ABSTRACT
The H1N1 Eurasian avian-like swine (EAsw) influenza viruses originated from an avian H1N1 virus. To characterize potential changes in the membrane fusion activity of the hemagglutinin (HA) during avian-to-swine adaptation of the virus, we studied EAsw viruses isolated in the first years of their circulation in pigs and closely related contemporary H1N1 viruses of wild aquatic birds. Compared to the avian viruses, the swine viruses were less sensitive to neutralization by lysosomotropic agent NH4Cl in MDCK cells, had a higher pH optimum of hemolytic activity, and were less stable at acidic pH. Eight amino acid substitutions in the HA were found to separate the EAsw viruses from their putative avian precursor; four substitutions—T492S, N722D, R752K, and S1132F—were located in the structural regions of the HA2 subunit known to play a role in acid-induced conformational transition of the HA. We also studied low-pH-induced syncytium formation by cell-expressed HA proteins and found that the HAs of the 1918, 1957, 1968, and 2009 pandemic viruses required a lower pH for fusion induction than did the HA of a representative EAsw virus. Our data show that transmission of an avian H1N1 virus to pigs was accompanied by changes in conformational stability and fusion promotion activity of the HA. We conclude that distinctive host-determined fusion characteristics of the HA may represent a barrier for avian-to-swine and swine-to-human transmission of influenza viruses.
IMPORTANCE Continuing cases of human infections with zoonotic influenza viruses highlight the necessity to understand which viral properties contribute to interspecies transmission. Efficient binding of the HA to cellular receptors in a new host species is known to be essential for the transmission. Less is known about required adaptive changes in the membrane fusion activity of the HA. Here we show that adaptation of an avian influenza virus to pigs in Europe in 1980s was accompanied by mutations in the HA, which decreased its conformational stability and increased pH optimum of membrane fusion activity. This finding represents the first formal evidence of alteration of the HA fusion activity/stability during interspecies transmission of influenza viruses under natural settings.
INTRODUCTION
Wild aquatic birds represent the major natural reservoir of influenza A viruses (1, 2). These viruses occasionally infect other avian and mammalian species, such as aquatic and terrestrial poultry, sea mammals, horses, and pigs. On rare occasions they adapt to and establish stable lineages in new species (2, 3). Documented transmissions of avian and swine influenza viruses to men are typically restricted to individual, often severe, cases of infection (for recent reviews, see references 4 and 5). However, four times within the last hundred years either whole animal viruses or their reassortants with contemporary human viruses acquired the ability to transmit between humans and initiated global pandemics (6, 7).
Despite the high impact of interspecies transmission of influenza viruses on animal and human health, host range restriction mechanisms and adaptive changes required for the virus to overcome the species barrier are not fully understood. Receptor-binding specificity of the viral hemagglutinin (HA) is the best-studied restriction factor. Receptor-dependent restriction is determined by distinctions in spectra of sialic acid (Sia) receptors in the target tissues of different species such as predominant expression of Sia2-3Gal-containing glycans in birds and horses and Sia2-6Gal-containing glycans in pigs and humans. It is believed that mutation of the HA that ensures alteration of the receptor specificity is critical for the adaptation of avian influenza viruses to humans and pigs (reviewed in references 8, 9, 10, and 11).
In addition to mediating attachment of the virus to cellular receptors, HA promotes membrane fusion, which is essential for the entry of viral RNPs into the cytoplasm. After internalization by receptor-mediated endocytosis, exposure of the virus to gradually decreasing pH in the endosomes triggers a series of structural rearrangements of the HA from its native “spring-loaded” high-energy conformation into a stable low-energy conformation. These rearrangements bring viral and endosomal membranes together and eventually lead to their fusion (12–15). Because both low pH and elevated temperature can trigger the same conformational transition of the HA and because in the absence of target membrane this transition causes irreversible virus inactivation, a higher pH optimum of HA-mediated membrane fusion correlates, in general, with a lower virus stability at reduced pH and high temperatures (reviewed in references 16, 17, and 18).
There is growing evidence that membrane fusion properties of the HA represent a host-range restriction factor (17–19). Thus, adaptation of human influenza viruses for efficient replication in mice typically results in the increase of viral pH optimum of fusion from pH 5.2 to 5.4 to pH 5.6 to 5.8. Highly pathogenic H5N1 poultry viruses fuse at a relatively high pH (>5.6); mutations that decrease pH of fusion were shown to facilitate replication and pathogenicity of H5N1 viruses in mice and ferrets (20–22). A clear-cut and striking effect was observed in two independent studies on adaptation of recombinant H5N1 viruses to ferrets (23, 24). In both studies, mutation in the HA that decreased pH of fusion and increased pH stability was shown to be indispensable for the virus transmission in ferrets by airborne droplets (24, 25).
Limited available data on fusion pH and stability of influenza viruses circulating in natural host species seem to indicate that human viruses fuse at a lower pH than avian and swine viruses (21, 26–30). However, different viral strains and distinctive assays were used by different authors, and pronounced subtype- and stain-dependent variation in the viral fusion activity was observed in these studies hampering solid conclusions. Thus, further systematic studies are needed to characterize host-specific differences in the membrane fusion properties of influenza viruses and potential alteration of these properties during interspecies transmission. To this end, we compared membrane fusion activity and pH stability of two panels of closely related avian and swine influenza viruses of the same subtype separated by a recent host switch event. We found that H1N1 avian-like swine viruses isolated in Europe from 1979 to 1981 have a higher pH optimum membrane fusion and cell entry and lower stability than their avian counterparts. We also identified mutations in the HA separating avian-like swine viruses from their putative avian precursor and inferred potential role of these mutations in the fusion activity change during avian-to-swine transmission.
MATERIALS AND METHODS
Cells.
MDCK and HeLa cells were propagated in Dulbecco′s modified Eagle medium (DMEM; Gibco, catalog no. 21969-035) supplemented with 10% fetal calf serum (Gibco), 100 IU ml−1 penicillin plus 100 μg ml−1 streptomycin (pen-strep), and 2 mM glutamine. Infection medium (IM) composed of DMEM containing 2 mM glutamine, pen-strep, and 0.1% bovine serum albumin (PAA Laboratories GmbH) was used for viral infections. Cells were grown and infections were performed at 37°C with 5% CO2.
Viruses and HA sequencing.
Viruses and their sources are listed in Table 1. All viruses were grown in MDCK cells in the presence of 1 μg ml−1 TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone) trypsin (Sigma), clarified by low-speed centrifugation, and stored in aliquots at −80°C. Viral stocks were titrated using single-cycle focus assay (31), and titers were expressed as focus-forming units (FFU) per ml. Total RNA was isolated from viral stocks using the QIAamp viral RNA minikit (Qiagen). The HA gene segment was amplified using a OneStep RT-PCR kit (Qiagen) with universal HA-specific primers (32) and Sanger sequenced.
TABLE 1.
Membrane fusion activity and stability of influenza virusesa
| Virusb | Infection inhibition by NH4Cl (IC50 [mM]) |
Hemolytic activity (pH50-hem) |
Inactivation at acidic pH (pH50-inact) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | 95% CI | P | Mean | 95% CI | P | Mean | 95% CI | P | |
| Avian viruses | |||||||||
| A/duck/Alberta/35/19761 | 0.26 | 0.011 | *** | 4.97 | 0.05 | 5.27 | 0.03 | ||
| A/duck/Bavaria/1/19772 | 0.50 | 0.007 | 5.20 | 0.06 | *** | 5.31 | 0.11 | ||
| A/duck/Bavaria/2/19772 | 0.31 | 0.014 | * | 5.03 | 0.05 | 5.43 | 0.03 | ** | |
| A/duck/Schleswig/21/19792 | 0.43 | 0.07 | 4.96 | 0.04 | 5.19 | 0.07 | |||
| A/coot/Schleswig/4/19792 | 0.55 | 0.07 | * | 5.05 | 0.03 | ** | |||
| A/coot/Schleswig/2/19802 | 0.50 | 0.07 | 5.10 | 0.021 | *** | ||||
| Eurasian avian-like swine viruses | |||||||||
| A/swine/Arnsberg/6554/19792 | 0.99 | 0.16 | *** | 5.07 | 0.07 | * | 5.75 | 0.017 | *** |
| A/swine/France/OLI/19802 | 1.04 | 0.21 | *** | 5.29 | 0.10 | *** | 5.64 | 0.03 | *** |
| A/swine/Marseille/2260/19802 | 1.13 | 0.13 | *** | 5.36 | 0.05 | *** | 5.91 | 0.11 | *** |
| A/swine/Italy/v147/19812 | 1.1 | 0.6 | * | 5.07 | 0.07 | * | |||
| A/swine/Germany/2/19812 | 0.95 | 0.16 | *** | 5.21 | 0.09 | ** | 5.75 | 0.20 | *** |
| A/swine/Germany/S27/19812 | 1.14 | 0.15 | *** | 5.13 | 0.07 | ** | 5.47 | 0.03 | ** |
| A/swine/Italy/215990-3/20053 c | 1.43 | 0.05 | *** | 5.39 | 0.024 | *** | |||
| A/swine/England/453/20064 | 0.83 | 0.15 | ** | ||||||
| A/swine/Italy/50175/20073 c | 1.80 | 0.10 | *** | 5.19 | 0.09 | ** | |||
| Swine viruses with classical swine HA isolated from humans | |||||||||
| A/Thailand/271/20055 c | 0.67 | 0.14 | * | 5.17 | 0.15 | * | |||
| A/Illinois/09/20076 c | 0.75 | 0.20 | * | ||||||
| A/South Dakota/03/20086 | 0.65 | 0.10 | * | ||||||
| A/Iowa/02/20096 c | 2.55 | 0.10 | *** | ||||||
| Swine viruses with human-like HA | |||||||||
| A/swine/Italy/30019-2/2007 (H1N2)3 c | 2.13 | 0.05 | *** | ||||||
| A/swine/Italy/50127/2007 (H3N2)3 c | 1.45 | 0.10 | *** | ||||||
| Human pandemic viruses | |||||||||
| A/Hong Kong/1/1968 (H3N2)7 | 0.61 | 0.05 | * | 5.08 | 0.07 | * | |||
| A/Hamburg/05/20091 c | 0.67 | 0.14 | * | 5.06 | 0.07 | * | |||
Viral phenotypes were studied using three assays described in Materials and Methods and in Fig. 2. The data show mean values and their 95% confidence intervals (CI). P values for the differences with respect to A/duck/Schleswig/21/1979 were calculated using an unpaired two-tailed Student t test (*, P < 0.05; **, P < 0.005; ***, P < 0.0005).
All viruses are H1N1, if not indicated otherwise. Superscript numbers following the strain name indicate the source of the virus as follows: 1, repository of the Institute of Virology, Philipps University, Marburg, Germany; 2, Christoph Scholtissek at the Institute of Medical Virology, Justus Liebig University, Giessen, Germany; 3, repository of the Istituto Zooprofilattico Sperimentale della Lombardia ed Emilia Romagna, Parma, Italy; 4, Sharon Brookes and Ian Brown, Animal and Plant Health Agency, Addlestone, Surrey, United Kingdom; 5, Ian Barr, WHO Collaborating Centre for Influenza, Melbourne, Victoria, Australia; 6, Alexander Klimov and Amanda Balish, Centers for Disease Control and Prevention, Atlanta, GA; 7, Earl Brown, University of Ottawa, Ottawa, Canada.
Viruses isolated and passaged solely in cell culture.
Inhibition of viral infection by NH4Cl.
Confluent monolayers of MDCK cells in 96-well plates (Greiner) were infected with 200 FFU of the viruses in 0.1 ml of IM containing variable concentrations of ammonium chloride. No trypsin was added to the medium to limit the infection to one replication cycle. After the cultures were incubated for 16 h at 37°C, the cells were fixed, and virus-infected cells were detected by immunostaining for viral nucleoprotein (NP) as described previously (31). Numbers of infected cells per culture were counted under the microscope and expressed in percentages with respect to the control cultures that were infected in the absence of NH4Cl. The dose-response curves were plotted, and concentrations of NH4Cl that inhibited infection by 50% (IC50) were determined for each replicate curve by linear interpolation as illustrated in Fig. 2a. Experiments were performed in triplicates on the same day and repeated at least twice on different days.
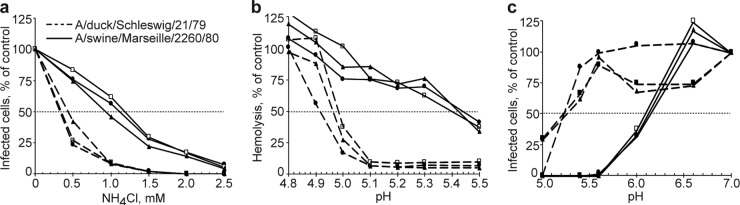
FIG 2.
Examples of the experiments used to characterize membrane fusion activity and stability of influenza viruses. (a) Inhibition of viral infection by NH4Cl. (b) pH dependence of virus-mediated hemolysis. (c) Virus inactivation at low pH. The experiments were performed as described in Materials and Methods. Each panel shows results of three replicate experiments performed either on the same day (a and c) or on different days (b) using the viruses A/swine/Marseille/2260/1980 (H1N1) (solid lines) and A/duck/Schleswig/21/1979 (H1N1) (dashed lines). For each replicate, the concentrations of NH4Cl and values of pH treatments that caused 50% effect were determined from the curve by interpolation. All individual values were averaged (Table 1).
Virus-mediated hemolysis.
Viruses were diluted with PBS to HA titer 64. Mixtures of 0.45 ml of viral suspension with 15 μl of 1% suspension of chicken red blood cells were incubated for 1 h on ice for virus adsorption. To account for nonspecific hemolysis at low pH (background control), phosphate-buffered saline (PBS) was used instead of the virus. The mixtures were vortex mixed to disperse erythrocytes, and 50-μl aliquots were dispensed in the wells of a 96-well plate (Greiner). Aliquots of either PBS (pH 7) or 100 mM sodium acetate buffers with pH from 4.8 to 5.5 were added to experimental and control wells at 75 μl per well. After incubation for 30 min at 37°C, the mixtures were neutralized by addition of 25 μl of 1 M Tris-HCl buffer (pH 7.3). As a control for 100% hemolysis, red blood cells were lysed by the addition of Triton X-100 to a 0.01% concentration. Nonlysed erythrocytes and cell debris were removed by centrifugation of the plate for 10 min at 1,500 rpm. Supernatants were transferred to an empty 96-well plate at 50 μl per well. The level of hemolysis was quantified by measuring the peroxidase activity of released hemaproteins (33) using 3,3′,5,5′-tetramethylbenzidine peroxidase substrate. The reaction with substrate was terminated by adding 5% sulfuric acid. Absorbance values at 450 nm were measured by using a microplate reader (Epoch; Biotek), corrected by subtracting the absorbencies of corresponding background control wells, and expressed as a percentage of absorbency with respect to the positive control. Absorbency-versus-pH curves were plotted, and pH values that corresponded to 50% hemolysis (pH50-hem) were determined by linear interpolation as illustrated in Fig. 2b. The experiments were performed at least twice on different days.
Virus inactivation at low pH.
Aliquots of virus stocks were diluted to concentration 200,000 FFU per ml in the buffers containing 100 mM MES, 150 mM NaCl, 0.9 mM CaCl2, and 0.5 mM MgCl2. The pH of the buffers varied from 5.0 to 7.0. After incubation for 15 min at 37°C, the mixtures were neutralized by adding a 100-fold excess of IM. Portions (100 μl) of the mixtures were inoculated into confluent monolayers of MDCK cells in 96-well plates. The cultures were incubated for 16 h, fixed, and immunostained for viral NP as described previously (31). The numbers of infected cells per culture were counted and expressed in percentages with respect to the cultures infected with the viruses that were treated with the MES buffer at pH 7. Infection-versus-pH curves were plotted, and pH values that corresponded to virus inactivation by 50% (pH50-inact) were determined by linear interpolation, as illustrated in Fig. 2c. Experiments were performed in triplicates and repeated at least twice on different days.
HA plasmids.
pHW2000 plasmid was kindly provided by Erich Hoffmann and Robert Webster (St. Jude Children's Research Hospital, Memphis, TN). pHW2000 plasmids containing HA genes of A/Hong Kong/1/1968 (H3N2) and A/Hamburg/5/2009 (H1N1) were described previously (31, 34). pHW2000 plasmid containing HA gene of A/swine/Marseille/2260/1980 (H1N1) was prepared as described previously (32). The full-length HA gene of A/Brevig Mission/1/1918 (H1N1) (GenBank accession no. AF250356) was synthesized commercially (Genscript Corporation, Piscataway, NJ). pHW2000 plasmid containing the HA gene of A/Singapore/1/1957 (H2N2) was kindly provided by Volker Czudai-Matwich (Institute of Virology, Philipps University, Marburg, Germany). For cellular expression and fusion assays, all HAs were subcloned into pCAGGS plasmid. The identity of all plasmids was confirmed by sequencing.
Syncytium formation assay.
The assay was performed as described previously (24) with some modifications. In brief, monolayers of 80% confluent HeLa cells grown in 12-well plates were transfected with 1 μg of pCAGGS-HA plasmids using Lipofectamine 2000 (Invitrogen). At 16 h posttransfection, TPCK trypsin was added to the culture medium (1 μg ml−1) for proteolytic activation of the HA, and the cells were incubated for 15 min at 37°C. The cells were exposed either to low-pH buffers (145 mM NaCl, 20 mM sodium acetate; pH 5.1 to 6.0) or to PBS (pH 7) for 5 min at 37°C. The buffers were replaced by IM; the cells were incubated for 3 h at 37°C, fixed with methanol, and stained with Giemsa stain (1:10 in water). Three randomly chosen fields in each well were photographed under the microscope equipped with digital camera at ×300 magnification. Cell nuclei were counted in each photograph, and syncytium formation was quantified as a percentage of nuclei in polykaryons with respect to the total number of nuclei in the same field. The experiments were performed at least twice on different days, and the results were averaged.
Analysis of HA sequences.
We compared HA sequences of EAsw viruses isolated between 1979 and 1981 in Europe with HA sequences of H1 avian viruses isolated around the world. All available full-length nonidentical nucleotide HA sequences of these viruses were downloaded from GenBank through the NCBI Influenza Virus Resource (35). Sequences of avian viruses with classical-swine-like- and human-like HA (15 sequences) were identified by phylogenetic analysis and removed from the data set. Six avian H1 HAs and six EAsw HAs were determined in this study. All sequences were combined, aligned and analyzed using BioEdit 7.1.11 (36). A final set included 455 avian and 22 EAsw sequences. The phylogenetic trees were generated for nucleotide sequences using MEGA6 (37) with the minimum-evolution method. The ancestral amino acid sequences were reconstructed using MEGA6 with the maximum-likelihood method under a Dayhoff matrix-based model.
Molecular modeling.
Location of amino acid substitutions on the X-structure of the HA molecule was analyzed with Pymol, version 1.7.4.1 (Schrödinger, LLC) using atomic coordinates of the HA of A/duck/Alberta/35/1976 (H1N1) (2WRH; protein Data Bank) (38).
Nucleotide sequence accession numbers.
The nucleotide sequences of the HA genes have been deposited in the GenBank database under accession numbers KT715445 to KT715456.
RESULTS
Selection of viruses for the study.
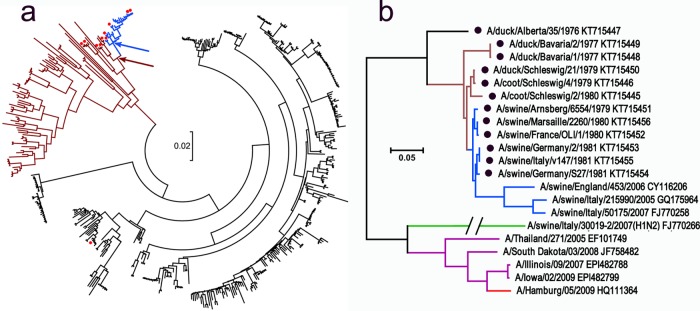
In 1979, novel H1N1 viruses were isolated from pigs in Belgium and Germany (39). These viruses were closely related to European H1N1 duck viruses (39–41), indicating a recent avian-to-swine transmission event (Fig. 1a). The viruses became endemic in Europe and Asia and form the so-called Eurasian avian-like swine lineage (EAsw) (42, 43). To assess potential changes in the viral membrane fusion characteristics during the avian-to-swine transmission, we focused on EAsw viruses isolated in the first years of their circulation in pigs (1979 to 1981) and on the closest available avian counterparts (Fig. 1, Table 1). For a comparison, we also tested a limited number of other swine and human viruses, among them, recent isolates of EAsw viruses, viruses with “classical” swine HA isolated from cases of zoonotic infections in Thailand and the United States (44, 45), swine H1N2 and H3N2 viruses with human-virus-like HAs (42, 43), and two pandemic human viruses (Fig. 1b, Table 1). The avian viruses and early EAsw viruses were originally isolated and passaged in embryonated hen's eggs, whereas recent swine and human viruses were isolated and propagated solely in cell culture. To exclude potential effects on viral phenotype of laboratory substrate used to grow the viruses, all viral stocks were prepared for this study in MDCK cells.
FIG 1.
Phylogenetic relationships between HAs of H1 influenza viruses. (a) Tree based on the sequences of 455 avian viruses from North America (black), Eurasia, Oceania, and Africa (brown) and avian-like swine viruses isolated in 1979 to 1981 in Europe (blue). Red dots depict virus strains that were tested in this study. Arrows show locations of hypothetical first swine virus (blue) and its putative avian precursor (brown). (b) Tree showing all of the viruses with H1 HA used in this study. Color coding of HA lineages: black, brown, and blue (same as in panel a); green, H1N2 swine viruses with human-like HA; purple, viruses with classical swine HA isolated from humans; red, H1N1/2009 pandemic virus. GenBank accession numbers are shown next to strain names. Two sequences (A/Illinois/09/2007 and A/Iowa/02/2009) (44) were obtained from GISAID EpiFlu Database (www.platform.gisaid.org/). Black dots depict sequences determined in the present study. The scale bars in both panels represent units of nucleotide substitutions per site.
pH dependence of viral membrane fusion activity and stability of the viruses at low pH.
HA-mediated fusion of the viral and cellular membranes is triggered by the low pH in the endosomes and is essential for viral infection. To compare requirements of the viruses for the acidic pH during cell entry, we studied inhibition of viral single-cycle infection in MDCK cells by the lysosomotropic agent ammonium chloride, which counteracts acidification of endosomes (46, 47). Concentrations of NH4Cl that reduced infection by 50% were determined from the dose-response curves, as illustrated in Fig. 2a. Values of 50% inhibitory concentration (IC50) for all tested viruses are shown in the Table 1. These values were significantly lower for avian viruses (IC50s from 0.26 to 0.55 mM NH4Cl) than for the Eurasian avian-like swine viruses (IC50s from 0.83 to 1.8 mM NH4Cl). Swine viruses of other lineages, similarly to EAsw viruses, were less sensitive to ammonium chloride than avian H1 viruses. It should be noticed that virus strains with classical swine HA isolated from zoonotic human infections were rather variable in their sensitivity (IC50s from 0.65 to 2.55 mM NH4Cl). Two pandemic virus strains were less sensitive than avian viruses and more sensitive than a majority of swine viruses.
These findings showed that swine viruses are less dependent on the level of endosomal acidification than avian viruses with the same HA subtype. We assumed that this effect is determined, at least in part, by a higher pH optimum of the HA-mediated membrane fusion of swine viruses. To test this hypothesis, we compared EAsw viruses and avian viruses using two additional assays. We studied the pH dependence of virus-mediated hemolysis (48), as illustrated in the Fig. 2b, and determined values of IC50-hem (Table 1). Viruses that were less sensitive to neutralization by ammonium chloride typically displayed higher values of IC50-hem, although this correlation was not perfect. This inconsistency may be due to different nature of the assays used. In general, EAsw viruses lysed red blood cells at a higher pH than did avian viruses (pH50-hem in the range of 5.07 to 5.39 for swine viruses and 4.96 to 5.20 for avian viruses; P = 0.019, two-sided unpaired t test). We also determined inactivation of the viral infectivity at acidic pH (Fig. 2c, Table 1) and found that swine viruses were less stable than avian viruses (pH50-inact in the range of 5.47 to 5.91 for swine viruses and 5.19 to 5.43 for avian viruses; P = 0.0037, two-sided unpaired t test).
Analysis of HA sequences.
Our assays suggested that the HAs of EAsw lineage undergo acid-induced conformational change and mediate fusion at higher pH values than the HAs of closely related avian viruses. To assess molecular changes in the HA that were responsible for these differences we compared HAs of the EAsw viruses isolated in 1979 to 1981 with HAs of all H1 avian viruses sequenced to date and reconstructed amino acid sequences at the nodes of phylogenetic tree separating EAsw viruses from the avian viruses (see Fig. 1a). This analysis revealed that the hypothetical first EAsw virus differed from the hypothetical avian precursor by eight amino acid substitutions (Table 2 and Fig. 3).
TABLE 2.
Amino acids at the HA positions that separate H1N1 Eurasian avian-like swine viruses from their putative avian precursora
| Virus | Amino acid (no. of sequences with the indicated amino acid) |
|||||||
|---|---|---|---|---|---|---|---|---|
| 126a [138]b | 155 [169] | 188 [202] | 190 [204] | 492 [393] | 722 [416] | 752 [419] | 1132 [457] | |
| Hypothetical avian precursor | S | T | A | E | T | N | R | S |
| Hypothetical first EAsw virus | N | I | T | D | S | D | K | F |
| Avian virusesc | N (285) | T (285) | T (370) | E (452) | T (452) | N | R (400) | S (454) |
| S (167) | I (164) | V (53) | Q (2) | I (3) | K (54) | F (1) | ||
| T (1) | V (4) | A (28) | X (1) | Q (1) | ||||
| X (2) | X (2) | I (3) | ||||||
| X (1) | ||||||||
Sequences of the first EAsw virus and its avian precursor were inferred based on the HA sequences of avian and avian-like swine influenza viruses as described in Materials and Methods. Amino acids are shown in a single-letter code; “X” depicts ambiguity at this position. Numbers in parentheses indicate the numbers of sequences with the indicated amino acid.
The first number is based on the H3 numbering system in accord with H3/H1 alignment of Nobusawa et al. (49). A separate numbering is used for HA1 and HA2 subunits; the subscript refers to HA2. The numbers in square brackets correspond to the numbers of codons within the complete coding sequence of the HA precursor (codons 1 to 17, signal peptide; codons 18 to 566, HA0).
Analysis of 455 full-length H1 HA sequences.
FIG 3.
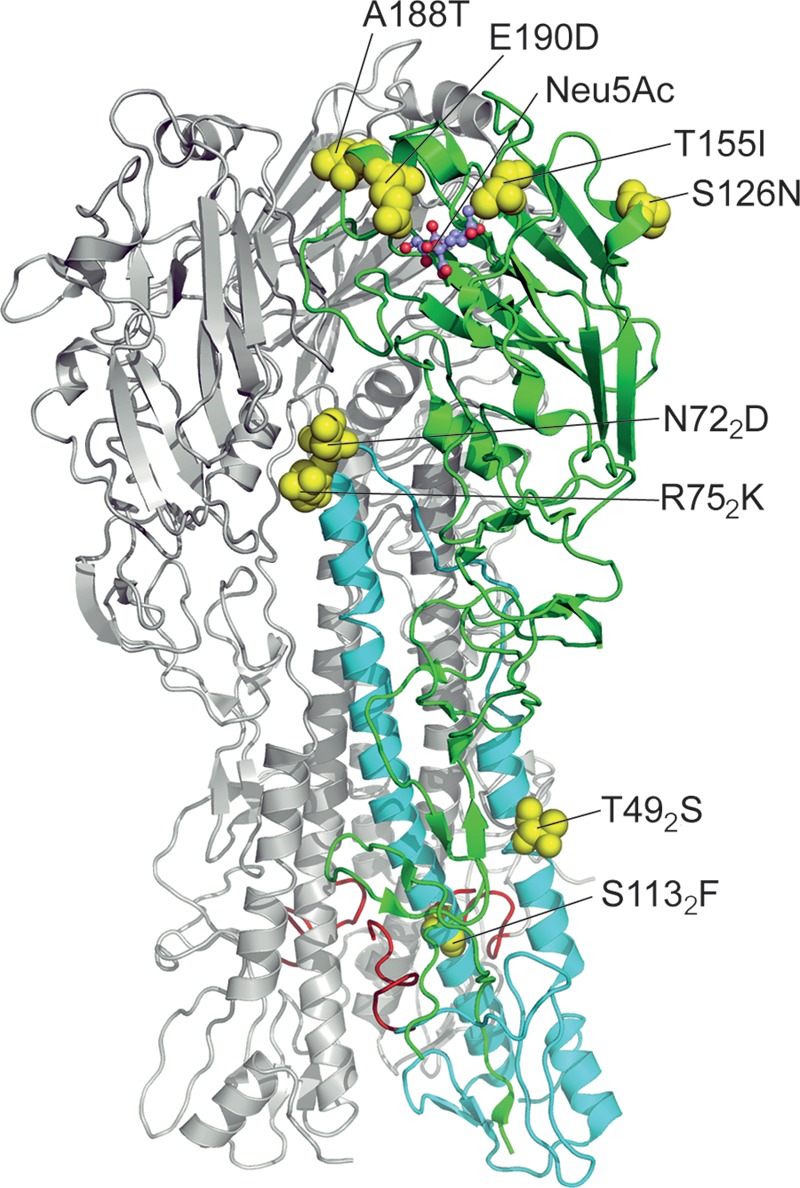
Eight amino acid substitutions separating HAs of EAsw viruses from their putative avian ancestor. The model is based on the X-ray structure of the HA of A/duck/Alberta/35/1976 (2WRH, Protein Data Bank). For clarity, two HA monomers are colored gray, and the third monomer is colored green (HA1) and cyan (HA2). Location of substitutions is shown on this monomer as yellow spaced-filled models; sialic acid (Neu5Ac) in the receptor-binding site is shown as ball-and-stick model. Amino acids are numbered using H3 numbering system, with the subscript referring to HA2. Ten N-terminal amino acid residues of the fusion peptides of all three monomers are depicted in red.
Four substitutions were located in the receptor-binding domain of the HA1 subunit. Amino acids in positions 155 and 190 (H3 numbering [49]) are in direct contact with sialic acid and play important role in receptor recognition (50, 51). Amino acid in position 188 is located at the rim of the receptor binding site in close proximity to position 190. A substitution in position 126a is about 20 Å away from the sialic acid in the binding site with the side chain of the amino acid being highly exposed to the solvent. Importantly, none of the four HA1 mutations was located in the structural regions of the HA1 known to modulate the low pH conformational transition, such as interface between HA1 monomers, interface between HA1 and HA2 monomers and interface between the receptor-binding and vestigial esterase subdomains (16–18). Thus, the potential effects, if any, of these four mutations in HA1 on pH optimum of fusion remain obscure.
Four other substitutions (T492S, N722D, R752K, and S1132F) were located in the HA2 subunit (Fig. 3). Amino acid residues in three out of four mentioned positions are conserved in the HAs of avian viruses (Table 2), implying that substitutions in these three positions could reduce virus fitness in birds by altering HA expression and/or functions. The amino acid 492 is located in the midsection of the smaller HA2 helix, and its side chain is exposed to solvent without making contacts with other parts of the HA. It is not likely, therefore, that conservative substitution T492S can significantly affect fusion activity of swine virus HA. The amino acids in positions 722 and 752 belong, respectively, to the loop connecting two helixes and the tip of the central large helix. Side chains of either residue forms close contacts with amino acids of the vestigial esterase subdomain of HA1. Mutations in this structural region are well known to affect the pH of the HA conformational transition (16–18). In particular, the mutation G752R in the HA of H3N2 human virus increased pH threshold for fusion by 0.4 U (52). Residue 1132 is located on the large helix with its side chain facing the internal hydrophobic pocket with buried fusion peptides. The residue contacts phenylalanine in position 32 of the fusion peptide of its own HA2 monomer and leucine in position 22 of the neighboring HA2 monomer. Mutations in the fusion peptide and in its pocket are known to alter HA conformational stability and fusion pH threshold (16–18). All but 1 of 455 avian HA sequences analyzed harbored S in position 1132, whereas one avian virus (GenBank accession number KC209515), similarly to the avian-like swine viruses, carried the nonconservative mutation S1132F. Remarkably, the mutation seemed to emerge after serial passaging of the original virus isolate A/mallard/Netherlands/10-Nmkt/1999 (H1N1) in the newborn pig trachea cell line NPTr (53).
In summary, based on their nature and location, from one to three substitutions in the HA2 (positions 72, 75, and 113) were likely responsible for the alteration of conformational stability and fusion pH optimum during the emergence of an avian-like swine virus from its avian ancestor. Studies are in progress in our lab to test this hypothesis and to identify specific set of mutations involved.
pH dependence of the fusion activity of pandemic virus HAs.
Based on our data (Table 1), the HAs of viruses from 1968 and 2009 pandemics had a lower pH optimum of fusion than the HAs of most swine viruses analyzed. To test this observation further, we compared the HAs of all four known pandemic viruses for their fusion activation at low pH. Because the whole 1918 pandemic virus was not available, the experiments were performed using cell-expressed HAs. The HA of A/swine/Marseille/1980 was included in the analysis since this virus displayed fusion characteristics typical of the majority of swine viruses tested (see Table 1). The 1918 HA required the lowest pH for fusion activation, since syncytium formation was only observed at pH 5.1 and below (Fig. 4). The 1957 and 1968 HAs started to cause weak but significant syncytium formation at pH 5.2. The HA of 2009 virus was able to induce pronounced cell-to-cell fusion at pH 5.4, whereas the HA of A/swine/Marseille/1980 was already active at pH 6.0. Thus, in this assay all four pandemic HAs required significantly lower pH for fusion activation compared to the swine virus HA.
FIG 4.
Syncytium formation in HeLa cells expressing HA proteins after exposure to low pH. Cells were transfected with plasmids encoding HAs of pandemic viruses A/Brevig Mission/1/1918 (H1N1), A/Singapore/1/1957 (H2N2), A/Hong Kong/1/1968 (H3N2), A/Hamburg/5/2009 (H1N1), and HA of A/swine/Marseille/2260/1980 (H1N1). The cells were exposed to different pHs 16 h posttransfection, incubated to allow syncytium formation, and contrasted using Giemsa stain. The numbers of nuclei in syncytia were counted and are expressed as percentages of the total nuclei in the same microscopic field. The data represent mean values; error bars show 95% confidence intervals.
DISCUSSION
Whereas receptor-binding specificity of influenza virus HA is a well-known factor determining viral host range (8–11), the role of the fusion-promotion characteristics of the HA in host range restriction is less well understood (17, 19). The pioneering studies of Scholtissek showed that stability of influenza viruses at low pH correlated, at least partially, with the viral host species (27, 54), implying that host-specific differences exist between the viruses in the pH optimum of HA conformational stability. Recently, this notion was confirmed by studies on highly pathogenic H5N1 viruses which showed that replication and pathogenesis of H5N1 viruses in experimentally infected ducks, chickens, mice, and ferrets were differently regulated by pH of HA activation (20–22, 24, 25, 55–57). These results highlight the necessity to characterize host-specific parameters of membrane fusion activity of influenza viruses with various HA subtypes from major natural host species, such as wild and domestic birds, pigs, horses, and humans.
In the present study, we focused on H1N1 viruses in aquatic birds and pigs. The H1N1 viruses are ubiquitous, they demonstrated a propensity for interspecies transmission, and they caused pandemics in 1918 and 2009. Furthermore, availability of EAsw viruses isolated at the very beginning of the European swine epizootic and closely related aquatic bird viruses isolated at the same time in the same geographical area (see Fig. 1) provided a unique opportunity to assess genotypic and phenotypic changes in the HA that accompanied avian-to-swine adaptation of the virus. The results of three phenotypic assays indicated that the HAs of the early EAsw virus strains undergo conformational transition and mediate fusion at a higher pH than HAs of the closest available avian counterparts (Table 1). These differences in pH optimum of HA activation were associated with amino acid substitutions in the conserved positions of HA2 subunit that separate early isolates of EAsw swine viruses from the avian precursor (Fig. 3). Several other tested swine viruses of the EAsw lineage and other lineages behaved similarly to the early EAsw virus isolates, suggesting that relatively high pH optimum of fusion is a typical trait of swine-adapted influenza viruses. Our results suggest that HA fusion characteristics of avian H1N1 viruses reduced viral fitness in pigs, and alteration of the pH optimum of fusion occurred during the avian-to-swine adaptation of the virus. It remains obscure whether these changes were essential for the initial adaptation of the virus to replicate and transmit in pigs or they were acquired during subsequent years of virus circulation in pigs prior to its detection and isolation in 1979 (41). This question can be answered by studying replication and transmission of wild type and recombinant viruses with relevant mutations in the HA in experimentally infected pigs as described previously (58, 59).
Hypothetical mechanisms by which pH optimum of HA-mediated fusion could affect virus fitness in distinct host species have been discussed (17–19, 22, 28, 54). First, the kinetics of acidification and the levels of pH in the endosomal compartment may vary depending on host species, target tissue, cell type, and differentiation and metabolic state of the cell. A balance between endosomal pH in the cells and pH optimum of HA conformational transition was shown to affect the efficiency of viral infection (20, 28, 60). Differences in sensitivity of viruses to ammonium chloride during cell entry (Table 1) well illustrate this phenomenon. Second, host-specific differences in the pH and temperature at mucosal surfaces in target tissues of different species may also play a role in determining pH optimum of HA fusion and stability. For example, in humans, the nasal pH varies from 5.2 to 8.0 (61, 62), the lower values being sufficiently acidic to can cause rapid virus inactivation. In ducks, the viruses replicate in the cells lining the lower intestinal tract (63, 64) with pH values between 6 and 8 (65, 66), although on its route to target cells the virus has to pass the highly acidic environment of the gizzard. Third, requirements for virus persistence in the environment may vary significantly depending on ecology of viral host species and mode of virus transmission (67, 68). For example, a higher pH stability and thermostability may be required for virus transmission by fecal-oral route through water in aquatic birds and by airborne droplets in humans compared to direct contact transmission in crowded domestic poultry flocks and pig herds. Thus, the optimal pH of fusion and stability of the HA in its host species is determined by a balance between several not yet well-defined selective pressures. Our results suggest that this balance differs between aquatic birds and pigs.
Unlike most avian influenza viruses, swine-adapted viruses preferentially bind to Sia2-6Gal-terminated (“human-type”) receptors (50, 69–72). It is believed, therefore, that adaptation of the HA of avian viruses to receptors in pigs increases viral zoonotic and pandemic potential. This theory gained strong support after emergence of the 2009 pandemic virus from a swine virus without significant changes in receptor specificity (73, 74). In contrast, our data suggest that the levels of conformational stability of the HA of swine-adapted viruses may not be optimal for viral circulation in humans. In our experiments, two pandemic viruses tested had more pH-stable HAs than a majority of swine viruses, as judged by viral sensitivity to ammonium chloride and pH optimum of hemolytic activity (Table 1). Interestingly, three of four swine viruses isolated in Thailand and United States from sporadic human cases of infection differed from other swine viruses and resembled pandemic virus strains with respect to their sensitivity to ammonium chloride. It remains obscure whether this effect represents natural strain-specific variation among viruses with “classical” swine HA or reflects ongoing adaptation of these swine viruses to humans. The distinctions between the pH stability of swine and pandemic virus HAs were further confirmed in experiments with expressed HA proteins (Fig. 4). The HA of the viruses from three previous pandemics displayed a similar pH of fusion activation that was significantly lower than that of the swine virus HA. Interestingly, the HA of the swine-origin pandemic virus A/Hamburg/05/2009 showed an intermediate phenotype. This virus was isolated in late April 2009, at the very beginning of the emerging pandemic. Notably, subsequent circulation of the virus in human population led to a selection of a fitter variant with mutation E472K in the HA that lowered pH of fusion and increased pH stability (75).
In summary, our finding that transmission of an avian H1N1 virus to pigs in Europe in late 1980s was accompanied by changes in conformational stability of the HA represents the first formal evidence of alteration of the HA fusion activity/stability during influenza virus adaptation to a new host species under natural settings. This finding prompts further detailed studies on membrane fusion characteristics of influenza viruses circulating in different host species, alterations of these characteristics during interspecies transmission, and their potential effects on viral zoonotic and pandemic potential.
ACKNOWLEDGMENTS
This study was supported by European Union's Seventh Framework Programme for Research, Technological Development, and Demonstration under grant agreements 258084-FLUPIG and 278433-PREDEMICS and by the German Research Foundation (SFB 1021, project B02).
We are very grateful to Christoph Scholtissek and other colleagues listed in Table 1, footnote b, for influenza viruses. We thank Robert Webster (St. Jude Children's Research Hospital, Memphis, TN) for the pHW2000 plasmid.
REFERENCES
- 1.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, Fouchier RA. 2006. Global patterns of influenza A virus in wild birds. Science 312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 2.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munoz O, De Nardi M, van der Meulen K, van Reeth K, Koopmans M, Harris K, von Dobschuetz S, Freidl G, Meijer A, Breed A, Hill A, Kosmider R, Banks J, Stark KD, Wieland B, Stevens K, van der Werf S, Enouf V, Dauphin G, Dundon W, Cattoli G, Capua I. 2015. Genetic adaptation of influenza A viruses in domestic animals and their potential role in interspecies transmission: a literature review. Ecohealth 29:29. [DOI] [PubMed] [Google Scholar]
- 4.Short KR, Richard M, Verhagen JH, van Riel D, Schrauwen EJA, van den Brand JMA, Mänz B, Bodewes R, Herfst S. 2015. One health, multiple challenges: the inter-species transmission of influenza A virus. One Health 1:1–13. doi: 10.1016/j.onehlt.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freidl GS, Meijer A, de Bruin E, de Nardi M, Munoz O, Capua I, Breed AC, Harris K, Hill A, Kosmider R, Banks J, von Dobschuetz S, Stark K, Wieland B, Stevens K, van der Werf S, Enouf V, van der Meulen K, van Reeth K, Dauphin G, Koopmans M, Consortium F. 2014. Influenza at the animal-human interface: a review of the literature for virological evidence of human infection with swine or avian influenza viruses other than A(H5N1). Euro Surveill 19:8–26. [DOI] [PubMed] [Google Scholar]
- 6.Kilbourne ED. 2006. Influenza pandemics of the 20th century. Emerg Infect Dis 12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morens DM, Taubenberger JK. 2011. Pandemic influenza: certain uncertainties. Rev Med Virol 21:262–284. doi: 10.1002/rmv.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Graaf M, Fouchier RA. 2014. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J 33:823–841. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulson JC, de Vries RP. 2013. H5N1 receptor specificity as a factor in pandemic risk. Virus Res 178:99–113. doi: 10.1016/j.virusres.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matrosovich MN, Gambaryan AS, Klenk H-D. 2008. Receptor specificity of influenza viruses and its alteration during interspecies transmission, p 134–155. In Klenk H-D, Matrosovich MN, Stech J (ed), Avian influenza, vol 27 Karger, Basel, Switzerland. [Google Scholar]
- 11.Neumann G, Kawaoka Y. 2015. Transmission of influenza A viruses. Virology 479-480:234–246. doi: 10.1016/j.virol.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruigrok RWH, Martin SR, Wharton SA, Skehel JJ, Bayley PM, Wiley DC. 1986. Conformational-changes in the hemagglutinin of influenza-virus which accompany heat-induced fusion of virus with liposomes. Virology 155:484–497. doi: 10.1016/0042-6822(86)90210-2. [DOI] [PubMed] [Google Scholar]
- 13.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 14.Carr CM, Chaudhry C, Kim PS. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci U S A 94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniels RS, Downie JC, Hay AJ, Knossow M, Skehel JJ, Wang ML, Wiley DC. 1985. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell 40:431–439. doi: 10.1016/0092-8674(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 16.Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 17.Russell CJ. 2014. Acid-induced membrane fusion by the hemagglutinin protein and its role in influenza virus biology. Curr Top Microbiol Immunol 385:93–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mair CM, Ludwig K, Herrmann A, Sieben C. 2014. Receptor binding and pH stability: how influenza A virus hemagglutinin affects host-specific virus infection. Biochim Biophys Acta 1838:1153–1168. doi: 10.1016/j.bbamem.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Long JS, Benfield CT, Barclay WS. 2015. One-way trip: Influenza virus' adaptation to gallinaceous poultry may limit its pandemic potential. Bioessays 37:204–212. doi: 10.1002/bies.201400133. [DOI] [PubMed] [Google Scholar]
- 20.Zaraket H, Bridges OA, Duan S, Baranovich T, Yoon SW, Reed ML, Salomon R, Webby RJ, Webster RG, Russell CJ. 2013. Increased acid stability of the hemagglutinin protein enhances H5N1 influenza virus growth in the upper respiratory tract but is insufficient for transmission in ferrets. J Virol 87:9911–9922. doi: 10.1128/JVI.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shelton H, Roberts KL, Molesti E, Temperton N, Barclay WS. 2013. Mutations in haemagglutinin that affect receptor binding and pH stability increase replication of a PR8 influenza virus with H5 HA in the upper respiratory tract of ferrets and may contribute to transmissibility. J Gen Virol 94:1220–1229. doi: 10.1099/vir.0.050526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krenn BM, Egorov A, Romanovskaya-Romanko E, Wolschek M, Nakowitsch S, Ruthsatz T, Kiefmann B, Morokutti A, Humer J, Geiler J, Cinatl J, Michaelis M, Wressnigg N, Sturlan S, Ferko B, Batishchev OV, Indenbom AV, Zhu R, Kastner M, Hinterdorfer P, Kiselev O, Muster T, Romanova J. 2011. Single HA2 mutation increases the infectivity and immunogenicity of a live attenuated H5N1 intranasal influenza vaccine candidate lacking NS1. PLoS One 6:e18577. doi: 10.1371/journal.pone.0018577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de, Munster WEVJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linster M, van, De BSGM, Schrauwen EJ, Lexmond P, Manz B, Bestebroer TM, Baumann J, van, Rimmelzwaan RDGF, Osterhaus AD, Matrosovich M, Fouchier RA, Herfst S. 2014. Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell 157:329–339. doi: 10.1016/j.cell.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beyer WE, Ruigrok RW, van Driel H, Masurel N. 1986. Influenza virus strains with a fusion threshold of pH 5.5 or lower are inhibited by amantadine: brief report. Arch Virol 90:173–181. doi: 10.1007/BF01314156. [DOI] [PubMed] [Google Scholar]
- 27.Scholtissek C. 1985. Stability of infectious influenza A viruses at low pH and at elevated temperature. Vaccine 3:215–218. doi: 10.1016/0264-410X(85)90109-4. [DOI] [PubMed] [Google Scholar]
- 28.Daidoji T, Watanabe Y, Ibrahim MS, Yasugi M, Maruyama H, Masuda T, Arai F, Ohba T, Honda A, Ikuta K, Nakaya T. 2015. Avian influenza virus infection of immortalized human respiratory epithelial cells depends upon a delicate balance between hemagglutinin acid stability and endosomal pH. J Biol Chem 290:10627–10642. doi: 10.1074/jbc.M114.611327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galloway SE, Reed ML, Russell CJ, Steinhauer DA. 2013. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog 9:e1003151. doi: 10.1371/journal.ppat.1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byrd-Leotis L, Galloway SE, Agbogu E, Steinhauer DA. 2015. Influenza hemagglutinin (HA) stem region mutations that stabilize or destabilize the structure of multiple HA subtypes. J Virol 89:4504–4516. doi: 10.1128/JVI.00057-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matrosovich M, Matrosovich T, Uhlendorff J, Garten W, Klenk HD. 2007. Avian-virus-like receptor specificity of the hemagglutinin impedes influenza virus replication in cultures of human airway epithelium. Virology 361:384–390. doi: 10.1016/j.virol.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 32.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 33.Montano RF, Morrison SL. 1999. A colorimetric-enzymatic microassay for the quantitation of antibody-dependent complement activation. J Immunol Methods 222:73–82. doi: 10.1016/S0022-1759(98)00181-1. [DOI] [PubMed] [Google Scholar]
- 34.Gerlach T, Kuhling L, Uhlendorff J, Laukemper V, Matrosovich T, Czudai-Matwich V, Schwalm F, Klenk HD, Matrosovich M. 2012. Characterization of the neuraminidase of the H1N1/09 pandemic influenza virus. Vaccine 30:7348–7352. doi: 10.1016/j.vaccine.2012.09.078. [DOI] [PubMed] [Google Scholar]
- 35.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. J Virol 82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall T. 2004. BioEdit, v7.0. 0. Distributed by the author. www.mBio.ncsu.edu/BioEdit/bioedit.html.
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, Gamblin SJ, Skehel JJ. 2009. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc Natl Acad Sci U S A 106:17175–17180. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pensaert M, Ottis K, Vandeputte J, Kaplan MM, Bachmann PA. 1981. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull World Health Organ 59:75–78. [PMC free article] [PubMed] [Google Scholar]
- 40.Scholtissek C, Burger H, Bachmann PA, Hannoun C. 1983. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology 129:521–523. doi: 10.1016/0042-6822(83)90194-0. [DOI] [PubMed] [Google Scholar]
- 41.Krumbholz A, Lange J, Sauerbrei A, Groth M, Platzer M, Kanrai P, Pleschka S, Scholtissek C, Buttner M, Durrwald R, Zell R. 2014. Origin of the European avian-like swine influenza viruses. J Gen Virol 95:2372–2376. doi: 10.1099/vir.0.068569-0. [DOI] [PubMed] [Google Scholar]
- 42.Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, Donis R, Culhane M, Hamilton K, Lewis N, Mumford E, Nguyen T, Parchariyanon S, Pasick J, Pavade G, Pereda A, Peiris M, Saito T, Swenson S, Van Reeth K, Webby R, Wong F, Ciacci-Zanella J. 2014. Review of influenza A virus in swine worldwide: a call for increased surveillance and research. Zoonoses Public Health 61:4–17. doi: 10.1111/zph.12049. [DOI] [PubMed] [Google Scholar]
- 43.Kuntz-Simon G, Madec F. 2009. Genetic and antigenic evolution of swine influenza viruses in Europe and evaluation of their zoonotic potential. Zoonoses Public Health 56:310–325. doi: 10.1111/j.1863-2378.2009.01236.x. [DOI] [PubMed] [Google Scholar]
- 44.Shu B, Garten R, Emery S, Balish A, Cooper L, Sessions W, Deyde V, Smith C, Berman L, Klimov A, Lindstrom S, Xu X. 2012. Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990-2010. Virology 422:151–160. doi: 10.1016/j.virol.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 45.Komadina N, Roque V, Thawatsupha P, Rimando-Magalong J, Waicharoen S, Bomasang E, Sawanpanyalert P, Rivera M, Iannello P, Hurt AC, Barr IG. 2007. Genetic analysis of two influenza A (H1) swine viruses isolated from humans in Thailand and the Philippines. Virus Genes 35:161–165. doi: 10.1007/s11262-007-0097-9. [DOI] [PubMed] [Google Scholar]
- 46.Koerner I, Matrosovich MN, Haller O, Staeheli P, Kochs G. 2012. Altered receptor specificity and fusion activity of the haemagglutinin contribute to high virulence of a mouse-adapted influenza A virus. J Gen Virol 93:970–979. doi: 10.1099/vir.0.035782-0. [DOI] [PubMed] [Google Scholar]
- 47.Matlin KS, Reggio H, Helenius A, Simons K. 1981. Infectious entry pathway of influenza virus in a canine kidney cell line. J Cell Biol 91:601–613. doi: 10.1083/jcb.91.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maeda T, Ohnishi S-i. 1980. Activation of influenza virus by acidic media causes hemolysis and fusion of erythrocytes. FEBS Lett 122:283–287. doi: 10.1016/0014-5793(80)80457-1. [DOI] [PubMed] [Google Scholar]
- 49.Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. 1991. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 50.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol 74:8502–8512. doi: 10.1128/JVI.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glaser L, Stevens J, Zamarin D, Wilson IA, Garcia-Sastre A, Tumpey TM, Basler CF, Taubenberger JK, Palese P. 2005. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol 79:11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakowitsch S, Wolschek M, Morokutti A, Ruthsatz T, Krenn BM, Ferko B, Ferstl N, Triendl A, Muster T, Egorov A, Romanova J. 2011. Mutations affecting the stability of the haemagglutinin molecule impair the immunogenicity of live attenuated H3N2 intranasal influenza vaccine candidates lacking NS1. Vaccine 29:3517–3524. doi: 10.1016/j.vaccine.2011.02.100. [DOI] [PubMed] [Google Scholar]
- 53.Bourret V, Croville G, Mariette J, Klopp C, Bouchez O, Tiley L, Guérin J-L. 2013. Whole-genome, deep pyrosequencing analysis of a duck influenza A virus evolution in swine cells. Infect Genet Evol 18:31–41. doi: 10.1016/j.meegid.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 54.Scholtissek C. 1987. Molecular aspects of the epidemiology of virus disease. Experientia 43:1197–1201. doi: 10.1007/BF01945523. [DOI] [PubMed] [Google Scholar]
- 55.Reed ML, Bridges OA, Seiler P, Kim JK, Yen HL, Salomon R, Govorkova EA, Webster RG, Russell CJ. 2010. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity and transmissibility in ducks. J Virol 84:1527–1535. doi: 10.1128/JVI.02069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reed ML, Yen HL, DuBois RM, Bridges OA, Salomon R, Webster RG, Russell CJ. 2009. Amino acid residues in the fusion peptide pocket regulate the pH of activation of the H5N1 influenza virus hemagglutinin protein. J Virol 83:3568–3580. doi: 10.1128/JVI.02238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zaraket H, Bridges OA, Russell CJ. 2013. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus replication and pathogenesis in mice. J Virol 87:4826–4834. doi: 10.1128/JVI.03110-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Poucke S, Uhlendorff J, Wang ZF, Billiau V, Nicholls J, Matrosovich M, Van Reeth K. 2013. Effect of receptor specificity of A/Hong Kong/1/68 (H3N2) influenza virus variants on replication and transmission in pigs. Influenza Other Respir Viruses 7:151–159. doi: 10.1111/j.1750-2659.2012.00376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brookes SM, Nunez A, Choudhury B, Matrosovich M, Essen SC, Clifford D, Slomka MJ, Kuntz-Simon G, Garcon F, Nash B, Hanna A, Heegaard PMH, Queguiner S, Chiapponi C, Bublot M, Garcia JM, Gardner R, Foni E, Loeffen W, Larsen L, Van Reeth K, Banks J, Irvine RM, Brown IH. 2010. Replication, pathogenesis and transmission of pandemic (H1N1) 2009 virus in non-immune pigs. PLoS One 5:e9068. doi: 10.1371/journal.pone.0009068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murakami S, Horimoto T, Ito M, Takano R, Katsura H, Shimojima M, Kawaoka Y. 2012. Enhanced growth of influenza vaccine seed viruses in Vero cells mediated by broadening the optimal pH range for virus membrane fusion. J Virol 86:1405–1410. doi: 10.1128/JVI.06009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.England RJ, Homer JJ, Knight LC, Ell SR. 1999. Nasal pH measurement: a reliable and repeatable parameter. Clin Otolaryngol Allied Sci 24:67–68. doi: 10.1046/j.1365-2273.1999.00223.x. [DOI] [PubMed] [Google Scholar]
- 62.Washington N, Steele RJ, Jackson SJ, Bush D, Mason J, Gill DA, Pitt K, Rawlins DA. 2000. Determination of baseline human nasal pH and the effect of intranasally administered buffers. Int J Pharm 198:139–146. doi: 10.1016/S0378-5173(99)00442-1. [DOI] [PubMed] [Google Scholar]
- 63.Webster RG, Yakhno M, Hinshaw VS, Bean WJ, Murti KG. 1978. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology 84:268–278. doi: 10.1016/0042-6822(78)90247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kida H, Yanagawa R, Matsuoka Y. 1980. Duck influenza lacking evidence of disease signs and immune response. Infect Immun 30:547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kararli TT. 1995. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharmaceutics Drug Disposition 16:351–380. [DOI] [PubMed] [Google Scholar]
- 66.Farner DS. 1942. The hydrogen ion concentration in avian digestive tracts. Poultry Sci 21:445–450. doi: 10.3382/ps.0210445. [DOI] [Google Scholar]
- 67.Stallknecht DE, Brown JD. 2009. Tenacity of avian influenza viruses. Rev Sci Tech 28:59–67. [DOI] [PubMed] [Google Scholar]
- 68.Reperant LA, Kuiken T, Osterhaus AD. 2012. Adaptive pathways of zoonotic influenza viruses: from exposure to establishment in humans. Vaccine 30:4419–4434. doi: 10.1016/j.vaccine.2012.04.049. [DOI] [PubMed] [Google Scholar]
- 69.Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, Kawaoka Y. 1998. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol 72:7367–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma W, Vincent AL, Gramer MR, Brockwell CB, Lager KM, Janke BH, Gauger PC, Patnayak DP, Webby RJ, Richt JA. 2007. Identification of H2N3 influenza A viruses from swine in the United States. Proc Natl Acad Sci U S A 104:20949–20954. doi: 10.1073/pnas.0710286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bateman AC, Busch MG, Karasin AI, Bovin N, Olsen CW. 2008. Amino acid 226 in the hemagglutinin of H4N6 influenza virus determines binding affinity for alpha2,6-linked sialic acid and infectivity levels in primary swine and human respiratory epithelial cells. J Virol 82:8204–8209. doi: 10.1128/JVI.00718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Byrd-Leotis L, Liu R, Bradley KC, Lasanajak Y, Cummings SF, Song X, Heimburg-Molinaro J, Galloway SE, Culhane MR, Smith DF, Steinhauer DA, Cummings RD. 2014. Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc Natl Acad Sci U S A 111:E2241–E2250. doi: 10.1073/pnas.1323162111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen LM, Rivailler P, Hossain J, Carney P, Balish A, Perry I, Davis CT, Garten R, Shu B, Xu X, Klimov A, Paulson JC, Cox NJ, Swenson S, Stevens J, Vincent A, Gramer M, Donis RO. 2011. Receptor specificity of subtype H1 influenza A viruses isolated from swine and humans in the United States. Virology 412:401–410. doi: 10.1016/j.virol.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bradley KC, Jones CA, Tompkins SM, Tripp RA, Russell RJ, Gramer MR, Heimburg-Molinaro J, Smith DF, Cummings RD, Steinhauer DA. 2011. Comparison of the receptor binding properties of contemporary swine isolates and early human pandemic H1N1 isolates (novel 2009 H1N1). Virology 413:169–182. doi: 10.1016/j.virol.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 75.Cotter CR, Jin H, Chen ZY. 2014. A single amino acid in the stalk region of the H1N1pdm influenza virus HA protein affects viral fusion, stability and infectivity. PLoS Pathog 10:e1003831. doi: 10.1371/journal.ppat.1003831. [DOI] [PMC free article] [PubMed] [Google Scholar]