of VZV genes (the promoter, body, and transcription termination site) was independent of mRNA abundance. In contrast, little to no enrichment of serine 3 phosphorylation of RNAP was detected at these virus gene regions. This is distinct from the findings for RNAP at highly regulated host genes, where RNAP S5P occupancy decreased and S2P levels increased as the polymerase transited through the gene. Overall, these results suggest that RNAP associates with human and virus transcriptional units through different mechanisms.
INTRODUCTION
Chromatin immunoprecipitation (ChIP) has been used to determine the association of proteins with DNA in cells of humans (1), mice (2), Drosophila melanogaster (3), Caenorhabditis elegans (4), and Saccharomyces cerevisiae yeast (5) and has shown that gene regulation requires the interaction of multiple nuclear proteins, including RNA polymerase II (RNAP), with various transcription factors across the genome. ChIP has also been used to explore the phosphorylation status of the C-terminal domain (CTD) of RNAP at different positions along a transcription unit and showed that in eukaryotes, phosphorylation of serine 5 (S5P) residues is enriched at the beginning of genes, whereas the ends of genes are enriched with RNAP phosphorylated on serine 2 (S2P) residues (6). ChIP assays can also map the genomic location of posttranslationally modified histone proteins and chromosomal insulator elements that influence host and virus gene transcription (7–9). ChIP is a multistep procedure that requires experimental optimization. Whether sensitivity is measured by sequencing, microarray analysis, or quantitative PCR (qPCR), the sensitivity of a ChIP experiment depends on enrichment of protein-bound DNA fragments among a background of unbound fragments. Thus, antibody quality and the immunoprecipitation procedure are critical parameters. While the number and quality of ChIP-grade antibodies are increasing and immunoprecipitation is becoming increasingly standardized (10), chromatin fragmentation is a critical step that is receiving increased attention (11). Chromatin is fragmented by enzyme digestion or sonication, with the approximate size range of DNA fragments being determined by agarose gel electrophoresis. Typically, shearing of formaldehyde-cross-linked chromatin produces random DNA fragments ranging from 100 to 1,000 bp in length. Such fragmentation is necessary to show increased enrichment of protein binding to specific sites over unbound genomic regions and is a critical parameter when mapping adjacent protein binding sites. In particular, determination of the size of DNA fragments is important when investigating small genomes with compact transcriptional units.
Varicella-zoster virus (VZV), a ubiquitous neurotropic herpesvirus, has the smallest genome among the human alphaherpesviruses (12). To date, 74 open reading frames (ORFs) have been identified within the 124,884-bp VZV genome. While transcripts (13–15) and proteins (16) mapping to these ORFs have been detected, few of their promoters have been identified (17) and even fewer associated transcription factors have been characterized (18). Distal VZV gene regulatory elements, including transcriptional enhancers, remain to be identified, and most identified transcription regulatory regions are located proximal to the virus ORFs. Each VZV ORF is separated by an average of 272 bp (19). Since 146 bp of DNA is packaged into a nucleosome core particle (20), regulation of VZV transcription may involve posttranslational modifications of histones enriched on the gene promoters. Histone proteins containing posttranslational modifications indicative of permissive chromatin are bound to promoters of VZV genes transcribed during both latent and lytic infection (21).
Due to the high density of the VZV genome coding sequence with a close proximity of regulatory regions to associated genes, it is important to accurately quantify DNA fragmentation efficiency during ChIP. Thus, we developed a qPCR assay to rapidly determine DNA shearing efficiency. Using this assay and ChIP-qPCR, we investigated changes in RNAP density and CTD phosphorylation during transcription of VZV genes 9, 51, and 66 in human fetal lung (HFL) fibroblasts harvested at 1 and 3 days postinfection (dpi).
Unlike RNAP CTD phosphorylation in mammalian gene transcription, we found that the RNAP CTD heptad repeat contained no S2P in VZV promoter regions, suggesting that serine 2 phosphorylation is not involved in the exit of polymerase from the promoter region. In contrast, RNAP near the promoter displayed high levels of S5P. However, unlike their levels during host gene transcription, RNAP S5P levels did not decrease as polymerase transcribed beyond the promoter region into the gene body but were constant throughout the length of the VZV gene region and independent of transcript levels. These results suggest that the dynamic changes in CTD phosphorylation patterns seen during the transcription of VZV genes are distinct from those seen during the transcription of human genes.
MATERIALS AND METHODS
Virus and cells.
The VZV Ellen strain was used to infect HFL cells (ATCC CCL-153) propagated in Dulbecco's modified Eagle's medium (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (HyClone, Logan, UT) (22).
Preparation of sheared chromatin from VZV-infected HFL cells.
VZV-infected HFL cells in 10-cm plates were fixed in 1% formaldehyde at room temperature for 10 min, quenched with 0.25 M glycine at room temperature for 5 min, washed with ice-cold phosphate-buffered saline (PBS) containing proteinase and phosphatase inhibitors (Pierce Biotechnology, Rockford, IL), scraped, pelleted, and stored at −80°C. For sonication, cell pellets were resuspended in 1 ml sonication buffer {5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 85 mM KCl, 10 μl/ml NP-40} supplemented with proteinase and phosphatase inhibitors. Total DNA from 2 × 106 cells (approximately 20 μg) was sonicated at −7°C (maintained by immersion in NaCl-saturated ice water) in 15 ml polymethylpentene conical tubes (Diagenode, Denville, NJ) using the microprobe, at a setting of 4 (40% maximal power output), of a Sonifer W140D cell disrupter (Heat Systems-Ultrasonics, Farmingdale, NY). After sonication, samples were centrifuged for 30 min at 40,000 × g and 4°C to eliminate nucleocapsids containing virus DNA (23).
DNA fragment size determination.
Formaldehyde cross-links were reversed by exposure to 65°C for 1.5 h in 0.2 M NaCl, and DNA was extracted by affinity chromatography (E.Z.N.A. tissue DNA kit; Omega Bio-Tek, Norcross, GA) and quantified by determination of the optical absorbance at 260 nm (NanoDrop spectrophotometer; Thermo Scientific). The size of the DNA was estimated using capillary gel electrophoresis (Bioanalyzer; Agilent Technologies, Santa Clara, CA). Equal amounts of sonicated DNA were analyzed by qPCR (7500 Fast PCR machine; Applied Biosystems, Foster City, CA) using virus- and cell-specific primers (Integrated DNA Technologies, Coralville, IA) (Table 1) with the following cycling conditions: 15 min at 95°C (activation), followed by 40 cycles at 95°C for 30 s (denaturation), 55°C for 30 s (annealing), and 72°C for 90 s (extension). Amplified DNA was resolved on 2.0% agarose gels, visualized with ethidium bromide, and imaged on a FluorChem Q imager (R&D Systems, Minneapolis, MN). The amount of PCR-amplifiable DNA at each region and each sonication time point was quantified by comparison to the amounts for dilutions of intact (unsonicated) DNA analyzed in the same PCR. The efficiency of VZV DNA shearing was calculated as follows: [(N0 − Nt)/N0] × 100, where N0 is the initial VZV DNA copy number and Nt is the VZV DNA copy number at time t.
TABLE 1.
Oligonucleotide primers
| Assay | Target | Primer namea | Region | Sequence | 5′ locationb |
|---|---|---|---|---|---|
| Long-range qPCR | VZV | ORF 9 LRPCR for | ORF 9 promoter | GCCCTTATTTCGTTAGCTTATCATGC | 10823 |
| ORF 9 short rev | ORF 9 TSSc | ACTGCATTAGAGCGACAAAGTC | 11041 | ||
| ORF 9 medium rev | ORF 9 center | CCGCCGCATTTTTAGTAAAAGATC | 11392 | ||
| ORF 9 long rev | ORF 9 terminus | AACCGTTACTGCGTAATTATATCCC | 11931 | ||
| ORF 51 LRPCR for | ORF 51 promoter | CGCTTCCACCTCGGGTGTC | 87808 | ||
| ORF 51 short rev | ORF 51 promoter | TACGACGGTTACCGGGCG | 88075 | ||
| ORF 51 medium rev | ORF 51 center | GCTGGATACGCGGTGTAGGC | 88312 | ||
| ORF 51 long rev | ORF 51 center | CCGGCCCATATGACATTGGC | 88946 | ||
| ChIP | VZV | 9.1 for | ORF 9 promoter | GGGCCCCTGTTTAATACGC | 10943 |
| 9.1 rev | ORF 9 promoter | GGGTCGTGAAATATCCAAACACGG | 10991 | ||
| 9.2 for | ORF 9 center | GCCAGCGGGAGACCAA | 11459 | ||
| 9.2 rev | ORF 9 center | GCGGTTTTTGGTGCAGTGC | 11502 | ||
| 9.3 for | ORF 9 terminus | GCATCAAGAACTGATACGCG | 11929 | ||
| 9.3 rev | ORF 9 terminus | GGGTAAACCGTTACTGCG | 11981 | ||
| 51.1 rev | ORF 51 promoter | GATCAGTGTATATACCGCCATG | 87511 | ||
| 51.1 for | ORF 51 promoter | TACTACACGGCTTTAACGGC | 87549 | ||
| 51.2 for | ORF 51 TSS | GCGTCCAGTACACAGC | 87920 | ||
| 51.2 rev | ORF 51 TSS | CCCTCCATATAACGCCCG | 87958 | ||
| 51.3 for | ORF 51 TSS | CGCGCGGGCGTTATAT | 87937 | ||
| 51.3 rev | ORF 51 TSS | CCACGAAACCAGATCCCC | 87973 | ||
| 51.4 for | ORF 51 center | CCTGTTCTCGCCAATGTTACTA | 89044 | ||
| 51.4 rev | ORF 51 center | TTGCACATCGTTGCCTAAATG | 89156 | ||
| 51.5 for | ORF 51 terminus | GCGCTATCGGAACTGTCC | 90320 | ||
| 51.5 rev | ORF 51 terminus | CGTTTATCCGCGGCCA | 90362 | ||
| 66.1 for | ORF 66 promoter | CCGACATTCTCACAAAGAGGTA | 112781 | ||
| 66.1 rev | ORF 66 promoter | GAGGGCGGAGATAATGACAAA | 112924 | ||
| 66.2 for | ORF 66 center | CGCGCGTTACAGTATCTTCATA | 113610 | ||
| 66.2 rev | ORF 66 center | CTCCAAAGTCTCCCACACAAA | 113715 | ||
| 66.3 for | ORF 66 terminus | CGTATAACCCGTTAAGGCAAATAG | 114266 | ||
| 66.3 rev | ORF 66 terminus | GGGCGTGACTCTGTATTAAGT | 114364 | ||
| HFL cells | ZFP154 for | Zinc finger protein 154 terminus | TGGATTTGGGGAAAATGGTA | Chr 19, position 2,784,893 | |
| ZFP154 rev | Zinc finger protein 154 terminus | GCCGTGTTGATTTTGTTGTG | Chr 19, position 2,785,192 | ||
| ZFP180 for | Zinc finger protein 180 terminus | TGATGCACAATAAGTCGAGCA | Chr 19, position 49,672,583 | ||
| ZFP180 for | Zinc finger protein 180 terminus | TGCAGTCAATGTGGGAAGTC | Chr 19, position 49,672,719 | ||
| GAPdH promoter | GAPDH promoter | Proprietary sequence | Catalog no. C17011001-50d | ||
| GAPdH TSS for | GAPDH TSS | CGGCTACTAGCGGTTTTACG | Chr 12, position 6,643,535 | ||
| GAPdH TSS rev | GAPDH TSS | GCTGCGGGCTCAATTTATAG | Chr 12, position 6,643,672 | ||
| GAPdH 2K DS for | GAPDH 2 kbp downstream of TSS | CGTAGCTCAGGCCTCAAGAC | Chr 12, position 6,645,635 | ||
| GAPdH 2K DS rev | GAPDH 2 kbp downstream of TSS | GTCGAACAGGAGGAGCAGAG | Chr 12, position 6,645,744 | ||
| GAPdH 4K DS for | GAPDH 4 kbp downstream of TSS | ACGCTGACTGGTTAGTGGAG | Chr 12, position 6,647,880 | ||
| GAPdH 4K DS rev | GAPDH 4 kbp downstream of TSS | GGCACAGAAAGCAATAGAGC | Chr 12, position 6,647,988 |
for, foward; rev, reverse.
The nucleotide location on the VZV (Dumas) genome (GenBank accession number X04370.1) or the position on the indicated chromosome (Chr) is indicated. For HFL cell (HG19) data, see https://genome.ucsc.edu/.
TSS, transcriptional start site.
Diagenode, Denville, NJ.
Chromatin immunoprecipitation.
Immunoprecipitations were carried out using a Pierce magnetic ChIP kit (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions and overnight antibody incubations with mouse anti-RNA polymerase II S2P IgG (Diagenode, Denville, NJ) or mouse anti-RNA polymerase II S5P IgG (Thermo Scientific, Rockford, IL). qPCR was used to determine the amount of VZV DNA immunoprecipitated, after normalization of the results to the amount of input DNA. The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene promoter served as a positive ChIP control for RNAP S5P enrichment, while two regions on HFL cell chromosome 19 known to lack RNAP S5P were used as negative controls (see Table 2). A region 4 kbp downstream from the transcriptional start site of the GAPDH gene was selected as a positive ChIP control for S2P enrichment on the basis of published Encode data from A549 (human lung carcinoma) cells and human umbilical vein endothelial cells (HUVEC) (http://genome.ucsc.edu). Further ChIP controls included normal mouse IgG (AbCam, Eugene, OR) and no IgG (beads alone). ChIP data were analyzed by analysis of variance (ANOVA) and multiple t tests using the Bonferroni correction to adjust for type 1 error.
TABLE 2.
RNAP S5P and S2P occupancy on VZV genes 9, 51, and 66 and GAPDH gene
| Target | Primer set | Region | Antibody | Day 1 |
Day 3 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold input |
Fold input |
Fold input |
na | Fold input |
||||||||||||
| Avg | SD | ChIP 1 | ChIP 2 | Avg | SD | ChIP 1 | ChIP 2 | ChIP 3 | ChIP 4 | ChIP 5 | ChIP 6 | |||||
| VZV | 9.1 | ORF 9 promoter | No Abb | NDc | 2.3E−05 | 5.0E−06 | 2 | 2.7E−05 | 2.0E−05 | |||||||
| Control Ig | 7.2E−04 | 5.7E−05 | 6.8E−04 | 7.6E−04 | 2.1E−04 | 2.2E−04 | 6 | 4.7E−05 | 1.3E−05 | 6.7E−05 | 5.0E−04 | 1.7E−04 | 4.7E−04 | |||
| RNAP S2P | 1.7E−03 | 3.1E−03 | 1.7E−03 | 1.7E−03 | 4.3E−04 | 2.7E−04 | 2 | 6.2E−04 | 2.4E−04 | |||||||
| RNAP S5P | 1.4E−02 | 3.3E−05 | 1.1E−02 | 1.6E−02 | 1.0E−02 | 1.1E−02 | 6 | 5.3E−03 | 5.8E−03 | 2.8E−03 | 1.2E−02 | 4.1E−03 | 3.2E−02 | |||
| 9.2 | ORF 9 center | No Ab | ND | 1.7E−04 | 2.0E−04 | 2 | 3.1E−04 | 2.2E−05 | ||||||||
| Control Ig | 6.8E−04 | 9.5E−05 | 6.1E−04 | 7.5E−04 | 2.9E−04 | 2.5E−04 | 6 | 6.1E−04 | 3.5E−05 | 4.9E−04 | 1.6E−04 | 4.3E−04 | 3.1E−05 | |||
| RNAP S2P | 1.7E−03 | 1.8E−03 | 1.8E−03 | 1.7E−03 | 3.8E−04 | 2.8E−04 | 2 | 5.8E−04 | 1.8E−04 | |||||||
| RNAP S5P | 1.1E−02 | 2.3E−05 | 1.0E−02 | 1.3E−02 | 1.1E−02 | 9.5E−03 | 6 | 2.7E−02 | 2.2E−03 | 1.2E−02 | 3.8E−03 | 1.6E−02 | 3.9E−03 | |||
| 9.3 | ORF 9 terminus | No Ab | ND | 1.2E−04 | 1.6E−04 | 2 | 2.3E−04 | 1.1E−05 | ||||||||
| Control Ig | 8.5E−04 | 2.1E−04 | 7.0E−04 | 1.0E−03 | 2.1E−04 | 2.3E−04 | 4 | 2.9E−04 | 4.1E−06 | 4.8E−05 | 5.0E−04 | |||||
| RNAP S2P | 2.0E−03 | 1.5E−03 | 2.5E−03 | 1.6E−03 | 4.3E−04 | 2.1E−04 | 2 | 5.8E−04 | 2.9E−04 | |||||||
| RNAP S5P | 1.8E−02 | 6.2E−04 | 1.7E−02 | 1.9E−02 | 8.6E−03 | 6.1E−03 | 4 | 1.7E−02 | 3.3E−03 | 5.2E−03 | 8.8E−03 | |||||
| 51.1 | ORF 51 promoter | No Ab | ND | 4.0E−05 | 2.8E−05 | 3 | 3.9E−05 | 1.2E−05 | 6.8E−05 | |||||||
| Control Ig | 5.9E−04 | 6.5E−05 | 5.4E−04 | 6.3E−04 | 3.6E−05 | 2.9E−05 | 3 | 6.2E−05 | 5.3E−06 | 4.0E−05 | ||||||
| RNAP S2P | 1.6E−03 | 4.8E−04 | 1.7E−03 | 1.5E−03 | 2.6E−04 | 7.9E−05 | 2 | 3.2E−04 | 2.1E−04 | |||||||
| RNAP S5P | 1.4E−02 | 3.5E−04 | 1.4E−02 | 1.5E−02 | 4.2E−03 | 2.5E−03 | 3 | 6.6E−03 | 4.3E−03 | 1.7E−03 | ||||||
| 51.2 | ORF 51 TSSd | No Ab | ND | 8.5E−05 | 7.8E−05 | 2 | 3.0E−05 | 1.4E−04 | ||||||||
| Control Ig | 7.6E−04 | 5.8E−04 | 3.5E−04 | 1.2E−03 | 6.6E−05 | 2.5E−05 | 3 | 3.8E−05 | 7.4E−05 | 8.6E−05 | ||||||
| RNAP S2P | 1.5E−03 | 1.1E−04 | 1.8E−03 | 1.3E−03 | 3.7E−04 | 1.9E−04 | 2 | 5.0E−04 | 2.4E−04 | |||||||
| RNAP S5P | 1.1E−02 | 3.7E−04 | 1.1E−02 | 1.2E−02 | 8.2E−03 | 4.8E−03 | 3 | 3.5E−03 | 1.3E−02 | 8.0E−03 | ||||||
| 51.3 | ORF 51 TSS | No Ab | ND | 3.4E−05 | 3.0E−05 | 3 | 3.0E−05 | 5.8E−06 | 6.5E−05 | |||||||
| Control Ig | 6.1E−04 | 1.9E−04 | 7.4E−04 | 4.8E−04 | 3.4E−05 | 2.5E−05 | 3 | 3.7E−05 | 8.1E−06 | 5.8E−05 | ||||||
| RNAP S2P | 1.9E−03 | 8.5E−04 | 2.0E−03 | 1.8E−03 | 3.8E−04 | 1.3E−04 | 2 | 4.7E−04 | 2.9E−04 | |||||||
| RNAP S5P | 1.2E−02 | 1.3E−04 | 1.2E−02 | 1.3E−02 | 3.4E−03 | 1.3E−03 | 3 | 3.9E−03 | 4.3E−03 | 1.9E−03 | ||||||
| 51.4 | ORF 51 center | No Ab | ND | 8.2E−05 | 1.0E−04 | 2 | 9.0E−06 | 1.6E−04 | ||||||||
| Control Ig | 5.9E−04 | 1.7E−04 | 4.6E−04 | 7.1E−04 | 1.0E−04 | 9.7E−05 | 3 | 4.2E−06 | 2.0E−04 | 9.9E−05 | ||||||
| RNAP S2P | 1.7E−03 | 3.5E−03 | 2.0E−03 | 1.4E−03 | 4.0E−04 | 8.7E−05 | 2 | 4.6E−04 | 3.4E−04 | |||||||
| RNAP S5P | 1.5E−02 | 4.0E−04 | 1.3E−02 | 1.8E−02 | 1.1E−02 | 7.7E−03 | 3 | 4.2E−03 | 1.9E−02 | 9.3E−03 | ||||||
| 51.5 | ORF 51 terminus | No Ab | ND | 8.2E−05 | 1.0E−04 | 3 | 2.0E−04 | 5.6E−06 | 4.0E−05 | |||||||
| Control Ig | 7.7E−04 | 2.7E−04 | 5.8E−04 | 9.6E−04 | 8.8E−05 | 1.2E−04 | 3 | 2.3E−04 | 5.6E−06 | 2.7E−05 | ||||||
| RNAP S2P | 1.2E−03 | 8.8E−04 | 8.7E−04 | 1.5E−03 | 3.6E−04 | 1.5E−04 | 2 | 4.6E−04 | 2.5E−04 | |||||||
| RNAP S5P | 8.5E−03 | 4.7E−04 | 7.8E−03 | 9.1E−03 | 5.4E−03 | 4.9E−03 | 3 | 1.1E−02 | 3.0E−03 | 2.2E−03 | ||||||
| 66.1 | ORF 66 promoter | No Ab | ND | ND | ||||||||||||
| Control Ig | 4.9E−04 | 1.4E−04 | 3.9E−04 | 5.9E−04 | 2.6E−04 | 2.0E−04 | 4 | 2.9E−05 | 4.3E−04 | 4.2E−04 | 1.6E−04 | |||||
| RNAP S2P | 2.7E−03 | 3.9E−03 | 3.1E−03 | 2.4E−03 | 5.1E−04 | 5.3E−05 | 2 | 5.4E−04 | 4.7E−04 | |||||||
| RNAP S5P | 2.6E−02 | 4.5E−04 | 2.4E−02 | 2.9E−02 | 1.0E−02 | 8.6E−03 | 4 | 5.5E−03 | 2.3E−02 | 8.6E−03 | 4.0E−03 | |||||
| 66.2 | ORF 66 center | No Ab | ND | ND | ||||||||||||
| Control Ig | 5.7E−04 | 2.3E−04 | 4.0E−04 | 7.3E−04 | 2.4E−04 | 1.6E−04 | 4 | 3.7E−05 | 3.0E−04 | 4.1E−04 | 2.1E−04 | |||||
| RNAP S2P | 1.9E−03 | 3.1E−03 | 1.9E−03 | 1.8E−03 | 5.0E−04 | 3.4E−05 | 2 | 4.8E−04 | 5.3E−04 | |||||||
| RNAP S5P | 2.6E−02 | 1.1E−04 | 2.4E−02 | 2.8E−02 | 8.8E−03 | 6.1E−03 | 4 | 3.2E−03 | 1.7E−02 | 1.0E−02 | 4.9E−03 | |||||
| 66.3 | ORF 66 terminus | No Ab | ND | ND | ||||||||||||
| Control Ig | 6.2E−04 | 1.9E−04 | 4.8E−04 | 7.5E−04 | 2.3E−04 | 1.6E−04 | 4 | 2.6E−05 | 3.4E−04 | 3.8E−04 | 1.7E−04 | |||||
| RNAP S2P | 2.4E−03 | 1.5E−03 | 2.8E−03 | 2.0E−03 | 4.0E−04 | 2.1E−04 | 2 | 5.5E−04 | 2.5E−04 | |||||||
| RNAP S5P | 2.4E−02 | 6.0E−04 | 2.3E−02 | 2.5E−02 | 9.9E−03 | 7.9E−03 | 4 | 5.3E−03 | 2.1E−02 | 9.5E−03 | 3.6E−03 | |||||
| HFL | GAPdH promoter | GAPDH promoter | No Ab | ND | 3.3E−04 | 1.8E−04 | 2 | 4.5E−04 | 2.0E−04 | |||||||
| Control Ig | 3.1E−03 | 8.7E−04 | 3.7E−03 | 2.4E−03 | 4.8E−04 | 4.1E−04 | 5 | 2.8E−04 | 2.3E−04 | 2.7E−04 | 1.2E−03 | 4.2E−04 | ||||
| RNAP S2P | 3.2E−03 | 3.1E−03 | 5.4E−03 | 9.8E−04 | 6.1E−03 | 5.4E−03 | 2 | 2.3E−03 | 9.9E−03 | |||||||
| RNAP S5P | 4.2E−02 | 2.5E−02 | 6.0E−02 | 2.4E−02 | 3.8E−02 | 3.1E−02 | 5 | 8.9E−03 | 1.1E−02 | 8.5E−02 | 4.9E−02 | 3.4E−02 | ||||
| ZFP154 | Zinc finger protein 154 terminus | No Ab | ND | 3.5E−05 | 3.9E−05 | 2 | 6.2E−05 | 7.5E−06 | ||||||||
| Control Ig | 9.4E−04 | 1.3E−03 | 4.3E−05 | 1.8E−03 | 1.9E−04 | 1.7E−04 | 2 | 7.0E−05 | 3.1E−04 | |||||||
| RNAP S2P | 2.4E−03 | 4.9E−04 | 2.0E−03 | 2.7E−03 | 9.8E−04 | 4.7E−04 | 2 | 6.5E−04 | 1.3E−03 | |||||||
| RNAP S5P | 3.4E−04 | 4.3E−04 | 2.9E−05 | 6.4E−04 | 1.6E−04 | 1.2E−04 | 2 | 7.1E−05 | 2.4E−04 | |||||||
| ZFP180 | Zinc finger protein 180 terminus | No Ab | ND | 5.5E−04 | 3.8E−04 | 2 | 2.8E−04 | 8.2E−04 | ||||||||
| Control Ig | 2.0E−03 | 5.0E−04 | 2.4E−03 | 1.7E−03 | 4.5E−04 | 1.9E−04 | 2 | 3.1E−04 | 5.8E−04 | |||||||
| RNAP S2P | 1.1E−03 | 1.1E−04 | 1.2E−03 | 1.0E−03 | 5.9E−04 | 1.3E−04 | 2 | 6.8E−04 | 5.0E−04 | |||||||
| RNAP S5P | 1.9E−03 | 4.1E−04 | 2.2E−03 | 1.6E−03 | 6.4E−04 | 5.2E−04 | 2 | 2.7E−04 | 1.0E−03 | |||||||
| GAPdH TSS | GAPDH TTSe | Control Ig | 5.8E−04 | 1.6E−04 | 4.6E−04 | 6.9E−04 | 1.3E−04 | 1.2E−04 | 2 | 2.1E−04 | 4.0E−05 | |||||
| RNAP S2P | 6.7E−03 | 8.5E−04 | 6.1E−03 | 7.3E−03 | 5.0E−03 | 1.9E−03 | 2 | 6.3E−03 | 3.7E−03 | |||||||
| RNAP S5P | 5.3E−02 | 2.7E−02 | 3.3E−02 | 7.2E−02 | 2.8E−02 | 4.5E−04 | 2 | 2.9E−02 | 2.8E−02 | |||||||
| GAPdH 2K DS | GAPDH 2 kbp downstream of TTS | Control Ig | 2.5E−05 | 9.8E−06 | 1.8E−05 | 3.2E−05 | 1.7E−03 | 2.2E−03 | 2 | 3.2E−03 | 1.6E−04 | |||||
| RNAP S2P | 9.4E−03 | 1.1E−04 | 9.5E−03 | 9.3E−03 | 9.8E−03 | 1.1E−03 | 2 | 1.1E−02 | 9.0E−03 | |||||||
| RNAP S5P | 4.0E−02 | 1.4E−02 | 3.0E−02 | 5.0E−02 | 2.4E−02 | 5.2E−03 | 2 | 2.0E−02 | 2.8E−02 | |||||||
| GAPdH 4K DS | GAPDH 4 kbp downstream of TTS | Control Ig | 1.2E−03 | 1.1E−03 | 4.6E−04 | 2.0E−03 | 5.0E−04 | 2.4E−04 | 2 | 3.3E−04 | 6.7E−04 | |||||
| RNAP S2P | 2.6E−02 | 1.2E−02 | 1.7E−02 | 3.4E−02 | 1.3E−02 | 2.0E−03 | 2 | 1.5E−02 | 1.2E−02 | |||||||
| RNAP S5P | 1.1E−02 | 4.2E−03 | 8.5E−03 | 1.4E−02 | 8.9E−03 | 3.1E−04 | 2 | 8.6E−03 | 9.1E−03 | |||||||
n, number of independent replicates.
Ab, antibody.
ND, not done.
TSS, transcriptional start site.
TTS, transcription termination site.
Quantitative RNA analysis.
Total RNA was extracted (TriPure isolation reagent; Roche, San Francisco, CA) from VZV-infected HFL cells at 1 dpi before a virus-induced cytopathic effect (CPE) and 3 dpi, when the CPE was at its height, and first-strand cDNA was synthesized (Transcriptor first-strand cDNA synthesis kit, Roche). Transcripts were quantified by qPCR as described above using VZV- and cell-specific primers (Table 1). Each VZV qPCR mixture contained dilutions of VZV DNA at known concentrations to determine the abundance of the virus transcripts (24).
RESULTS
Efficiency of VZV DNA shearing.
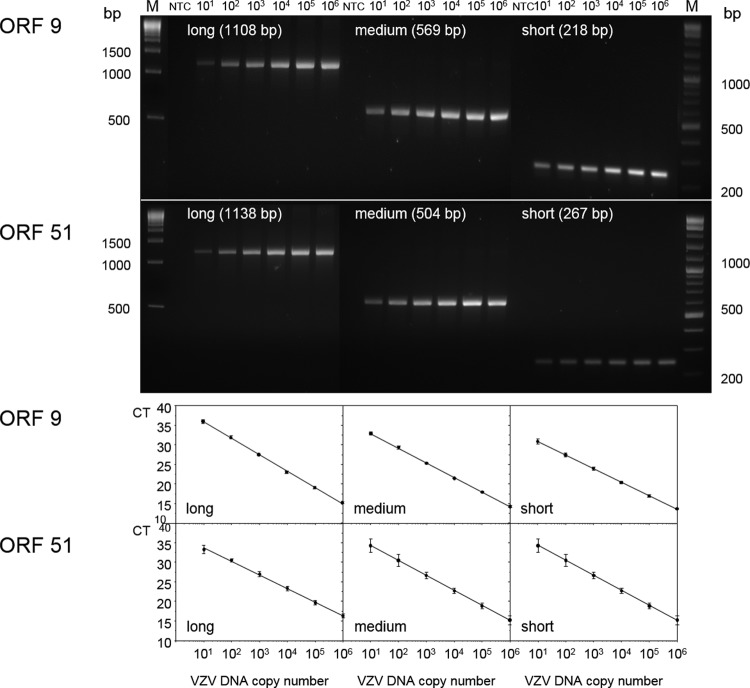
Initial experiments showed that most sheared cell DNA averaged ∼300 bp in size, but ChIP routinely demonstrated a high nonspecific background (data not shown). While a high ChIP background can be attributed to many factors, we reasoned that virus DNA was insufficiently sheared, since shorter DNA fragments would be expected to have fewer sites for nonspecific binding. Thus, PCR primers that spanned 100% of VZV ORF 9 and 45% of ORF 51 were used to determine the extent of VZV DNA shearing by sonication (Fig. 1). For VZV ORF 9 (Fig. 1B) or ORF 51 (Fig. 1C), one forward primer and three reverse primers were selected to yield nested PCR products of ∼250 bp (short fragments), 550 bp (fragments of medium length), and 1,100 bp (long fragments) (Table 1). VZV ORFs 9 and 51 represent two distinct regions of the virus genome (Fig. 1A) separated by ∼77 kbp. To ensure that the SYBR-based quantitative PCR conformed to the minimum information for publication of quantitative real-time PCR experiments (MIQE guidelines for PCR specificity, sensitivity, and efficiency) (25), PCR amplification of known amounts of VZV DNA revealed single bands of the expected size for each primer pair (Fig. 2, top), and each PCR assay detected as few as 10 copies of VZV DNA with little variation in 6 independent trials (Fig. 2, bottom).
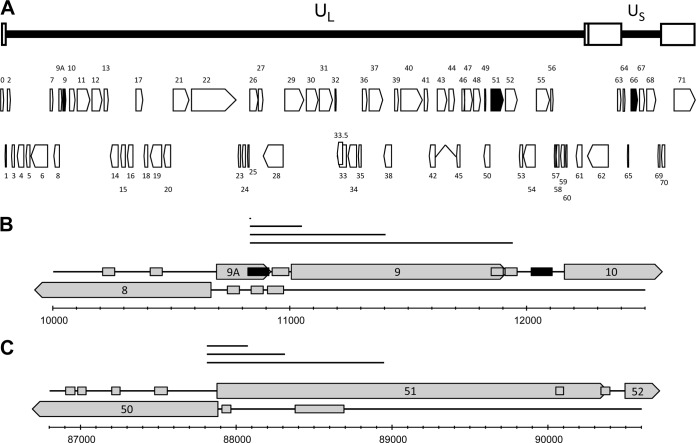
FIG 1.
Locations of oligonucleotide primers on the VZV transcription map. (A) The VZV genome consists of unique long (UL) and unique short (US) DNA segments, each of which is bounded by inverted repeat regions (open boxes). A total of 74 ORFs, 39 on the top DNA strand and 35 on the bottom DNA strand, were identified. Filled arrows show the locations of VZV ORFs 9, 51, and 66. (B) VZV DNA between 10 kbp and 12.5 kbp begins within ORF 8, ends within ORF 10, and contains experimentally identified promoters for ORF 9 and ORF 10 (black boxes), predicted promoter regions for ORFs 8, 9A, 9, and 10 (gray boxes), and PCR fragments generated by long-range PCR (solid lines). (C) VZV DNA between 86.8 kbp and 90.6 kbp begins within ORF 50, ends within ORF 52, and contains predicted promoter regions for ORF 50, 51, and 52 (gray boxes) and PCR fragments generated by long-distance PCR (solid lines).
FIG 2.
Long-range qPCR. (Top) Log10 dilutions of VZV DNA (106 to 101 virus DNA copies) were amplified with PCR primers that generated long (1,108 bp for ORF 9 and 1,138 bp for ORF 51), medium-length (569 bp for ORF 9 and 504 bp for ORF 51), and short (218 bp for ORF 9 and 267 bp for ORF 51) VZV DNA fragments. (Bottom) qPCR of the same VZV DNA dilutions with the same primer sets resulted in highly reproducible linear relationships between DNA yield (the cycle threshold [CT] value) and initial VZV DNA copy number for all targets in ORF 9 and ORF 51. Each graph represents the mean ± standard deviation from 6 independent amplifications. Lanes M, DNA size markers; lanes NTC, no-template control.
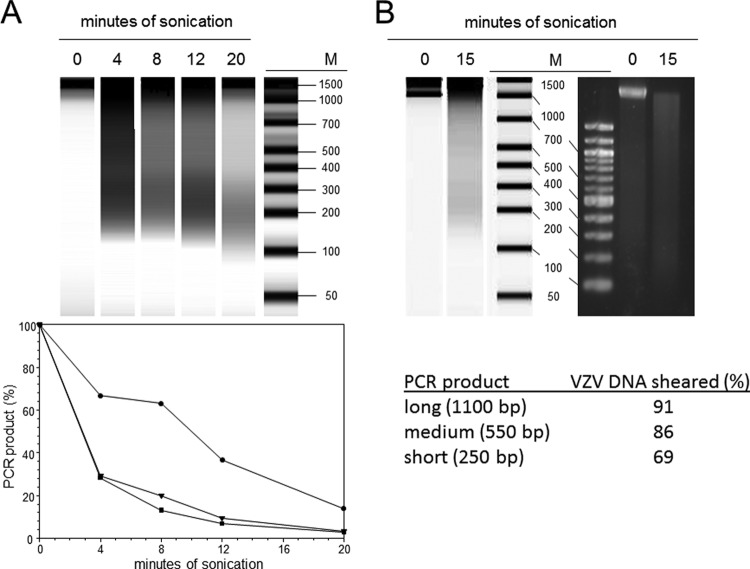
The efficiency of VZV DNA shearing was determined by analysis of formaldehyde-cross-linked chromatin from virus-infected HFL cells after sonication for 0, 4, 8, 12, and 20 min. The amount of host DNA with lengths of between 100 and 1,000 bp increased with the time of sonication and peaked at ∼200 bp after 20 min of sonication (Fig. 3A). Bioanalyzer analysis provided a convenient and sensitive estimate of the DNA fragmentation pattern within the size range of the capillary gel. DNA fragments larger than the capillary exclusion limit (1,500 bp) were concentrated at the top of the gel and obscured by the 1,500-bp DNA marker added to each sample. While Southern blotting of sheared DNA probed with labeled VZV DNA could be used to determine the size range of the virus DNA fragments, we used long-range qPCR to determine the efficiency of shearing of the virus DNA among the background of host DNA fragments. The amount of VZV DNA remaining following sonication was determined by long-range PCR with both ORF 9- and ORF 51-specific primer sets, and the results from two independent biological replicates were averaged and showed that nucleocapsid-free VZV DNA was sensitive to sonication (Fig. 3A, bottom). After 4 min, 30% of virus DNA fragments of <∼550 bp remained intact. However, a 20-min sonication was required to reduce the amount of VZV DNA fragments of <250 bp that remained intact to <20% of the starting quantity. A 15-min sonication was selected for subsequent ChIP experiments, since 6 independent trials showed that this amount of time was sufficient to reduce the amount of VZV DNA fragments of <1,100 bp to <5% of the initial quantity and VZV DNA fragments of <550 bp to <10% of the initial quantity by PCR amplification. An example of the DNA fragmentation achieved following 15 min of sonication of formaldehyde-cross-linked VZV-infected HFL cell DNA is shown in Fig. 3B. The Bioanalyzer analysis (Fig. 3B, top, left) showed an increase in the amount of DNA fragments of <1,000 bp obtained following 15 min of sonication. Similar results were observed when the same DNA was analyzed by standard electrophoresis on 2% agarose gels (Fig. 3B, top, right). The percentage of VZV DNA sheared was calculated from the amount of PCR product remaining, as described in Materials and Methods, and indicated that 91% of the VZV DNA was sheared to fragments of <1,100 bp, 86% of the VZV DNA was sheared to fragments of <550 bp, and 69% of the VZV DNA was sheared to fragments of <250 bp (Fig. 3B, bottom).
FIG 3.
Sheared chromatin size. (A) Formaldehyde-fixed DNA from VZV-infected HFL cells was sonicated for 0, 4, 8, 12, and 20 min. (Top) The abundance of DNA fragments with lengths of between 50 bp and 1,500 bp was determined by capillary gel electrophoresis after heat reversal of the formaldehyde-induced protein-DNA cross-links. DNA fragments of >1,500 bp were retained at the top of the gel and comigrated with the added 1,500-bp internal standard. (Bottom) The amount of short (circles), medium-length (triangles), and long (squares) VZV DNA fragments generated by increasing the sonication time was determined by qPCR using primers specific for regions within ORF 9 and ORF 51. The percentage of VZV DNA sheared was compared to that obtained with no sonication (0 min). Results reflect the averages from two independent assays. (B) Formaldehyde-fixed DNA from VZV-infected HFL cells was sonicated for 15 min and resolved by capillary gel electrophoresis along with agarose gel electrophoresis. The efficiency of VZV DNA shearing was determined by long-distance PCR. Lanes M, DNA size markers (in base pairs).
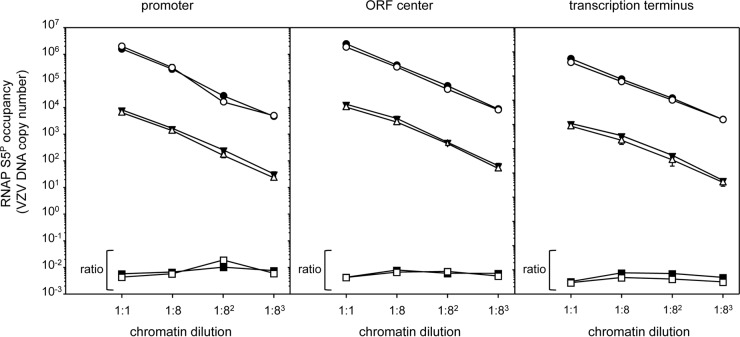
After optimization of VZV DNA fragmentation, ChIP was conducted to ensure that excess antibody was present. Serial 8-fold dilutions of fragmented chromatin from duplicate biological replicates were immunoprecipitated with 5 μg purified anti-RNAP S5P antibody. qPCR amplification of input VZV DNA with VZV ORF 9- and ORF 51-specific primers reflected the 8-fold dilutions (Fig. 4). The amount of VZV DNA immunoprecipitated with anti-RNAP S5P antibody also resulted in similar 8-fold reductions. For all dilutions and at all 3 locations within both VZV ORF 9 and VZV ORF 51, the same ratio of input VZV DNA to immunoprecipitated virus DNA was obtained (ORF 9, 6.5% ± 1.7%; ORF 51, 5.0% ± 1.1%). Overall, 5.77% ± 1.57% of input VZV DNA was immunoprecipitated with anti-RNAP S5P IgG. On the basis of the results presented above, antibody excess was clearly evident.
FIG 4.
ChIP under conditions of excess antibody. Serial log8 dilutions of sonicated chromatin from formaldehyde-fixed, VZV-infected HFL cells were immunoprecipitated with equal amounts of mouse anti-RNAP S5P-specific IgG. The amount of VZV DNA added to each assay mixture and immunoprecipitation was determined by qPCR with primers specific for VZV ORF 9 (closed symbols) and ORF 51 (open symbols). The horizontal lines reflect the ratio (squares) of the amount of input VZV DNA (circles) to the amount of immunoprecipitated VZV DNA (triangles) for ORF 9 and ORF 51 and indicate that ChIP conditions were independent of the input DNA concentration under conditions of antibody excess. Results reflect the averages from two biological replicates.
RNAP S5P and RNAP S2P enrichment on VZV genes at height of CPE.
qPCR primers were designed to amplify regions within the promoter, the approximate ORF center, and the terminus of VZV ORFs 9, 51, and 66 (Table 1) and used to determine RNAP S5P and RNAP S2P occupancy (Fig. 5, top). All primer pairs, regardless of product length (38 to 143 bp), detected as few as 10 copies of VZV DNA and produced a single discrete band on agarose gels, as well as sharp melting temperature profiles (data not shown).
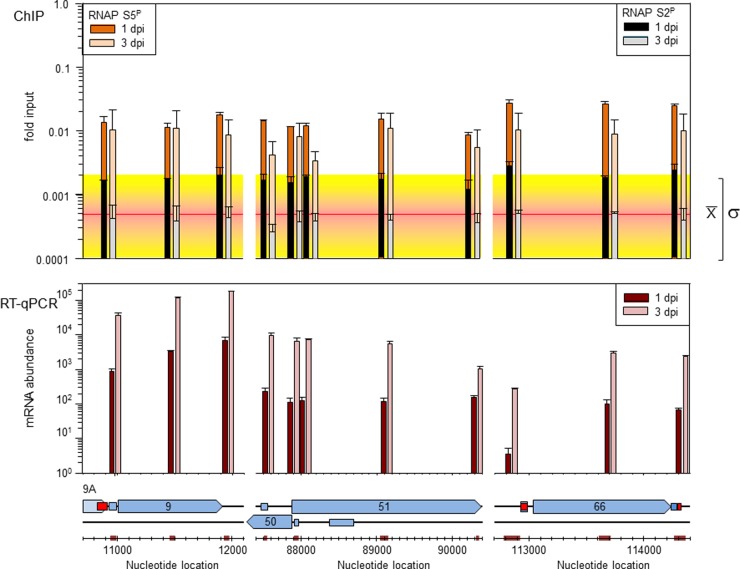
FIG 5.
Comparison of RNAP S5P and RNAP S2P occupancy to VZV mRNA abundance at 1 and 3 days postinfection. VZV ORFs 9, 51, and 66 were investigated for RNAP S5P and RNAP S2P occupancy by ChIP (top), and mRNA abundance was determined by RT-qPCR at 1 and 3 dpi (middle). (Bottom) Predicted (blue boxes) and experimentally determined (red boxes) promoters for ORFs 9, 50, 51, and 66 are shown, as are the location and relative size of PCR products (colored boxes on the nucleotide scale). (Top) The amount of VZV DNA immunoprecipitated with mouse anti-RNAP S5P and mouse anti-RNAP S2P IgG compared to the amount of input VZV DNA (fold input) was determined by qPCR for each location. The average background (isotype control IgG) (red line) and standard deviation (yellow region) for all ChIP reactions are shown. (Middle) At 1 and 3 dpi, the average mRNA abundance was determined by RT-qPCR. The RT-qPCR results reflect those from two independent biological replicates, while the number of independent replicates for each immunoprecipitation is listed in Table 2.
Table 2 lists all individual ChIP data along with the overall results obtained for each location in VZV genes 9, 51, and 66 as well as three cellular regions (the GAPDH gene promoter, ZFP154, and ZFP180). No statistically significant difference (ANOVA, P = 0.92) in RNAP S5P occupancy in the promoter, ORF center, or transcription terminus was detected for VZV genes 9, 51, and 66. However, the amount of VZV DNA immunoprecipitated with anti-RNAP S5P antibody was significantly greater (t test, P < 0.001) than that recovered using an isotype control antibody or when antibody was omitted from the reaction mixture (background). With respect to the host control regions, significant enrichment of RNAP S5P on the GAPDH gene promoter compared to that for the isotype control was detected (ANOVA, P = 0.05; t test, P = 0.05), while no significant RNAP S5P enrichment (ANOVA, P = 0.56) at the two host negative-control regions relative to the amount seen in the ChIP control immunoprecipitations was detected (Table 2). Since both ChIP assays for determination of the amount of background (assays with nonspecific, isotype control IgG and assays in which IgG was omitted altogether) gave the same results (ANOVA, P = 0.55), subsequent assays used only the nonspecific, isotype antibody as the ChIP background control.
No significant difference in the occupancy of RNAP S2P at the promoter, ORF center, or transcription terminus was detected for VZV genes 9, 51, and 66 in virus-infected HFL cells harvested at 1 and 3 dpi (ANOVA, P = 0.93) and with respect to that obtained with nonspecific isotype control IgG (ANOVA, P = 0.06) (Table 2).
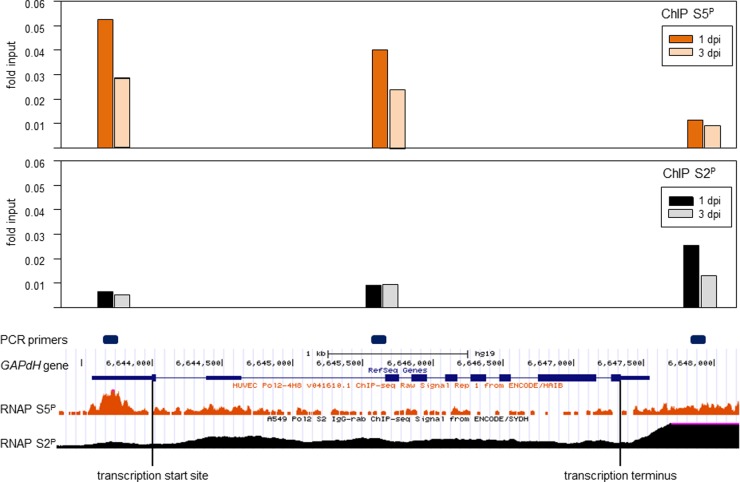
With respect to the results obtained with the host control regions (the GAPDH gene promoter and the two zinc finger protein genes), no significant amounts of RNAP S2P above the amount of isotypic control IgG were immunoprecipitated (ANOVA, P = 0.98). Since these regions were specifically selected for their RNAP S5P binding characteristics, Encode ChIP-DNA sequencing data were searched for phosphorylated RNAP binding, and combination of the information for human umbilical endothelial cells (HUVEC) with that for human lung carcinoma (A549) cells provided three regions within the GAPDH gene that showed differences in RNAP S5P and RNAP S2P occupancy (Fig. 6 bottom).
FIG 6.
Comparison of RNAP S5P and RNAP S2P occupancy on the GAPDH gene. DNA from VZV-infected HFL cells was harvested at 1 and 3 dpi. Formaldehyde-cross-linked, sheared chromatin was immunoprecipitated with mouse anti-RNAP S5P IgG (top) and mouse anti-RNAP S2P IgG (middle) and compared to the amount of input DNA (fold input) at the promoter, gene body, and transcription termination site of the GAPDH gene. (Bottom) Purple boxes represent GAPDH-specific primer locations. The occupancy of RNAP S5P and RNAP S2P on the GAPDH gene from VZV-infected HFL cells was compared to that of RNAP S5P (orange histogram) and RNAP S2P (black histogram) at previously published locations (http://genome.ucsc.edu). The GAPDH transcription start site, termination site, exons (blue boxes), and introns (thin blue lines) are shown.
Before quantitative ChIP analysis, each new GAPDH gene-specific primer pair was shown to amplify HFL cell DNA with a similar efficiency and specificity and produced a single discrete amplicon (data not shown). For all GAPDH gene-specific primers and at both 1 and 3 dpi, RNAP S2P and RNAP S5P occupancy on HFL DNA was significantly more than that of the isotype control antibody (t test, P < 0.01). Within the GAPDH gene, RNAP S5P occupancy decreased from the promoter to the transcription termination site in HFL cell chromatin harvested at both 1 and 3 dpi. More RNAP S5P was present at each location at 1 dpi than at 3 dpi. However, RNAP S2P occupancy on the GAPDH gene was maximal at the transcription termination site at 1 dpi. At 3 dpi, the RNAP S2P occupancy at the GAPDH gene promoter was reduced by 48%. The RNAP S2P occupancy was similar (0.08-fold ± 0.002-fold the input amount) at the GAPDH gene promoter and gene body but 8-fold above that for the respective background (isotype) controls (Fig. 6, top).
RNAP S5P and RNAP S2P enrichment on VZV genes early during VZV infection.
Since RNAP S2P occupancy at the GAPDH transcription termination site decreased from 1 to 3 dpi (Fig. 6) and at 3 dpi no significant RNAP S2P enrichment was detected on VZV genes 9, 51, and 66 (Fig. 5, top), posttranslationally modified RNAP occupancy on VZV DNA was investigated in virus-infected HFL cells harvested early (1 dpi). No significant difference (ANOVA, P = 0.06) in the occupancy of RNAP S2P at the promoter, ORF center, and transcription terminus of VZV genes 9, 51, and 66 was detected; however, the amount of VZV DNA immunoprecipitated at these regions was 2.8-fold above the background, which was a significant increase (t test, P < 0.01).
While the amount of RNAP S5P specifically immunoprecipitated was significantly above the background level (ANOVA, P < 0.01; t test, P < 0.01), the occupancy of RNAP S5P at the promoter, ORF center, and transcription terminus was the same within each gene (ANOVA, P < 0.01; t test, P < 0.01). Since RNAP S5P co-occupancy within each gene was the same at each location tested, each gene pair was compared. Gene 66 contained the most RNAP S5P (t test, P < 0.01), and genes 9 and 51 contained the same amount of RNAP S5P (t test, P = 0.12).
Steady-state VZV mRNA abundance.
The steady-state mRNA levels present at the promoter, ORF center, and transcription terminus of VZV ORFs 9, 51, and 66 RNA, as determined by reverse transcription-qPCR (RT-qPCR), was 46-fold ± 16-fold more at 3 dpi than at 1 dpi for all sites except the ORF 51 transcription termination site, whereas at 3 dpi it was only 6-fold more than that at 1 dpi (Fig. 5, middle section, top). Using gene body PCR results as an indication of gene transcript abundance, the levels of the ORF 9 transcripts were increased 30- and 40-fold over those of the ORF 51 transcripts at 1 and 3 dpi, respectively, which are similar to the 29- and 21-fold increases in the levels of the ORF 9 transcripts over those of the ORF 51 and 66 transcripts at 3 dpi, respectively. The abundance of ORF 51 and ORF 66 transcripts was the same at 1 dpi (t test, P = 0.27), while at 3 dpi, the amount of ORF 66 transcripts was twice that of the ORF 51 transcripts, which is a significant difference (t test, P < 0.001). The compactness of VZV transcriptional units was shown by the qPCR data and suggested continuation of ORF 9A into ORF 9 and overlap of the 5′ region of VZV ORF 51 with the 5′ untranslated region of ORF 50. In addition, the qPCR data indicated that ORF 66 mRNA terminates at least 200 nucleotides (nt) beyond the termination codon for ORF 66.
DISCUSSION
ChIP is a powerful tool used to decipher transcription regulation in alphaherpesviruses. Indeed, ChIP has shown that during productive herpes simplex virus 1 (HSV-1) infection, virus gene transcription is regulated, in part, by binding of virus (26) or host (27) proteins to specific DNA sequences within gene promoters or to protein components of the transcriptional apparatus (28). Importantly, ChIP analyses of HSV-1-infected neurons have shown that posttranslational modification of histone proteins is a critical factor that maintains the unique transcriptional pattern of virus genes during latency. Most of the ∼80 HSV-1 genes expressed during productive infection are transcriptionally silent during latent infection, and their promoters are associated with nucleosomes containing posttranslationally modified histones indicative of transcriptionally silent heterochromatin. Conversely, the promoter of the single HSV-1 gene transcribed during latency is associated with histones containing posttranslational modifications indicative of actively transcribed euchromatin (29, 30). ChIP has shown that transcriptionally distinct regions of the virus genome are maintained by chromosomal insulators, most notably, the CCCTC-binding factor, CTCF (31). During HSV-1 reactivation, these chromatin boundary elements are released from the virus DNA (32, 33). Since the release of CTCF from the virus genome is an attractive mechanism connecting external stress to HSV-1 reactivation (34, 35) and the critical CCCTC-binding sites are separated by at least 3,300 nt (31), the HSV-1 DNA fragment size is not a critical aspect of these ChIP analyses. However, determination of the efficiency of virus DNA fragmentation is critical when locating transcription factors on the virus genome where promoters are closely linked to the genes that they control. Although computer algorithms that analyze DNA sequence data obtained by ChIP are continually updated to better locate protein binding domains on the genome (36), our long-distance PCR assay is a quick, convenient, and sensitive means to determine the efficiency of size fractionations of virus DNA within a large background of cell chromatin. The long-distance PCR assay can also be applied to determine the DNA fragmentation generated by single-strand-specific endo-exonuclease (micrococcal nuclease [MNase]) or DNase I, the well-established “gold standard” method for probing chromatin accessibility (37). Additionally, Southern blot analysis requires >2 days and significant amounts of sample, and it is difficult to accurately quantify the results. The <4-h PCR-based assay requires minimal sample (<10 ng input DNA), does not require an isotope, and is completed in a time frame that permits further sonication, if required.
In tissue culture and at the single-cell level, VZV productive infection takes 9 to 12 h, during which time immediate early, early, and late proteins are sequentially synthesized (38). However, VZV is a highly cell-associated virus, and titers of cell-free virus sufficient to infect cultures at multiplicities of >1 are unattainable. Consequently, in vitro infections are initiated by cocultivation of virus-infected cells with uninfected cells. Under these conditions, the CPE progresses slowly and transcription analysis does not reveal the sequential synthesis of individual classes of virus transcripts but, rather, reveals an overall increase in the number of transcripts of all classes (13). This was seen in the uniform increase in ORF 9 (late), ORF 51 (early), and ORF 66 (late) transcripts from 1 to 3 dpi. Also, since high-multiplicity VZV infections are not feasible, assignment of virus genes into kinetic classes (immediate early, early, and late) is done by analogy to the genes of herpes simplex virus 1, the prototype alphaherpesvirus. Transcripts for all annotated VZV genes have been detected by RT-qPCR (15) and quantified on PCR-based arrays (13) and long-oligonucleotide-based arrays (14), as well as strand-specific cDNA deep sequencing (22). These studies do not reveal the sequential transcription of different classes of virus genes, as is seen in other herpesviruses (39), but they do show a variation in the steady-state abundance of VZV transcripts. For example, the ratio of VZV ORF 9 to ORF 66 transcripts in virus-infected HFL cells at 3 dpi determined in this study was 21:1 and compared favorably to previous findings for VZV-infected HFL cells (30:1) (22) and virus-infected malignant melanoma cells (22:1) (13). Both VZV ORF 9 and ORF 66 transcripts have similar decay constants, indicating that mRNA stability does not account for the difference in ORF 9 and ORF 66 abundance (40). Instead, we reasoned that the number of VZV DNA templates involved in transcribing these genes differed and used RNAP ChIP analysis to determine polymerase occupancy.
RNAP CTD serves as a large flexible docking site for the coordinated assembly of factors involved in pre-mRNA capping, splicing, and 3′ polyadenylation at different stages in transcription of nascent RNA molecules (41). In eukaryotes ranging from yeast to humans, the phosphorylation of serines 2 and 5 of the RNAP CTD heptad repeat coordinates distinct phases of the polymerase complex assembly. Accordingly, CTD undergoes a cycle of phosphorylation and dephosphorylation during transcription initiation, elongation, and termination (42). During transcription initiation, cyclin-dependent kinase 7 (CDK7) phosphorylates serine 5 residues at gene promoters (43–45) which enhance the association and activity of mRNA capping complexes (46, 47). RNAP is released from its promoter-paused position by pTEFb-dependent phosphorylation of RNAP at serine 2 (48). As RNAP transits the gene body, during transcriptional elongation, CDK9 further phosphorylates serine 2 (49, 50), while phosphatases progressively dephosphorylate S5P (41).
Consequently, RNAP phosphorylation on serine 5 generally decreases and is replaced by serine 2 phosphorylation as the polymerase transits the gene body (51). While the RNAP S5P-to-S2P switch typically present during eukaryotic transcription (52) was seen on the HFL cell GAPDH gene, RNAP S5P levels on VZV genes 9, 51, and 66 remained the same at the promoter, ORF center, and transcription terminus at both 1 and 3 dpi. Additionally, RNAP S2P levels did not differ within VZV genes 9, 51, and 66 at 1 dpi and showed no enrichment above the background at 3 dpi. RNAP S2P is inhibited during HSV-1 infection (26, 53), suggesting that herpesvirus transcription differs from the typical progression of RNAP C-terminal domain phosphorylation patterns seen in eukaryotic cells, a notion consistent with our finding regarding RNAP S2P. A possible explanation is that since RNA splicing is rare in VZV genes, the lack of RNAP S2P could reduce splicing and favor virus gene transcription; RNAP S2P is a signal for splicing factor accretion (54). In addition, the steady-state amounts of the VZV gene 9 and 66 transcripts are independent of the amount of RNAP S5P and RNAP S2P present at any location within these virus genes, suggesting that the rate of transcription may be a key component in virus transcript accumulation.
Overall, we have developed an accurate and sensitive PCR-based assay to quantify the efficiency of VZV DNA shearing. The assay showed significant and similar occupancy of RNAP S5P at three distinct regions within three virus genes whose steady-state mRNA abundance differs by ∼20-fold. Determining the factors involved in maintaining mRNA steady-state levels is critical to a full understanding of virus gene transcription. While RNAP occupancy is a predictor of mRNA abundance (55), our findings indicate that mRNA degradation, RNAP S5P, or the transition to RNAP S2P occupancy does not determine VZV transcript abundance during productive infection. Future studies will investigate RNAP processivity as a factor influencing VZV mRNA accumulation.
ACKNOWLEDGMENTS
We thank Wenhua Ren and Bifeng Gau in the Genomics and Microarray Core at the University of Colorado School of Medicine for technical assistance, Marina Hoffman for editorial assistance, and Cathy Allen for manuscript preparation.
Funding Statement
This work was supported in part by Public Health Service grants AG032958 (D.G. and R.J.C.), NS093716 (D.G.), and NS082228 (R.J.C.) from the National Institutes of Health. H.B. and N.L.B. were supported by training grant NS007321 to D.G. from the National Institutes of Health.
REFERENCES
- 1.Raney BJ, Cline MS, Rosenbloom RK, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, Suh BB, Hinrichs AS, Clawson H, Zweig AS, Kirkup V, Fujita PA, Rhead B, Smith KE, Pohl A, Kuhn RM, Karolchik D, Haussler D, Kent WJ. 2011. ENCODE whole-genome data in the UCSC genome browser (2011 update). Nucleic Acids Res 39:D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L, Huebner N, Mann M, Stunnenberg HG, Buchholz F. 2011. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS One 6:e19470. doi: 10.1371/journal.pone.0019470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim SJ, Boyle PJ, Chinen M, Dale RK, Lei EP. 2013. Genome-wide localization of exosome components to active promoters and chromatin insulators in Drosophila. Nucleic Acids Res 41:2963–2980. doi: 10.1093/nar/gkt037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD. 2008. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet 4:e1000187. doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee HS, Pugh BF. 2011. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komarnitsky P, Cho EJ, Buratowski S. 2000. Differential phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herold M, Bartkuhn M, Renkawitz R. 2012. CTCF: insights into insulator function during development. Development 139:1045–1057. doi: 10.1242/dev.065268. [DOI] [PubMed] [Google Scholar]
- 8.Tempera I, Lieberman PM. 2014. Epigenetic regulation of EBV persistence and oncogenesis. Semin Cancer Biol 26:22–29. doi: 10.1016/j.semcancer.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teves SS, Weber CM, Henikoff S. 2014. Transcribing through the nucleosome. Trends Biochem Sci 39:577–586. doi: 10.1016/j.tibs.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Gasper WC, Marinov GK, Pauli-Behn F, Scott MT, Newberry K, DeSalvo G, Ou S, Myers RM, Vielmetter J, Wold BJ. 2014. Fully automated high-throughput chromatin immunoprecipitation for ChIP-seq: identifying ChIP-quality p300 monoclonal antibodies. Sci Rep 4:5152. doi: 10.1038/srep05152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Browne JA, Harris A, Leir SH. 2014. An optimized protocol for isolating primary epithelial cell chromatin for ChIP. PLoS One 9:e100099. doi: 10.1371/journal.pone.0100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruyechan WT. 2010. Roles of cellular transcription factors in VZV replication. Curr Top Microbiol Immunol 342:43–65. doi: 10.1007/82_2010_42. [DOI] [PubMed] [Google Scholar]
- 13.Cohrs RJ, Hurley MP, Gilden DH. 2003. Array analysis of viral gene transcription during lytic infection of cells in tissue culture with varicella-zoster virus. J Virol 77:11718–11732. doi: 10.1128/JVI.77.21.11718-11732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy PG, Grinfeld E, Craigon M, Vierlinger K, Roy D, Forster T, Ghazal P. 2005. Transcriptomal analysis of varicella-zoster virus infection using long oligonucleotide-based microarrays. J Gen Virol 86:2673–2684. doi: 10.1099/vir.0.80946-0. [DOI] [PubMed] [Google Scholar]
- 15.Nagel MA, Gilden D, Shade T, Gao B, Cohrs RJ. 2009. Rapid and sensitive detection of 68 unique varicella zoster virus gene transcripts in five multiplex reverse transcription-polymerase chain reactions. J Virol Methods 157:62–68. doi: 10.1016/j.jviromet.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenac Roviš T, Bailer SM, Pothineni VR, Ouwendijk WJ, Simic H, Babic M, Miklic K, Malic S, Verweij MC, Baiker A, Gonzalez O, von Brunn A, Zimmer R, Fruh K, Verjans GM, Jonjic S, Haas J. 2013. Comprehensive analysis of varicella-zoster virus proteins using a new monoclonal antibody collection. J Virol 87:6943–6954. doi: 10.1128/JVI.00407-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandri-Goldin RM. 2007. Initiation of transcription and RNA synthesis, processing and transport in HSV and VZV infected cells, p 128–137. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 18.Cohrs RJ, Koelle DM, Schuette MC, Mehta S, Pierson D, Gilden DH, Hill JM. 2009. Asymptomatic alphaherpesvirus reactivation, p 133–166. In Gluckman TR. (ed), Herpesviridae: viral structure, life cycle and infections. Nova Biomedical, New York, NY. [Google Scholar]
- 19.Davison AJ, Scott JE. 1986. The complete DNA sequence of varicella-zoster virus. J Gen Virol 67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 20.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 21.Gary L, Gilden DH, Cohrs RJ. 2006. Epigenetic regulation of varicella-zoster virus open reading frames 62 and 63 in latently infected human trigeminal ganglia. J Virol 80:4921–4926. doi: 10.1128/JVI.80.10.4921-4926.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baird NL, Bowlin JL, Cohrs RJ, Gilden D, Jones KL. 2014. Comparison of varicella-zoster virus RNA sequences in human neurons and fibroblasts. J Virol 88:5877–5880. doi: 10.1128/JVI.00476-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilden DH, Shtram Y, Friedmann A, Wellish M, Devlin M, Cohen A, Fraser N, Becker Y. 1982. Extraction of cell-associated varicella-zoster virus DNA with Triton X-100-NaCl. J Virol Methods 4:263–275. doi: 10.1016/0166-0934(82)90073-8. [DOI] [PubMed] [Google Scholar]
- 24.Cohrs RJ, Gilden DH. 2007. Prevalence and abundance of latently transcribed varicella-zoster virus genes in human ganglia. J Virol 81:2950–2956. doi: 10.1128/JVI.02745-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 26.Guo L, Wu WJ, Liu LD, Wang LC, Zhang Y, Wu LQ, Guan Y, Li QH. 2012. Herpes simplex virus 1 ICP22 inhibits the transcription of viral gene promoters by binding to and blocking the recruitment of P-TEFb. PLoS One 7:e45749. doi: 10.1371/journal.pone.0045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Figliozzi RW, Chen F, Balish M, Ajavon A, Hsia SV. 2014. Thyroid hormone-dependent epigenetic suppression of herpes simplex virus-1 gene expression and viral replication in differentiated neuroendocrine cells. J Neurol Sci 346:164–173. doi: 10.1016/j.jns.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaborowska J, Baumli S, Laitem C, O'Reilly D, Thomas PH, O'Hare P, Murphy S. 2014. Herpes simplex virus 1 (HSV-1) ICP22 protein directly interacts with cyclin-dependent kinase (CDK) 9 to inhibit RNA polymerase II transcription elongation. PLoS One 9:e107654. doi: 10.1371/journal.pone.0107654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubat NJ, Tran RK, McAnany P, Bloom DC. 2004. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J Virol 78:1139–1149. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A 102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amelio AL, McAnany PK, Bloom DC. 2006. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J Virol 80:2358–2368. doi: 10.1128/JVI.80.5.2358-2368.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Q, Lin L, Smith S, Huang J, Berger SL, Zhou J. 2007. CTCF-dependent chromatin boundary element between the latency-associated transcript and ICP0 promoters in the herpes simplex virus type 1 genome. J Virol 81:5192–5201. doi: 10.1128/JVI.02447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ertel MK, Cammarata AL, Hron RJ, Neumann DM. 2012. CTCF occupation of the herpes simplex virus 1 genome is disrupted at early times postreactivation in a transcription-dependent manner. J Virol 86:12741–12759. doi: 10.1128/JVI.01655-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bloom DC, Giordani NV, Kwiatkowski DL. 2010. Epigenetic regulation of latent HSV-1 gene expression. Biochim Biophys Acta 1799:246–256. doi: 10.1016/j.bbagrm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy PG, Rovnak J, Badani H, Cohrs RJ. 2015. A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation. J Gen Virol 96(Pt 7):1581–1602. doi: 10.1099/vir.0.000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y, Fear J, Hu J, Haecker I, Zhou L, Renne R, Bloom D, McIntyre LM. 2014. Leveraging biological replicates to improve analysis in ChIP-seq experiments. Comput Struct Biotechnol J 9:e201401002. doi: 10.5936/csbj.201401002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsompana M, Buck MJ. 2014. Chromatin accessibility: a window into the genome. Epigenetics Chromatin 7:33. doi: 10.1186/1756-8935-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reichelt M, Brady J, Arvin AM. 2009. The replication cycle of varicella-zoster virus: analysis of the kinetics of viral protein expression, genome synthesis, and virion assembly at the single-cell level. J Virol 83:3904–3918. doi: 10.1128/JVI.02137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eshleman E, Shahzad A, Cohrs RJ. 2011. Varicella zoster virus latency. Future Virol 6:341–355. doi: 10.2217/fvl.10.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azarkh Y, Dolken L, Nagel M, Gilden D, Cohrs RJ. 2011. Synthesis and decay of varicella zoster virus transcripts. J Neurovirol 17:281–287. doi: 10.1007/s13365-011-0029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsin JP, Manley JL. 2012. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu H, Zawel L, Fisher L, Egly JM, Reinberg D. 1992. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature 358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 44.Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell 2:43–53. doi: 10.1016/S1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 45.He Y, Fang J, Taatjes DJ, Nogales E. 2013. Structural visualization of key steps in human transcription initiation. Nature 495:481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho EJ, Takagi T, Moore CR, Buratowski S. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev 11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho CK, Shuman S. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell 3:405–411. doi: 10.1016/S1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 48.Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, Varzavand K, Cooper JJ, Hu X, Gnatt A, Young RA, Price DH. 2012. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol Cell 45:38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marshall NF, Price DH. 1995. Purification of P-TEBb, a transcription factor required for the transition into productive elongation. J Biol Chem 270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 50.Marshall NF, Peng J, Xie Z, Price DH. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 51.Bowman EA, Kelly WG. 2014. RNA polymerase II transcription elongation and Pol II CTD Ser2 phosphorylation: a tail of two kinases. Nucleus 5:224–236. doi: 10.4161/nucl.29347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heidemann M, Hintermair C, Voss K, Eick D. 2013. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta 1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 53.Fraser KA, Rice SA. 2007. Herpes simplex virus immediate-early protein ICP22 triggers loss of serine 2-phosphorylated RNA polymerase II. J Virol 81:5091–5101. doi: 10.1128/JVI.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu B, Eick D, Bensaude O. 2013. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res 41:1591–1603. doi: 10.1093/nar/gks1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. 2013. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell 50:212–222. doi: 10.1016/j.molcel.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]