ABSTRACT
Parechoviruses are human pathogens that cause diseases ranging from gastrointestinal disorders to encephalitis. Unlike those of most picornaviruses, parechovirus capsids are composed of only three subunits: VP0, VP1, and VP3. Here, we present the structure of a human parechovirus 1 (HPeV-1) virion determined to a resolution of 3.1 Å. We found that interactions among pentamers in the HPeV-1 capsid are mediated by the N termini of VP0s, which correspond to the capsid protein VP4 and the N-terminal part of the capsid protein VP2 of other picornaviruses. In order to facilitate delivery of the virus genome into the cytoplasm, the N termini of VP0s have to be released from contacts between pentamers and exposed at the particle surface, resulting in capsid disruption. A hydrophobic pocket, which can be targeted by capsid-binding antiviral compounds in many other picornaviruses, is not present in HPeV-1. However, we found that interactions between the HPeV-1 single-stranded RNA genome and subunits VP1 and VP3 in the virion impose a partial icosahedral ordering on the genome. The residues involved in RNA binding are conserved among all parechoviruses, suggesting a putative role of the genome in virion stability or assembly. Therefore, putative small molecules that could disrupt HPeV RNA-capsid protein interactions could be developed into antiviral inhibitors.
IMPORTANCE Human parechoviruses (HPeVs) are pathogens that cause diseases ranging from respiratory and gastrointestinal disorders to encephalitis. Recently, there have been outbreaks of HPeV infections in Western Europe and North America. We present the first atomic structure of parechovirus HPeV-1 determined by X-ray crystallography. The structure explains why HPeVs cannot be targeted by antiviral compounds that are effective against other picornaviruses. Furthermore, we found that the interactions of the HPeV-1 genome with the capsid resulted in a partial icosahedral ordering of the genome. The residues involved in RNA binding are conserved among all parechoviruses, suggesting an evolutionarily fixed role of the genome in virion assembly. Therefore, putative small molecules disrupting HPeV RNA-capsid protein interactions could be developed into antiviral inhibitors.
INTRODUCTION
Human parechoviruses (HPeVs) belong to the family Picornaviridae, which contains many vertebrate and human pathogens. Parechoviruses mainly cause mild gastrointestinal diseases in neonates and young children (1–4). Occasionally, however, parechovirus infections progress to serious and debilitating illnesses, including pneumonia, flaccid paralysis, encephalitis, sepsis, and meningitis (5–7). After their discovery (8, 9), HPeVs were placed in the same genus as human enteroviruses, exemplified by polioviruses and rhinoviruses (10). In the early 1990s, a separate genus, Parechovirus, was designated, which now includes HPeVs and Ljungan viruses (11). Parechoviruses exhibit high genetic variability, with at least 16 different types (12, 13). HPeV-1, HPeV-3, and HPeV-6 are the most prevalent clinically diagnosed types (14, 15).
HPeVs are small, nonenveloped icosahedral viruses. Their virions have an outer diameter of about 300 Å and contain a positive-sense single-stranded RNA (ssRNA) genome approximately 7,500 nucleotides long (16). HPeV capsids are built out of 60 copies of each of three viral proteins, VP0, VP1, and VP3. These three capsid proteins are co- and posttranslationally cleaved from a single polyprotein and constitute a protomer—the elementary building block of the capsid. In most picornaviruses, the capsid protein VP0 is cleaved into fragments VP2 and VP4 after virion assembly (17). The RNA genome has been proposed to have a role in cleavage (18). However, in parechoviruses, VP0 proteins remain intact, even in the mature virions (11, 19).
A distinctive feature of the capsid surface of many picornaviruses is a depression encircling the icosahedral 5-fold axes, called the “canyon” (20). For many picornaviruses, the canyon is the binding site of receptors with an immunoglobulin fold (21–23). Integrins αVβ3 and αVβ6 were proposed to be receptors for HPeVs that possess the integrin recognition sequence arginine-glycine-aspartate (RGD) in the C terminus of VP1, including HPEV-1, -2,- 4, -5, and -6 (24, 25). The binding of receptors within the canyon induces the release of a pocket factor from a hydrophobic pocket immediately below the surface of the canyon (26). The shape of the electron density and the hydrophobic environment of the pocket suggest that the pocket factor is a lipid (27–30). The expulsion of the pocket factor is associated with a decrease in virion stability and leads to genome release.
Genome release from picornavirus virions requires structural rearrangements of the capsid (31, 32). In enteroviruses, the capsid proteins change their positions relative to each other, resulting in the opening of pores at icosahedral 2-fold symmetry axes (33–35). VP1 N termini become exposed at the particle surface, and VP4s are released from the virion. Finally, the genomic RNA leaves the empty capsids. The standard in vitro procedure to induce the genome release of picornaviruses by heating to 56°C (34) results in disruption of the HPeV-1 virions to pentamers (36). Thus, the in vitro experiments provide putative evidence that the genome release mechanism of parechoviruses might be different from that of the other enteroviruses.
Pocket-binding antipicornavirus compounds have been developed that overstabilize the capsids and thus prevent genome release (30, 37–40). The capsid-binding inhibitors targeting rhinoviruses demonstrated a moderate level of success in human clinical trials (41, 42); however, they were not effective against parechovirus infections (43).
Single-particle cryo-electron microscopy reconstructions of HPeV-1 and its complex with the integrin receptor were reported previously at a resolution of 8.5 Å (25). However, despite the impact of HPeVs on human health, the atomic-level structural details of their virions are unknown. Here, we report the crystal structure of the virion of HPeV-1 (strain Harris) determined to a resolution of 3.1 Å. We show that specific interactions of the RNA genome with the capsid proteins result in a partial icosahedral ordering of the genome. This indicates a possible role of the genome in the stability or assembly of the HPeV-1 virion.
MATERIALS AND METHODS
Virus preparation.
Human parechovirus (strain Harris; ATTC VR-52) was propagated in A549 human lung carcinoma cells (ATCC CCL-185). For a typical preparation, fifty 140-cm2 tissue culture plates were infected with HPeV-1 at a multiplicity of infection (MOI) of 0.1 at 90% confluence, and the infection was allowed to proceed for 72 h at 37°C, at which point 90% of the cells exhibited cytopathic effects. The supernatant was harvested, and any remaining attached cells were removed from the plates using cell scrapers. The supernatant was centrifuged at 7,500 × g for 15 min, and the resulting pellet was resuspended in 10 ml of resuspension buffer (0.25 M HEPES-HCl, pH 7.5, 0.25 M NaCl). This fraction was subjected to three rounds of freeze-thawing by sequential transfer between −80°C and 37°C and homogenized with a Dounce tissue grinder to lyse the remaining cells. Cell debris was separated from the supernatant by low-speed centrifugation at 7,500 × g for 15 min. The resulting supernatant was combined with that obtained during the first low-speed centrifugation step. Viral particles were precipitated by adding polyethylene glycol (PEG) 8000 and NaCl to final concentrations of 15% (wt/vol) and 0.5 M, respectively, and incubating at 4°C with mild shaking (60 rpm) overnight. The following day, the solution was spun down at 10,000 × g for 20 min, and the visible white precipitate was resuspended in 12 ml of the resuspension buffer. MgCl2 was added to a final concentration of 5 mM, and the sample was subjected to trypsin (80 μg/ml), DNase (10 μg/ml), and RNase (10 μg/ml) treatment for 30 min at 22°C. EDTA (pH 9.5) was added to a final concentration of 15 mM, and a nonionic detergent, Nonidet P-40 (Sigma-Aldrich Inc.), was added to a final concentration of 1%. The solution was incubated for an additional 20 min at 22°C and centrifuged at 3,500 × g, and the supernatant was spun down through a 30% (wt/wt) sucrose cushion in 30 mM Tris-HCl, pH 8.0, 250 mM NaCl at 200,000 × g using a Ti50.2 rotor (Beckman Coulter). The pellet was resuspended in approximately 1 ml of cold resuspension buffer and added to 10 ml of 60% (wt/wt) CsCl solution in an ultracentrifuge tube. Gradient ultracentrifugation was allowed to proceed for at least 12 h at 100,000 × g in a SW40Ti rotor (Beckman Coulter). The opaque virus band was extracted with an 18-gauge needle on a 3-ml disposable syringe. The virus was transferred to the resuspension buffer by multiple rounds of centrifugation using a centrifugal-filter device with a 100-kDa molecular mass cutoff (Centricon, Millipore Inc.). The yield was approximately 100 μg of purified virus.
Crystallization, data collection, and data processing.
Purified HPeV-1 at a concentration of 3.5 mg/ml was subjected to sparse-matrix screening using a number of commercially available crystallization screens. The initial hits were obtained at room temperature using 0.1 M Tris-HCl, pH 8.0, 1 M ammonium sulfate in a sitting-drop format. The crystals measured about 30 μm in the largest dimension and diffracted X rays to a resolution of 3.1 Å in the Diamond Light Source beamline I23. In an effort to obtain larger crystals, optimization of the original crystallization conditions centered around testing various salts as precipitants and various divalent cations as additives. The best crystals were obtained in 0.1 M Tris-HCl, pH 8.0, 0.6 M ammonium sulfate, 0.1 M MgCl2, 5% (wt/vol) glycerol with growth at room temperature for about 3 weeks in a 2.0-μl hanging drop consisting of 0.5 μl reservoir solution and 1.5 μl of purified virus at a concentration of 3.5 mg/ml. Two independent data sets from two different crystals were collected in the Synchrotron Soleil Proxima 1 beamline and processed to a resolution of 3.1 Å using the XDS software package in space group P6322 (44). The two data sets were scaled together using the program Aimless from the CCP4 suite (45, 46).
Phasing, model building, and refinement.
HPeV-1 crystallized in space group P6322. Plots of the 2-fold, 3-fold, and 5-fold self-rotation function calculated using the program GLRF had shown that virions crystallized with one of the icosahedral 3-fold axes aligned with the crystallographic 3-fold axis and icosahedral 2-fold axes aligned with the crystallographic 2-fold axes (47). Reflections between 5 Å and 3.8 Å were used for the calculations. The radius of integration was set to 140 Å. Thus, icosahedral symmetry had to be rotated (φ = 90°, φ = 90°, κ = 60°) according to the XYK polar-angle convention relative to the standard icosahedral orientation as defined by Rossmann and Blow (48). One sixth of a virus particle occupied a crystallographic asymmetric unit. The only possible way to place the particle center in the crystallographic unit cell was at the intersection of the crystallographic 3-fold and 2-fold axes [x = a/2, y = (atg30°), z = c/4] (Table 1).
TABLE 1.
Diffraction data and structure quality indicators
| Parameter | Value |
|---|---|
| Space group | P6322 |
| Cell parameters | |
| a, b, c (Å) | 399.5, 399.5, 332.9 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution shella | 65.0–3.1 (3.15–3.10) |
| No. of observationsa | 1,187,478 (5,621) |
| No. of unique reflectionsa | 233,083 (4,306) |
| Observation multiplicitya | 5.4 (1.3) |
| Completenessa | 78.2 (31.8) |
| Rmerge (%)a,b | 0.354 (0.904) |
| I/σ(I)a | 5.0 (0.9) |
| Rfactora | 0.29 (0.41) |
| No. of protein atomsc | 5,283 |
| No. of RNA atomsc | 119 |
| Average B factor protein (Å2) | 50 |
| Average B factor RNA (Å2) | 71 |
| Ramachandran plot statistics | |
| Preferred regions (%)d | 90 |
| Allowed regions (%)d | 9.41 |
| Disallowed regions (%)d | 0.59 |
| RMSDe bond angles (°) | 0.005 |
| RMSD bond lengths (Å) | 1.16 |
Statistics for the highest-resolution shell are shown in parentheses.
Rmerge = ΣhΣj|lhj − <lh>|/ΣΣ|lhj|.
Statistics are given for one icosahedral asymmetric unit.
As calculated by Molprobity (79).
RMSD, root mean square deviation.
A Protein Data Bank [PDB] model of bovine enterovirus 1 (BeV1) (PDB code 1BEV) converted to polyalanine was used for molecular replacement (28). The model was rotated and positioned in the unit cell and used to calculate the initial phases for reflections up to a resolution of 10 Å in the program CNS (49, 50). The phases were refined by 15 cycles of real-space electron density averaging in the program AVE, using 10-fold noncrystallographic symmetry (NCS) (51). A mask for electron density averaging was generated using the program Mama from the Uppsala Software Factory program package by including all voxels within 5 Å of any atoms of the PDB model of the BeV1 icosahedral asymmetric unit (52). Phase extension was applied in order to obtain phases for higher-resolution reflections (53). The addition of a small fraction of higher-resolution data (one index along the a axis at a time) was followed by three cycles of averaging. This procedure was repeated until phases were obtained for all the reflections up to a resolution of 3.1 Å. The program SigmaA from the CCP4 package was used to include weights in the electron density calculations (45, 54). Inspection of the resulting electron density map indicated that the molecular-averaging mask was too large. Thus, a new mask was prepared based on a correlation map calculated by comparing electron density distributions among the 10 NCS-related icosahedral asymmetric units. The correlation map was calculated using the program Coma from Uppsala Software Factory (55). A cutoff value of 0.7 was used for the inclusion of voxels in the mask. The phase extension procedure was repeated using the new mask. The resulting map was of sufficient quality to enable model building. The program Buccaneer was used for automated building of the protein part of the model, utilizing the 10-fold NCS present in the crystallographic asymmetric unit (56, 57). The program Nautilus from the CCP4 suite (45) was used for the automated building of RNA chains and was able to build 3 nucleotides, while the remaining 3 nucleotides were built manually using the program Coot (58). The combined protein-RNA model from the automated building was about 80% complete with the assigned amino acid sequence. This initial model was subjected to manual rebuilding using the programs Coot and O (59) and to coordinate and B-factor refinement using the program CNS (49, 50). No water molecules were added, due to the limited resolution of the diffraction data. All the measured reflections were used in the refinement.
Data analysis.
The volumes of the particles were calculated using the programs Mama and Voidoo from Uppsala Software Factory (60). Average radii of virions were calculated using the program Moleman2 from Uppsala Software Factory (55). Multiple-sequence alignments were carried out using the ClustalW server (http://www.ebi.ac.uk/Tools/msa/clustalw2/) (61). Figures were generated using the programs UCSF Chimera (62), PyMOL (PyMOL Molecular Graphics System, version 1.7.4; Schrödinger, LLC), and RiverM (63). Structure-based pairwise alignments of biological protomers of various picornaviruses were prepared using the program Gr-Align (64). The similarity score provided by Gr-Align was used as an evolutionary distance to construct a nexus format matrix file, which was converted into the phylogenetic tree and visualized with the program SplitsTree (65).
Protein structure accession number.
The HPeV-1 model, structure factor amplitudes, and phases derived by phase extension have been deposited in the Protein Data Bank with PDB code 4Z92.
RESULTS AND DISCUSSION
Quality of diffraction data and HPeV-1 structure.
The structure of HPeV-1 was determined by X-ray crystallography at a resolution of 3.1 Å. The electron density map resulting from 10-fold NCS averaging enabled the HPeV-1 capsid proteins to be built, except for residues 1 to 31 and 289 of VP0, 1 to 24 and 217 to 235 of VP1, and 1 to 14 of VP3. Identifying the sequences of the individual residues was straightforward, due to the good quality of the electron density map. Six nucleotides corresponding to the RNA genome were modeled per icosahedral asymmetric unit. Water molecules could not be modeled because of the limited resolution of the available diffraction data. If calculated, the Rfree value would have been very close to the Rwork value due to the 10-fold NCS (66). Thus, all measured reflections were used in the crystallographic refinement. The basic crystallographic structure quality indicators are listed in Table 1.
Structures of HPeV-1 capsid proteins and virion.
The icosahedral asymmetric unit of HPeV-1 consists of subunits VP0, VP1, and VP3 (Fig. 1). The core of each of the capsid proteins is a jelly roll β-sandwich composed of two β-sheets, each containing four antiparallel β-strands. The β-strands are conventionally named B to I, and the two β-sheets contain strands BIDG and CHEF, respectively. The C termini of the capsid proteins are located on the virion surface, while the extended N termini mediate interactions among the capsid proteins and with the RNA genome on the inner surface of the capsid.
FIG 1.
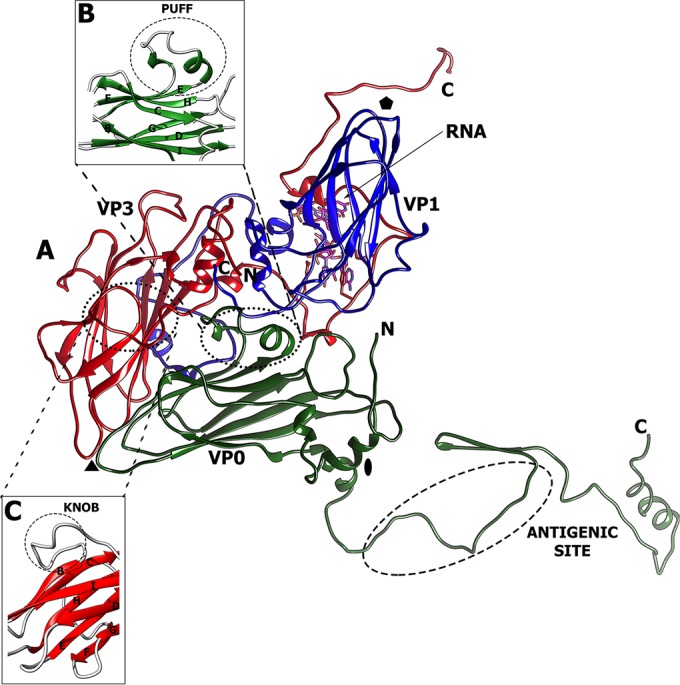
Icosahedral asymmetric unit of HPeV-1. (A) Cartoon representation of major capsid proteins: VP0 (green), VP1 (blue), and VP3 (red). The locations of the 5-fold, 3-fold, and 2-fold symmetry axes are indicated by a pentagon, a triangle, and an oval, respectively. (B and C) The puff (B) and knob (C) loops constituting the major capsid surface features.
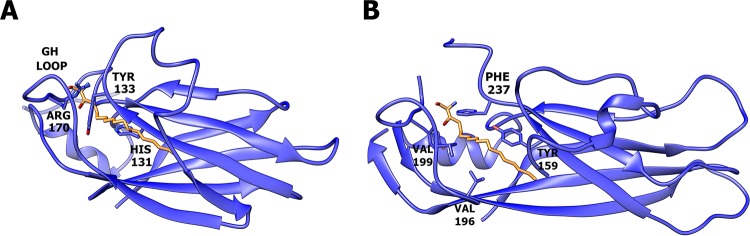
A pocket in the capsid protein VP1, which is present in most picornaviruses, can be targeted by capsid-binding inhibitors. This pocket is not formed in HPeV-1 (Fig. 2). Bulky amino acid side chains of His-131, Tyr-133, and Arg-170 fill the equivalent volume of the cavity, and the mouth of the pocket is occluded by the main-chain atoms of the GH loop (Fig. 2A). This explains the previous findings that the capsid-binding inhibitors that inhibit enteroviruses and rhinoviruses are not effective against parechoviruses (43).
FIG 2.
VP1 of HPeV-1 does not contain a hydrophobic pocket. The VP1 proteins of HPeV-1 (A) and poliovirus type 1 (B) are shown as cartoon representations. The pocket factor in poliovirus type 1 is shown as a stick model in orange. The side chains of residues that interact with the pocket factor are also shown as sticks. In panel A, the poliovirus type 1 pocket factor was superimposed onto the HPeV-1 structure. However, the pocket is not formed, and the side chains of several residues clash with the pocket factor.
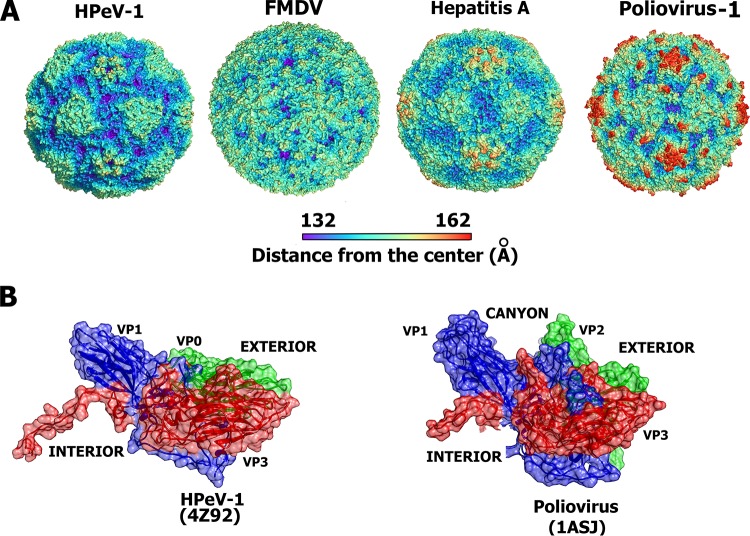
Of the picornaviruses that have been structurally characterized, HPeV-1 has the smallest observed virion, with an average diameter of 247 Å (Table 2). The surface of the HPeV-1 virion is relatively flat in comparison to the other picornaviruses because no canyon is formed (Fig. 3A). This is due to the relative shortening of features on the capsid surface that form the borders of the canyon. The HI loop of VP1, which forms protrusions around a 5-fold axis, is shortened from 12 residues in poliovirus type 1 (PDB code 1ASJ) to 6 residues in HPeV-1, resulting in a decrease in the height of the “northern rim” of the canyon. The “southern rim” of the canyon is mostly formed by two loops called the “puff” and the “knob.” The puff is a loop between βE and βF of VP0/VP2 and in HPeV-1 contains two short 3.10 helices connected by a 10-residue loop (Fig. 1B). The knob is a loop preceding βB of VP3 and in HPeV-1 contains 10 residues (Fig. 1C). The knob and puff of HPeV-1 are 2 and 65 residues shorter than those found in poliovirus type 1. In addition, the residues forming the puff and knob do not protrude away from the viral surface, as observed in poliovirus type 1 (Fig. 3B).
TABLE 2.
Physical parameters of selected picornaviruses
| Virusa/PDB code | Diameter (Å)b | Virion vol (106 Å3) | Genome size (nucleotides) | RNA density (Å3/nucleotide) |
|---|---|---|---|---|
| HPeV-1 | 247 | 6.88 | 7,321 | 10.6 |
| Poliovirus type 1/1ASJ | 256 | 8.70 | 7,433 | 8.6 |
| HRV-16/1AYM | 258 | 8.44 | 7,124 | 8.4 |
| BeV1/1BEV | 259 | 8.14 | 7,414 | 8.0 |
| FMDV/1BTT | 258 | 7.19 | 8,176 | 11.3 |
| HAV/4QPI | 254 | 7.45 | 7,478 | 10.0 |
HRV-16, human rhinovirus type 16.
Determined as the distance between the center of mass of the capsid protein protomer and the center of the virion.
FIG 3.
Comparison of virion surface features of selected picornaviruses. (A) Molecular surfaces of selected picornaviruses rainbow colored according to the distance from the particle center. (B) Comparison of side views of biological protomers of HPeV-1 and poliovirus type 1 showing that the canyon is not formed in the HPeV-1 structure. The subunits VP0 and VP2 are shown in green, VP1 in blue, and VP3 in red.
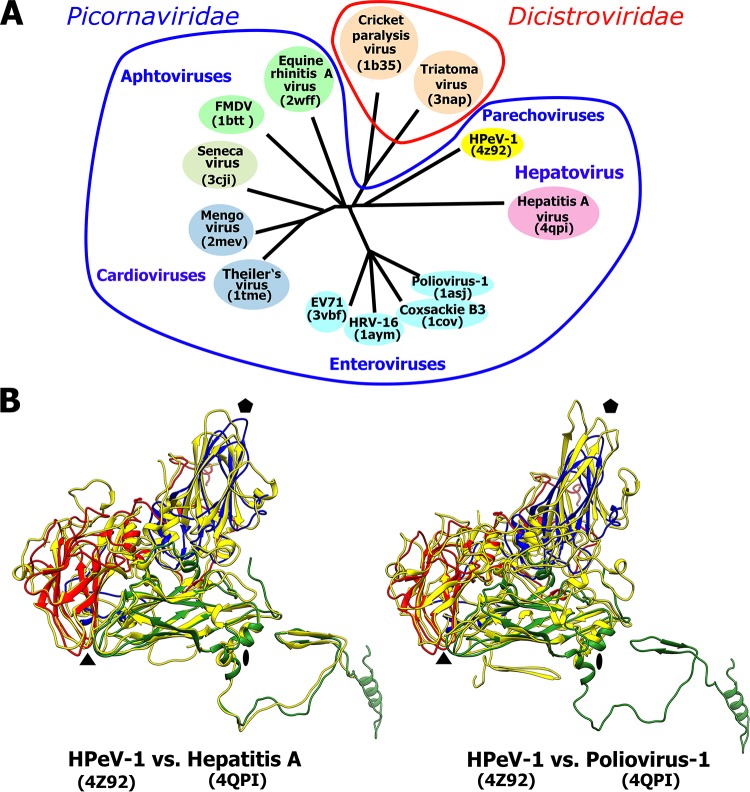
The availability of the HPeV-1 virion structure enabled the construction of a structure-based phylogenetic tree comparing HPeV-1 to 12 other viruses from the order Picornavirales (Fig. 4A). The phylogenetic analysis shows that the HPeV-1 capsid is most similar to that of hepatitis A virus (HAV) from the family Picornaviridae and to those of cricket paralysis virus and triatoma virus from the family Dicistroviridae (Fig. 4A). The relatively close evolutionary relationship between HPeV-1 and HAV is further indicated by the similar positions of the structured N-terminal arms of HPeV-1 VP0 and of HAV VP2 (Fig. 4B). The structured parts of the N termini of the two viruses mediate interactions among the capsid protein protomers along a line connecting the icosahedral 2-fold and 3-fold axes of the capsid (Fig. 4B). The N-terminal arm of VP2 of poliovirus type 1 has the same function; however, in poliovirus type 1, the structured part of the N-terminal arm of VP2 mediates interactions among protomers different than those in the virions of HPeV-1 and HAV (Fig. 4B). This is an example of domain swapping where part of a protein retains its function; however, its location in the quaternary complex is different. The closer similarity of the HPeV-1 capsid to those of viruses from the family Dicistroviridae than to those of viruses from the family Picornaviridae, with the exception of HAV, might be due to the differences in the processing of the polyprotein precursor of capsid proteins. The amino acid sequence of the VP4 subunit is located between VP2 and VP3 in viruses from the family Dicistroviridae while it is located before the VP2 sequence in viruses from the family Picornaviridae. In contrast, parechovirus capsids do not contain VP4, and this might be reflected in the structural organization of the capsid. Thus, HPeVs might represent an evolutionary link between picornaviruses and other virus families in the order Picornavirales, as was previously suggested for HAV (67).
FIG 4.
Evolutionary relationship among viruses from the families Picornaviridae and Dicistroviridae based on structural alignment of capsid proteins. (A) Phylogenetic tree based on structural similarity of icosahedral asymmetric units of the indicated viruses. For details on the construction of the diagram, see Materials and Methods. (B) Icosahedral asymmetric unit of HPeV-1 (VP0, green; VP1, blue; VP3, red) superimposed on those of HAV and poliovirus type 1 (all capsid proteins, yellow). Pentagons, triangles, and ovals indicate the positions of the icosahedral 5-fold, 3-fold, and 2-fold symmetry axes, respectively.
Integrin receptor binding site of HPeV-1.
The integrin αVβ6 is a cellular receptor for HPeV-1 (24, 25). The integrin binds to an integrin recognition motif, RGD, in the C terminus of VP1. However, the RGD residues are not visible in the HPeV-1 electron density map, probably due to the flexibility of the loop that contains the sequence. The last structured residue of VP1 is Thr-216, which is 6 residues before the integrin-binding sequence (Fig. 5). Seitsonen et al. used cryo-electron microscopy to calculate a three-dimensional reconstruction of the HPeV-1–integrin complex (25). Rigid-body fitting of the HPeV-1 virion into the map of the complex enabled us to estimate the location of the RGD motif to be approximately in the middle between the icosahedral 5-fold and 3-fold axes immediately above the last structured residue of VP1 (Fig. 5; see Fig. S1 in the supplemental material). In contrast to HPeV-1, the integrin-binding site in foot and mouth disease virus (FMDV) is located above the core of subunit VP2 in the GH loop of VP1 (Fig. 5) (68). In coxackievirus A9 (CV-A9), the integrin-binding site is next to the icosahedral 5-fold axis (Fig. 5) (69). Similar to HPeV-1, the RGD residues are not visible in the CV-A9 crystal structure (70). Thus, the location of the RGD sequence in the flexible part of the capsid protein might be required for binding to the integrin receptor. The distinct integrin-binding sites in HPeV1, FMDV, and CV-A9 indicate a convergent evolution in which the different viruses independently acquired the ability to utilize the integrins as receptors for cell entry.
FIG 5.
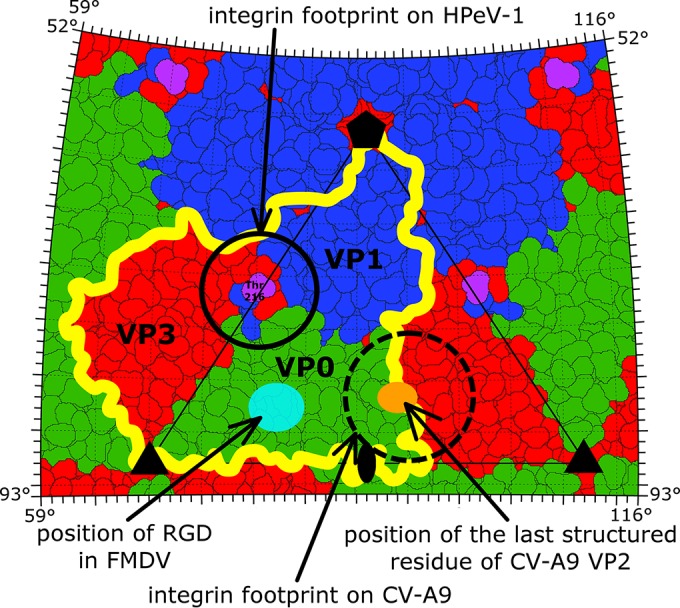
Comparison of αVβ6 integrin receptor binding sites of HPeV-1, FMDV, and CV-A9. Shown is a stereographic projection of HPeV-1 surface residues, with subunits VP0, VP1, and VP3 in green, blue, and red, respectively. The yellow line shows the border of a selected protomer. Features of FMDV and CV-A9 were plotted onto the HPeV-1 surface. The last structured C-terminal residue of HPeV-1 VP1 is highlighted in magenta, while that of CV-A9 is shown as an orange oval. The integrin-binding site in HPeV-1 as determined previously by cryo-electron microscopy is encircled by a solid line. The location of the footprint of the integrin receptor on CV-A9 (EMD-5512) is encircled by a dashed line. The conserved RGD motif in FMDV (PDB code 1FOD) is shown as a light-blue oval. The positions of the icosahedral-symmetry axes are indicated by a pentagon (5-fold), triangles (3-fold), and an oval (2-fold).
HPeV-1 virion-antibody interactions.
An intravenous immunoglobulin infusion containing large amounts of HPeV-1-neutralizing antibodies proved to be an efficient treatment for parechovirus-induced cardiomyopathy in an infant (3). One of the strongest antigenic sites of HPeV-1 consists of residues 82 to 94 in the N-terminal arm of VP0, which form an extended loop connecting β3 and α2 on the inside of the capsid (Fig. 6A) (71, 72). A possible explanation for the high immunogenicity of a sequence that is inside the capsid is the dynamic motions of virions referred to as “capsid breathing.” Even residues that are located on the inside of the capsid can be temporarily exposed on the virion surface and may be accessible to antibodies. Similar targeting of internal peptides by antibodies has been described in other picornaviruses (73, 74). In poliovirus type 1, antibodies can bind to residues 34 to 53 of VP1 located on the interior of the particle (Fig. 6B) (73, 75). The buried antigenic sites of poliovirus type 1 and HPeV-1 are located at the interpentamer boundary (Fig. 6B). The immunological reactivity of the buried epitopes suggests that they might be temporarily exposed on the capsid surface without disrupting the integrity of the virion.
FIG 6.
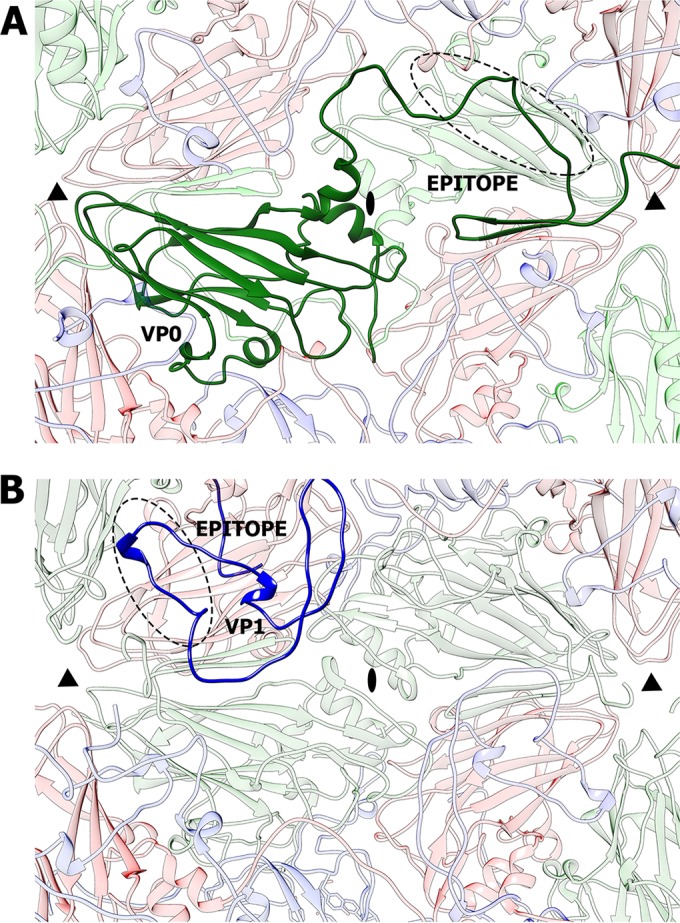
Comparison of internal immunogenic epitopes of HPeV-1 and poliovirus type 1 located at the interpentamer boundary. (A) Interior view of an HPeV-1 particle. A selected VP0 subunit is shown in dark green. Residues 82 to 94, targeted by the neutralizing antibodies, are highlighted with a dashed oval. (B) Interior view of a poliovirus type 1 particle. The N-terminal arm of a selected VP1 subunit is shown in dark blue. The immunogenic epitope, consisting of residues 33 to 54 of VP1, is highlighted with a dashed oval. The positions of the 2-fold and 3-fold icosahedral-symmetry axes are indicated by ovals and triangles, respectively.
Role of N termini of VP0 subunits in mediating interpentamer contacts within the HPeV-1 capsid.
The N-terminal part of HPeV-1 VP0 corresponds to the residues of VP4 and of the N terminus of VP2 in enteroviruses, according to the positions of the amino acids in the polyprotein precursors of the capsid proteins. However, the locations of the structured residues of the N terminus of VP0 in the HPeV-1 capsid are different from those of VP2 and VP4 in poliovirus type 1 and other enteroviruses while they are similar to those of HAV (Fig. 4). In HPeV-1, the structured N-terminal part of VP0 forms a tentacle-like extension that interacts with capsid proteins from two neighboring pentamers (Fig. 7A). Residues from the VP0 N terminus following helix α1 form stabilizing interactions with other parts of the capsid, including the following: (i) residues between β3 and α2 of VP0 form hydrogen bonds to Ser-67 and Arg-71 of VP3 and Asp-38 of VP1 subunits in the neighboring pentamer (Fig. 7B) and (ii) β1 of VP0 forms a two-stranded antiparallel β-sheet with β2 of VP0 in another adjacent pentamer (Fig. 7C). The N-terminal part of parechovirus VP0 might be homologous to VP4 of enteroviruses, which has to be released from the virion in order to ensure delivery of the virus genome across the endosomal membrane into the cytoplasm. Therefore, upon cell entry, the N terminus of VP0 might be required for interaction with the host cell membranes. Since the VP0 N termini are involved in interpentamer interactions, alteration of their structure leads to capsid disruption. Thus, the role of the VP0 N-terminal arm in mediating interactions among the pentamers within the HPeV-1 capsid might explain why the parechovirus capsids disassemble into pentamers after genome release in vitro (36). The first N-terminal residues of VP0 that are visible in the HPeV-1 electron density map form helix α1, which contains predominantly hydrophobic amino acids. The side chains of residues from helix α1 interact with the polar side chains of the residues from βG of subunit VP0 in the adjacent pentamer (Fig. 7D). The unfavorable nature of α1-βG interactions indicates that they are not required for capsid stability. The average B factor of α1 atoms is 51 Å2, while it is 40 Å2 for the whole capsid. The elevated B factor shows that the helix is more mobile and might be ready for exposure at the virion surface and interaction with membranes. However, it is possible that the part of the VP0 N terminus that is not visible in the structure (the first 30 residues) is responsible for interaction with the host membranes. In that case, the disruption of the HPeV-1 capsids into pentamers would not be required to ensure transfer of the ssRNA genome across the biological membrane.
FIG 7.
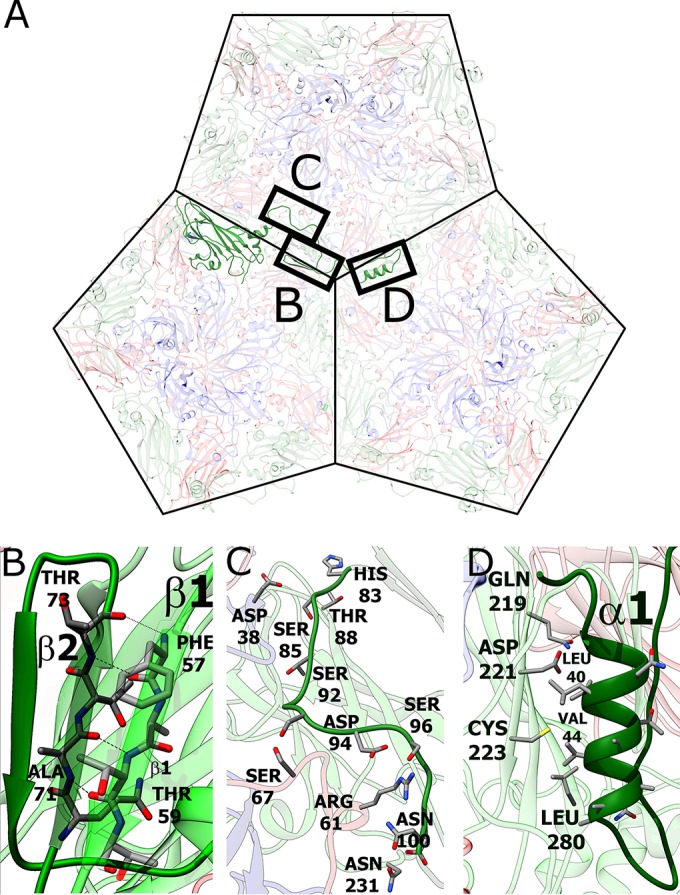
Role of the N-terminal arm of VP0 in interpentamer interactions. (A) The N-terminal arm of VP0 mediates contacts among three pentamers in the HPeV-1 virion. Shown is a view from inside the virion. (B) Interactions between antiparallel β-strands β1 and β2 of VP0 proteins from two adjacent pentamers. (C) Residues 83 to 100 of VP1 interact with residues from all three subunits, VP0, VP1, and VP3. (D) Interactions of α1 with βG of VP0 from another, neighboring pentamer.
The putative role of the HPeV-1 genome in HPeV-1 virion stability and assembly.
The HPeV-1 virion has an inner volume of 6.9 × 106 Å3 (Table 2). This is 20% smaller than the interior particle volumes of both poliovirus type 1 and rhinovirus type 16. However, the size of the genome of HPeV-1 is similar to that of poliovirus type 1, leading to a higher RNA density than in enteroviruses but similar to that in FMDV and HAV (Table 2). Furthermore, the internal surface of the HPeV-1 capsid around the 5-fold axes is more positively charged than the corresponding areas in other picornaviruses (see Fig. S2 in the supplemental material). This is probably required to neutralize the negative charge of the tightly packed RNA.
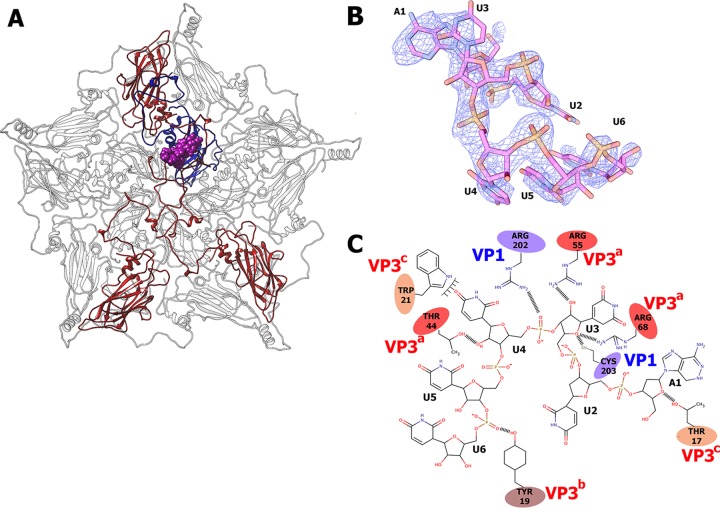
An electron density corresponding to an RNA hexanucleotide associated with each icosahedral asymmetric unit is located close to the 5-fold icosahedral axis on the inside of the HPeV-1 capsid (Fig. 8). The HPeV-1 genome is a linear RNA molecule that forms unique interactions with the inner surface of the capsid. However, the RNA does not affect the packing of particles within the crystal or the measured diffraction data, and therefore, it contains information about the icosahedrally averaged RNA structure. Thus, the observed RNA density corresponds to an averaged nucleotide sequence. The shapes of the electron densities of the individual bases indicate that the first nucleotide is a purine, while the following 5 nucleotides are pyrimidines. The RNA was modeled as an adenosine followed by five uridines (Fig. 8B). The average B factor of the RNA is 71 Å2, while that of the protein part of the capsid is 40 Å2, indicating higher mobility of the RNA. A crystallographic refinement resulted in 94% occupancy of the RNA, showing that the RNA is bound to nearly all the available positions in the icosahedral capsid. Each hexanucleotide forms extensive interactions with one VP1 and three VP3 subunits belonging to different protomers from one pentamer (Fig. 8). Overall, the 60 copies of the ordered hexanucleotides represent 5% of the 7,500-nucleotide genome.
FIG 8.
Interactions of HPeV-1 genomic RNA with the capsid. (A) Location of an RNA hexanucleotide (shown as a space-filling model in magenta) within the pentamer of capsid protein protomers as seen from inside the virion. Subunits in contact with RNA are shown in color (VP1, blue, and VP3s, red). (B) Electron density map of RNA hexanucleotide contoured at 2σ. (C) Two-dimensional representation of RNA-protein interactions. Hydrophobic interactions and hydrogen bonds are shown. VP3 subunits from different icosahedral asymmetric units are distinguished by the label colors and superscript a, b, and c.
The HPeV-1 virion contains more ordered RNA than is seen in other picornaviruses, in which only 1 or 2 nucleotides are observed (27, 76, 77). For example, in enteroviruses, a nucleotide base makes stacking interactions with a conserved tryptophan residue of VP1 located close to the icosahedral 2-fold axis (76). However, the partly ordered genomic RNA has been previously observed in a bean pod mottle virus (BPMV) from the family Secoviridae of the order Picornavirales (78). In BPMV, the RNA interacts with the capsid proteins close to the icosahedral 3-fold axis, while in HPeV-1, the RNA binds close to the 5-fold axis (see Fig. S3 in the supplemental material). The differences in the RNA-protein interactions between the two viruses indicate that the two viruses might differ in the structures of their packaged genomes. It is likely that the genome organization in HPeV-1 virions is also different from that in enteroviruses, which may be due to the necessity to package the genome with a higher density. The complete conservation of the RNA binding residues among HPeVs, together with the nearly complete occupancy of the RNA, indicates that the binding might have a role in virion assembly and perhaps also in ensuring the selective packaging of HPeV genomes into capsids. Small molecules that interfere with the genomic-RNA–capsid protein interactions could therefore be developed into antiviral compounds preventing HPeV virion assembly.
Supplementary Material
ACKNOWLEDGMENTS
We thank Charles Sabin and Synchrotron Soleil Proxima 1/Proxima 2A and beamline scientists Martin Slavko and Beatriz Guimaraes for their help with crystal screening and data collection. We thank the synchrotron Diamond beamline scientists for their help with preliminary crystal screening and data collection. We also thank Noel Malod-Dognin and Daniel Lundin for their help with structure-based phylogenetic analysis.
The research leading to these results received funding from the European Research Council under the European Union's Seventh Framework Programme, (FP/2007-2013)/ERC grant agreement no. 355855′, and from EMBO installation grant no. 3041 to Pavel Plevka.
L.P. optimized and conducted large-scale virus purifications. S.K. performed crystallization, model building, refinement, and data analysis and participated in manuscript preparation. P.P. designed the study, performed structure determination, directed and participated in data analysis, and wrote the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02346-15.
REFERENCES
- 1.Boivin G, Abed Y, Boucher FD. 2005. Human parechovirus 3 and neonatal infections. Emerg Infect Dis 11:103–105. doi: 10.3201/eid1101.040606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuffenecker I, Javouhey E, Gillet Y, Kugener B, Billaud G, Floret D, Lina B, Morfin F. 2012. Human parechovirus infections, Lyon, France, 2008-10: evidence for severe cases. J Clin Virol 54:337–341. doi: 10.1016/j.jcv.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 3.Wildenbeest JG, Wolthers KC, Straver B, Pajkrt D. 2013. Successful IVIG treatment of human parechovirus-associated dilated cardiomyopathy in an infant. Pediatrics 132:e243–e247. doi: 10.1542/peds.2012-1136. [DOI] [PubMed] [Google Scholar]
- 4.Stanway G, Joki-Korpela P, Hyypia T. 2000. Human parechoviruses—biology and clinical significance. Rev Med Virol 10:57–69. [DOI] [PubMed] [Google Scholar]
- 5.Esposito S, Rahamat-Langendoen J, Ascolese B, Senatore L, Castellazzi L, Niesters HG. 2014. Pediatric parechovirus infections. J Clin Virol 60:84–89. doi: 10.1016/j.jcv.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Verboon-Maciolek MA, Groenendaal F, Hahn CD, Hellmann J, van Loon AM, Boivin G, de Vries LS. 2008. Human parechovirus causes encephalitis with white matter injury in neonates. Ann Neurol 64:266–273. doi: 10.1002/ana.21445. [DOI] [PubMed] [Google Scholar]
- 7.Levorson RE, Jantausch BA, Wiedermann BL, Spiegel HM, Campos JM. 2009. Human parechovirus-3 infection: emerging pathogen in neonatal sepsis. Pediatr Infect Dis J 28:545–547. doi: 10.1097/INF.0b013e318194596a. [DOI] [PubMed] [Google Scholar]
- 8.Wigand R, Sabin AB. 1961. Properties of ECHO types 22, 23 and 24 viruses. Arch Gesamte Virusforsch 11:224–247. doi: 10.1007/BF01241688. [DOI] [PubMed] [Google Scholar]
- 9.Anonymous. 1955. ENTERIC cytopathogenic human orphan (ECHO) viruses. Science 122:1187–1188. doi: 10.1126/science.122.3181.1187. [DOI] [PubMed] [Google Scholar]
- 10.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed). 2012. Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, Waltham, MA. [Google Scholar]
- 11.Stanway G, Kalkkinen N, Roivainen M, Ghazi F, Khan M, Smyth M, Meurman O, Hyypia T. 1994. Molecular and biological characteristics of echovirus 22, a representative of a new picornavirus group. J Virol 68:8232–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun G, Wang Y, Tao G, Shen Q, Cao W, Chang X, Zhang W, Shao C, Yi M, Shao S, Yang Y. 2012. Complete genome sequence of a novel type of human parechovirus strain reveals natural recombination events. J Virol 86:8892–8893. doi: 10.1128/JVI.01241-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams CH, Panayiotou M, Girling GD, Peard CI, Oikarinen S, Hyoty H, Stanway G. 2009. Evolution and conservation in human parechovirus genomes. J Gen Virol 90:1702–1712. doi: 10.1099/vir.0.008813-0. [DOI] [PubMed] [Google Scholar]
- 14.Sedmak G, Nix WA, Jentzen J, Haupt TE, Davis JP, Bhattacharyya S, Pallansch MA, Oberste MS. 2010. Infant deaths associated with human parechovirus infection in Wisconsin. Clin Infect Dis 50:357–361. doi: 10.1086/649863. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Victoria J, Kapoor A, Naeem A, Shaukat S, Sharif S, Alam MM, Angez M, Zaidi SZ, Delwart E. 2009. Genomic characterization of novel human parechovirus type. Emerg Infect Dis 15:288–291. doi: 10.3201/eid1502.080341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyypia T, Horsnell C, Maaronen M, Khan M, Kalkkinen N, Auvinen P, Kinnunen L, Stanway G. 1992. A distinct picornavirus group identified by sequence analysis. Proc Natl Acad Sci U S A 89:8847–8851. doi: 10.1073/pnas.89.18.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnold E, Luo M, Vriend G, Rossmann MG, Palmenberg AC, Parks GD, Nicklin MJ, Wimmer E. 1987. Implications of the picornavirus capsid structure for polyprotein processing. Proc Natl Acad Sci U S A 84:21–25. doi: 10.1073/pnas.84.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basavappa R, Syed R, Flore O, Icenogle JP, Filman DJ, Hogle JM. 1994. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 A resolution. Protein Sci 3:1651–1669. doi: 10.1002/pro.5560031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanway G, Hyypia T. 1999. Parechoviruses. J Virol 73:5249–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossmann MG. 1989. The canyon hypothesis. Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J Biol Chem 264:14587–14590. [PubMed] [Google Scholar]
- 21.He Y, Chipman PR, Howitt J, Bator CM, Whitt MA, Baker TS, Kuhn RJ, Anderson CW, Freimuth P, Rossmann MG. 2001. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat Struct Biol 8:874–878. doi: 10.1038/nsb1001-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao C, Bator CM, Bowman VD, Rieder E, He Y, Hebert B, Bella J, Baker TS, Wimmer E, Kuhn RJ, Rossmann MG. 2001. Interaction of coxsackievirus A21 with its cellular receptor, ICAM-1. J Virol 75:2444–2451. doi: 10.1128/JVI.75.5.2444-2451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson NH, Kolatkar PR, Oliveira MA, Cheng RH, Greve JM, McClelland A, Baker TS, Rossmann MG. 1993. Structure of a human rhinovirus complexed with its receptor molecule. Proc Natl Acad Sci U S A 90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boonyakiat Y, Hughes PJ, Ghazi F, Stanway G. 2001. Arginine-glycine-aspartic acid motif is critical for human parechovirus 1 entry. J Virol 75:10000–10004. doi: 10.1128/JVI.75.20.10000-10004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seitsonen J, Susi P, Heikkila O, Sinkovits RS, Laurinmaki P, Hyypia T, Butcher SJ. 2010. Interaction of alphaVbeta3 and alphaVbeta6 integrins with human parechovirus 1. J Virol 84:8509–8519. doi: 10.1128/JVI.02176-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossmann MG, He Y, Kuhn RJ. 2002. Picornavirus-receptor interactions. Trends Microbiol 10:324–331. doi: 10.1016/S0966-842X(02)02383-1. [DOI] [PubMed] [Google Scholar]
- 27.Filman DJ, Syed R, Chow M, Macadam AJ, Minor PD, Hogle JM. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J 8:1567–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smyth M, Tate J, Hoey E, Lyons C, Martin S, Stuart D. 1995. Implications for viral uncoating from the structure of bovine enterovirus. Nat Struct Biol 2:224–231. doi: 10.1038/nsb0395-224. [DOI] [PubMed] [Google Scholar]
- 29.Smyth M, Pettitt T, Symonds A, Martin J. 2003. Identification of the pocket factors in a picornavirus. Arch Virol 148:1225–1233. doi: 10.1007/s00705-002-0974-4. [DOI] [PubMed] [Google Scholar]
- 30.Plevka P, Perera R, Yap ML, Cardosa J, Kuhn RJ, Rossmann MG. 2013. Structure of human enterovirus 71 in complex with a capsid-binding inhibitor. Proc Natl Acad Sci U S A 110:5463–5467. doi: 10.1073/pnas.1222379110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakker SE, Groppelli E, Pearson AR, Stockley PG, Rowlands DJ, Ranson NA. 2014. Limits of structural plasticity in a picornavirus capsid revealed by a massively expanded equine rhinitis A virus particle. J Virol 88:6093–6099. doi: 10.1128/JVI.01979-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuchs R, Blaas D. 2012. Productive entry pathways of human rhinoviruses. Adv Virol 2012:826301. doi: 10.1155/2012/826301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garriga D, Pickl-Herk A, Luque D, Wruss J, Caston JR, Blaas D, Verdaguer N. 2012. Insights into minor group rhinovirus uncoating: the X-ray structure of the HRV2 empty capsid. PLoS Pathog 8:e1002473. doi: 10.1371/journal.ppat.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy HC, Bostina M, Filman DJ, Hogle JM. 2010. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J Virol 84:4426–4441. doi: 10.1128/JVI.02393-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bostina M, Levy H, Filman DJ, Hogle JM. 2011. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J Virol 85:776–783. doi: 10.1128/JVI.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shakeel S. 2014. Human picornaviruses: uncoating, assembly and interaction with cellular receptors. University of Helsinki, Helsinki, Finland. [Google Scholar]
- 37.De Palma AM, Vliegen I, De Clercq E, Neyts J. 2008. Selective inhibitors of picornavirus replication. Med Res Rev 28:823–884. doi: 10.1002/med.20125. [DOI] [PubMed] [Google Scholar]
- 38.Thibaut HJ, De Palma AM, Neyts J. 2012. Combating enterovirus replication: state-of-the-art on antiviral research. Biochem Pharmacol 83:185–192. doi: 10.1016/j.bcp.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 39.Smith TJ, Kremer MJ, Luo M, Vriend G, Arnold E, Kamer G, Rossmann MG, McKinlay MA, Diana GD, Otto MJ. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286–1293. doi: 10.1126/science.3018924. [DOI] [PubMed] [Google Scholar]
- 40.Grant RA, Hiremath CN, Filman DJ, Syed R, Andries K, Hogle JM. 1994. Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design. Curr Biol 4:784–797. [DOI] [PubMed] [Google Scholar]
- 41.Rotbart HA. 2000. Antiviral therapy for enteroviruses and rhinoviruses. Antivir Chem Chemother 11:261–271. doi: 10.1177/095632020001100402. [DOI] [PubMed] [Google Scholar]
- 42.Rotbart HA, Webster AD, Pleconaril Treatment Registry Group . 2001. Treatment of potentially life-threatening enterovirus infections with pleconaril. Clin Infect Dis 32:228–235. doi: 10.1086/318452. [DOI] [PubMed] [Google Scholar]
- 43.van de Ven AA, Douma JW, Rademaker C, van Loon AM, Wensing AM, Boelens JJ, Sanders EA, van Montfrans JM. 2011. Pleconaril-resistant chronic parechovirus-associated enteropathy in agammaglobulinaemia. Antivir Ther 16:611–614. doi: 10.3851/IMP1792. [DOI] [PubMed] [Google Scholar]
- 44.Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 47.Tong L, Rossmann MG. 1997. Rotation function calculations with GLRF program. Methods Enzymol 276:594–611. doi: 10.1016/S0076-6879(97)76080-4. [DOI] [PubMed] [Google Scholar]
- 48.Rossmann MG, Blow DM. 1962. Detection of sub-units within crystallographic asymmetric unit. Acta Crystallogr 15:24–31. doi: 10.1107/S0365110X62000067. [DOI] [Google Scholar]
- 49.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54:905–921. [DOI] [PubMed] [Google Scholar]
- 50.Brunger AT. 2007. Version 1.2 of the crystallography and NMR system. Nat Protoc 2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 51.Kleywegt GJ, Read RJ. 1997. Not your average density. Structure 5:1557–1569. doi: 10.1016/S0969-2126(97)00305-5. [DOI] [PubMed] [Google Scholar]
- 52.Kleywegt GJ, Jones TA. 1999. Software for handling macromolecular envelopes. Acta Crystallogr D Biol Crystallogr 55:941–944. doi: 10.1107/S0907444999001031. [DOI] [PubMed] [Google Scholar]
- 53.Rossmann MG. 1995. Ab initio phase determination and phase extension using non-crystallographic symmetry. Curr Opin Struct Biol 5:650–655. doi: 10.1016/0959-440X(95)80058-1. [DOI] [PubMed] [Google Scholar]
- 54.Read RJ. 1986. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr A 42:140–149. doi: 10.1107/S0108767386099622. [DOI] [Google Scholar]
- 55.Kleywegt GJ. 1999. Experimental assessment of differences between related protein crystal structures. Acta Crystallogr D Biol Crystallogr 55:1878–1884. doi: 10.1107/S0907444999010495. [DOI] [PubMed] [Google Scholar]
- 56.Cowtan K. 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 57.Cowtan K. 2008. Fitting molecular fragments into electron density. Acta Crystallogr D Biol Crystallogr 64:83–89. doi: 10.1107/S0907444907033938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47:110–119. doi: 10.1107/S0108767390010224. [DOI] [PubMed] [Google Scholar]
- 60.Kleywegt GJ, Jones TA. 1994. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr 50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 61.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 63.Xiao C, Rossmann MG. 2007. Interpretation of electron density with stereographic roadmap projections. J Struct Biol 158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malod-Dognin N, Przulj N. 2014. GR-Align: fast and flexible alignment of protein 3D structures using graphlet degree similarity. Bioinformatics 30:1259–1265. doi: 10.1093/bioinformatics/btu020. [DOI] [PubMed] [Google Scholar]
- 65.Huson DH. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 66.Kleywegt GJ, Brunger AT. 1996. Checking your imagination: applications of the free R value. Structure 4:897–904. doi: 10.1016/S0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- 67.Wang X, Ren J, Gao Q, Hu Z, Sun Y, Li X, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. 2015. Hepatitis A virus and the origins of picornaviruses. Nature 517:85–88. doi: 10.1038/nature13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Logan D, Abu-Ghazaleh R, Blakemore W, Curry S, Jackson T, King A, Lea S, Lewis R, Newman J, Parry N, Rowlands D, Stuart D, Fry E. 1993. Structure of a major immunogenic site on foot-and-mouth disease virus. Nature 362:566–568. doi: 10.1038/362566a0. [DOI] [PubMed] [Google Scholar]
- 69.Shakeel S, Seitsonen JJ, Kajander T, Laurinmaki P, Hyypia T, Susi P, Butcher SJ. 2013. Structural and functional analysis of coxsackievirus A9 integrin alphavbeta6 binding and uncoating. J Virol 87:3943–3951. doi: 10.1128/JVI.02989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hendry E, Hatanaka H, Fry E, Smyth M, Tate J, Stanway G, Santti J, Maaronen M, Hyypia T, Stuart D. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7:1527–1538. doi: 10.1016/S0969-2126(00)88343-4. [DOI] [PubMed] [Google Scholar]
- 71.Joki-Korpela P, Roivainen M, Lankinen H, Poyry T, Hyypia T. 2000. Antigenic properties of human parechovirus 1. J Gen Virol 81:1709–1718. doi: 10.1099/0022-1317-81-7-1709. [DOI] [PubMed] [Google Scholar]
- 72.Alho A, Marttila J, Ilonen J, Hyypia T. 2003. Diagnostic potential of parechovirus capsid proteins. J Clin Microbiol 41:2294–2299. doi: 10.1128/JCM.41.6.2294-2299.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roivainen M, Piirainen L, Rysa T, Narvanen A, Hovi T. 1993. An immunodominant N-terminal region of VP1 protein of poliovirion that is buried in crystal structure can be exposed in solution. Virology 195:762–765. doi: 10.1006/viro.1993.1427. [DOI] [PubMed] [Google Scholar]
- 74.Park KB, Lim BK, Ye MB, Chung SY, Nam JH. 2012. A peptide vaccine based on a B-cell epitope on the VP1 protein of enterovirus 70 induces a strong antibody response. Acta Virol 56:337–342. doi: 10.4149/av_2012_04_337. [DOI] [PubMed] [Google Scholar]
- 75.Lin J, Lee LY, Roivainen M, Filman DJ, Hogle JM, Belnap DM. 2012. Structure of the Fab-labeled “breathing” state of native poliovirus. J Virol 86:5959–5962. doi: 10.1128/JVI.05990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hadfield AT, Lee W, Zhao R, Oliveira MA, Minor I, Rueckert RR, Rossmann MG. 1997. The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle. Structure 5:427–441. doi: 10.1016/S0969-2126(97)00199-8. [DOI] [PubMed] [Google Scholar]
- 77.Arnold E, Rossmann MG. 1990. Analysis of the structure of a common cold virus, human rhinovirus 14, refined at a resolution of 3.0 A. J Mol Biol 211:763–801. doi: 10.1016/0022-2836(90)90076-X. [DOI] [PubMed] [Google Scholar]
- 78.Chen ZG, Stauffacher C, Li Y, Schmidt T, Bomu W, Kamer G, Shanks M, Lomonossoff G, Johnson JE. 1989. Protein-RNA interactions in an icosahedral virus at 3.0 A resolution. Science 245:154–159. doi: 10.1126/science.2749253. [DOI] [PubMed] [Google Scholar]
- 79.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.