Abstract
The GP64 envelope fusion protein is a hallmark of group I alphabaculoviruses. However, the Diatraea saccharalis granulovirus genome sequence revealed the first betabaculovirus species harboring a gp64 homolog (disa118). In this work, we have shown that this homolog encodes a functional envelope fusion protein and could enable the infection and fusogenic abilities of a gp64-null prototype baculovirus. Therefore, GP64 may complement or may be in the process of replacing F protein activity in this virus lineage.
TEXT
Members of the Baculoviridae are a family of insect viruses with double-stranded DNA genomes. The family is currently divided into four genera, two of which, Alphabaculovirus and Betabaculovirus, contain members that are infective to the larval stages of moths and butterflies. During a complete infection cycle, viruses from both genera produce two virion phenotypes, (i) the occlusion-derived virus (ODV), which is surrounded by a crystalline protein matrix, the occlusion body (OB), and is responsible for interhost oral primary infection, and (ii) the budded virion (BV), responsible for intrahost systemic infection (1). GP64 is the major envelope fusion protein (EFP) found in the BVs of all group I alphabaculoviruses (G1-α) (1). It was probably acquired by the ancestor of G1-α, likely from an insect retrovirus-like element (2, 3), and is clearly related to the glycoprotein found in the genus Thogotovirus (from Orthomyxoviridae, a single-stranded RNA [ssRNA] negative-strand segmented virus family) (4). GP64 is a class III integral membrane glycoprotein (5) that plays essential roles in host cell receptor binding (6), low-pH-triggered viral membrane fusion (7), and systemic infection of the host insect (8). Other baculoviruses, including those from group II alphabaculovirus (G2-α) and betabaculovirus, share a GP64 analog called F protein as the major BV EFP (5).
The betabaculovirus Diatraea saccharalis granulovirus (DisaGV) was isolated from one of the most devastating insect pests of sugarcane and other cultures in Brazil. After complete genome sequencing, both a gp64 homolog, disa118, and an F protein-encoding gene, disa025, were found. The disa118 gene encoding the putative EFP clustered with genes from alphabaculovirus group I instead of with orthomyxovirus homologs, which confirms that gp64 was acquired first by alphabaculovirus and then transferred to either DisaGV or a related ancestor (D. M. P. Ardisson-Araújo, F. L. Melo, R. J. Clem, J. L. C. Wolff, and B. M. Ribeiro, unpublished data). The predicted amino acid sequence of the protein encoded by disa118 presented 505 amino acid (aa) residues, and both the signal peptide (SP; residues 1 to 21) and the transmembrane domain (TMD; residues 481 to 503) were present. The protein showed the highest pairwise identity with the Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) GP64 sequence (72.8%) and the lowest pairwise identity with AgMNPV protein (66.6%). In this work, we investigated whether the gp64 (disa118) homolog found in DisaGV encodes a functional EFP.
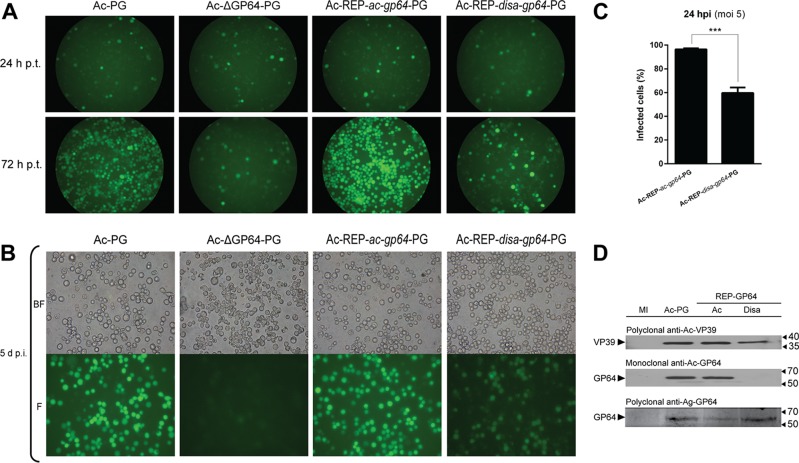
To examine the function of the protein encoded by the DisaGV gp64 homolog, we generated a gp64-null Autographa californica multiple nucleopolyhedrovirus (Ac-Δgp64-PG) bacmid (9) pseudotyped with the disa118 gene (called disa-gp64 here). Both disa-gp64 and the native gene (ac-gp64) used to repair the mutant bacmid were under the transcriptional control of the AcMNPV gp64 promoter region (10). The pseudotyped Ac-REP-disa-gp64-PG virus was able to infect and spread upon transfection into Spodoptera frugiperda cell line 9 (Sf9) (Fig. 1A). To confirm that infectious BVs were being produced after transfection, we transferred the supernatants from the transfection to healthy Sf9 cell cultures. Ac-REP-disa-gp64-PG was able to cause infection (Fig. 1B). However, the efficiency was lower than that seen with the control viruses in a controlled infection assay (triplicate infection at a multiplicity of infection [MOI] for 5 and 6 h with rocking) (Fig. 1C). In a previous study, a gp64-null virus expressing the EFP of vesicular stomatitis virus G was able to produce infection, replicate, and propagate in Sf9 cells despite the cell-to-cell propagation being delayed in comparison to the parental virus (11). Although Disa-GP64 showed a pairwise identity of about 73% with Ac-GP64, a commercial monoclonal antibody against Ac-GP64 (AcV1 from eBioscience) was unable to recognize Disa-GP64. This can be explained by the variability of the region to which the commercial monoclonal antibody binds (12). However, a rat polyclonal antibody raised against His tag-purified Anticarsia gemmatalis multiple nucleopolyhedrovirus GP64 (Ag-GP64) (lacking the signal peptide and the transmembrane domain and expressed in bacteria) recognized both Disa-GP64 and Ac-GP64 (Fig. 1D). We also carried out a fusogenic activity assay to verify whether Disa-GP64 could mediate low-pH-triggered membrane fusion. We found that cells infected with both Ac-REP-disa-gp64-PG and Ac-REP-ac-gp64-PG mediated membrane fusion and syncytium formation when exposed to low pH (Fig. 2, left panels and right panels, respectively). The efficiency of syncytium formation was apparently much lower than that of the Ac-REP-ac-gp64-GP positive control. Moreover, no syncytium formation was observed when the cells were mock infected and treated with low pH (data not shown).
FIG 1.
Disa-GP64 is a functional envelope fusion protein. (A) Transfection assay of Ac-PG (used as a positive control); this is an AcMNPV bacmid containing both the polyhedrin [P] gene under the control of its natural promoter and the green florescence protein [G] gene under the control of the AcMNPV immediate early-1 gene promoter, Ac-ΔGP64-PG (negative control), Ac-REP-ac-gp64-PG (repaired virus), and Ac-REP-disa-gp64-PG (pseudotyped virus). A 1-μg volume of DNA from each virus was transfected into Sf9 cells. The cells were photographed at 24 and 72 h posttransfection (h p.t.). (B) Infection assay of transfection supernatants, Ac-REP-disa-gp64-PG transfection supernatant is infective to Sf9 cells. At 5 days posttransfection, clarified supernatants were used to infect Sf9 cells. The cells were photographed at 5 days postinfection (d p.i.). (C) The infection efficiency of the pseudotyped Ac-REP-disa-gp64-PG was reduced compared to that of the repaired Ac-REP-ac-gp64-PG. Cells were infected at an MOI of 5 (determined by endpoint dilution) and photographed at 24 h postinfection (hpi). (D) Disa-GP64 was detected only with a polyclonal antibody raised against GP64 of an alphabaculovirus (anti-Ag-GP64). Anti-Ac-VP39 antibody was used as a baculovirus infection control. Cells were mock infected (MI) or infected with (i) Ac-PG, (ii) Ac-REP-ac-gp64-PG (Ac), or (iii) Ac-REP-disa-gp64-PG (Disa) at an MOI of 5 for 72 hpi. Cells were harvested, and the total proteins were extracted, resolved on SDS-12% PAGE gels, and analyzed by immunoblotting with polyclonal anti-Ac-VP39, monoclonal anti-Ac-GP64, or polyclonal anti-Ag-GP64 antibody.
FIG 2.
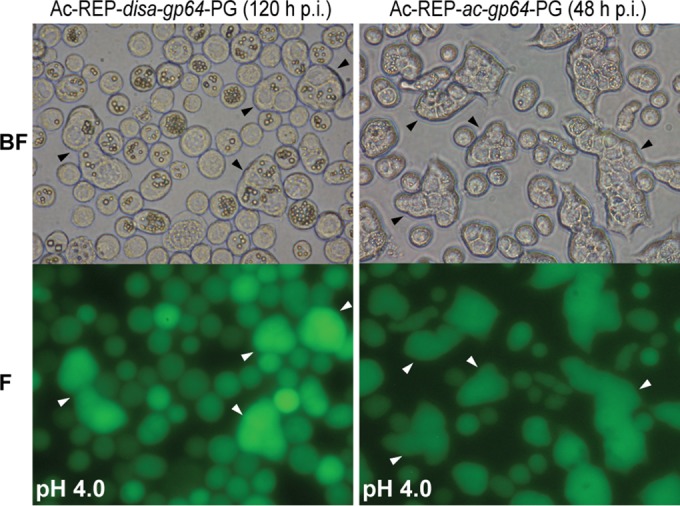
Syncytium formation mediated by recombinant baculovirus infections. Sf9 cells were infected with either AcRep-Disa-gp64-PG or AcRep-Ac-gp64-PG at an MOI of 1. The infected cells were then incubated with low-pH TC100 media (pH 4.0) for 10 min at 48 or 120 hpi, as indicated. After 10 min, the media were replaced by media at pH 6.0. Syncytium formation was observed and photographed at 4 h after treatment. Multinucleated cells are indicated by arrowheads. The absence of OBs is due to the different times postinfection used for the repaired virus. BF, bright field; F, fluorescence.
To understand this reduction in virus infectivity, spread, and syncytium formation efficiency, we mapped functionally important amino acid residues in Disa-GP64 based on previous reports and protein alignment (Fig. 3). Two main regions were analyzed which included the signal peptide (SP) and the ectodomain (ED; the region between the SP and the transmembrane domain [TMD]). First, when the entire proteins of AcMNPV, AgMNPV, and DisaGV were aligned by the MAFFT method (13), the mean pairwise identity was 71%, suggesting that these proteins are well conserved. Second, by mapping the protein on the basis of previous reports, we found that GP64 SPs across baculovirus species are variable per se, with a pairwise identity of 39.8% (Fig. 3A), which is not an exclusive feature in GP64 alone. Other baculoviral envelope proteins and secreted enzymes present highly variable SP sequences as well (e.g., per os infectivity factors and ecdysteroid UDP glucosyltransferase [EGT]) (data not shown). These amino acid substitutions could be related to host adaptation and might be responsible for the efficiency reduction displayed by the pseudotyped virus, since DisaGV and AcMNPV were found infecting caterpillars from different lepidopteran families, i.e., Crambidae and Noctuidae, respectively. On the other hand, the ED has been shown to present important regions for the functions of GP64 (14–16). Using the same alignment method cited above, we found that most of the previously mapped ED regions and sites, such as intramolecular disulfide bonds, which are critical in membrane fusion, are highly conserved in Disa-GP64 (17) (not shown). Nevertheless, three of four glycosylation sites identified in Ac-GP64 ED (N198, N355, N385, and N426) (18) and conserved in all other G1-α GP64 orthologs are maintained in Disa-GP64; only N355 underwent a substitution (Fig. 3B). We believe that there could be a connection between the putative Disa-GP64 glycosylation site mutation and the reduced infectivity and fusogenicity observed. However, next to N355, there is another asparagine residue that could work as a glycosylation site as well. Therefore, this change may have no particular functional significance; further investigation is needed for confirmation. In previous work, cell surface expression, assembly into infectious BV, and fusogenic activity did not require N-linked oligosaccharide processing; however, the removal of one or more N-glycosylation sites in Ac-GP64 impaired binding of budded virus to the cell, indicating that this modification is necessary for optimal GP64 function (18, 19). Interestingly, both the production of infectious BV and the fusion activity were reduced when glycosylation of GP64 was inhibited in Bombyx mori nucleopolyhedrovirus (20).
FIG 3.
MAFFT alignment of the predicted amino acid sequences of Disa-GP64 and related baculoviruses. (A) The entire protein was aligned with two orthologs from AcMNPV and AgMNPV. All the predicted signal peptides (SP), glycosylation sites (N198, N355, N385, and N426), and transmembrane domains (TMD) are shown in red boxes. SPs and TMDs were predicted by the SignalP 4.1 and TMHMM v. 2.0 servers, respectively. The region recognized by the commercial AcV1 monoclonal antibody is shown in yellow. (B) Signal peptide region alignment. The last residue shown (glutamate) is the predicted beginning of the soluble portion of the protein. (C) Alignment of part of the soluble portion, revealing the substitution in DisaGV from N355 to I355 compared to the other alphabaculovirus species (star). This residue has been experimentally shown to be a N-linked glycosylation site in AcMNPV. Strictly conserved amino acid residues are shown in black boxes and partially conserved residues in gray boxes. PlxyMNPV, Plutella xylostella multiple nucleopolyhedrovirus; BomaNPV-S2, Bombyx mandarina nucleopolyhedrovirus-S2; AnfaNPV, Anagrapha falcifera nucleopolyhedrovirus; RoMNPV, Rachiplusia ou nuclear polyhedrosis virus; BmNPV, Bombyx mori nuclear polyhedrosis virus; ThorNPV, Thysanoplusia orichalcea nucleopolyhedrovirus; MaviMNPV, Maruca vitrata (F.) multinucleopolyhedrovirus; DekiNPV, Dendrolimus kikuchii Matsumura nucleopolyhedrovirus; AgMNPV, Anticarsia gemmatalis multiple nucleopolyhedrovirus; AnpeNPV-L2, Antheraea pernyi nucleopolyhedrovirus-L2; PhcyNPV, Philosamia cynthia nucleopolyhedrovirus; CfDEFMNPV, Choristoneura fumiferana defective MNPV; CfMNPV, Choristoneura fumiferana multinucleocapsid nuclear polyhedrovirus; ChocNPV, Choristoneura occidentalis; ChmuNPV, Choristoneura murinana NPV; Choristoneura rosaceana nucleopolyhedrovirus, ChroNPV; OpNPV, Orgyia pseudotsugata nuclear polyhedrosis virus; HycuNPV, Hyphantria cunea nucleopolyhedrovirus; EppoNPV, Epiphyas postvittana nucleopolyhedrovirus.
The main question here is this: why has gp64 been fixed into DisaGV? Fixation of gp64 is responsible for improvement of both fusion and binding activities (21–23) and possibly led to replacement of the F protein in G1-α (3). In fact, G1-α viruses also contain a remnant F protein homolog in their genomes that is unable to compensate for gp64 loss (8, 11) and that plays a role in virus pathogenicity (24). Previous experimental analysis showed that the incorporation of GP64 into a G2-α enhanced virus infectivity in vivo and in vitro (22). Since D. saccharalis is an insect borer during the larval stage and presents a very short time of virus exposure between the egg hatching and the insect penetration into the host plants, which include sugar cane, rice, and other monocots, it is reasonable to propose that a novel gene acquisition event occurred that allowed the virus to improve its spread within the host and more effectively establish infection.
In summary, GP64 of DisaGV is a functional EFP that is able to pseudotype a gp64-null AcMNPV, although with lower efficiency in spreading the infection and in fusogenic activity. The lack of one conserved glycosylation site and the possible adaptation to a different lepidoptera-family cell machinery could explain this reduction. We are constructing different mutants of disa-gp64 to test those hypotheses. Importantly, in research for a submitted work describing the DisaGV genome, we found several early transcriptional motifs upstream of the gp64 start codon; however, it is not clear whether DisaGV expresses the gp64 homolog and uses it as a functional EPF. We can only speculate that GP64 could complement or may even be in the process of replacing F protein activity in this betabaculovirus lineage.
Nucleotide sequence accession number.
The nucleotide sequence of DisaGV was submitted to GenBank under accession number KP296186.
ACKNOWLEDGMENTS
We thank the Brazilian agencies Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Apoio à Pesquisa do Distrito Federal (FAPDF), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for financial support.
We thank Ana Lorena Passarelli for providing the gp64-null AcMNPV bacmid from the laboratory of Gary Blissard.
REFERENCES
- 1.Rohrmann GF. 2013. Baculovirus molecular biology, 3rd [Internet] ed National Center for Biotechnology Information, Bethesda, MD: http://www.ncbi.nlm.nih.gov/books/NBK49500/. [PubMed] [Google Scholar]
- 2.Rohrmann GF, Karplus PA. 2001. Relatedness of baculovirus and gypsy retrotransposon envelope proteins. BMC Evol Biol 1:1. doi: 10.1186/1471-2148-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang M, Wang J, Yin F, Tan Y, Deng F, Chen X, Jehle JA, Vlak JM, Hu Z, Wang H. 2014. Unraveling the entry mechanism of baculoviruses and its evolutionary implications. J Virol 88:2301–2311. doi: 10.1128/JVI.03204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morse MA, Marriott AC, Nuttall PA. 1992. The glycoprotein of Thogoto virus (a tick-borne orthomyxo-like virus) is related to the baculovirus glycoprotein GP64. Virology 186:640–646. doi: 10.1016/0042-6822(92)90030-S. [DOI] [PubMed] [Google Scholar]
- 5.Garry CE, Garry RF. 2008. Proteomics computational analyses suggest that baculovirus GP64 superfamily proteins are class III penetrenes. Virol J 5:28. doi: 10.1186/1743-422X-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hefferon KL, Oomens AG, Monsma SA, Finnerty CM, Blissard GW. 1999. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology 258:455–468. doi: 10.1006/viro.1999.9758. [DOI] [PubMed] [Google Scholar]
- 7.Kingsley DH, Behbahani A, Rashtian A, Blissard GW, Zimmerberg J. 1999. A discrete stage of baculovirus GP64-mediated membrane fusion. Mol Biol Cell 10:4191–4200. doi: 10.1091/mbc.10.12.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monsma SA, Oomens AG, Blissard GW. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J Virol 70:4607–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lung O, Westenberg M, Vlak JM, Zuidema D, Blissard GW. 2002. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J Virol 76:5729–5736. doi: 10.1128/JVI.76.11.5729-5736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen YR, Zhong S, Fei Z, Hashimoto Y, Xiang JZ, Zhang S, Blissard GW. 2013. The transcriptome of the baculovirus Autographa californica multiple nucleopolyhedrovirus in Trichoplusia ni cells. J Virol 87:6391–6405. doi: 10.1128/JVI.00194-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangor JT, Monsma SA, Johnson MC, Blissard GW. 2001. A GP64-null baculovirus pseudotyped with vesicular stomatitis virus G protein. J Virol 75:2544–2556. doi: 10.1128/JVI.75.6.2544-2556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Blissard GW. 2006. Mapping the conformational epitope of a neutralizing antibody (AcV1) directed against the AcMNPV GP64 protein. Virology 352:427–437. doi: 10.1016/j.virol.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katou Y, Yamada H, Ikeda M, Kobayashi M. 2010. A single amino acid substitution modulates low-pH-triggered membrane fusion of GP64 protein in Autographa californica and Bombyx mori nucleopolyhedroviruses. Virology 404:204–214. doi: 10.1016/j.virol.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Blissard GW. 2009. The pre-transmembrane domain of the Autographa californica multicapsid nucleopolyhedrovirus GP64 protein is critical for membrane fusion and virus infectivity. J Virol 83:10993–11004. doi: 10.1128/JVI.01085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Blissard GW. 2008. Identification of a GP64 subdomain involved in receptor binding by budded virions of the baculovirus Autographica californica multicapsid nucleopolyhedrovirus. J Virol 82:4449–4460. doi: 10.1128/JVI.02490-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Blissard GW. 2010. Baculovirus GP64 disulfide bonds: the intermolecular disulfide bond of Autographa californica multicapsid nucleopolyhedrovirus GP64 is not essential for membrane fusion and virion budding. J Virol 84:8584–8595. doi: 10.1128/JVI.00264-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis DL, Wills L, Burow G, Bohlmeyer DA. 1998. Mutational analysis of the N-linked glycans on Autographa californica nucleopolyhedrovirus gp64. J Virol 72:9459–9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvis DL, Garcia A Jr. 1994. Biosynthesis and processing of the Autographa californica nuclear polyhedrosis virus gp64 protein. Virology 205:300–313. doi: 10.1006/viro.1994.1646. [DOI] [PubMed] [Google Scholar]
- 20.Rahman MM, Gopinathan KP. 2003. Characterization of the gene encoding the envelope fusion glycoprotein GP64 from Bombyx mori nucleopolyhedrovirus. Virus Res 94:45–57. doi: 10.1016/S0168-1702(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 21.Liang C, Song J, Chen X. 2005. The GP64 protein of Autographa californica multiple nucleopolyhedrovirus rescues Helicoverpa armigera nucleopolyhedrovirus transduction in mammalian cells. J Gen Virol 86:1629–1635. doi: 10.1099/vir.0.80857-0. [DOI] [PubMed] [Google Scholar]
- 22.Shen S, Gan Y, Wang M, Hu Z, Wang H, Deng F. 2012. Incorporation of GP64 into Helicoverpa armigera nucleopolyhedrovirus enhances virus infectivity in vivo and in vitro. J Gen Virol 93:2705–2711. doi: 10.1099/vir.0.046458-0. [DOI] [PubMed] [Google Scholar]
- 23.Yu IL, Lin YC, Robinson JH, Lung O. 2009. Transduction of vertebrate cells with Spodoptera exigua multiple nucleopolyhedrovirus F protein-pseudotyped gp64-null Autographa californica multiple nucleopolyhedrovirus. J Gen Virol 90:2282–2287. doi: 10.1099/vir.0.012138-0. [DOI] [PubMed] [Google Scholar]
- 24.Lung OY, Cruz-Alvarez M, Blissard GW. 2003. Ac23, an envelope fusion protein homolog in the baculovirus Autographa californica multicapsid nucleopolyhedrovirus, is a viral pathogenicity factor. J Virol 77:328–339. doi: 10.1128/JVI.77.1.328-339.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]