ABSTRACT
Chikungunya virus is a positive-stranded RNA alphavirus. Structures of chikungunya virus-like particles in complex with strongly neutralizing antibody Fab fragments (8B10 and 5F10) were determined using cryo-electron microscopy and X-ray crystallography. By fitting the crystallographically determined structures of these Fab fragments into the cryo-electron density maps, we show that Fab fragments of antibody 8B10 extend radially from the viral surface and block receptor binding on the E2 glycoprotein. In contrast, Fab fragments of antibody 5F10 bind the tip of the E2 B domain and lie tangentially on the viral surface. Fab 5F10 fixes the B domain rigidly to the surface of the virus, blocking exposure of the fusion loop on glycoprotein E1 and therefore preventing the virus from becoming fusogenic. Although Fab 5F10 can neutralize the wild-type virus, it can also bind to a mutant virus without inhibiting fusion or attachment. Although the mutant virus is no longer able to propagate by extracellular budding, it can, however, enter the next cell by traveling through junctional complexes without being intercepted by a neutralizing antibody to the wild-type virus, thus clarifying how cell-to-cell transmission can occur.
IMPORTANCE Alphaviral infections are transmitted mainly by mosquitoes. Chikungunya virus (CHIKV), which belongs to the Alphavirus genus, has a wide distribution in the Old World that has expanded in recent years into the Americas. There are currently no vaccines or drugs against alphaviral infections. Therefore, a better understanding of CHIKV and its associated neutralizing antibodies will aid in the development of effective treatments.
INTRODUCTION
Chikungunya virus (CHIKV) is the causative agent of an emerging disease. The first outbreaks occurred in the 1950s in Tanzania (1). The virus later reemerged in late 2005 on the Reunion islands and, subsequently, also in several Southeast Asian countries (2). However, by 2013 large epizootics/epidemics of CHIKV had spread into the Caribbean and by 2014 into South America as well as the United States (3). CHIKV is a member of the Togaviridae family, Alphavirus genus. Some alphaviruses, such as Sindbis virus (SINV), generally cause only mild disease symptoms in humans (4). Although mortality rates from CHIKV infections are low, CHIKV and Venezuelan equine encephalitis virus (VEEV) can be lethal or permanently disabling (2, 3). Furthermore, there are currently no treatments or vaccines for alphaviral infections.
The structures of a number of alphaviruses have been determined to resolutions better than 10 Å, including the structures of chikungunya virus-like particles (CHIK-VLPs), which have been determined to 5.3-Å resolution (5–7), and of VEEV, determined to 4.6-Å resolution (6). Alphaviruses have an external diameter of about 700 Å and are icosahedral with quasi-T=4 symmetry. They have a nucleocapsid core that is completely surrounded by a lipid envelope, derived from a host membrane, into which is embedded an icosahedral array of glycoproteins (8). A single virus particle contains 240 copies each of the E1 and E2 glycoproteins, which form 20 “i3” spikes situated on the icosahedral 3-fold axes and 60 quasi-3-fold “q3” spikes at general positions. There are 240 copies of the capsid protein on the internal cytoplasmic side of the viral lipid membrane, arranged as 12 pentamers about the 5-fold vertices and 30 hexamers about the icosahedral 2-fold vertices, consistent with the T=4 symmetry of the glycoprotein on the external side of the membrane.
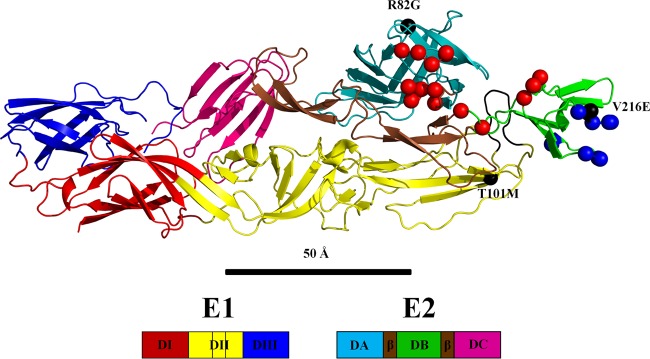
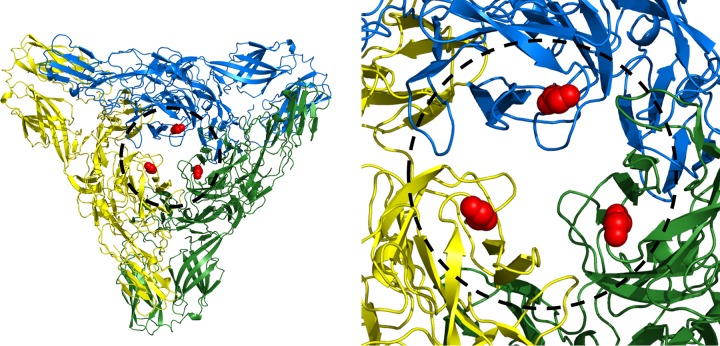
Alphaviruses have a positive-sense single-stranded RNA (ssRNA) genome that is about 11 kb in length and codes for 9 proteins. Four nonstructural proteins (NSP1 to -4) are coded at the 5′ end of the genome. The 3′ end of the genome is transcribed for subsequent translation into a polyprotein precursor containing the three structural proteins PE2 (the precursor of E3 and E2), E1, and the capsid protein. The E1 glycoprotein of CHIKV consists of 442 amino acids that form three β-barrel domains (I, II, and III). The CHIKV E2 glycoprotein consists of 423 amino acids arranged into three immunoglobulin-like domains, A, B, and C. Domain A contains the receptor binding site (9–12), domain B is at the distal end of each spike protecting the fusion loop on DII of E1, and domain C is situated closest to the viral membrane. Domain B is connected to domains A and C by a β ribbon (Fig. 1). The crystal structures of the E1E2 heterodimer of CHIKV (13, 14) and the E1E2 trimer of heterodimers from SINV (14) have been determined to near-atomic resolution.
FIG 1.
Structure of the E1E2 heterodimer of chikungunya virus (PDB 3N42). The amino and carboxy E2 β-ribbon connectors to domain B are colored brown, and the E1 fusion loop is colored black. Red and blue spheres represent the amino acids in the Fab 8B10 and 5F10 footprints, respectively. Gray spheres represent escape mutants.
The E1 glycoprotein contains a hydrophobic fusion loop that is responsible for membrane fusion with an endosomal membrane when initiating infection. The E2 protein can bind to cellular receptors and protects the E1 fusion loop at neutral pH. During the initial stages of infection, the host cellular receptor is recognized by the surface glycoprotein E2. Once the virus has been internalized, the low pH of the endosome causes the virion to undergo an irreversible conformational change resulting in the disassociation of E2 from E1 and the formation of E1 trimers (14). Upon E2 disassociation, the fusion loop then binds to and fuses with the endosomal membrane. Finally, the viral genome is released into the host cytoplasm, where replication of new viral particles can begin.
Antibody neutralization of alphaviruses can occur through various mechanisms. Most antibodies against alphaviruses target the E2 glycoprotein, since it covers much of E1 and is therefore far more exposed. The two most common mechanisms for neutralizing alphaviruses are by binding to the A domain of E2 and therefore blocking receptor attachment (9–12) and by preventing exposure of the E1 fusion loop by stabilizing the E2 B domain to cover and protect the fusion loop in the mature virus (9, 12). This can be achieved by binding the flexible B domain to the rigid A domain via a Fab fragment (8, 12).
The CHIKV-neutralizing antibodies 8B10 and 5F10 had been previously isolated from CD-40-activated B cells of infected patients (15) and have been shown to protect mice from CHIKV infection (16). However, CHIKV with a V216E or R82G mutation on E2 were also able to propagate in the presence of antibody 5F10 (16). Similarly, CHIKV with mutations T12I or R82G on domain A of E2 or T101M on domain II of E1 (16) were able to propagate in the presence of antibody 8B10. When mutant CHIKV containing these escape mutations to antibody 8B10 were propagated in the presence of 8B10, the virus particles migrated to cellular junctions (15), indicative of direct cell-to-cell transmission. Thus, antibody 8B10 enhances the direct transmission of mutant CHIKV between cells.
Here we report cryo-electron microscopy (cryo-EM) structures of CHIK-VLPs in complex with Fab fragments of monoclonal antibodies (MAbs) 8B10 and 5F10. These structures suggest the probable mechanism of neutralization and cell-to-cell transmission of the virus.
MATERIALS AND METHODS
CHIK-VLPs were produced and purified as described previously (17). The use of CHIK-VLPs instead of the live infectious virus allowed studies to be performed under nonbiosafety conditions.
Antibody production and purification.
Procedures for producing MAbs 8B10 and 5F10 have been previously reported (15, 16). Fab fragments were purified from the IgG after papain digestion using a protein A column and then further purified over a Superdex 200 16/600 column.
Complex formation, cryo-EM data collection, and single-particle reconstructions.
CHIK-VLP particles were incubated with Fabs at room temperature and pH 7.6 for 1 h at a 3-to-1 molar ratio of Fab to E2. For the prefusion complexes, the pH was lowered to 6.0 by addition of a 1/10-volume dilution with 1 M sodium citrate (pH 5.5). Aliquots of 3 μl for each complex were flash-frozen in liquid ethane on 400-mesh Cu Holey Carbon grids using a CP3 robotic plunger. Images of the frozen complexes were recorded on a 4k × 4k charge-coupled device (CCD) camera using an FEI Titan Krios electron microscope operating at 300 kV. Images were collected at a magnification of ×59,000 with an electron dose of ∼25 e−/Å2. Particles were boxed using e2boxer in the EMAN2 software package (18). The images were corrected for the contrast transfer function using CTFIT from the EMAN package (19). To initiate the orientation search, a cryo-EM reconstruction of native CHIK-VLP was used. Euler angles for each raw particle image were determined by projection matching of the raw particles into classes. The class averages were then used to generate a new three-dimensional (3D) model by using a Fourier reconstruction routine. The 3D model was then iteratively refined until the Fourier shell correlation no longer improved. After rejecting about 7% of the particles, the final number of particles used for the reconstruction of the CHIK-VLP/Fab 8B10 complex at pH 7.6 was 1,582. For the reconstruction of the CHIK-VLP/Fab 5F10 complexes, 1,621 and 1,428 particles were finally used at pH 7.6 and 6.0, respectively. This gave final resolutions of 17.6 Å for the 8B10 complex at pH 7.6 and 18.1 Å and 17.3 Å for Fab complexes 5F10, respectively. The resolution of the cryo-EM reconstructions was determined as the Fourier shell correlation at a 0.5 threshold (20) using the “gold standard” complete separation of two equally sized data sets.
X-ray crystal structure of Fab 5F10.
The purified Fab fragments were concentrated to ∼10 mg/ml. The concentrated Fab solution was used to set up crystallization trials using Hampton Crystal Screen I & II and Hampton Index HT (21). Promising crystallization conditions were optimized for buffer, precipitant, and pH at room temperature. The best-formed crystals were needle-shaped (100 μm by 30 μm), grown in 10 to 20% 2-propanol, 20 to 25% polyethylene glycol (PEG) 4000, and 50 mM HEPES buffer, pH 7.5. The exceedingly radiation-sensitive crystals were screened at the Advanced Photon Source, GM/CA, beamline 23 ID-B. Partial data sets were collected using an oscillation angle of 1° with an exposure time of 3 s with reflections extending to a 2.7-Å resolution (Table 1).
TABLE 1.
X-ray data and crystallographic information
| Parameter (unit) | Value(s) |
|---|---|
| Data collection | |
| X-ray wavelength (Å) | 1.03 |
| Space group | P212121 |
| Cell dimensions | a = 40.2 Å, b = 66.6 Å, c = 168.9 Å |
| α = β = γ = 90° | |
| Resolution (Å) | 2.75 (2.81–2.75)a |
| Rmergeb | 0.192 (0.692) |
| <I/ΔI> | 8.67 (2.15) |
| Completeness (%) | 95.3 (94.0) |
| Avg multiplicity | 5.0 (4.6) |
| Refinement | |
| No. of reflections | 11,899 |
| R-work/R-free | 0.253 (0.291) |
| Average B factor | 47.25 |
| RMSd deviations | |
| Bond length (Å) | 0.0053 |
| Bond angles (degrees) | 1.066 |
| Ramachandran plot (%)c | |
| Favored | 94.5 |
| Allowed | 5.3 |
| Outliers | 0.2 |
Values in parentheses represent the highest-resolution shell.
Rmerge = ∑hklΣi | Iihkl − <I(hkl)> | /ΣhklΣiIi(hkl).
According to the criteria of Molprobity (41).
RMS, root mean square.
The data sets were indexed and scaled using HKL2000 (22). The crystals belong to space group P212121 with one Fab molecule in the asymmetric unit. The final Rmerge value was 19.2% with a completeness of 95.3%. Using the sequence of Fab 5F10, a BLAST search was performed for Fab molecules in the Protein Data Bank (PDB) that had the highest sequence homology to antibody 5F10 (patent number EP 2374816A1). These molecules (PDB ID 3V0V, 1HZH, and 4D9L) were superimposed and morphed together to make an initial molecular replacement search model using the Phenix Ensembler (23). Initial phases were determined by molecular replacement using Phaser (24). The resultant molecular replacement solution was refined using Phenix (25) and modeled with Coot (26). The final working and free R values were 25 and 29%, respectively.
Homology model of Fab 8B10.
Using the sequence of Fab 8B10, a homology model was determined using the Swiss-Model server (27, 28) together with the crystal structure of Fab 5F10 as a template.
Structural analysis of the VLP/Fab complex.
The initial step in the interpretation of the cryo-EM electron density maps of the Fab-CHIK-VLP complexes was to fit either the homology model (8B10) or crystal structure (5F10) of the Fab fragment as a rigid body, assuming a quasi-T=4 icosahedral symmetry using the EMfit program (29). The approximate initial position of the Fab was given to the EMfit program. The program then performed a complete three-dimensional orientational search and limited three-dimensional positional search taking into account the icosahedral symmetry of the virus and the quasi-T=4 symmetry within the icosahedral asymmetric unit (29). The quality of each trial was measured by maximizing the average density heights of the fitted atoms and minimizing the clashes between symmetry-related structures as well as minimizing the number of atoms in low or negative density. Finally, the top 25 results were further refined with local, fine-increment, six-dimensional searches. Many of these fits converged to the same best result. The other fits were exceedingly poor based on the defined fitting criteria. All map pixels within 3.0 Å of a fitted atom were then set to zero to produce a modified cryo-EM map of each complex.
The second step in the interpretation was to fit the crystal structure of the E1E2 CHIKV heterodimer (PDB 3N42) into the modified cryo-EM maps. This procedure avoids clashes between the Fab molecule and the E1E2 glycoproteins, as the fitting maximizes the height of the average density at all fitted atoms, thus avoiding the regions of zero density where the Fab had been fitted (15, 16). This gave a starting position and orientation to perform independent fitting of individual domains (30). Glycoprotein E1 was split into domain I (residues 1 to 36, 132 to 168, and 273 to 293), domain II (residues 37 to 131 and 169 to 272), and domain III (residues 294 to 393). E2 was divided into domain A (residues 16 to 134), domain B (residues 173 to 231), domain C (residues 269 to 342), and the β-ribbon connector (residues 7 to 15, 135 to 172, and 232 to 268) (Fig. 1). While fitting the independent domains, the C terminus of each domain was restrained to be within 3.8 Å from the N terminus of the next domain (Table 2).
TABLE 2.
Fitting of E-glycoprotein and Fab structures into cryo-EM reconstructions
| Structure | pH | sumfa | Clashb (%) | −Denc (%) | θ1d | θ2 | θ3 | cxe | cy | cz |
|---|---|---|---|---|---|---|---|---|---|---|
| 8B10 | ||||||||||
| E1E2 | 7.6 | 36.2 | 0.0 | 2.8 | 184.5 | 80.2 | 175.0 | 21.9 | 88.4 | 277.3 |
| Fab | 7.6 | 31.4 | 0.0 | 2.1 | 200.0 | 10.3 | 200.0 | 22.7 | 94.3 | 337.8 |
| 5F10 | ||||||||||
| E1E2 | 7.6 | 37.2 | 0.0 | 1.9 | 185.0 | 80.5 | 178.0 | 23.6 | 88.7 | 275.7 |
| 6.0 | 36.8 | 0.0 | 1.5 | 185.8 | 80.0 | 179.2 | 23.9 | 89.2 | 274.1 | |
| Fab | 7.6 | 29.7 | 0.0 | 9.6 | 98.0 | 359.3 | 260.8 | 14.3 | 103.2 | 294.8 |
| 6.0 | 30.3 | 0.0 | 9.8 | 97.1 | 360.5 | 260.7 | 14.0 | 101.4 | 294.3 | |
Average density for all atomic positions normalized to the highest density in the map (set to 100).
Percentage of clashes between symmetry-related atoms.
Percentage of atoms positioned in negative density.
θ1, θ2, and θ3 are the Eulerian angles (°) that rotate the coordinates from their initial to fitted positions.
cx, cy, and cz (Å) are the final center positions of the molecules after fitting.
Protein structure accession numbers.
The crystal structure of Fab 5F10 has been deposited in the Protein Data Bank (www.pdb.org) (PDB code for the 5F10 crystal structure, 5BQ7); cryo-EM maps of CHIK-VLP in complex with Fabs 8B10 and 5F10 have been deposited in the Electron Microscopy Data Bank (www.emdatabank.org) (EMB codes EMD-6356 and EMD-6368, respectively).
RESULTS AND DISCUSSION
Fitting the Fab fragments into the cryo-EM densities.
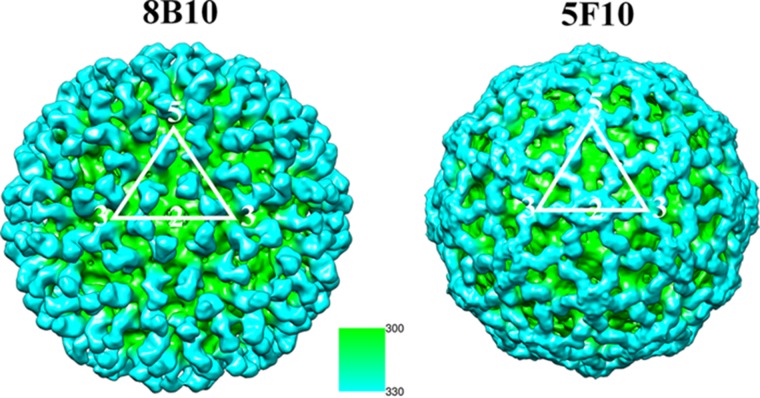
The cryo-EM maps of the complexes had a resolution ranging from 17 Å to 18 Å (Fig. 2), consistent with similar studies of many other viruses (e.g., dengue virus, West Nile virus [WNV], Venezuelan equine encephalomyelitis [VEEV] EV71, HIV). The reconstructions of the CHIK-VLPs in complex with 8B10 showed that each trimeric spike had three Fab fragments, extending radially outwards from the spike's outer surface. Thus, there are four bound 8B10 Fab fragments per icosahedral asymmetric unit and, therefore, 240 Fab fragments per VLP. The heights of the densities of the 8B10 Fab fragments and the virus were similar, indicating that the 8B10 Fab binding sites were fully occupied on the virus (Table 3). The 8B10 Fab light chain complementary determining region (CDR) loops 1 and 2 and the 8B10 Fab heavy chain CDR loop 2 were bound to the E2 A and B domains as well as the β-ribbon connector (Fig. 3 and Table 4).
FIG 2.
Cryo-EM reconstructions of CHIK-VLP in complex with Fab fragments of neutralizing antibodies looking down a 2-fold axis. One icosahedral asymmetric unit is outlined in white. The surface of the complex is colored according to its distance from the center of the virus as given by the inset scale in angstroms. Thus, the Fab fragment bound to the surface of the virus is blue, since it is at a greater distance from the center of the virus.
TABLE 3.
Average density heights for Cα positions (sumf, as defined in Table 2) upon fitting the individual domains from the CHIKV E1E2 heterodimer and the Fab structures into the cryo-EM map at four quasiequivalent positionsa
| Complex and domain | Avg density height upon: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Independent domain fitting |
T=4 fitting |
|||||||||
| #1 | #2 | #3 | #4 | Avg | #1 | #2 | #3 | #4 | Avg | |
| 8B10 complex at pH 7.6 | ||||||||||
| E1 I | 35.7 | 34.9 | 35.2 | 34.5 | 35.1 | 34.4 | 35.1 | 34.0 | 34.2 | 34.4 |
| E1 II | 36.9 | 35.3 | 35.1 | 35.0 | 35.6 | 35.9 | 35.6 | 34.8 | 34.2 | 35.1 |
| E1 III | 34.9 | 35.7 | 35.1 | 34.7 | 35.1 | 34.4 | 35.2 | 35.0 | 34.8 | 34.9 |
| E2 A | 37.4 | 38.1 | 37.2 | 37.1 | 37.5 | 36.8 | 37.3 | 36.6 | 36.3 | 36.8 |
| E2 B | 29.7 | 28.9 | 28.4 | 28.0 | 28.8 | 28.2 | 27.9 | 28.5 | 27.2 | 28.0 |
| E2 C | 36.0 | 36.7 | 37.1 | 35.9 | 36.4 | 35.7 | 36.0 | 35.9 | 35.3 | 35.7 |
| Fab V | 31.1 | 30.5 | 30.7 | 30.9 | 30.8 | 30.7 | 29.3 | 29.1 | 29.5 | 29.7 |
| Fab C | 30.2 | 31.5 | 29.9 | 29.6 | 30.3 | 29.5 | 29.1 | 28.7 | 28.1 | 28.9 |
| 5F10 complex at pH 7.6 | ||||||||||
| E1 I | 36.2 | 37.1 | 37.4 | 36.0 | 36.7 | 35.7 | 35.9 | 34.8 | 34.5 | 35.2 |
| E1 II | 35.9 | 36.3 | 35.2 | 34.9 | 35.6 | 35.1 | 34.7 | 35.8 | 34.3 | 35.0 |
| E1 III | 35.5 | 35.2 | 34.9 | 34.3 | 35.0 | 35.6 | 35.1 | 35.0 | 34.7 | 35.1 |
| E2 A | 34.6 | 34.0 | 33.9 | 33.7 | 34.1 | 33.4 | 33.0 | 32.9 | 32.3 | 32.9 |
| E2 B | 37.3 | 37.1 | 36.8 | 36.2 | 36.8 | 36.6 | 37.1 | 36.6 | 35.8 | 36.5 |
| E2 C | 35.9 | 35.5 | 35.7 | 35.1 | 35.6 | 34.5 | 34.9 | 33.8 | 33.4 | 34.2 |
| Fab V | 28.1 | 27.3 | 27.9 | 27.1 | 27.6 | 28.0 | 28.3 | 27.6 | 27.2 | 27.8 |
| Fab C | 27.0 | 27.7 | 27.0 | 26.9 | 27.2 | 26.7 | 27.0 | 26.1 | 26.0 | 26.5 |
| 5F10 complex at pH 6.0 | ||||||||||
| E1 I | 37.1 | 36.2 | 36.9 | 36.4 | 36.7 | 36.3 | 35.7 | 35.2 | 35.0 | 35.6 |
| E1 II | 35.2 | 35.7 | 35.2 | 34.0 | 35.0 | 34.5 | 34.8 | 34.1 | 34.1 | 34.4 |
| E1 III | 35.3 | 36.1 | 35.0 | 34.9 | 35.3 | 35.5 | 35.0 | 34.7 | 34.4 | 34.9 |
| E2 A | 35.6 | 34.8 | 34.8 | 34.2 | 34.9 | 36.0 | 36.2 | 36.9 | 35.7 | 36.2 |
| E2 B | 38.0 | 38.3 | 37.2 | 37.0 | 37.6 | 37.9 | 37.4 | 38.2 | 37.0 | 37.6 |
| E2 C | 35.4 | 35.3 | 35.1 | 35.0 | 35.2 | 34.1 | 34.9 | 33.8 | 34.3 | 34.3 |
| Fab V | 26.2 | 26.9 | 25.4 | 25.2 | 26.0 | 29.7 | 30.5 | 30.2 | 28.7 | 29.8 |
| Fab C | 26.0 | 25.8 | 26.1 | 25.6 | 25.9 | 28.7 | 30.9 | 31.6 | 29.4 | 30.2 |
Fab V, Fab variable; Fab C, Fab constant.
FIG 3.
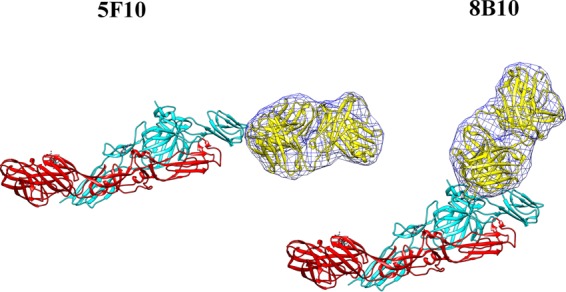
Fit of the Fab structures (yellow) into the cryo-EM density difference maps (blue mesh) that were calculated by performing a complete three-dimensional orientational search and a limited three-dimensional positional search using the program EMfit. The E1 glycoprotein is colored in red, and E2 is colored cyan.
TABLE 4.
Contacts between Fab 8B10 or 5F10 and glycoprotein E2d
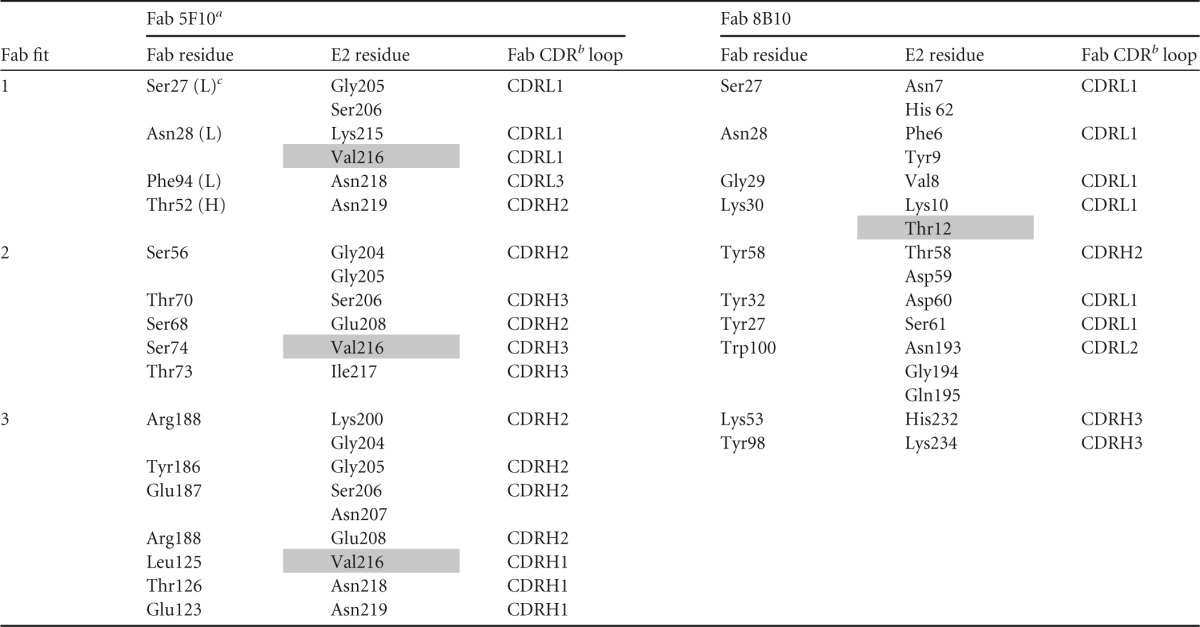
For Fab 3B4C-4, CD1 and CD2 are the Fab constant domains.
CDR is the complementary determining region on the Fabs.
(H) and (L) represent the heavy and light chains.
Shaded residues are escape mutants.
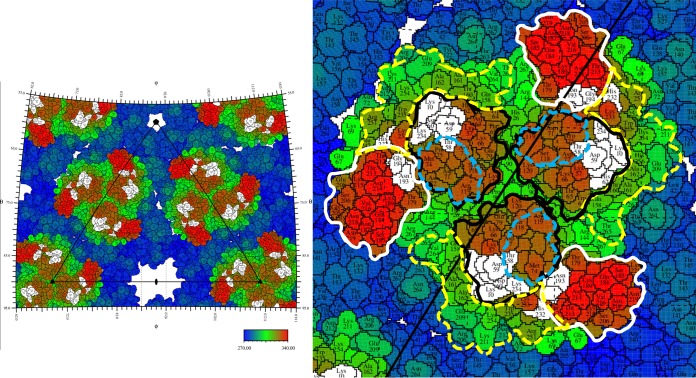
A construct containing the B domain and the β ribbon produced neutralizing antibodies in mice (31). Furthermore, a peptide of domain A fused with domain B, and the β ribbon induced potent antibodies. These observations suggest that both domains A and B are likely involved with inhibiting attachment, consistent with the footprint of Fab 8B10 on the virus. Thus, Fab 8B10 binds primarily to domain A on E2, which includes residue Thr58, which was identified to be in the putative receptor-binding region (Fig. 4) (32–34), and neutralizes the viral activity by inhibiting attachment of the virus to a cell.
FIG 4.
Stereographic projection showing a roadmap of CHIK-VLP with Fab 8B10 binding sites. The right-hand panel is a close-up of the leftmost trimeric glycoprotein spike. The amino acid colors represent the distance from the center of the virus as defined by the color scale bar. Domains A and B and the β-ribbon connector are bounded by black, white, and dashed yellow lines, respectively. Blue dashes represent the boundaries of the putative receptor-binding region. White-shaded residues represent where the Fab binds on E2. Positions on the surface of the virus can be identified by their latitude and longitude as is usual for locations on Earth. The edges of the left-hand figure correspond to lines of latitude and longitude.
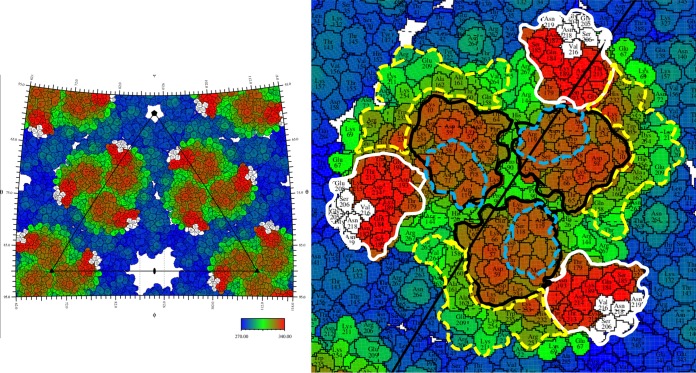
In contrast to the radial positioning of the 8B10 Fab molecules on the (E1E2)3 trimeric spikes, the 5F10 Fab molecules were bound to the edge of the spikes and are oriented tangentially to the virus surface and away from the receptor attachment site (Fig. 3 and 5) (32–34). This orientation gives rise to numerous steric clashes between Fabs bound to the i3 spikes and Fabs bound to the q3 spikes as well as clashes between Fabs bound to the q3 spikes around the pentameric vertices. Thus, not all of the symmetry-related sites can be occupied. Furthermore, the distribution of occupied sites is likely to be different for each particle. Therefore, the resultant cryo-EM reconstruction will represent an average of all these different structures.
FIG 5.
Stereographic projection showing a roadmap of CHIK-VLP with Fab 5F10 binding sites. The right-hand panel is a close-up of the leftmost trimeric glycoprotein spike. The amino acid colors represent the distance from the center of the virus as defined by the color scale bar. Domains A and B and the β-ribbon connector are bounded by black, white, and dashed yellow lines, respectively. Blue dashes represent the boundaries of the putative receptor-binding region. White-shaded residues represent where the Fab binds on E2.
Inhibition of fusion.
Attempts at determining the structure of the virus in complex with 8B10 at pH 6.0 failed because the virus aggregated. However, the virus in complex with 5F10 did not aggregate at pH 6.0, allowing the structure of this complex to be determined at both neutral and low pH.
The structures of CHIK-VLPs and also of other alphaviruses had shown that the B domain of glycoprotein E2 covers and protects the fusion loop in domain II of E1 (6, 7, 12). However, the B domain is exceedingly flexible (8, 15) as observed in the crystal structure of the E1E2 heterodimer (14) or the (E1E2)3 trimer (15). This allows the fusion loop to be exposed at low pH on the viral surface. Therefore, the ability of the 5F10 Fab fragments to stop aggregation at low pH implies that the B domains are rigidly fixed and prevented the exposure of the fusion loops. When the 5.3-Å resolution cryo-EM density of the native VLPs was fitted with the crystal structure of the E1E2 heterodimer, the average density height of the atoms in the B domain were about one-half of the average density in all other domains at all four quasiequivalent positions (15). Similarly, for the VLP complex with the 8B10 Fabs, the B domains have a significantly lower density than the other glycoprotein domains (Table 3). However, for the VLP complex with the 5F10 Fabs, the average density heights of the atoms in the B domain are slightly greater than the densities in all the other domains, demonstrating that the 5F10 Fab fragments have rigidly fixed the B domain, preventing the exposure of the fusion loops. Furthermore, there was no measurable difference between the structures of the VLP/Fab 5F10 complexes at pH 7.0 and at pH 6.0, showing that the reduction of pH did not cause any transition to a fusogenic conformation. The rigid B domain in the 5F10 complex is presumably caused by the tight packing of the 5F10 Fab molecules between the i3 and q3 spikes (Fig. 5). To accommodate 5F10 Fab binding, the B domains shifted 12 Å and rotated 5°.
Impact of escape mutations.
Locating the site of mutations on mutant viruses that escape neutralization by specific antibodies has been a standard and verified procedure for determining the binding site of the antibodies on the wild-type virus (15, 35). Escape mutations V216E and R82G from antibody 5F10, both in glycoprotein E2, as well as escape mutation T101M in glycoprotein E1 and T12I in glycoprotein E2 from antibody 8B10, were previously identified (15, 16). However, only mutations T12I and V216E in glycoprotein E2 were in the footprints of the corresponding antibodies (Table 4; Fig. 1 and 4). Nevertheless, the escape mutation T101M is within ∼20 Å of the corresponding Fab footprint. Thus, presumably this mutation causes a local conformational change that propagates to the antibody-binding site, which then abrogates antibody 8B10 binding and neutralization of the virus. As the binding site of antibody 8B10 stretches over both domain A and domain B, the 8B10 binding site would be distorted and, therefore, inhibit Fab 8B10 from binding or abrogate cellular attachment.
In contrast, escape mutation R82G in E2 is more than 45 Å away from the binding site of antibody 5F10 and therefore unlikely to have a direct impact on 5F10 binding (Fig. 1). However, the R82G escape mutation is situated in the central cavity of the trimeric spike (E1E2)3 and is about 10 Å from the two other 3-fold related arginine residues (Fig. 6). Thus, replacing this residue with a glycine would remove the repulsive forces between the 3-fold related arginine residues in the structure of the trimeric spikes. This will relieve crowding of the bound 5F10 Fab molecules and allow domain B to have greater freedom of movement, abrogating the strong inhibition of fusion that Fab 5F10 has on the wild-type virus (36). Thus, although Fab 5F10 could still bind to the mutant virus, there would be less restriction on the movement of the B domain than in the wild-type virus. In contrast, the other escape mutation to Fab 5F10 (V216E in E2) is in the footprint of the Fab and therefore inhibits binding of 5F10, thereby rendering the antibody ineffective.
FIG 6.
Overview of a single trimeric spike showing positions of residue Arg82 (red spheres). Each E1E2 heterodimer is colored separately for clarity.
Cell-to-cell transmission of the virus.
Certain mutations can render a virus incapable of budding from the plasma membrane, preventing the infection of another cell. The escape mutation R82G in domain A of E2 was generated under the neutralizing pressure of MAb 5F10 and was previously shown to promote the transmission of CHIKV isolates by direct cell-to-cell transfer of the virus (16). Although cell-to-cell transmission of viruses has been observed for many lipid-enveloped viruses (36–38), little is known about the exact mechanism. Nevertheless, it is known that cell-to-cell transmission is often accompanied by the increased concentration of molecules that can act as receptors (for instance, decay-accelerating factor) (39) for the free virus near cell junctions (39). Hence, presumably cell-to-cell transmission can occur when the virus buds near a cell junction and can then recognize receptor molecules on the neighboring cell. In the case of 5F10, the escape mutation abrogates the ability of the antibody to inhibit fusion, yet the antibody can still bind to the virus (16). As the location of the 5F10 binding site is on the edge of the B domain, far from the normal cellular receptor-binding site, the virus remains free to enter another cell at a cell-to-cell junction. In brief, although Fab 5F10 can neutralize the wild-type virus, it can also bind to the mutant virus without inhibiting fusion or attachment. Thus, the mutant virus would then be able to enter the next cell without being intercepted by what would be a neutralizing antibody to the wild-type virus.
Conclusions.
The two neutralizing anti-CHIKV antibodies 8B10 and 5F10 function very differently. The former primarily inhibits attachment, while the latter inhibits fusion. MAb 8B10 binds to the outside of the virus, whereas MAb 5F10 lies on the surface of the virus. The former inhibits by covering the cellular receptor-binding site, and the latter inhibits by crowding around and restricting the movement of the B domain, but leaving the cellular receptor binding site vacant for use in cell-to-cell transmission. Although the requirements of cell-to-cell transmission are roughly known for quite a few membrane-enveloped viruses (36–38, 40), this currently is the most detailed knowledge of the mechanism of this process.
ACKNOWLEDGMENTS
We thank Valorie Bowman for providing technical help and advice with cryo-EM sample preparation and data collection. We also thank Chia-Yin Lee (SIgN, A*STAR) for her technical assistance in generating the antibodies. We are also thankful to Andrei Fokine for useful discussions in interpreting the EM maps and to Sheryl Kelly for administrative aid in preparing the manuscript.
Funding Statement
This work was supported by National Institutes of Health (NIH) grants R01AI095366 to M.G.R. and F32AI106149 to J.P. and core grants provided by the Biomedical Research Council, A*STAR. We thank Purdue University for their support of the EM facility and the Advanced Photon Source for staff support and the use of beamline 23 ID-B. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) by Argonne National Laboratory, was supported by the U.S. DOE under contract no. DE-AC02-06CH11357. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The authors declare no conflict of interest.
REFERENCES
- 1.Johansson MA. 2015. Chikungunya on the move. Trends Parasitol 31:43–45. doi: 10.1016/j.pt.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paquet C, Quatresous I, Solet JL, Sissoko D, Renault P, Pierre V, Cordel H, Lassalle C, Thiria J, Zeller H, Schuffnecker I. 2006. Chikungunya outbreak in Reunion: epidemiology and surveillance, 2005 to early January 2006. Euro Surveill 11:2891 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=2891. [DOI] [PubMed] [Google Scholar]
- 3.Figueiredo ML, Figueiredo LT. 2014. Emerging alphaviruses in the Americas: Chikungunya and Mayaro. Rev Soc Bras Med Trop 47:677–683. doi: 10.1590/0037-8682-0246-2014. [DOI] [PubMed] [Google Scholar]
- 4.Laine M, Luukkainen R, Toivanen A. 2004. Sindbis viruses and other alphaviruses as cause of human arthritic disease. J Intern Med 256:457–471. doi: 10.1111/j.1365-2796.2004.01413.x. [DOI] [PubMed] [Google Scholar]
- 5.Kostyuchenko VA, Jakana J, Liu X, Haddow AD, Aung M, Weaver SC, Chiu W, Lok SM. 2011. The structure of Barmah Forest virus as revealed by cryo-electron microscopy at a 6-angstrom resolution has detailed transmembrane protein architecture and interactions. J Virol 85:9327–9333. doi: 10.1128/JVI.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, Baker ML, Weaver SC, Chiu W. 2011. 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J 30:3854–3863. doi: 10.1038/emboj.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun S, Xiang Y, Akahata W, Holdaway H, Pal P, Zhang X, Diamond MS, Nabel GJ, Rossmann MG. 2013. Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. eLife 2:e00435. doi: 10.7554/eLife.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng RH, Kuhn RJ, Olson NH, Rossmann MG, Choi HK, Smith TJ, Baker TS. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roehrig JT, Day JW, Kinney RM. 1982. Antigenic analysis of the surface glycoproteins of a Venezuelan equine encephalomyelitis virus (TC-83) using monoclonal antibodies. Virology 118:269–278. doi: 10.1016/0042-6822(82)90346-4. [DOI] [PubMed] [Google Scholar]
- 10.Hunt AR, Frederickson S, Hinkel C, Bowdish KS, Roehrig JT. 2006. A humanized murine monoclonal antibody protects mice either before or after challenge with virulent Venezuelan equine encephalomyelitis virus. J Gen Virol 87:2467–2476. doi: 10.1099/vir.0.81925-0. [DOI] [PubMed] [Google Scholar]
- 11.Smith TJ, Cheng RH, Olson NH, Peterson P, Chase E, Kuhn RJ, Baker TS. 1995. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc Natl Acad Sci U S A 92:10648–10652. doi: 10.1073/pnas.92.23.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porta J, Jose J, Roehrig JT, Blair CD, Kuhn RJ, Rossmann MG. 2014. Locking and blocking the viral landscape of an alphavirus with neutralizing antibodies. J Virol 88:9616–9623. doi: 10.1128/JVI.01286-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. 2010. Structural changes of envelope proteins during alphavirus fusion. Nature 468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee CY, Kam YW, Fric J, Malleret B, Koh EG, Prakash C, Huang W, Lee WW, Lin C, Lin RT, Renia L, Wang CI, Ng LF, Warter L. 2011. Chikungunya virus neutralization antigens and direct cell-to-cell transmission are revealed by human antibody-escape mutants. PLoS Pathog 7:e1002390. doi: 10.1371/journal.ppat.1002390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warter L, Lee CY, Thiagarajan R, Grandadam M, Lebecque S, Lin RT, Bertin-Maghit S, Ng LF, Abastado JP, Despres P, Wang CI, Nardin A. 2011. Chikungunya virus envelope-specific human monoclonal antibodies with broad neutralization potency. J Immunol 186:3258–3264. doi: 10.4049/jimmunol.1003139. [DOI] [PubMed] [Google Scholar]
- 17.Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, Lewis MG, Higgs S, Rossmann MG, Rao S, Nabel GJ. 2010. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat Med 16:334–338. doi: 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 20.Harauz G, Fong-Lochovsky A. 1989. Automatic selection of macromolecules from electron micrographs by component labelling and symbolic processing. Ultramicroscopy 31:333–344. doi: 10.1016/0304-3991(89)90331-8. [DOI] [PubMed] [Google Scholar]
- 21.Feil SC, Tang J, Hansen G, Gorman MA, Wiktelius E, Stenberg G, Parker MW. 2009. Crystallization and preliminary X-ray analysis of glutathione transferases from cyanobacteria. Acta Crystallogr Sect F Struct Biol Crystallogr Commun 65:475–477. doi: 10.1107/S1744309109011634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Snoeyink J. 2008. Defining and computing optimum RMSD for gapped and weighted multiple-structure alignment. IEEE/ACM Trans Comput Biol Bioinform 5:525–533. doi: 10.1109/TCBB.2008.92. [DOI] [PubMed] [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 28.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. 2009. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 29.Rossmann MG, Bernal R, Pletnev SV. 2001. Combining electron microscopic with X-ray crystallographic structures. J Struct Biol 136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- 30.Tang J, Jose J, Chipman P, Zhang W, Kuhn RJ, Baker TS. 2011. Molecular links between the E2 envelope glycoprotein and nucleocapsid core in Sindbis virus. J Mol Biol 414:442–459. doi: 10.1016/j.jmb.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber C, Buchner SM, Schnierle BS. 2015. A small antigenic determinant of the Chikungunya virus E2 protein is sufficient to induce neutralizing antibodies which are partially protective in mice. PLoS Negl Trop Dis 9:e0003684. doi: 10.1371/journal.pntd.0003684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernard KA, Klimstra WB, Johnston RE. 2000. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276:93–103. doi: 10.1006/viro.2000.0546. [DOI] [PubMed] [Google Scholar]
- 33.Dubuisson J, Rice CM. 1993. Sindbis virus attachment: isolation and characterization of mutants with impaired binding to vertebrate cells. J Virol 67:3363–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klimstra WB, Ryman KD, Johnston RE. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol 72:7357–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez R, Paredes A, Brown DT. 2008. Sindbis virus conformational changes induced by a neutralizing anti-E1 monoclonal antibody. J Virol 82:5750–5760. doi: 10.1128/JVI.02673-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malbec M, Porrot F, Rua R, Horwitz J, Klein F, Halper-Stromberg A, Scheid JF, Eden C, Mouquet H, Nussenzweig MC, Schwartz O. 2013. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. J Exp Med 210:2813–2821. doi: 10.1084/jem.20131244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mateo M, Generous A, Sinn PL, Cattaneo R. 2015. Connections matter—how viruses use cell-cell adhesion components. J Cell Sci 128:431–439. doi: 10.1242/jcs.159400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Z, Waeckerlin R, Urbanowski MD, van Marle G, Hobman TC. 2012. West Nile virus infection causes endocytosis of a specific subset of tight junction membrane proteins. PLoS One 7:e37886. doi: 10.1371/journal.pone.0037886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timpe JM, Stamataki Z, Jennings A, Hu K, Farquhar MJ, Harris HJ, Schwarz A, Desombere I, Roels GL, Balfe P, McKeating JA. 2008. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 47:17–24. [DOI] [PubMed] [Google Scholar]
- 40.Bergelson JM. 2009. Intercellular junctional proteins as receptors and barriers to virus infection and spread. Cell Host Microbe 5:517–521. doi: 10.1016/j.chom.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]