Abstract
Human herpesvirus 6A (HHV-6A) U14 is a virion protein with little known function in virus propagation. Here, we elucidated its function by constructing and analyzing U14-mutated viruses. We found that U14 is essential for HHV-6A propagation. We then constructed a mutant virus harboring dysfunctional U14. This virus showed severely reduced growth and retarded maturation. Taken together, these data indicate that U14 plays an important role during HHV-6A maturation.
TEXT
The herpesvirus virion has a proteinaceous layer (tegument proteins) between the envelope and capsid, making its structure strikingly different from that of other enveloped viruses. The functions of these proteins have been extensively investigated. Studies show that they function in virus infection by providing an optimum environment for virus infection, by facilitating viral gene expression, and by aiding viral packing and egress (1–9). Here, we focused on a predicted tegument protein with little known function in virus propagation, U14, belonging to human herpesvirus 6A (HHV-6A).
HHV-6A is a member of the betaherpesvirus subfamily (10) and is closely related to human herpesvirus 6B (HHV-6B) (11–18). HHV-6A was first isolated from patients with lymphoproliferative diseases (19). Although the virus may be associated with several diseases (20–25), the molecular mechanism underlying disease pathogenesis is unclear, partially because the functions of many HHV-6A genes, including the gene coding for U14, are rarely known. The gene products of HHV-6A-U14 positional homologs in HCMV (UL25, ∼25% identity) and in HHV-7 (U14, ∼52% identity) are reported to be tegument proteins and to be the major antigens for the production of antibody to these viruses (26, 27). HCMV UL25 is dispensable for the virus growth of the AD and Towne strains (28, 29). We previously showed that both HHV-6A U14 and -6B U14 interact with p53, and U14 and p53 are incorporated into the HHV-6B virion (not tested in HHV-6A); however, the detailed functions of U14 in virus propagation are unclear (30).
We previously used a bacterial artificial chromosome (BAC) and recombination systems in Escherichia coli to examine the function of HHV-6A genes (31–34). Theoretically, any of the HHV-6A genes can be deleted from the viral genome using these systems. It is also possible to define the function of a HHV-6A gene by analyzing the phenotype of mutant viruses from which the target gene has been deleted (if the gene is not essential for virus propagation). Because no studies have examined whether U14 is essential for virus propagation, we first deleted the U14 gene from the HHV-6A genome using a two-step Red recombination system in E. coli, as described previously (35). We then tried to reconstitute infectious virus from the genome. However, deletion of the U14 gene abolished the ability of the mutant viral genome to reconstitute infectious virus (as shown by these experiments, which were done independently by two researchers), suggesting that U14 is essential for HHV-6A propagation.
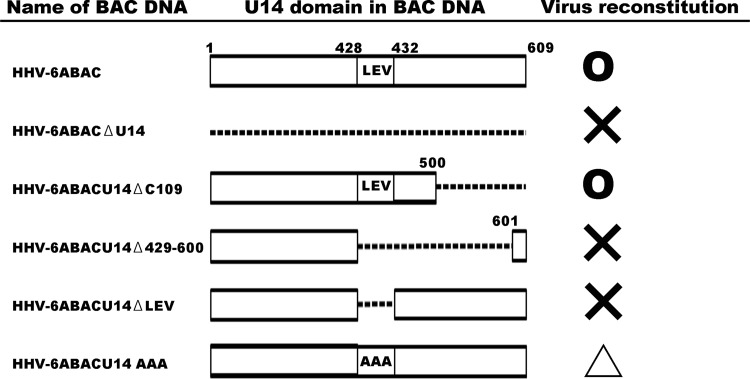
To further examine U14 gene function, we partially deleted the U14 gene, expecting that partial deletion would affect, but not completely abolish, U14 function. Thus, the resulting U14-mutated virus could be reconstituted and its phenotype investigated. Therefore, we deleted the U14 gene from its C terminus in the HHV-6A BAC genome to construct several U14-mutated HHV-6A genomes (Fig. 1) and tested whether they could be used to reconstitute infectious viruses. We found the following. (i) Infectious virus could be reconstituted when the C terminus of U14 (amino acids [aa] 501 to 609) was deleted, and (ii) infectious virus could not be reconstituted when aa 429 to 600 were deleted. Because we think that critical amino acids are positioned between aa 429 and 500, to identify the critical amino acid residues, first we tried to delete first 3 aa residues (aa 429 to 431) in aa 429 to 500. (iii) As a result, the virus could not be reconstituted when 3 aa residues (aa 429 to 431) were deleted (Fig. 1).
FIG 1.
Reconstitution of viruses from BAC DNAs. The name of each BAC DNA is listed on the left. The U14 domain within each HHV-6ABACDNA is shown in the middle. The infectious virus reconstitution results are shown on the right. ○, normally reconstituted; ×, not reconstituted; △, reconstituted but with a severe growth defect.
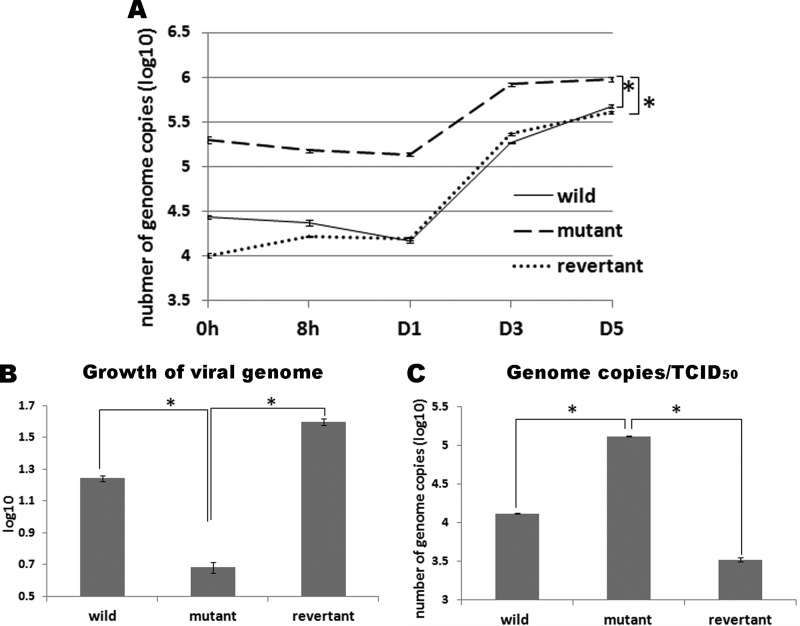
Therefore, we focused on aa 429 to 431 within U14. We replaced these three residues with alanine and tested whether infectious virus could be reconstituted. Infectious virus could be reconstituted when all three residues were replaced with alanine (Fig. 1), although the reconstituted virus (rHHV-6ABACU14AAA) showed slower growth than the wild-type virus (rHHV-6ABAC). Because this mutated U14 is partially functional during HHV-6A infection, it might help us elucidate the detailed function of U14 by enabling examination of the phenotype of the mutant virus. We first prepared virus stocks and tested virus growth in umbilical cord blood mononuclear cells (CBMCs), which were provided by H. Yamada (Kobe University Graduate School of Medicine, Kobe, Japan) or purchased from the Cell Bank of the RIKEN BioResource Center, Tsukuba, Japan. The use of CBMCs was approved by the Ethics Committee of the Kobe University Graduate School of Medicine. Compared with the titer of the wild-type or revertant virus stock, the titer of the mutant virus stock that could be prepared was relatively low. The titers of each virus stock used in this study are as follows: wild type, 9.3 × 103 50% tissue culture infective doses (TCID50)/ml; revertant, 6.3 × 104 TCID50/ml; and mutant, 3.6 × 102 TCID50/ml. CBMCs were infected with these viruses at a multiplicity of infection of ∼0.01. The number of green fluorescent protein (GFP)-positive cells increased smoothly in wild-type or revertant virus-infected cells, while mutant virus-infected cells showed a retarded increase in expression of GFP. We harvested the infected cells by centrifugation and monitored virus growth by measuring the viral genome copy number in these cells, as described previously (35). Wild-type and revertant viruses showed similar growth kinetics, although the initial viral genome copy numbers were different (Fig. 2A). This may be due to the fact that viruses were harvested at different time points after infection for preparation of virus stocks. Although the genome copy number of the mutant virus was initially higher than those of the wild type (105.30 versus 104.44, respectively) (Fig. 2A) and revertant (105.30 versus 104.01, respectively) (Fig. 2A), the increase per initial virus genome was low (100.68 versus 101.24compared with the wild type and 100.68 versus 101.60 compared with the revertant) (Fig. 2B). Since HHV-6A U14 is a tegument protein, it is unlikely that it would affect virus entry into host cells; however, it is highly likely that the mutant virus stock contained more virus particles than the wild-type stock, even though it showed the same TCID50. There could be a number of explanations for the higher initial genome copy numbers in the mutant. If the particle/TCID50 ratio is higher in the mutant, those additional defective particles could have bound but not been internalized or been internalized but not gone on to productive infection. It is also likely that replication of the mutant virus in CBMCs was severely reduced.
FIG 2.
(A) Growth kinetics of rHHV-6ABACU14AAA. CBMCs were infected with wild-type (rHHV-6ABAC), rHHV-6ABACU14AAA, or revertant virus at a multiplicity of infection of 0.01. The cells were harvested at 0 and 8 h and 1, 3, and 5 days postinfection and washed three times, and DNA was extracted from the cells as described previously (35). The viral genome copy number in each sample was quantitated by real-time PCR. (B) Comparison of the growth of each virus genome. Growth of the viral genome was expressed as viral genome copy number at day 5 divided by that at 0 h. (C) Comparison of genome copy number at the same TCID50. Total DNA was extracted from each virus stock at the same TCID50. The viral genome copy number was quantitated by real-time PCR. The t test was used for statistical analysis. *, P < 0.01.
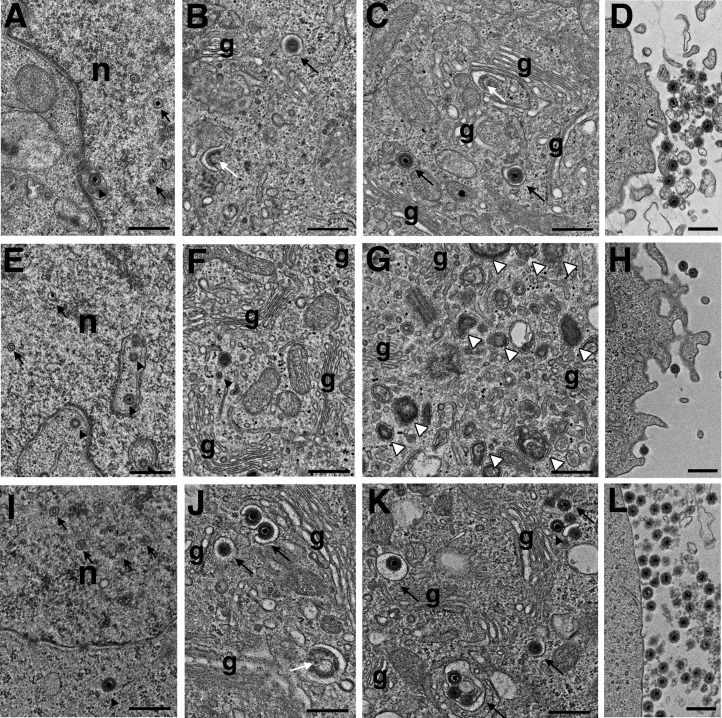
Thus, we next measured the number of genome copies in virus stocks with the same TCID50. We found that the copy number of rHHV-6ABACU14AAA was higher than that of the wild-type or revertant viruses (Fig. 2C). To examine the role of U14 during virus propagation, we observed CBMCs infected by rHHV-6ABACU14AAA, its revertant, and wild-type virus by electron microscopy (EM), as previously described (36). Although similar virus particles (viral capsid) were observed in the nuclei of wild-type- and rHHV-6ABACU14AAA-infected cells (Fig. 3A, E, and I), the viral particles in the cytosol were completely different. Cells infected with wild-type virus harbored enveloped virions in vacuoles near the Golgi complex (Fig. 3B, C, J, and K, black arrows). Furthermore, electron-dense tegument-like materials accumulated along the cytosolic face of some tubulovacuoles near the Golgi complex for virus budding (white arrows in Fig. 3B and J). Also, numerous virions, along with small internal vesicles, were released at the cell surface (Fig. 3D and L). The Golgi complex and tubulovesicular structures accumulated in cells infected with rHHV-6ABACU14AAA. Enveloped virions or electron-dense tegument-like materials that accumulated at tubulovacuoles for virus budding were rarely detected around them (Fig. F and G). Capsids were detected in the cytoplasm, but they were not enveloped (Fig. 3F; arrowhead). Lysosome-like organelles containing electron-dense materials accumulated in some cells (white arrowheads in Fig. 3G); these were not observed in cells infected with wild-type HHV-6A. Virions, along with small internal vesicles, were released from the cells (Fig. 3D and L) but were a few in number (Fig. 3H). These data strongly suggest that U14 plays a role during the postnuclear maturation process of HHV-6A infection.
FIG 3.
Electron microscopic observation of cells infected with wild-type rHHV-6ABAC (A to D) or rHHV-6ABACU14AAA (E to H) or its revertant virus (I to L). Cells were analyzed by transmission electron microscopy as previously described (36). Two or 3 fields in 30 cells in each group were observed. Arrows and arrowheads in panels A, E, and I indicate intranuclear and cytosolic capsids, respectively, in these three lines of infected cells. Black and white arrows in panels B, C, J, and K indicate enveloped virions in vacuoles near the Golgi complex (g) and accumulation of electron-dense tegument-like materials along the cytosolic face of some tubulovacuoles near the Golgi complex, respectively. n, nucleus. Scale bars, 0.5 μm.
In summary, we found that U14 is essential for HHV-6A propagation and that three amino acid residues (aa 429 to 431) are important for U14 function. Infectious virus could not be reconstituted without these residues. Replacement of these residues with alanine allowed reconstitution of infectious virus, but it exhibited severe growth defects. The mutant virus provides clues regardingU14 gene function. The EM data clearly show that the mutant virus harbors a marked maturation defect during postnuclear processes. The presence of fewer enveloped virus particles may be one of the reasons why the stock had a low titer and why fewer mature viral particles were released from infected cells. Another finding we found interesting was the presence of novel, amorphous, high-density (dense body) particles observed in rHHV-6ABACU14AAA-infected cells (Fig. 3G). We cannot explain the orientation of these structures or how they form. Since U14 is predicted to be a tegument protein (27, 30), any dysfunction may affect the tegumentation process during virus maturation, resulting in the accumulation of viral proteins in the cytosol of infected cells.
ACKNOWLEDGMENTS
We thank Gregory A. Smith (Department of Microbiology-Immunology, Northwestern University, Chicago, IL) for providing E. coli GS1783, Nikolaus Osterrieder (Institut für Virologie, Freie Universität Berlin, Berlin, Germany) for providing the pEP-KanS plasmid, Ulrich H. Koszinowski (Max von Pettenkofer-Institute, Ludwig-Maximilians-University, Munich, Germany) for providing the pHA-2 plasmid, and Hideto Yamada (Department of Obstetrics and Gynecology, Kobe University Graduate School of Medicine) for providing the CBMCs.
This study was supported in part by a Grant-in-Aid for Scientific Research (B) from the Japan Society for the Promotion of Science (JSPS).
REFERENCES
- 1.Guo H, Shen S, Wang L, Deng H. 2010. Role of tegument proteins in herpesvirus assembly and egress. Protein Cell 1:987–998. doi: 10.1007/s13238-010-0120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalejta RF. 2008. Functions of human cytomegalovirus tegument proteins prior to immediate early gene expression. Curr Top Microbiol Immunol 325:101–115. [DOI] [PubMed] [Google Scholar]
- 3.Kelly BJ, Bauerfeind R, Binz A, Sodeik B, Laimbacher AS, Fraefel C, Diefenbach RJ. 2014. The interaction of the HSV-1 tegument proteins pUL36 and pUL37 is essential for secondary envelopment during viral egress. Virology 454-455:67–77. doi: 10.1016/j.virol.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Le Sage V, Jung M, Alter JD, Wills EG, Johnston SM, Kawaguchi Y, Baines JD, Banfield BW. 2013. The herpes simplex virus 2 UL21 protein is essential for virus propagation. J Virol 87:5904–5915. doi: 10.1128/JVI.03489-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasdeloup D, McElwee M, Beilstein F, Labetoulle M, Rixon FJ. 2013. Herpesvirus tegument protein pUL37 interacts with dystonin/BPAG1 to promote capsid transport on microtubules during egress. J Virol 87:2857–2867. doi: 10.1128/JVI.02676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shu M, Taddeo B, Zhang W, Roizman B. 2013. Selective degradation of mRNAs by the HSV host shutoff RNase is regulated by the UL47 tegument protein. Proc Natl Acad Sci U S A 110:E1669–E1675. doi: 10.1073/pnas.1305475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strunk U, Saffran HA, Wu FW, Smiley JR. 2013. Role of herpes simplex virus VP11/12 tyrosine-based motifs in binding and activation of the Src family kinase Lck and recruitment of p85, Grb2, and Shc. J Virol 87:11276–11286. doi: 10.1128/JVI.01702-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tandon R, Mocarski ES. 2012. Viral and host control of cytomegalovirus maturation. Trends Microbiol 20:392–401. doi: 10.1016/j.tim.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xing J, Wang S, Lin R, Mossman KL, Zheng C. 2012. Herpes simplex virus 1 tegument protein US11 downmodulates the RLR signaling pathway via direct interaction with RIG-I and MDA-5. J Virol 86:3528–3540. doi: 10.1128/JVI.06713-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roizman B, Desrosiers RC, Fleckenstein B, Lopez C, Minson AC, Studdert MJ. 1992. The family Herpesviridae: an update. The Herpesvirus Study Group of the International Committee on Taxonomy of Viruses. Arch Virol 123:425–449. [DOI] [PubMed] [Google Scholar]
- 11.Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, Diluca D, Flamand L, Frenkel N, Gallo R, Gompels UA, Hollsberg P, Jacobson S, Luppi M, Lusso P, Malnati M, Medveczky P, Mori Y, Pellett PE, Pritchett JC, Yamanishi K, Yoshikawa T. 2014. Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol 159:863–870. doi: 10.1007/s00705-013-1902-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aubin JT, Collandre H, Candotti D, Ingrand D, Rouzioux C, Burgard M, Richard S, Huraux JM, Agut H. 1991. Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. J Clin Microbiol 29:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campadelli-Fiume G, Guerrini S, Liu X, Foa-Tomasi L. 1993. Monoclonal antibodies to glycoprotein B differentiate human herpesvirus 6 into two clusters, variants A and B. J Gen Virol 74:2257–2262. doi: 10.1099/0022-1317-74-10-2257. [DOI] [PubMed] [Google Scholar]
- 14.Dominguez G, Dambaugh TR, Stamey FR, Dewhurst S, Inoue N, Pellett PE. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J Virol 73:8040–8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gravel A, Ablashi D, Flamand L. 2013. Complete genome sequence of early passaged human herpesvirus 6A (GS strain) isolated from North America. Genome Announc 1:e00012-13. doi: 10.1128/genomeA.00012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isegawa Y, Mukai T, Nakano K, Kagawa M, Chen J, Mori Y, Sunagawa T, Kawanishi K, Sashihara J, Hata A, Zou P, Kosuge H, Yamanishi K. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J Virol 73:8053–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tweedy J, Spyrou MA, Donaldson CD, Depledge D, Breuer J, Gompels UA. 2015. Complete genome sequence of the human herpesvirus 6A strain AJ from Africa resembles strain GS from North America. Genome Announc 3:e01498-14. doi: 10.1128/genomeA.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyatt LS, Balachandran N, Frenkel N. 1990. Variations in the replication and antigenic properties of human herpesvirus 6 strains. J Infect Dis 162:852–857. doi: 10.1093/infdis/162.4.852. [DOI] [PubMed] [Google Scholar]
- 19.Salahuddin SZ, Ablashi DV, Markham PD, Josephs SF, Sturzenegger S, Kaplan M, Halligan G, Biberfeld P, Wong-Staal F, Kramarsky B, Gallo RC. 1986. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 20.Bigalke B, Klingel K, May AE, Kandolf R, Gawaz MG. 2007. Human herpesvirus 6 subtype A-associated myocarditis with “apical ballooning”. Can J Cardiol 23:393–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chi J, Gu B, Zhang C, Peng G, Zhou F, Chen Y, Zhang G, Guo Y, Guo D, Qin J, Wang J, Li L, Wang F, Liu G, Xie F, Feng D, Zhou H, Huang X, Lu S, Liu Y, Hu W, Yao K. 2012. Human herpesvirus 6 latent infection in patients with glioma. J Infect Dis 206:1394–1398. doi: 10.1093/infdis/jis513. [DOI] [PubMed] [Google Scholar]
- 22.Clark DA. 2002. Human herpesvirus 6 and human herpesvirus 7: emerging pathogens in transplant patients. Int J Hematol 76(Suppl 2):246–252. doi: 10.1007/BF03165124. [DOI] [PubMed] [Google Scholar]
- 23.Fremont M, Metzger K, Rady H, Hulstaert J, De Meirleir K. 2009. Detection of herpesviruses and parvovirus B19 in gastric and intestinal mucosa of chronic fatigue syndrome patients. In Vivo 23:209–213. [PubMed] [Google Scholar]
- 24.Garcia-Montojo M, De Las Heras V, Dominguez-Mozo M, Bartolome M, Garcia-Martinez MA, Arroyo R, Alvarez-Lafuente R. 2011. Human herpesvirus 6 and effectiveness of interferon beta1b in multiple sclerosis patients. Eur J Neurol 18:1027–1035. doi: 10.1111/j.1468-1331.2011.03410.x. [DOI] [PubMed] [Google Scholar]
- 25.Ogata M, Satou T, Kadota J, Saito N, Yoshida T, Okumura H, Ueki T, Nagafuji K, Kako S, Uoshima N, Tsudo M, Itamura H, Fukuda T. 2013. Human herpesvirus 6 (HHV-6) reactivation and HHV-6 encephalitis after allogeneic hematopoietic cell transplantation: a multicenter, prospective study. Clin Infect Dis 57:671–681. doi: 10.1093/cid/cit358. [DOI] [PubMed] [Google Scholar]
- 26.Lazzarotto T, Varani S, Gabrielli L, Pignatelli S, Landini MP. 2001. The tegument protein ppUL25 of human cytomegalovirus (CMV) is a major target antigen for the anti-CMV antibody response. J Gen Virol 82:335–338. doi: 10.1099/0022-1317-82-2-335. [DOI] [PubMed] [Google Scholar]
- 27.Stefan A, Secchiero P, Baechi T, Kempf W, Campadelli-Fiume G. 1997. The 85-kilodalton phosphoprotein (pp85) of human herpesvirus 7 is encoded by open reading frame U14 and localizes to a tegument substructure in virion particles. J Virol 71:5758–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takemoto M, Koike M, Mori Y, Yonemoto S, Sasamoto Y, Kondo K, Uchiyama Y, Yamanishi K. 2005. Human herpesvirus 6 open reading frame U14 protein and cellular p53 interact with each other and are contained in the virion. J Virol 79:13037–13046. doi: 10.1128/JVI.79.20.13037-13046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang H, Hayashi M, Maeki T, Yamanishi K, Mori Y. 2011. Human herpesvirus 6 glycoprotein complex formation is required for folding and trafficking of the gH/gL/gQ1/gQ2 complex and its cellular receptor binding. J Virol 85:11121–11130. doi: 10.1128/JVI.05251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang H, Kawabata A, Yoshida M, Oyaizu H, Maeki T, Yamanishi K, Mori Y. 2010. Human herpesvirus 6 encoded glycoprotein Q1 gene is essential for virus growth. Virology 407:360–367. doi: 10.1016/j.virol.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 33.Tang H, Mahmoud NF, Mori Y. 2015. Maturation of human herpesvirus 6A glycoprotein O requires coexpression of glycoprotein H and glycoprotein L. J Virol 89:5159–5163. doi: 10.1128/JVI.00140-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 35.Oyaizu H, Tang H, Ota M, Takenaka N, Ozono K, Yamanishi K, Mori Y. 2012. Complementation of the function of glycoprotein H of human herpesvirus 6 variant A by glycoprotein H of variant B in the virus life cycle. J Virol 86:8492–8498. doi: 10.1128/JVI.00504-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mori Y, Koike M, Moriishi E, Kawabata A, Tang H, Oyaizu H, Uchiyama Y, Yamanishi K. 2008. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 9:1728–1742. doi: 10.1111/j.1600-0854.2008.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]