ABSTRACT
Hepatitis C virus (HCV) is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma in humans. We showed previously that HCV induces autophagy for viral persistence by preventing the innate immune response. Knockdown of autophagy reduces extracellular HCV release, although the precise mechanism remains unknown. In this study, we observed that knockdown of autophagy genes enhances intracellular HCV RNA and accumulates infectious virus particles in cells. Since HCV release is linked with the exosomal pathway, we examined whether autophagy proteins associate with exosomes in HCV-infected cells. We observed an association between HCV and the exosomal marker CD63 in autophagy knockdown cells. Subsequently, we observed that levels of extracellular infectious HCV were significantly lower in exosomes released from autophagy knockdown cells. To understand the mechanism for reduced extracellular infectious HCV in the exosome, we observed that an interferon (IFN)-stimulated BST-2 gene is upregulated in autophagy knockdown cells and associated with the exosome marker CD63, which may inhibit HCV assembly or release. Taken together, our results suggest a novel mechanism involving autophagy and exosome-mediated HCV release from infected hepatocytes.
IMPORTANCE Autophagy plays an important role in HCV pathogenesis. Autophagy suppresses the innate immune response and promotes survival of virus-infected hepatocytes. The present study examined the role of autophagy in secretion of infectious HCV from hepatocytes. Autophagy promoted HCV trafficking from late endosomes to lysosomes, thus providing a link with the exosome. Inhibition of HCV-induced autophagy could be used as a strategy to block exosome-mediated virus transmission.
INTRODUCTION
Hepatitis C virus (HCV) establishes chronic infection in more than 70% of infected individuals worldwide, and over 170 million people are currently infected with HCV. Persistent HCV infection is associated with a chronic inflammatory disease that ultimately leads to hepatic fibrosis, cirrhosis, and hepatocellular carcinoma (HCC). HCV is a member of the genus Hepacivirus, which belongs to the family Flaviviridae. The virus is enveloped, and the single-stranded positive-sense RNA genome contains an open reading frame flanked by untranslated regions (1). Translation of the single open reading frame is driven by an internal ribosomal entry site (IRES) sequence present within the 5′ untranslated region (UTR), resulting in synthesis of a polyprotein approximately 3,000 amino acids in length. This polyprotein is processed by cellular and viral proteases into its individual viral proteins. The nonstructural proteins NS3, NS4A, NS4B, NS5A, and NS5B are sufficient to support efficient HCV RNA replication in the membranous compartments of the cytosol (1, 2). In infected individuals, HCV particles circulate as low-density lipoprotein (LDL)–virus complexes that are rich in triglycerides and contain HCV RNA, core protein, and apolipoproteins B and E (apoB and apoE), which are components of the beta-lipoproteins (very-low-density lipoproteins [VLDL] and LDL). The precise mechanisms of HCV assembly, budding, and release are currently under investigation.
There are different mechanisms by which cells release large biomolecules into the extracellular space, with the most common process being exocytosis (3). Exosomes are extracellular vesicles secreted upon fusion of endosomal multivesicular bodies (MVBs) with the plasma membrane (4). Endosomal sorting complex required for transport (ESCRT) proteins are involved in the generation of secretory MVBs. The ESCRT-0 component hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) and the ESCRT-I protein TSG101 are involved early in cargo recruitment. VPS4B is an ATPase that provides energy for the dissolution of ESCRT-III complexes at membrane fission, whereas apoptosis-linked gene 2-interacting protein X (ALIX) contributes to exosome biogenesis (5, 6). MVBs can either fuse with lysosomes to release vesicle contents for degradation or fuse with the plasma membrane to release extracellular exosomes (7). Virus budding is usually coupled tightly to virion assembly, and most viruses, therefore, use their structural proteins to recruit the ESCRT pathway. Enveloped viruses commonly assemble and release at the plasma membrane, although some are released into internal compartments (8, 9). In the latter case, the internal compartment must ultimately fuse with the plasma membrane to release the virus from the cell (8, 9).
The role of exosomes in HCV infection was first demonstrated by the presence of viral RNA in exosomes isolated from plasma of HCV-infected patients (10). The components required for MVB biogenesis, Vps4 and the ESCRT-III complex, are required for release of infectious HCV particles (11). Subsequently, an ESCRT-0 component and Hrs were reported to be involved in HCV release through the exosomal pathway (12). Further, the functional role of hepatocyte-derived exosomes in carrying viral RNA and transferring it to plasmacytoid dendritic cells was suggested for the activation and production of alpha interferon (IFN-α) (13). Exosome-mediated transmission of HCV also establishes productive infection in hepatocytes (14). Apart from HCV, the existence of infectious hepatitis A virus particles within extracellular vesicles has been observed and shown to be dependent on ESCRT-III-associated proteins, ALIX, and Vps4B of the MVB pathway (15). An interaction of ALIX with the ESCRT-III protein CHMP4B has been reported to facilitate the budding of human immunodeficiency virus (16).
The autophagy pathway, an evolutionarily conserved membrane system, is known to engulf damaged organelles and maintains cellular homeostasis through the formation of double-membrane vesicles called autophagosomes (17, 18). Closed autophagosomes undergo a maturation process as they subsequently fuse with endosomes and lysosomes. Autophagosomes fuse with both early and late endosomes to form amphisomes prior to the lysosomal fusion step. The autophagosomal system extensively overlaps the endosomal system. The endosomal system provides membrane components for autophagosome biogenesis, and the amphisomes acquire machinery required for lysosomal fusion following the fusion of nascent autophagosomes with endosomes. Host cellular molecules, such as LC3, p62, and Rab7, are the substrates of autophagic degradation and may partly reflect the rate of endolysosomal degradation. LC3-I undergoes a posttranslational modification to form lipidated LC3-II, an autophagosomal marker. p62 (SQTSM1), is an adaptor of LC3-II and serves as a substrate for autophagic degradation. Rab7, a small GTPase, contributes to the progression of the autophagy pathway and regulates trafficking of MVBs to lysosomes. We have shown previously that inhibition of autophagy machinery decreases the production of infectious virus in the extracellular medium. Induction of autophagy is required to sustain the survival of virus-infected cells, a crucial aspect of virus-mediated chronic infection (19, 20).
In the present study, we examined the role of autophagy in the release of infectious HCV from infected cells. We observed that knockdown of autophagy protein, Beclin1 or ATG7, inhibits the release of infectious HCV particles into the extracellular medium by accumulating the virus in infected cells. Our study also suggested that autophagy intersects with the exosomal pathway to help the exosome-mediated HCV release process.
MATERIALS AND METHODS
Cell culture, transfection, and HCV infection.
Immortalized human hepatocytes (IHH) (21) or Huh7.5 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin G, and 100 μg/ml streptomycin at 37°C in a 5% carbon dioxide atmosphere. For infection, IHH (3 × 105) were incubated with HCV genotype 1a (clone H77) or HCV genotype 2a (clone JFH1) in a minimum volume of medium. DMEM supplemented with 5% heat-inactivated fetal bovine serum was added after 8 h of virus adsorption on hepatocytes. Green fluorescent protein (GFP)-tagged HCV JFH1 (JFH1-GFP), in which the V3 region of domain III of NS5A was replaced with an enhanced green fluorescent protein (EGFP) gene, was provided by Curt H. Hagedorn (22). Hepatocytes transfected with control (scrambled) or Beclin1 (BCN1) small interfering RNA (siRNA), ATG7 siRNA, or Rab27a siRNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) were similarly infected with cell culture-grown HCV. The siRNAs used in this study are mixtures of three oligonucleotides and were purchased from Santa Cruz (the sequences are not available from the supplier).
RNA quantitation.
IHH or Huh7.5 cells were infected with HCV, and RNA was isolated with TRIzol (Invitrogen) at different days postinfection. cDNA was synthesized using the Superscript III reverse transcriptase kit (Invitrogen). Real-time PCR was performed using cDNA with the TaqMan gene expression PCR master mix and 6-carboxyfluorescein (FAM)-labeled reporter minor groove binding (MGB) probes for BCN1, CD9, HCV, bone marrow stromal cell antigen 2 (BST-2), and 18S RNA (Applied Biosystems).
Intracellular and extracellular virus infectivity assay.
Infectious HCV titers were determined by measuring the 50% tissue culture infective dose (TCID50) in serial dilutions. Control siRNA-, BCN1 siRNA-, or Rab27a siRNA-treated cells were infected with HCV. The supernatant of the infected cells was collected at day 3 postinfection for extracellular-virus titer determination. To harvest cell-associated virus, the infected cells were washed with phosphate-buffered saline (PBS) three times, collected into a new tube, and resuspended in 500 μl DMEM. The cells were freeze-thawed three times. Both extracellular and cell-associated supernatants were sedimented at 14,000 rpm and 4°C for 5 min to remove cell debris. The viral titer was determined by TCID50 assay. Briefly, naive Huh7.5 cells were seeded in 96-well plates. A serial dilution of virus stock was added to the cells and incubated for 8 h at 37°C. The diluted virus supernatant was removed from the cells, the cells were washed with PBS, and the medium was replaced with fresh medium. At day 3 postinfection, the infected cells were washed once with PBS and fixed with cold methanol. The level of HCV infection in the cells was analyzed by using a mouse monoclonal antibody against HCV NS5A (9E14, kindly provided by Charles M. Rice) at 4°C overnight and an Alexa Fluor 488-conjugated goat anti-mouse antibody (Molecular Probes) at room temperature for 2 h. The HCV-expressing cells were counted using a fluorescence microscope, and the viral titer was calculated using the TCID50.
Exosome isolation.
Exosome-depleted serum was prepared by ultracentrifugation of FBS at 110,000 × g at 4°C for 16 h using an SW41 rotor (Beckman Coulter), followed by filtration through a 0.22-μm filter (Nunc) as described previously (23). For exosome isolation, virus-infected cells were washed 3 times with PBS and then supplemented with DMEM containing 2% exosome-depleted serum and incubated for 3 days. The culture supernatant was collected and centrifuged at 300 × g at 4°C for 5 min, followed by centrifugations at 2,000 × g at 4°C for 10 min, 26,500 × g at 4°C for 30 min, and 110,000 × g at 4°C for 90 min. The exosome pellet was washed 2 times with PBS by centrifugation at 110,000 × g at 4°C for 60 min and finally resuspended in PBS.
Immunofluorescence assay.
IHH or Huh7.5 cells were seeded in a 4-well chamber slide (Nunc) and transfected with control siRNA or BCN1 siRNA, followed by infection with JFH1-GFP at a multiplicity of infection (MOI) of 0.1 (22, 24). Three days postinfection, the cells were washed with PBS, fixed with 3.7% formaldehyde for 20 min at room temperature, and blocked with 3% bovine serum albumin (BSA) for 1 h. The fixed cells were permeabilized with 0.2% Triton X-100 for 5 min at room temperature. Subsequently, the cells were incubated with CD63 specific mouse antibody (Santa Cruz) overnight at 4°C. The cells were washed and incubated with anti-mouse Ig conjugated with Alexa 594 (Molecular Probes) secondary antibody for 1 h at room temperature. Finally, the cells were washed and mounted for confocal microscopy (Olympus FV1000). Images were superimposed digitally for fine comparisons. For lysosome staining, the control siRNA- or BCN1 siRNA-treated virus-infected cells were treated with 1 μM LysoTracker Red DND-99 (Invitrogen) at 37°C for 30 min, as previously described (20). GFP-tagged viral protein and LysoTracker red images were collected using the sequential-scanning mode (488-nm and 543-nm excitation and 522-nm and 595-nm emission, respectively) of the Olympus FV1000 confocal system. CD63 was stained with a mouse monoclonal antibody and anti-mouse immunoglobulin conjugated to Alexa Fluor 647 (Molecular Probes). Images were superimposed digitally for fine comparisons.
Western blotting.
Mock-infected, HCV-infected, or control siRNA- or BCN1 siRNA-treated virus-infected cells were lysed in a sample buffer, subjected to SDS-PAGE, and transferred onto a nitrocellulose membrane. The membrane was probed with specific antibodies to HCV core (C7-50; ThermoFisher); BST-2 (Novus Biologicals); Hsp70 (StressMarq; BD Biosciences); p62 (Abnova); and Beclin1, CD63, and Rab27a (Santa Cruz). The membrane was reprobed with actin or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) for comparison of the protein loads. Proteins were visualized using an enhanced chemiluminescence (ECL) Western blot substrate (Pierce) and subjected to densitometric scanning by using an image analyzer and Quantity One software (Bio-Rad).
RESULTS
HCV promotes a complete autophagy maturation process.
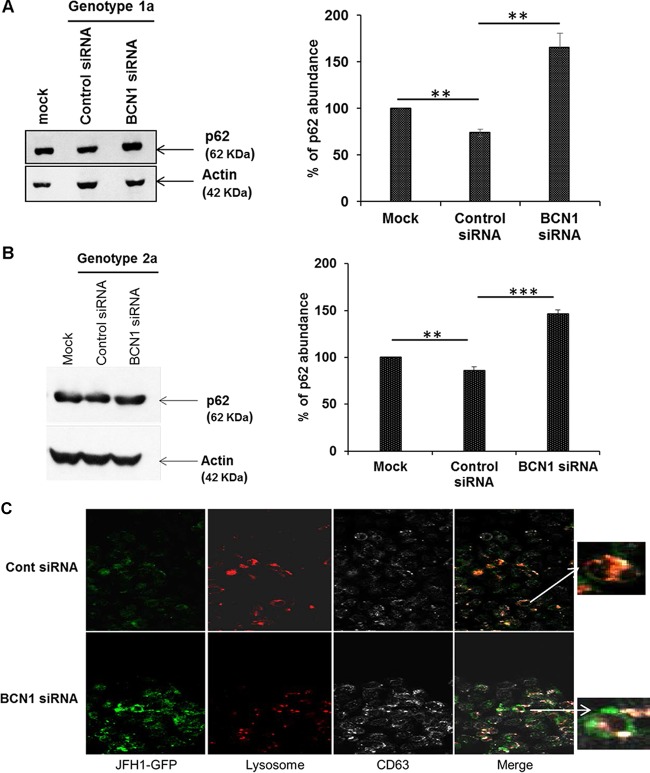
Autophagy is a dynamic multistep process and can be modulated at several levels. Accumulation of autophagosomes can either reflect an increase of autophagy flux, through the formation of de novo autophagosomes, or a reduced turnover of autophagosome recycling due to inhibition of their fusion with lysosomes. p62 (SQTSM1) serves as a substrate for autophagic degradation. We examined the expression of p62 in mock-infected and HCV (either genotype 1a or genotype 2a)-infected IHH or Huh7.5 cells. We observed lower expression of p62, suggesting that the lysosomal degradation process was not inhibited in HCV-infected cells (Fig. 1A and B). When we inhibited autophagy by gene silencing of either BCN1 or ATG7, followed by virus infection, we observed an accumulation of p62, suggesting impairment of the lysosomal degradation process. Similar observations were noted in autophagy knockdown HCV genotype 2a (JFH1)-infected Huh7.5 cells. Thus, HCV-infected cells undergo complete autophagy maturation, which is in agreement with previous reports (25, 26). We examined virus-infected cells for association of late endosomes and lysosomes and observed colocalization of CD63 and lysosomes in the cells. On the other hand, reduced colocalization was noted in BCN1 knockdown virus-infected cells (Fig. 1C). Our results suggest that knockdown of the autophagy pathway can cause lysosomal dysfunction and may impair delivery of late endosome/exosome cargo to the lysosome in virus-infected cells.
FIG 1.
HCV infection induces autophagosome-lysosome fusion in hepatocytes. (A and B) IHH were treated with control or BCN1 siRNA and infected with HCV genotype 1a (A) or genotype 2a (B) at an MOI of 0.1. After 3 days of infection, total cell lysates were collected for Western blot analysis. The Western blots were probed for p62-specific antibody and reprobed with an antibody to actin for comparison of protein loads. Silencing of BCN1 induced an accumulation of p62 in HCV-infected cells compared to control siRNA-treated virus-infected cells. Densitometry scanning for quantitation is shown on the right. **, P < 0.01; ***, P < 0.001. (C) IHH were treated with control (Cont) or BCN1 siRNA, infected with GFP-tagged HCV JFH1, and incubated for 72 h. Knockdown of BCN1 in HCV-infected IHH displayed reduced colocalization of CD63 and lysosome. Green, HCV; red, lysotracker red dye; white, late endosome/exosome marker CD63. Merged images (with enlarged areas) are shown on the right.
Autophagy in hepatocytes regulates secretion of infectious HCV.
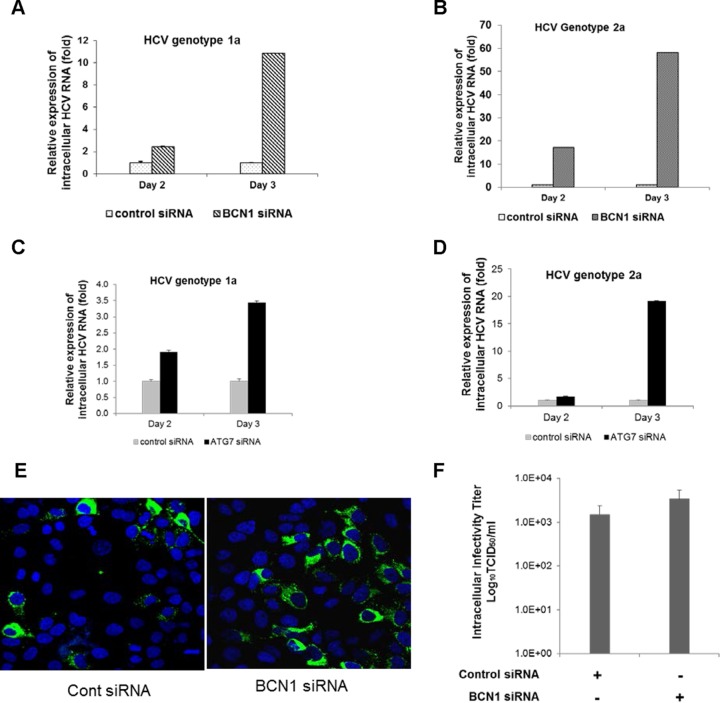
To investigate whether autophagy is involved in the late stage of the HCV life cycle, IHH or Huh7.5 cells were transfected with siRNA against BCN1 or ATG7 and then infected with HCV. The infected cells and culture supernatants were collected at different time points postinfection. We observed increased expression of intracellular viral RNA in BCN1 knockdown virus-infected cells compared to control siRNA-treated virus-infected cells (Fig. 2A and B). Upregulation of intracellular viral RNA in ATG7 knockdown virus-infected cells was observed compared to control siRNA-treated virus-infected cells (Fig. 2C and D). We observed an increase in the expression of HCV protein in autophagy knockdown virus-infected cells compared to parallel control siRNA-treated cells, indicating more virus may be present in autophagy knockdown infected cells (Fig. 2E). We also analyzed the infectivity of isolated intracellular viral particles from either control or BCN1 siRNA-treated virus-infected cells after repeated freeze-thawing and measured the viral titer on naive Huh7.5 cells. Upregulation of the infectivity of intracellular viral particles in BCN1 knockdown virus-infected cells was observed compared to control siRNA-treated virus-infected cells (Fig. 2F), suggesting HCV particles are retained in autophagy knockdown hepatocytes.
FIG 2.
Knockdown of BCN1 or ATG7 induces accumulation of intracellular viral particles. (A and B) IHH were treated with control siRNA or BCN1 siRNA and infected with HCV genotype 1a (H77) or genotype 2a (JFH1) at an MOI of 0.1. Cells were collected at different time points to measure the levels of intracellular HCV RNA and normalized to GAPDH by quantitative reverse transcription (qRT)-PCR. (C and D) IHH were treated with control siRNA or ATG7 siRNA and infected with HCV genotype 1a or genotype 2a at an MOI of 0.1. The intracellular HCV RNA level was measured by qRT-PCR at different time points and normalized with GAPDH. All experiments were performed three times, and the results are presented as the means of the results of the three experiments plus standard deviations. (E) IHH were treated with control siRNA or BCN1 siRNA and infected with GFP-tagged HCV genotype 2a (JFH1-GFP) at an MOI of 0.1. Representative photographs were taken 3 days postinfection. (F) IHH were treated with control siRNA or BCN1 siRNA and infected with HCV genotype 2a at an MOI of 0.1. Cells were collected 3 days postinfection, and infectious virus particles were isolated by repeated freezing and thawing. The titers of intracellular HCV particles were determined in naive Huh7.5 cells. HCV-infected cells were stained for NS5A protein, and the viral titer was calculated by TCID50. The results are presented in log scale.
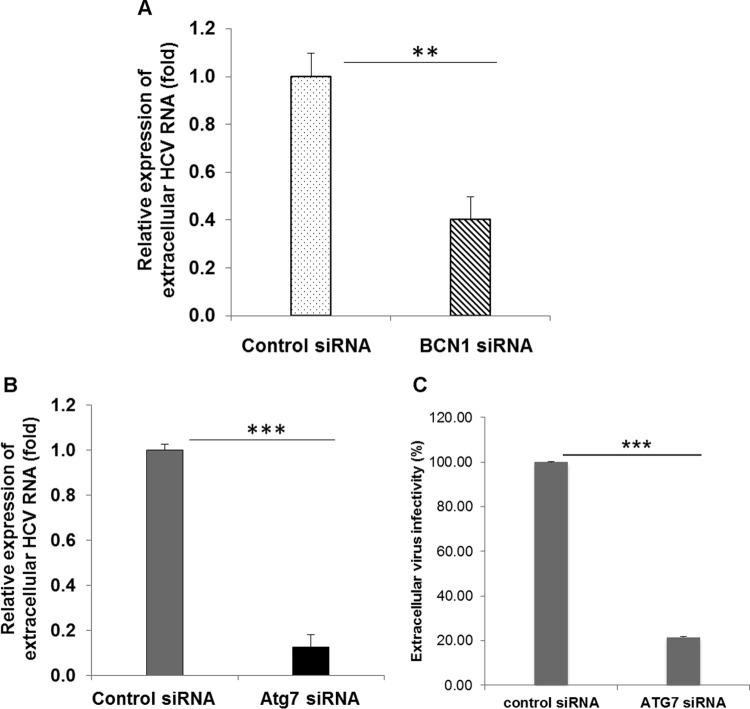
We also measured viral RNA in culture supernatants of autophagy knockdown virus-infected cells compared with control siRNA-treated HCV-infected cells. As expected, we observed a decrease in extracellular viral RNA in BCN1 or ATG7 knockdown virus-infected cells compared to control siRNA-treated virus-infected cells (Fig. 3A and B). We also examined the infectivity of virus particles in the culture supernatant by measuring the virus titer in naive Huh7.5 cells. We observed reduced infectivity of extracellular HCV particles generated from ATG7 knockdown virus-infected cells compared to those from control siRNA-treated virus-infected cells (Fig. 3C). We have shown previously that BCN1 knockdown impairs the infectivity of extracellular viral particles (19). A similar result was also observed in ATG7 knockdown HCV-infected Huh7.5.1 cells (27), although intracellular HCV RNA expression remained unchanged in that study. The discrepancy with the latter observation may be due to the use of a different cell line or subclone. The reduced infectivity in the supernatants and concomitant increased infectivity of intracellular virus in autophagy knockdown virus-infected cells compared to parallel controls suggested that secretion of virion particles was inhibited upon silencing of the autophagy machinery. Collectively, these results suggest that autophagy is required for HCV secretion into the cell culture medium.
FIG 3.
Knockdown of BCN1 or ATG7 reduces the extracellular viral RNA level. (A) IHH were treated with control siRNA or BCN1 siRNA and infected with HCV genotype 1a at an MOI of 0.1. At 3 days postinfection, the culture supernatant was collected to measure the extracellular HCV RNA by qRT-PCR. (B) IHH were treated with control siRNA or ATG7 siRNA and infected with HCV genotype 1a at an MOI of 0.1. RNA from the culture supernatant was collected, and extracellular HCV RNA was measured by qRT-PCR. (C) The extracellular HCV titer was measured from the culture supernatant of control siRNA- or ATG7 siRNA-treated HCV genotype 2a cells by TCID50. All experiments were performed three times, and the results are presented as the means of the results of the three experiments plus standard deviations. **, P < 0.01; ***, P < 0.001.
Silencing autophagy impairs exosome-associated HCV release.
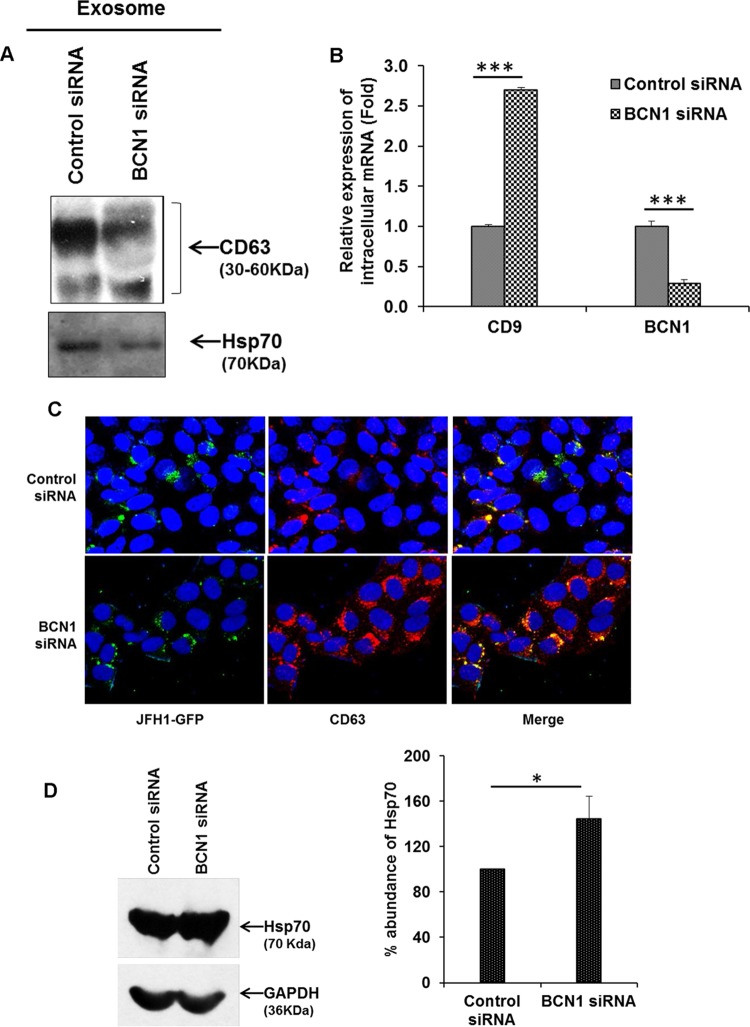
HCV utilizes the exosome pathway for its release from host cells (13, 14). To examine whether autophagy is required for exosome-mediated viral release, we first isolated the exosomes from culture supernatants of control siRNA- or BCN1 siRNA-treated HCV-infected cells using a standard ultracentrifugation protocol (23, 28). Reduced expression levels of CD63 and Hsp70 (exosome markers) were observed in exosomes secreted from BCN1 knockdown HCV-infected cell culture supernatants compared to a mock-treated control (Fig. 4A). We did not observe expression of the early endosome marker EEA1 when used as a negative control, as expected (data not shown). Interestingly, we detected a higher level of expression of intracellular CD9 mRNA (an exosome marker) in autophagy knockdown virus-infected cells in comparison to the control (Fig. 4B). As expected, Beclin1 expression was reduced in cells treated with BCN1 siRNA compared to cells treated with control siRNA. However, intracellular CD9 expression was not altered in BCN1 knockdown naive hepatocytes (data not shown). We next examined the cellular localization of exosomes in the control or autophagy knockdown HCV-infected cells. We observed a significant increase in CD63 expression and higher colocalization of CD63 with HCV particles in BCN1 knockdown virus-infected cells than in control siRNA-treated virus-infected cells (Fig. 4C). In addition, Hsp70 expression was higher in BCN1 knockdown HCV-infected cell lysates than in control siRNA-treated virus-infected cells (Fig. 4D), suggesting that HCV particles were retained inside the exosome in autophagy-impaired virus-infected cells. Together, our results demonstrate that exosome-associated HCV release is linked with autophagy in infected cells.
FIG 4.
Silencing of BCN1 accumulates exosomes within infected cells. (A) IHH were treated with control siRNA or BCN1 siRNA, followed by infection with HCV genotype 2a at an MOI of 0.1. After incubation with virus for 8 h, the cells were washed and incubated with medium containing exosome-depleted serum. At 3 days postinfection, the culture supernatants were collected to isolate exosomes. Western blot analysis for CD63 or Hsp70 was performed with exosomes isolated from culture supernatants of control siRNA- or BCN1 siRNA-treated HCV-infected cells. (B) Total RNA was isolated from control siRNA- or BCN1 siRNA-treated HCV-infected IHH. Intracellular CD9 or BCN1 was measured by qRT-PCR, and GAPDH was used for normalization. All experiments were performed three times, and the results are presented as the means of the results of the three experiments plus standard deviations. (C) Control siRNA- or BCN1 siRNA-treated IHH were infected with HCV genotype 2a (JFH1-GFP) at an MOI of 0.1. At 3 days postinfection, the cells were fixed, permeabilized, and stained with CD63 antibody (red). Nuclei were stained with DAPI (blue). Colocalization of CD63 and viral protein was examined by superimposing images in the Merge column by confocal microscopy, and representative images are shown. (D) Western blot analysis for Hsp70 was performed using specific antibody in cell lysates from control siRNA- or BCN1 siRNA-treated HCV-infected cells. The blots were reprobed with GAPDH to normalize the equal protein loads. Silencing of BCN1 enhanced Hsp70 protein in HCV-infected IHH in comparison to control siRNA-treated virus-infected cells. Densitometry scanning for quantitation is shown on the right. *, P < 0.05; ***, P < 0.001.
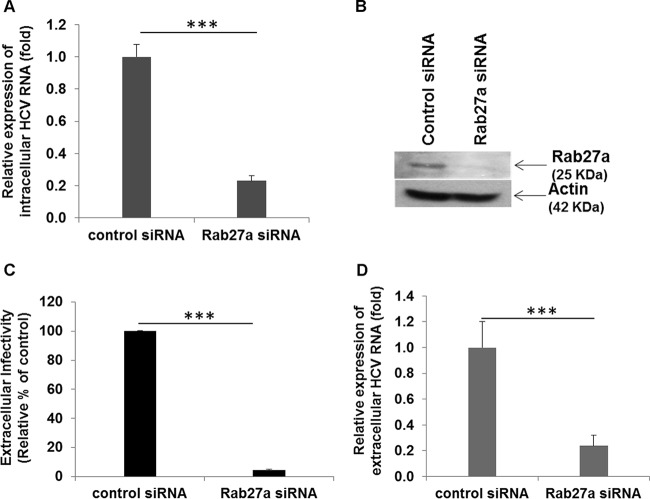
Knockdown of Rab27a inhibits intracellular and extracellular HCV RNA expression.
Rab27a regulates exosome secretion by docking MVB at the plasma membrane (29). To examine the role of Rab27a in HCV infection, we treated cells with control siRNA or Rab27a siRNA and then infected them with HCV. Intracellular HCV RNA levels were significantly lower in Rab27a siRNA-treated cells than in control siRNA-treated virus-infected cells (Fig. 5A). Cell lysates treated with control siRNA or Rab27a siRNA were subjected to Western blot analysis for expression of Rab27a protein. As expected, Rab27a protein expression was inhibited in specific siRNA-treated cells (Fig. 5B). Interestingly, we did not observe a difference in expression of the exosomal marker CD9 at the mRNA level or of CD63 at the protein level (data not shown). We also observed that extracellular HCV RNA or infectivity was significantly lower (Fig. 5C and D) in Rab27a knockdown virus-infected cells than in control siRNA-treated virus-infected cells. Rab27a knockdown blocks HCV-induced autophagy (data not shown), although the precise mechanism remains to be determined. While our article was in preparation, Chen et al. (30) reported similar observations, although the relationship of Rab27a with exosome secretion in HCV-infected cells needs further investigation.
FIG 5.
Knockdown of Rab27a reduces HCV replication and secretion. (A) Huh7.5 cells were treated with control or Rab27a siRNA, followed by infection with HCV genotype 2a at an MOI of 0.1. At 3 days postinfection, RNA was isolated from the cell lysates to determine intracellular HCV RNA levels. Intracellular HCV RNA levels were measured by qRT-PCR and normalized with GAPDH. (B) Cell lysates transfected with control or Rab27a siRNA were subjected to Western blot analysis for expression of Rab27a protein. Rab27a protein expression was inhibited in specific siRNA-treated cells. (C) Culture supernatants were collected from control siRNA- or Rab27a siRNA-treated HCV-infected cells 3 days postinfection and used to infect naive Huh7.5 cells for virus titer determination. The results are presented as percentages relative to control cells (100%). (D) Extracellular HCV RNA was measured by qRT-PCR from control siRNA- or Rab27a siRNA-treated virus-infected cells as described for panel A. ***, P < 0.001. The error bars represent standard deviations.
Knockdown of autophagy enhances BST-2 expression and inhibits HCV release.
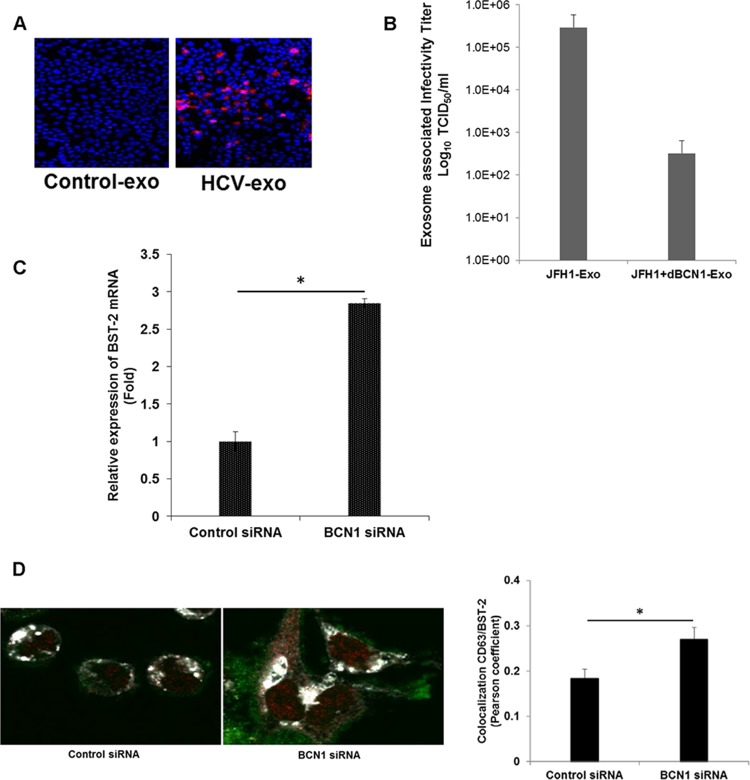
We next examined the presence of infectious HCV in exosomes. For this, exosomes isolated from mock- or HCV-infected cells were incubated with naive Huh7.5 cells, and HCV core protein was stained with a specific antibody to examine HCV infection. We observed the presence of HCV infectivity in Huh7.5 cells, as evident from viral protein expression. A representative image is shown in Fig. 6A, although we could not detect HCV protein in extracellular exosomes. We further measured viral RNA and infectivity of virus particles in exosomes isolated from control siRNA- or BCN1 siRNA-treated virus-infected cells. We observed variable HCV RNA levels in exosomes; however, the infectivity of HCV particles was significantly lower in exosomes isolated from autophagy knockdown virus-infected cells than in control siRNA-treated virus-infected cells (Fig. 6B). These results suggested that autophagy is required for exosome-mediated release of viral particles. BST-2 (also known as tetherin, CD317, or HM1.24) is encoded by an interferon-inducible gene that blocks the release of enveloped viruses, including retroviruses, filoviruses, arenaviruses, and herpesviruses, as well as paramyxoviruses and rhabdoviruses (31). We have previously shown that knockdown of autophagy enhances interferon-stimulated genes and the innate immune response in HCV-infected cells (19). To further elucidate the mechanism for restriction of exosome-mediated HCV release from the autophagy knockdown cells, we analyzed the expression of the BST-2/tetherin gene in autophagy knockdown HCV-infected cells. We observed increased expression of BST-2/tetherin in autophagy knockdown virus-infected cells compared to parallel controls (Fig. 6C). This result is in agreement with the recent finding that BST-2 inhibits HCV release (32). We next examined the intracellular location of BST-2 in the absence of BCN1 expression in HCV-infected cells. Immunofluorescence analysis suggested colocalization of BST-2 with CD63 in HCV-infected cells (Fig. 6D). Accumulation of BST-2 with CD63 was significantly higher in BCN1 knockdown HCV-infected cells. CD63-BST2 colocalization was assessed by calculating the Pearson correlation coefficient on three images. Together, our results suggested that HCV particles traffic to the exosome with the help of autophagy machinery to be released from the cells.
FIG 6.
Knockdown of BCN1 inhibits secretion of exosome-associated HCV. (A) Exosomes (exo) isolated from mock- or HCV-infected cell supernatants were incubated with naive Huh7.5 cells, and 3 days postinfection, viral infection was examined. HCV core protein (red) was stained with a specific antibody to determine the presence of virus. (B) Exosomes were isolated from the culture supernatant of control siRNA- or BCN1 siRNA-treated HCV-infected cells. The titer of exosome-associated HCV particles was determined by infecting naive Huh7.5 cells, and after 3 days of infection, the cells were stained for HCV protein-specific monoclonal antibody and Alexa Fluor anti-mouse 488 secondary antibody. NS5A-positive cells were counted, and the viral titer was calculated using the TCID50. The results are presented in log scale. (C) BST-2 expression was determined by qRT-PCR from RNA isolated from HCV-infected control or autophagy knockdown cells and normalized with GAPDH. Experiments were performed three times, and the results are presented as the means of the results of the three experiments plus standard deviations. (D) Cells were treated with control siRNA or BCN1 siRNA and then infected with HCV (JFH1-GFP). After 48 h of infection, the cells were stained with BST2 (red) and CD63 (white). Merged images are shown. CD63-BST2 colocalization was assessed (right) by calculating the Pearson correlation coefficient for three images per condition using Fluoview (FV10ASW1.7) software. *, P < 0.05.
DISCUSSION
In our present study, we demonstrated that autophagy knockdown in HCV-infected cells displays (i) higher intracellular HCV RNA and protein accumulation; (ii) increased intracellular exosomes, as evident by higher expression status of CD9, Hsp70, and CD63; and (iii) lower levels of infectious HCV in secreted (extracellular) exosomes. We also observed that the autophagy pathway regulates exosome-mediated virus transmission and promotes virus trafficking from late endosomes to lysosomes.
Many cell types continuously secrete a large number of exosomes. They have a diameter of approximately 50 to 150 nm and a buoyant density between 1.08 g/ml and 1.22 g/ml. Exosomes are released into the extracellular space from late endosomes/MVBs after fusion with the plasma membrane. Previous studies have indicated that cell culture (33–35) and patient serum-derived (36, 37) HCV particles have a broad range of buoyant densities (between 1.01 g/ml and 1.17 g/ml). Further, the low-density (1.1-g/ml) fraction displays exosome-like structures and also infectivity (34). HCV can also spread to neighboring cells with the help of exosomes carrying replication-competent viral RNA (38). Furthermore, exosomes carrying virus particles can establish productive infection in naive hepatocytes (14). Recently, both NS5A and core protein, along with the exosomal proteins CD63 and CD81, were detected in the low-density (1.083- to 1.098-g/ml) infective HCV particles. Exosomal markers and NS5A or core protein were found to be colocalized at the plasma membrane, from which virus release occurs (39). We have also observed colocalization of HCV with the exosome marker protein CD63 in virus-infected cells, confirming the presence of virus particles in the exosomes. However, cross talk between autophagy and exosome release in HCV infection still remained unclear.
The endosomal and autophagy pathways are functionally connected. Several studies implicated the components of endosomal trafficking, including ESCRT complexes, Rab GTPases (40), and vesicle fusion machinery SNAREs, in autophagosome maturation (41, 42). Recently, physical interaction between the core autophagy component ATG12-ATG3 and the ESCRT-associated protein ALIX was established (43). Autophagosomes fuse with MVBs before delivery to the lysosome. Autophagy components regulate MVB formation and exosome release in neurodegenerative diseases. Toxic alpha-synuclein oligomers can be secreted through exosomes when the autophagic machinery fails (44). On the other hand, in age-related disorders, the decline in autophagy-related proteins leads to the accumulation of deleterious molecules, promoting exosome secretion to remove harmful materials (45). Autophagy impairment is linked with lysosomal dysfunction, and we observed accumulation of p62 in autophagy knockdown HCV-infected cells. Virus infection goes through a complete autophagy maturation process, as reported by us and others (20, 25, 26). However, in the absence of autophagy proteins, the number of autolysosomes is reduced, leading to accumulation of virus particles within the exosomes.
Exosomes facilitate the budding of human immunodeficiency virus and hepatitis A virus (15, 16, 46). Infectious hepatitis A virus particles were observed within extracellular vesicles and were dependent on host proteins associated with endosomal sorting complexes required for transport (ESCRT), namely, VPS4B and ALIX (15). Interaction of ALIX with the ESCRT-III protein CHMP4B has been reported to facilitate the budding of human immunodeficiency virus (16). We observed that silencing of the autophagy pathway impairs the exosome-mediated release of infectious HCV particles. We also observed that knockdown of Rab27a inhibits generation of intracellular and extracellular infectious HCV particles. Silencing of Rab27a decreases exosome release in the culture supernatant without altering the exosome protein content (29), thus implying a role in exosome-mediated HCV secretion. Increased expression of the BST-2/tetherin gene in autophagy knockdown HCV-infected cells was also noted. BST-2 is a transmembrane protein that contains a short N-terminal cytoplasmic domain, a membrane-spanning alpha-helix, a coiled-coil ectodomain, and a C-terminal glycosylphosphatidylinositol (GPI) anchor (31). This antiviral protein localizes at the plasma membrane, as well as the membranes of multiple intracellular vesicles, including endosomes and the trans-Golgi network. The overexpression of BST-2 did not affect the efficiency of dengue virus (DENV) infection and intracellular replication but significantly reduced the virion yield from infected cells (47). A recent study demonstrated that BST-2/tetherin and HCV core protein colocalize on lipid droplets, where the assembly and maturation of HCV particles take place (32). The IFN-induced BST-2/tetherin protein inhibits hepatitis B virus (HBV) release from cells, and the colocalization of BST-2/tetherin and HBV particles on MVBs has been noted (48, 49). We have observed a large accumulation of BST2 with CD63 in BCN1 knockdown HCV-infected cells. Therefore, we postulate that this could be one of the possible mechanisms of BST-2/tetherin-mediated restriction of infectious HCV release from autophagy knockdown cells. Further study is necessary to understand the cross talk between autophagy and exosomes in HCV infection and the role of BST-2/tetherin in restricting HCV release. Exosomes containing HCV might fuse with uninfected cells in a mechanism independent of viral envelope proteins and may contribute to natural infection. Our studies provide the role of exosomes in the spread of HCV infection, which can be blocked by the use of autophagy inhibitors.
ACKNOWLEDGMENTS
We thank Curt H. Hagedorn for providing GFP-tagged JFH1 virus and Charles Rice for the HCV NS5A antibody.
This work was supported by research grants DK081817 (R.B.R.) and DK080812 (R.R.) from the National Institutes of Health.
REFERENCES
- 1.Moradpour D, Penin F, Rice CM. 2007. Replication of hepatitis C virus. Nat Rev Microbiol 5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 2.Lindenbach BD, Rice CM. 2013. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol 11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wurdinger T, Gatson NN, Balaj L, Kaur B, Breakefield XO, Pegtel DM. 2012. Extracellular vesicles and their convergence with viral pathways. Adv Virol 2012:767694. doi: 10.1155/2012/767694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simons M, Raposo G. 2009. Exosomes: vesicular carriers for intercellular communication. Curr Opin Cell Biol 21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Bissig C, Gruenberg J. 2014. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol 24:19–25. doi: 10.1016/j.tcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raposo G, Stoorvogel W. 2013. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorizate M, Kräusslich HG. 2011. Role of lipids in virus replication. Cold Spring Harb Perspect Biol 3:a004820. doi: 10.1101/cshperspect.a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Votteler J, Sundquist WI. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe 14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masciopinto F, Giovani C, Campagnoli S, Galli-Stampino L, Colombatto P, Brunetto M, Yen TS, Houghton M, Pileri P, Abrignani S. 2004. Association of hepatitis C virus envelope proteins with exosomes. Eur J Immunol 34:2834–2842. doi: 10.1002/eji.200424887. [DOI] [PubMed] [Google Scholar]
- 11.Corless L, Crump CM, Griffin SD, Harris M. 2010. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol 91:362–372. doi: 10.1099/vir.0.017285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamai K, Shiina M, Tanaka N, Nakano T, Yamamoto A, Kondo Y, Kakazu E, Inoue J, Fukushima K, Sano K, Ueno Y, Shimosegawa T, Sugamura K. 2012. Regulation of hepatitis C virus secretion by the Hrs-dependent exosomal pathway. Virology 422:377–385. doi: 10.1016/j.virol.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Dreux M, Garaigorta U, Boyd B, Décembre E, Chung J, Whitten-Bauer C, Wieland S, Chisari FV. 2012. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe 12:558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JA, Stalin Raj V, Jenster G, Kwekkeboom J, Tilanus HW, Haagmans BL, Baumert TF, van der Laan LJ. 2013. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A 110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. 2011. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb CA, Yoshimori T, Tooze SA. 2013. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol 14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 18.Abada A, Elazar Z. 2014. Getting ready for building: signaling and autophagosome biogenesis. EMBO Rep 15:839–852. doi: 10.15252/embr.201439076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shrivastava S, Raychoudhuri A, Steele R, Ray R, Ray RB. 2011. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology 53:406–414. doi: 10.1002/hep.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shrivastava S, Bhanja Chowdhury J, Steele R, Ray R, Ray RB. 2012. Hepatitis C virus upregulates Beclin1 for induction of autophagy and activates mTOR signaling. J Virol 86:8705–8712. doi: 10.1128/JVI.00616-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanda T, Basu A, Steele R, Wakita T, Ryerse JS, Ray R, Ray RB. 2006. Generation of infectious hepatitis C virus in immortalized human hepatocytes. J Virol 80:4633–4639. doi: 10.1128/JVI.80.9.4633-4639.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Chen R, Hagedorn CH. 2014. Direct visualization of hepatitis C virus-infected Huh7.5 cells with a high titre of infectious chimeric JFH1-EGFP reporter virus in three-dimensional Matrigel cell cultures. J Gen Virol 95:423–433. doi: 10.1099/vir.0.055772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Théry C, Amigorena S, Raposo G, Clayton A. 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3:Unit 3.22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 24.Meyer K, Kwon Y-C, Liu S, Hagedorn CH, Ray RB, Ray R. 2015. Interferon-α inducible protein 6 impairs EGFR activation by CD81 and inhibits hepatitis C virus infection. Sci Rep 5:9012. doi: 10.1038/srep09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Tian Y, Ou JH. 2015. HCV induces the expression of Rubicon and UVRAG to temporally regulate the maturation of autophagosomes and viral replication. PLoS Pathog 11:e1004764. doi: 10.1371/journal.ppat.1004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ke PY, Chen SS. 2011. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest 121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. 2009. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy 5:937–945. doi: 10.4161/auto.5.7.9243. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Liu K, Liu Y, Xu Y, Zhang F, Yang H, Liu J, Pan T, Chen J, Wu M, Zhou X, Yuan Z. 2013. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat Immunol 14:793–803. doi: 10.1038/ni.2647. [DOI] [PubMed] [Google Scholar]
- 29.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. 2010. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 30.Chen TC, Hsieh CH, Sarnow P. 2015. Supporting role for GTPase Rab27a in hepatitis C virus RNA replication through a novel miR-122-mediated effect. PLoS Pathog 11:e1005116. doi: 10.1371/journal.ppat.1005116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauter D. 2014. Counteraction of the multifunctional restriction factor tetherin. Front Microbiol 5:163. doi: 10.3389/fmicb.2014.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amet T, Byrd D, Hu N, Sun Q, Li F, Zhao Y, Hu S, Grantham A, Yu Q. 2014. BST-2 expression in human hepatocytes is inducible by all three types of interferons and restricts production of hepatitis C virus. Curr Mol Med 14:349–360. doi: 10.2174/1566524013666131118111719. [DOI] [PubMed] [Google Scholar]
- 33.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gastaminza P, Dryden KA, Boyd B, Wood MR, Law M, Yeager M, Chisari FV. 2010. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J Virol 84:10999–11009. doi: 10.1128/JVI.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merz A, Long G, Hiet MS, Brügger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. 2011. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem 286:3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prince AM, Huima-Byron T, Parker TS, Levine DM. 1996. Visualization of hepatitis C virions and putative defective interfering particles isolated from low-density lipoproteins. J Viral Hepat 3:11–17. doi: 10.1111/j.1365-2893.1996.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 37.André P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Bréchot C, Paranhos-Baccalà G, Lotteau V. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol 76:6919–6928. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longatti A, Boyd B, Chisari FV. 2015. Virion-independent transfer of replication-competent hepatitis C virus RNA between permissive cells. J Virol 89:2956–2961. doi: 10.1128/JVI.02721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai CK, Saxena V, Tseng CH, Jeng KS, Kohara M, Lai MM. 2014. Nonstructural protein 5A is incorporated into hepatitis C virus low-density particle through interaction with core protein and microtubules during intracellular transport. PLoS One 9:e99022. doi: 10.1371/journal.pone.0099022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gutierrez MG, Munafó DB, Berón W, Colombo MI. 2004. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci 117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 41.Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, Baba M, McNew JA, Jiang X, Reggiori F, Melia TJ, Klionsky DJ. 2011. SNARE proteins are required for macroautophagy. Cell 146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Itakura E, Kishi-Itakura C, Mizushima N. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 43.Murrow L, Malhotra R, Debnath J. 2015. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat Cell Biol 17:300–310. doi: 10.1038/ncb3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Danzer KM, Kranich LR, Ruf WP, Cagsal-Getkin O, Winslow AR, Zhu L, Vanderburg CR, McLean PJ. 2012. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol Neurodegener 7:42. doi: 10.1186/1750-1326-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J. 2010. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proc Natl Acad Sci U S A 107:14164–14169. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sette P, Mu R, Dussupt V, Jiang J, Snyder G, Smith P, Xiao TS, Bouamr F. 2011. The Phe105 loop of Alix Bro1 domain plays a key role in HIV-1 release. Structure 19:1485–1495. doi: 10.1016/j.str.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan XB, Han JC, Cong X, Wei L. 2012. BST2/tetherin inhibits dengue virus release from human hepatoma cells. PLoS One 7:e51033. doi: 10.1371/journal.pone.0051033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lv M, Zhang B, Shi Y, Han Z, Zhang Y, Zhou Y, Zhang W, Niu J, Yu XF. 2015. Identification of BST-2/tetherin-induced hepatitis B virus restriction and hepatocyte-specific BST-2 inactivation. Sci Rep 5:11736. doi: 10.1038/srep11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan R, Zhao X, Cai D, Liu Y, Block TM, Guo JT, Guo H. 2015. The interferon-inducible protein tetherin inhibits hepatitis B virus virion secretion. J Virol 89:9200–9212. doi: 10.1128/JVI.00933-15. [DOI] [PMC free article] [PubMed] [Google Scholar]