ABSTRACT
Promyelocytic leukemia protein nuclear bodies (PML-NBs) are subnuclear domains implicated in cellular antiviral responses. Despite the antiviral activity, several nuclear replicating DNA viruses use the domains as deposition sites for the incoming viral genomes and/or as sites for viral DNA replication, suggesting that PML-NBs are functionally relevant during early viral infection to establish productive replication. Although PML-NBs and their components have also been implicated in the adenoviral life cycle, it remains unclear whether incoming adenoviral genome complexes target PML-NBs. Here we show using immunofluorescence and live-cell imaging analyses that incoming adenovirus genome complexes neither localize at nor recruit components of PML-NBs during early phases of infection. We further show that the viral DNA binding protein (DBP), an early expressed viral gene and essential DNA replication factor, independently targets PML-NBs. We show that DBP oligomerization is required to selectively recruit the PML-NB components Sp100 and USP7. Depletion experiments suggest that the absence of one PML-NB component might not affect the recruitment of other components toward DBP oligomers. Thus, our findings suggest a model in which an adenoviral DNA replication factor, but not incoming viral genome complexes, targets and modulates PML-NBs to support a conducive state for viral DNA replication and argue against a generalized concept that PML-NBs target incoming viral genomes.
IMPORTANCE The immediate fate upon nuclear delivery of genomes of incoming DNA viruses is largely unclear. Early reports suggested that incoming genomes of herpesviruses are targeted and repressed by PML-NBs immediately upon nuclear import. Genome localization and/or viral DNA replication has also been observed at PML-NBs for other DNA viruses. Thus, it was suggested that PML-NBs may immediately sense and target nuclear viral genomes and hence serve as sites for deposition of incoming viral genomes and/or subsequent viral DNA replication. Here we performed a detailed analyses of the spatiotemporal distribution of incoming adenoviral genome complexes and found, in contrast to the expectation, that an adenoviral DNA replication factor, but not incoming genomes, targets PML-NBs. Thus, our findings may explain why adenoviral genomes could be observed at PML-NBs in earlier reports but argue against a generalized role for PML-NBs in targeting invading viral genomes.
INTRODUCTION
Viruses are intracellular parasites and utilize and/or divert cellular mechanisms for their propagation. To eliminate invading viruses and suppress viral replication, cells have evolved intracellular antiviral defense mechanisms. A prominent example is the antiviral activity of the promyelocytic leukemia protein nuclear body (PML-NB) (1–3). PML-NBs can be observed as punctate dots in the nucleus in immunofluorescence (IF) analyses and have been shown to occupy stable positions in the nucleus over time (1, 2). Interferon promotes PML-NB formation, and several interferon-responsive factors, including PML, Sp100, and Daxx, are known to localize at PML-NBs (1, 2) but differ significantly in their average residing times (4). Maul et al. were the first to show that incoming genomes of several nuclear replicating DNA viruses, such as herpes simplex virus 1 (HSV-1), simian virus 40 (SV40), and adenovirus (Ad), reside in and then start DNA replication at PML-NBs (5). Later similar observations were made for other members of the herpesvirus family, including human cytomegalovirus (HCMV) (6) and Epstein-Barr virus (EBV) (7), as well as papillomavirus (8). Thus, it is speculated that this subnuclear domain is a general site for deposition of incoming viral genomes and/or viral DNA replication (9, 10), although whether this association occurs through active targeting of existing PML-NBs or via de novo formation on the genome remains uncertain. HSV-1 is the best-studied model of the involvement of PML-NBs (9, 10). ICP0, an immediate early gene product of HSV-1 encoding an E3 ubiquitin ligase (11), promotes viral replication by degrading several host proteins, including PML (12). Everett et al. showed that immediately after nuclear entry of HSV-1 genomes, PML-NB components are recruited onto viral genomes at the nuclear periphery, suggesting that PML-NB-like structures form de novo (13). Since depletion of PML or other PML-NB components can rescue the replication defect of ICP0-null mutant viruses (14–16), the recruitment of the components onto incoming HSV-1 genomes has been thought to be a cellular antiviral response against infection (9, 10). This idea is partly supported by the report of HCMV showing PML-NB localization of a viral protein IE2, a possible marker for viral genomes, in immediate early phases of infection (6). However, since few spatiotemporal analyses for incoming genomes of other DNA viruses have been reported (9), it remains to be determined whether the encounter of PML-NB components is a general cellular defense against invading DNA viruses. In addition, the effects of depletion of PML-NB components have different effects in different viral systems: knockdown or knockout of certain PML-NB components resulted in enhancement, no effect, or suppression of viral propagation (8, 17–23), suggesting distinct responses against each virus. Furthermore, although colocalization has been observed, it is not clear how incoming viral genomes and/or viral DNA replication activities are connected to PML-NBs.
Ad is a nonenveloped virus with a linear double-stranded DNA genome. The Ad genome forms a chromatin-like structure with viral basic core proteins in the virion (24). The major DNA binding protein VII forms irregularly spaced nucleosomes on the genome (24, 25), which remain associated with the viral genome at least during the first hours of infection (26, 27). Protein V appears to be lost before the nuclear import of the genomes (28), while the fate of polypeptide X/Mu is unclear. Thus, for the first hours after nuclear import, protein VII marks viral genome complexes in cells (29). An early report suggested that incoming Ad genomes localize at PML-NBs (5). In this report, however, the genome localization at PML-NBs was observed at 4 but not at 1.5 h postinfection (hpi) (5). This is somewhat different from HSV-1, where genomes recruit PML-NB components at the nuclear periphery immediately upon nuclear entry (13). Ishov and Maul also reported that Ad DNA replication occurs at sites juxtaposed to PML-NBs and that the PML-NB resident protein Sp100 is specifically relocalized into viral DNA replication centers (5), which can be visualized by immunostaining of DBP, a viral single-strand DNA binding protein involved in DNA replication (30, 31). This observation is further supported by the recent works of Dobner et al. showing that USP7, another PML-NB component, and the specific isoforms of Sp100 are recruited into Ad DNA replication centers (18, 32). In contrast, PML is not recruited into viral DNA replication centers but is observed to be closely associated with newly formed replication centers (5). In summary, it remains open if Ad genome complexes associate with PML-NBs immediately upon nuclear entry, similar to HSV-1, or if the association with PML-NBs occurs later (e.g., when viral DNA replication takes place).
In this study, we sought to clarify the interplay between incoming Ad genome complexes and PML-NBs during early phases of infection. Using imaging analyses, including a recently developed live-cell imaging system (29), we show that PML-NBs are not immediate deposition sites for incoming Ad genome complexes. Furthermore, we found that DBP alone is sufficient to target PML-NBs and recruit Sp100 and USP7 through oligomerization. Taken together, our findings suggest that the Ad DNA replication factor DBP, but not incoming viral genome complexes themselves, targets PML-NBs, which may explain the earlier observation of the “delayed” genome localization at PML-NBs and argue against a general role for PML-NBs in the recognition of incoming viral genomes.
MATERIALS AND METHODS
Cell and viruses.
U2OS (ATCC HTB-96), H1299 (ATCC CRL-5803), and HEK293 cells (ATCC CRL-1573) were maintained in Dulbecco's modified Eagle's medium (DMEM)-Glutamax (Life Technologies) supplemented with 10% fetal calf serum (FCS). Human foreskin fibroblasts (HFFs) were obtained from J. Dechanet (CIRID University of Bordeaux) and maintained as described above. Recombinant replication-competent human adenovirus type 5 (Ad5), replication-deficient E1-deleted green fluorescent protein (GFP)-expressing Ad5 vector (Ad5-GFP), and Ad5-GFP-M1, in which the PPxY motif of protein VI is mutated (33), were amplified and purified as described previously (33, 34). Ad infection was carried out at a multiplicity of infection (MOI) of 100 PFU/cell. The transfection of plasmids was done using Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol.
pLKO.1-based lentiviral vectors expressing control short hairpin RNAs (shRNAs) (pLKO1-puro-shCtrl; kindly provided by the Plateforme de Vectorologie, Université de Bordeaux) and validated shRNAs against the PML and USP7 genes (pLKO.1-puro-shPML [NM_002675.x-1501s1c1] and pLKO.1-puro-shUSP7 [NM_003470.x-2618s1c1]; Sigma-Aldrich) were prepared and titrated by the Plateforme de Vectorologie (Université de Bordeaux). For knockdown experiments, cells were infected with lentiviral vectors at 5 infectious particles/cell and subjected to puromycin selection at 4 days post-lentiviral transduction.
Antibodies.
The antibodies used in this study are as follows: rat anti-protein VII (27), mouse anti-Daxx (ab9091 [Abcam]), rabbit anti-Daxx (07-471 [Millipore]), rabbit anti-ATRX (sc-15408 [Santa Cruz Biotechnology]), mouse anti-PML (sc-966 [Santa Cruz Biotechnology]), rabbit anti-PML (sc-5621 [Santa Cruz Biotechnology], and NB100-59787 [Novus Biologicals]), rabbit antihemagglutinin (anti-HA) (sc-805 [Santa Cruz Biotechnology]), and rat anti-HA (3F10 [Roche Life Science]).
Rabbit anti-Ad5, mouse anti-DBP, rat anti-USP7, and rabbit anti-Sp100 antibodies were kind gifts provided by R. Iggo (Institut Bergonié), T. Dobner (Heinrich-Pette-Institute), and T. Sternsdorf (Research Institute Children's Cancer Center Hamburg), respectively.
Plasmids.
The expression vectors for enhanced green fluorescent protein (EGFP)-TAF-Iβ, HA-DBP, and HA-DBPΔC (pEGFP-C1-TAF-Iβ, pCHA-puro-DBP, and pCHA-puro-DBPΔC) are described elsewhere (29, 35). For the construction of the Daxx expression vector, the cDNA fragment for Daxx was amplified by PCR, digested with BamHI and EcoRI, and inserted into the pCHA-puro vector (35) (pCHA-puro-Daxx) and subsequently combined with a N-terminal insertion of the FLAG-mCherry tag. The expression vector for mCherry-TAF-Iβ (pCHA-puro-FLAG-mCherry-TAF-Iβ) was constructed by first inserting the cDNA for TAF-Iβ into the BamHI/EcoRI site of the pCHA-puro vector (pCHA-puro-TAF-Iβ) and then inserting the FLAG-mCherry cDNA fragment into the BamHI site of the resultant plasmid. The expression vector for EGFP-DBP was constructed as follows. The cDNA fragment for DBP was obtained from pCHA-puro-DBP by digestion with BamHI and EcoRI and inserted into the BglII/EcoRI site of the pEGFP-C1 vector (cloning details provided upon request).
The expression vector for mCherry-tagged PML (pcDNA3-PML-mCherry) containing the cDNA for PML-IIA/isoform 11 of PML was obtained from MGC Montpellier Genomic Collections (Institut de Génétique Moléculaire de Montpellier). The expression vector for EGFP-tagged ATRX (pEGFP-C2-ATRX) is a generous gift from D. Pickets (Ottawa Hospital Research Institute) (36).
For the preparation of cells stably expressing EGFP-tagged and mCherry-tagged TAF-Iβ, U2OS cells were transfected with either pEGFP-C1-TAF-Iβ or pCHA-puro-FLAG-mCherry-TAF-Iβ and cultured for 2 weeks in the presence of 2 mg/ml G418 or 2 μg/ml puromycin, respectively.
Immunofluorescence and live-cell imaging analysis.
Indirect immunofluorescence (IF) and live-cell imaging analyses were carried out as described previously (29). IF samples were analyzed by a Leica SP5 confocal microscope. Confocal stacks were taken every 0.3 μm, and images were processed using ImageJ and presented as maximum intensity projections. For live-cell imaging, cells were seeded in Ibidi μ-slide VI0.4 (Ibidi), and images were acquired using a Leica spinning-disk microscopy system (×100 objective) equipped with an incubation chamber at 37°C. Frames were taken every 3 s for each color channel and assembled into movies using MetaMorph software.
Production and detection of BrdU-labeled viruses.
To produce bromodeoxyuridine (BrdU)-labeled viruses, HEK293 cells were infected with Ad5, and 10 μM BrdU was added to the culture medium at 16 hpi. At 24 hpi, cells were extensively washed with phosphate-buffered saline (PBS) to remove unincorporated BrdU, resuspended in fresh DMEM, and subjected to five freeze-thaw cycles to release progeny virions. Supernatants were cleared by centrifugation and collected as progeny virus solution. U2OS cells were infected with the progeny virus solution and at 2 hpi subjected to IF analyses as well as BrdU detection using a Roche IF-compatible BrdU labeling and detection kit according to the manufacturer's protocol. Samples were analyzed by microscopy as described above.
RESULTS
Incoming Ad genome complexes do not localize at PML-NBs during the first hours of infection.
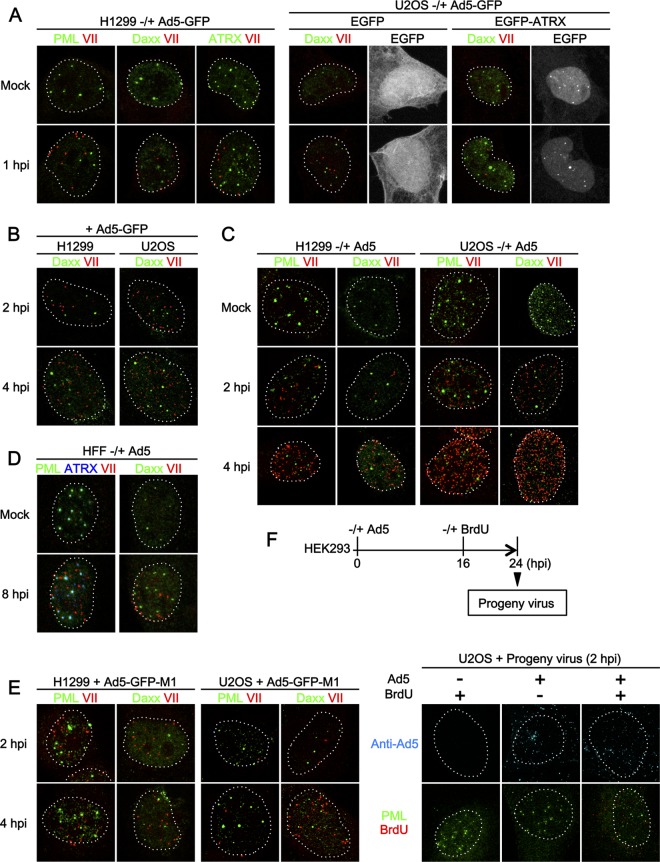
The fate of incoming Ad genome complexes after nuclear import is an open question. To examine whether incoming Ad genome complexes localize at PML-NBs, we performed IF analyses using antibodies against PML-NB components and protein VII, a marker for viral genome complexes (29) (Fig. 1). First, we wanted to investigate if incoming genome complexes target PML-NBs independently of viral gene expression. To this end, we used the replication-deficient Ad vector (Ad5-GFP), in which the E1 gene is replaced with the GFP-expressing cassette. H1299 cells were either mock infected or infected with Ad5-GFP and at 1 hpi subjected to IF analyses (Fig. 1A, left panels). We did not observe any specific colocalization between protein VII foci and the different PML-NB components, PML, Daxx, and ATRX. Similar analyses were carried out using U2OS cells (Fig. 1A, right panels). In the U2OS cell, ATRX is not expressed due to the deletion of the gene (37). Recently it has been reported that ATRX functions together with Daxx as a negative regulator in Ad gene expression and that the expression of EGFP-tagged ATRX can reconstitute the functional Daxx/ATRX complex in U2OS cells (38). Therefore, U2OS cells were first transfected with the expression vectors for EGFP alone or EGFP-ATRX and then infected with Ad5-GFP and subjected to IF analyses. Again no colocalization between protein VII foci and Daxx regardless of the ATRX expression was observed (Fig. 1A, right panel). Colocalization between protein VII and Daxx was also not observed up to 4 hpi in both H1299 and U2OS cells (Fig. 1B). Lack of colocalization was not due to the absence of the E1 gene, as similarly we did not observe any colocalization between viral genome complexes and PML or Daxx up to 4 hpi when using the replication-competent wild-type viruses (Fig. 1C, Ad5). In this study, we neither performed synchronized infection nor removed unbound viruses, allowing unsynchronized, continuous infection events during incubation periods. Consequently, the number of protein VII foci was generally greater in later time points (e.g., Fig. 1C, compare 4 hpi with 2 hpi). This increment was not due to de novo synthesis of protein VII, as it was also observed with Ad5-GFP (Fig. 1B). Even in cells exceptionally showing an excess amount of protein VII foci (e.g., Fig. 1C, 4 hpi), protein VII-free PML-NBs were still observed, with only a limited number occasionally overlapping, further suggesting the absence of specific colocalization or targeted recruitment. Likewise, we observed no colocalization between a marker for incoming Ad genome complexes and two other PML-NB-resident proteins, Sp100 and USP7 (data not shown). The absence of colocalization was not due to our choice of cell models, as we confirmed our observation in a primary cell model using human foreskin fibroblasts (HFFs) (Fig. 1D). HFFs have been shown to be susceptible to Ad infection but allow its replication only at a very low rate (39). PML-NB components did not show track-like localization (induced by E4orf3 [see Discussion]) even at 8 hpi, confirming that cells were still in immediate early stages of infection. Thus, we were not able to observe the localization of incoming Ad genome complexes at PML-NBs, regardless of immediate early gene expression, cell types, and ATRX expression.
FIG 1.
Incoming Ad genomes are not colocalized with PML-NBs. (A) IF analyses with Ad5-GFP. H1299 cells were either mock infected (first row) or infected with replication-deficient Ad5-GFP (second row) and at 1 hpi subjected to IF analyses using antibodies against PML-NB components (green) and protein VII (red, left panels). Dashed lines indicate the shapes of the nuclei indicated by DAPI (4′,6-diamidino-2-phenylindole) staining (not shown). For U2OS cells, cells were first transfected with the expression vectors for either EGFP alone or EGFP-ATRX (gray), and at 24 h posttransfection (hpt), IF analyses were performed as described above (right panels). (B) IF analyses with later time points. H1299 (left) or U2OS (right panels) cells were infected with Ad5-GFP and at 2 hpi (first row) and 4 hpi (second row) subjected to IF analyses. (C) IF analyses with Ad5. H1299 (left) or U2OS (right panels) cells were either mock infected (first row) or infected with replication-competent Ad5 and at 2 hpi (second row) and 4 hpi (third row) subjected to IF analyses. (D) IF analyses with human foreskin fibroblasts (HFFs). HFFs were either mock infected (first row) or infected with Ad5 (second row) and at 8 hpi subjected to IF analyses using antibodies against PML-NB components (green and blue) and protein VII (red). (E) IF analyses with Ad5-GFP-M1. H1299 (left panels) or U2OS (right panels) cells were infected with Ad5-GFP-M1, in which the PPxY motif of protein VI is mutated, and at 2 (first row) and 4 (second row) hpi subjected to IF analyses with the indicated antibodies. (F) IF analyses using BrdU-labeled viruses. To produce BrdU-labeled viruses, HEK293 cells were infected with Ad5, and 10 μM BrdU was added to the culture medium at 16 hpi. At 24 hpi, progeny viruses were released from infected cells. U2OS cells were infected with progeny viruses produced under the indicated conditions and at 2 hpi subjected to IF analyses using either anti-Ad5 (upper panels, cyan) or anti-PML (green) and anti-BrdU (red) antibodies (lower panels).
In addition to PML-NBs, centromeric heterochromatin is reported to associate with foreign DNAs delivered by polyomavirus-like particles (40). Thus, we next examined if protein VII foci associate with CENP-A, a histone H3 variant specific for centromeres (data not shown). Again, we did not observe any specific colocalization between protein VII foci and CENP-A, suggesting that centromeric heterochromatin is also not the site where incoming Ad genome complexes are deposited.
Previously we reported that protein VI, a component of incoming virions, may counteract Daxx to activate viral gene expression and that the conserved PPxY motif of protein VI could be important for this action (34). To examine the involvement of protein VI in the localization of protein VII foci, we performed IF analyses using the protein VI PPxY-mutated virus (33) (Fig. 1E, Ad5-GFP-M1). In both H1299 and U2OS cells, however, colocalization between protein VII foci and PML-NB components was not observed, as was the case for Ad5-GFP, suggesting that the PPxY-motif of protein VI is unlikely the cause of the lack of Ad genome association with PML-NBs.
To further strengthen our data, we also directly visualized intracellular viral genomes to confirm their lack of PML-NB targeting. To this end, we used BrdU for labeling of viral genomes (Fig. 1F). To produce BrdU-labeled viruses, cells were infected with Ad5, and BrdU was added to the culture medium at 16 hpi. Progeny virions released from infected cells at 24 hpi were collected and used for the next round of infection with U2OS cells. While progeny virus infection was observed irrespective of BrdU addition during the initial virus production step (Fig. 1F, anti-Ad5), BrdU signals were highly specific for progeny viruses that were produced in the presence of BrdU (Fig. 1F, BrdU), indicating the specific incorporation of BrdU into progeny viral genomes. Again we observed no colocalization between viral genomes (BrdU signals) and PML, consistent with our observations obtained using anti-protein VII antibody.
Incoming Ad genome complexes do not recruit PML-NB components in living cells.
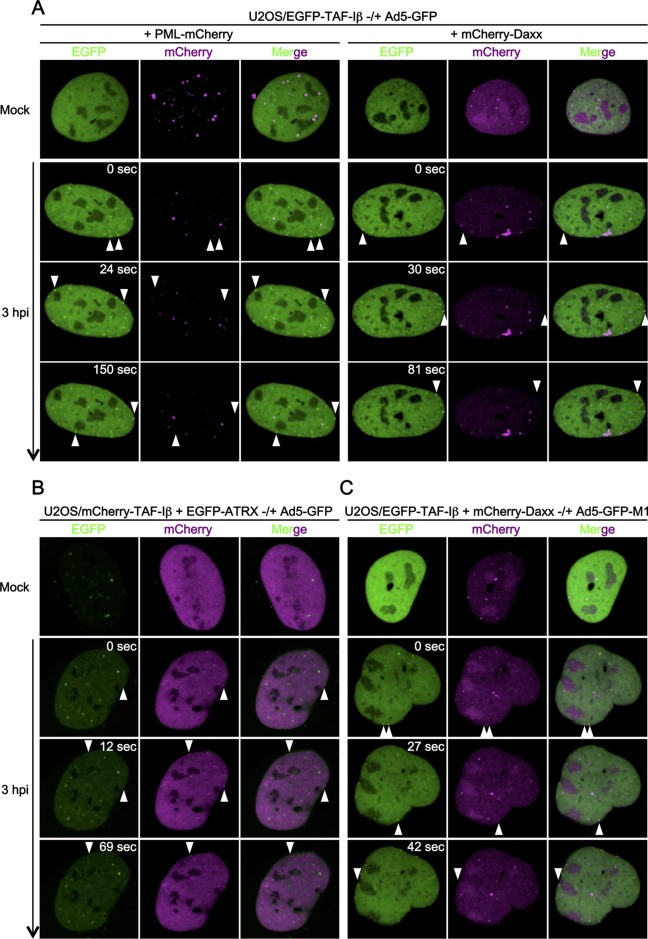
Although we could not observe colocalization between protein VII foci and PML-NB components in IF analyses (Fig. 1), it remained possible that incoming Ad genome complexes only transiently localize at PML-NBs and/or recruit its components—for instance, upon nuclear import, as reported for HSV-1 (13). To test this possibility, we next tested the association of incoming Ad genome complexes with PML-NBs in living cells using our recently developed live-cell imaging system (29). The analysis is based on the use of fluorescently labeled TAF-I, a cellular chromatin protein binding to protein VII upon Ad infection (41). Because EGFP-tagged or mCherry-tagged TAF-I forms complexes with genome-bound protein VII upon nuclear import of Ad genome complexes, it can be used as a marker depicting the localization of viral genome complexes in living cells (29). U2OS cells stably expressing EGFP-TAF-Iβ (U2OS/EGFP-TAF-Iβ cells) were transiently transfected with the expression vectors for either mCherry-tagged PML (Fig. 2, left; see Movie S1 in the supplemental material) or mCherry-tagged Daxx (Fig. 2, right panels; see Movie S2 in the supplemental material) and infected with Ad5-GFP for imaging. In both cases, TAF-I foci started to form ∼1 hpi, when Ad genome import initiated, and could be observed at the nuclear periphery for over 4 hpi (data not shown). No colocalization between TAF-I foci and PML or Daxx was observed in living cells, confirming our results in fixed cells (Fig. 2, arrowheads). Similarly, EGFP-tagged ATRX (when expressed in U2OS cells) was not colocalized with mCherry-TAF-I foci (Fig. 2B; see Movie S3 in the supplemental material). Because no recruitment of Daxx was also observed when using the protein VI mutant virus Ad5-GFP-M1 (Fig. 2C; see Movie S4 in the supplemental material), this was independent of the PPxY motif in protein VI. These results suggest that unlike HSV-1, incoming Ad genomes do not recruit PML-NB components immediately after nuclear entry. Taken together, our data from IF and live-cell imaging analyses strongly suggest that incoming Ad genome complexes neither stably reside at PML-NBs during the first hours of infection nor recruit its components at the nuclear periphery upon nuclear import.
FIG 2.
Incoming Ad genomes neither recruit components of nor are colocalized with PML-NBs in living cells. (A) Live-cell imaging using EGFP-TAF-Iβ. U2OS cells stably expressing EGFP-tagged TAF-Iβ (U2OS/EGFP-TAF-Iβ cells [green]) were first transiently transfected with the expression vectors for either mCherry-tagged PML (left panels) or Daxx (right panels, magenta) and then either mock infected (first row) or infected with Ad5-GFP (second through fourth rows) for live-cell imaging. Frames were taken every 3 s for 2 min (for mock-infected cells) or 3 min (for infected cells); snapshots from the movies are shown. Arrowheads indicate infection-specific TAF-I foci at the nuclear periphery. Full movies are provided as Movies S1 and S2 in the supplemental material. (B) Live-cell imaging using EGFP-ATRX. U2OS/mCherry-TAF-Iβ (magenta) cells were transiently transfected with the expression vector for EGFP-tagged ATRX (green) and then either mock infected or infected with Ad5-GFP for live-cell imaging. Full movies are provided as Movie S3 in the supplemental material. (C) Live-cell imaging using Ad5-GFP-M1. U2OS/EGFP-TAF-Iβ cells (green) were transiently transfected with the expression vector for mCherry-Daxx (magenta) and then either mock infected or infected with Ad5-GFP-M1 for live-cell imaging. Full movies are provided as Movie S4 in the supplemental material.
DBP targets PML-NBs and recruits its components in the absence of any other viral factors.
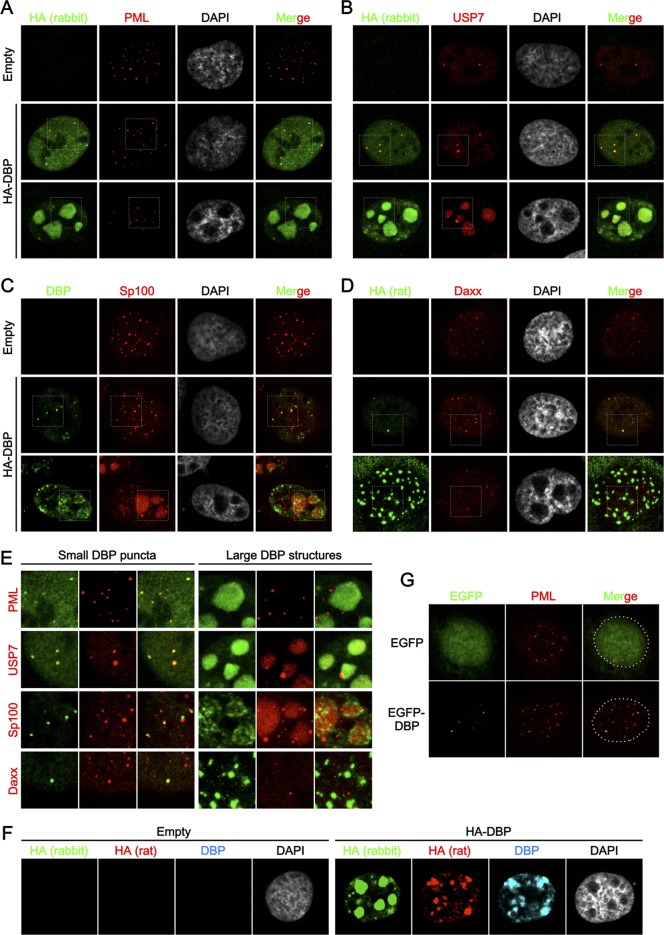
It is important to note that Ad gene expression resumes within the first hour of infection and is well on its way at 4 hpi (26, 34, 41). Because we did not see any increase in association with PML-NBs of Ad genome complexes up to 4 hpi, it is unlikely that early viral gene expression is a major contributor to PML-NB association. Thus, we next sought to investigate the involvement of PML-NBs in viral DNA replication. Since no colocalization between incoming Ad genome complexes and PML-NBs was observed, we speculated that PML-NBs and/or its components might recruit or be recruited to viral DNA replication components/compartments independently of viral genomes. Previously we reported that transiently expressed DBP forms subnuclear structures through its oligomerization in the absence of any other viral components and proposed that by forming these structures, DBP establishes an environment conducive for viral DNA replication (35). Here we hypothesized that DBP itself may also have the ability to associate with and/or modulate PML-NBs and/or its components. To test this, we first carried out IF analyses using cells transiently expressing HA-tagged DBP (Fig. 3). In a population of cells, HA-DBP formed large structures in nuclei as reported previously (35) (Fig. 3A to D, third rows), while some cells showed small puncta of DBP (second rows). The formation of either small puncta or large structures likely depended on the relative expression levels of DBP; higher expression levels of the protein tended to form large structures, while cells showing very low expression levels sometimes exhibited only diffuse nuclear localization without forming any foci (not shown). When costained with anti-PML antibody, some but not all of the small DBP foci were observed at or juxtaposed with PML-NBs (Fig. 3A, second row, and E), as was previously observed for the localization of DBP in infected cells (5). Furthermore, we observed close association between PML dots and large structures of DBP (Fig. 3A, third row, and E), although we cannot formally exclude the possibility of random association due to the large size of the structures. We next performed the same IF assays using antibodies against the PML-NB components USP7 and Sp100, both of which have been reported to be relocalized into Ad DNA replication centers in infected cells (Fig. 3B and C). Similar to PML, colocalization and/or close association between small DBP puncta and USP7 or Sp100 was observed (Fig. 3B and C, second rows, and D). In cells showing large DBP structures, both USP7 and Sp100 were recruited into the structures (Fig. 3B and C, third rows, and E), as also observed in infected cells (5, 18, 32). We examined the localization of Daxx in the presence of HA-DBP and observed colocalization and/or association with small DBP dots but no recruitment into large DBP structures (Fig. 3D and E), similar to PML. We noted that the rabbit anti-HA antibody could depict large DBP structures much better than mouse anti-DBP and rat anti-HA antibodies under our experimental conditions (Fig. 3F). These phenomena were also observed when using EGFP-tagged DBP (Fig. 3G) or exogenously expressed PML and USP7 (data not shown), confirming the findings presented above. Taken together, these results suggest that DBP can target PML-NBs and recruit USP7 and Sp100 into large structures in the absence of any additional viral factors, including viral genomes.
FIG 3.
DBP targets PML-NBs in the absence of additional viral factors. U2OS cells were transfected with either an empty vector (first rows) or the vector for HA-DBP (second and third rows) and at 24 hpt subjected to IF analyses. HA-DBP was detected using either rabbit anti-HA (A and B, first columns, green), anti-DBP (C), or rat anti-HA (D) antibodies, while PML-NB components were stained with specific antibodies (second columns, red, PML, USP7, Sp100, and Daxx for panels A, B, C, and D, respectively). DAPI staining (gray) and merged images are shown in the third and fourth columns, respectively. Cells with small DBP puncta and large DBP structures are shown in the second and third rows, respectively. (E) Details for DBP structures. Higher-magnification images marked by squares are shown. (F) IF analyses using antibodies against HA and DBP. U2OS cells were transfected with either an empty vector (left) or the vector for HA-DBP (right panels) and at 24 hpt subjected to IF analyses using rabbit anti-HA (green, first column), rat anti-HA (red, second column), and mouse anti-DBP (cyan, third column) antibodies. DAPI staining is shown in the fourth columns (gray). (G) IF analyses using EGFP-tagged DBP. U2OS cells were transfected with the expression vectors for EGFP alone (first row, green) or EGFP-DBP (second row) and at 24 hpt subjected to IF analyses using anti-PML antibody (red).
PML-NB components are independently recruited into DBP structures upon oligomerization.
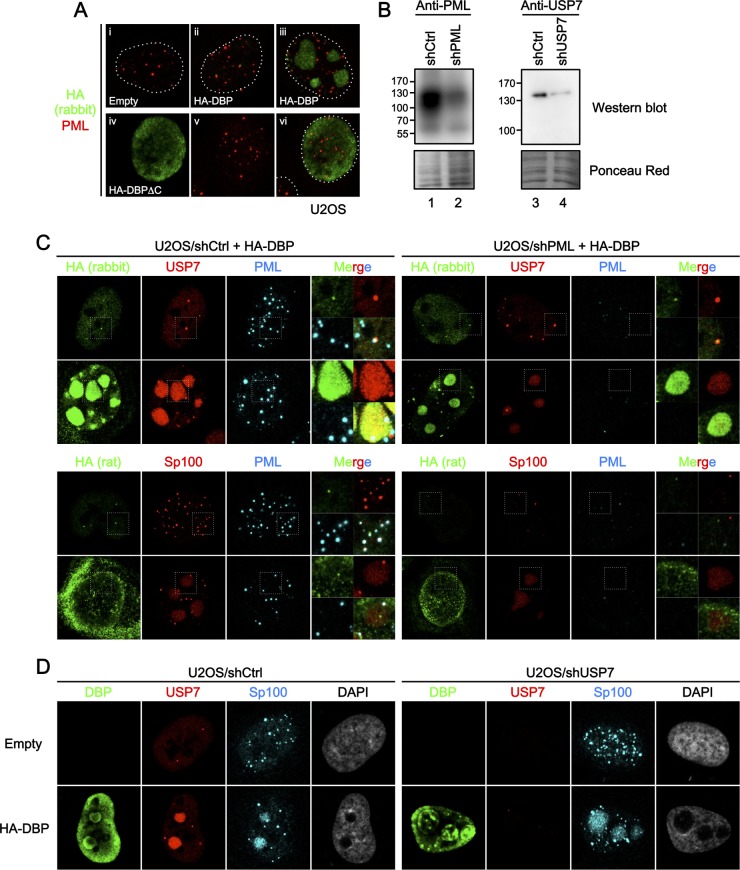
Next we sought to examine the underlying mechanisms of how PML-NB components recruit and/or are recruited into DBP structures. In the previous study, we showed that deletion of the C-terminal extension of DBP, which is necessary for its oligomerization (31), results in loss of the formation of the subnuclear structures in cells (35). Consistent with the previous observation, the deletion mutant of DBP (DBPΔC) localized only diffusely in the nucleus, independently of its expression levels, and did not show any specific association with PML-NBs (Fig. 4A), suggesting that PML-NB targeting or recruitment of DBP is dependent on its oligomerization.
FIG 4.
Depletion of one PML-NB component does not impair the recruitment of other factors into DBP structures. (A) IF analyses using HA-DBPΔC. U2OS cells were transfected with an empty vector (i), the vectors for HA-DBP (ii and iii), or HA-DBPΔC (iv to vi) and at 24 hpt subjected to IF analyses using anti-HA (green) and anti-PML (red) antibodies. (B) Western blotting with shRNA-treated cells. Cell lysates were prepared from U2OS cells transduced with either control shRNA-expressing (shCtrl, lanes 1 and 3), shPML-expressing (lane 2), or shUSP7-expressing (lane 4) lentiviral vectors and subjected to Western blot analyses using either anti-PML (left) or anti-USP7 (right panels) antibodies. Ponceau red staining is shown below as a loading control. (C) IF analyses with shPML-treated cells. U2OS/shCtrl (left) and U2OS/shPML (right panels) cells were transfected with an expression vector for HA-DBP, and IF analyses were carried out at 24 hpt using the indicated antibodies. Higher-magnification images marked by squares and merged images are also shown. (D) IF analyses with shUSP7-treated cells. U2OS/shCtrl (left panels) and U2OS/shUSP7 (right panels) cells were transfected with either an empty vector (first row) or a vector for HA-DBP (second row). At 24 hpt, IF analyses were carried out using the indicated antibodies.
Next we prepared knockdown cells for PML and USP7 using shRNA-expressing lentiviral vectors (Fig. 4B), to examine whether knockdown of one PML-NB component affected the recruitment of other components into DBP structures. When HA-DBP was expressed in shPML-treated cells, USP7 remained colocalizing with DBP structures (Fig. 4C, U2OS/shPML, first and second rows). Likewise, Sp100 was recruited into large DBP structures in the absence of PML (Fig. 4C, U2OS/shPML, fourth row). We next wanted to know if the recruitment of USP7 and Sp100 into DBP structures was linked. When we depleted cells of USP7 (Fig. 4D), we found that USP7 knockdown did not affect the recruitment of Sp100 into large DBP structures (U2OS/shUSP7, second row). We also performed small interfering RNA (siRNA)-mediated knockdown of Sp100 and found no effect on the recruitment of USP7 into DBP structures (data not shown). Thus, our results suggest that the absence of one PML-NB component is unlikely to impair the recruitment of other components into DBP structures.
In summary our results show that the Ad replication factor DBP when overexpressed autonomously forms subnuclear structures through oligomerization resembling viral DNA replication compartments and selectively recruits USP7 and Sp100 into the structures.
DBP alone is not sufficient to recruit viral genomes into PML-NBs.
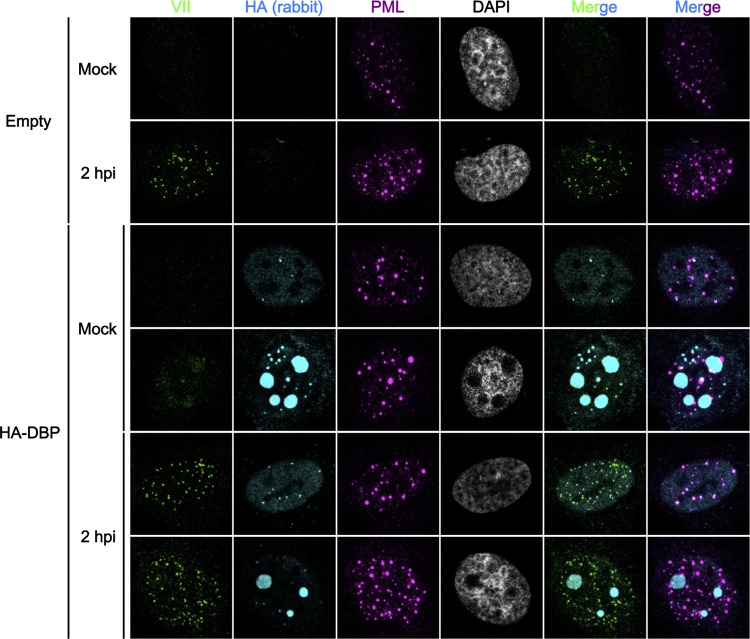
Our findings so far suggested that DBP is a major contributing factor to PML-NB targeting of viral genomes during Ad infection. Thus, we finally examined whether DBP alone is sufficient to recruit incoming viral genome complexes into DBP structures and/or PML-NBs (Fig. 5). U2OS cells were first transfected with the expression vector for HA-DBP and then infected with Ad5. However, we observed no specific colocalization between protein VII foci and PML, even in the presence of preexpressed DBP (Fig. 5). This suggests that DBP is not sufficient to mediate the association between PML-NB components and viral genome complexes, but other viral factors and/or active DNA replication may be needed to recruit viral genomes (see Discussion).
FIG 5.
Exogenously expressed DBP is not sufficient to recruit incoming viral genome complexes into PML-NBs. U2OS cells were transfected with either an empty vector (first and second rows) or the vector for HA-DBP (second through sixth rows) and at 24 hpt either mock infected (first, third, and fourth rows) or infected with Ad5 (second, fifth, and sixth rows). At 2 hpi, cells were subjected to IF analyses using anti-protein VII (green, first column), anti-HA (cyan, second column), and anti-PML (magenta, third column) antibodies. DAPI staining (gray) and merged images are shown in the fourth to sixth columns.
DISCUSSION
It has been proposed that the recruitment of PML-NB components onto viral genomes is a common cellular response against nuclear replicating DNA viruses (3, 9, 10). In this study, however, we observed neither the genome localization at PML-NBs nor recruitment of the components toward viral genomes during the first hours of Ad infection. Our observation thus opens the question of whether the recruitment of the PML-NB components upon genome delivery is unique for HSV-1 or if Ad evolved specific mechanisms to prevent antiviral activities of PML-NBs at this stage. There are several differences between HSV-1 and Ad genomes: the HSV-1 genome is more than 150 kbp in length and possibly unprotected immediately after nuclear entry, while the Ad genome is smaller (about 35 kbp) and chromatinized with protein VII. These features might be critical for how cells sense and respond to incoming viral genomes.
Previously we reported that an Ad capsid component, protein VI, targets PML-NBs and may interact with and counteract Daxx to ensure viral gene expression (34). In this study, using the virus harboring protein VI mutated in the PPxY motif (33), which is critical for counteracting Daxx (34), we did not find evidence of altered genome localization with respect to PML-NBs (Fig. 1E and 2C). Therefore, in contrast to HSV-1, the genomes of which are globally occupied and repressed by PML-NB components, it appears that incoming Ad genomes are not targeted by PML-NB components irrespective of protein VI. However, the results obtained here do not exclude the possibility that Daxx plays a role in local repression of specific regions on the Ad genome (e.g., promoter regions), as we proposed previously (34).
In contrast to our observation, Ishov and Maul observed Ad genome localization at PML-NBs at 4 hpi (5). To address this contradiction, we investigated the involvement of viral factors involved in DNA replication and found that DBP alone can target PML-NBs even in the absence of viral genomes. Thus, our study suggests a model in which viral genomes do not localize at PML-NBs immediately after nuclear entry but become associated with the domains only after DBP is expressed and DNA replication initiated. This scenario would be in line with the fact that Ad genomes were observed at PML-NBs at 4 but not 1.5 hpi in the previous report and that viral replication centers are associated with PML-NBs (5). However, DBP alone is not sufficient to recruit incoming viral genome complexes into DBP structures and/or PML-NBs (Fig. 5). Accordingly, it is possible that in the infection context, DBP oligomerizes on viral genomes during ongoing DNA replication, which is on one hand essential for the replication process (31) and as we show in this article required for PML-NB targeting (Fig. 4A), promoting the association with PML-NBs and/or the recruitment of the components. It remains unclear how DBP recruits and/or associates with PML-NBs and its components. In our immunoprecipitation analyses, both HA-DBP and HA-DBPΔC failed to coprecipitate endogenous PML, Sp100, and USP7 (data not shown), suggesting that the association is unlikely to be simply mediated by protein-protein interactions, nor does the recruitment of individual PML-NB components by DBP seem to be linked. Further studies are needed to address this point.
What is the biological/virological significance for the association between PML-NBs and DBP or Ad DNA replication centers? The consequence of the recruitment of PML-NB component Sp100 or USP7 into viral DNA replication centers is unclear, because knockdown of the components has been shown to either promote (Sp100) or inhibit (USP7) progeny virus production (18, 32). Furthermore, our knockdown experiments suggest that depletion of one PML-NB component does not affect the recruitment of other components into DBP structures (Fig. 4). In addition, it is well documented that PML-NBs are transformed into track forms by a viral early gene product, E4orf3 (42, 43), suggesting a complex and distinct regulation for each component in infected cells.
Although we investigated the localization of endogenous PML in infected cells using specific antibodies (Fig. 1), only one isoform of PML (PML-IIA/isoform 11) was examined in living cells (Fig. 2A). Given the different dynamics and specific functions of PML isoforms (4), it remains possible that specific isoforms of PML, as well as other PML-NB components that have not been tested in this study, may exhibit distinct behavior upon Ad infection. Furthermore, most recently it has been reported using live-cell imaging analyses that PML and Daxx respond to incoming HSV-1 with distinct dynamics within single cells (44), suggesting a need for further detailed analyses for PML-NB components in living Ad-infected cells.
In sum, our study demonstrates that an early expressed and essential Ad DNA replication factor, but not the incoming viral genome complex itself, contributes to PML-NB targeting by Ad, providing a rationale for how Ad genomes associate with PML-NBs. Our findings differ from the HSV-1 (and possibly HCMV) case and argue against the conceptual view that incoming genomes of nuclear replicating DNA viruses are immediately encountered by PML-NBs and/or its components. Thus, to separate special case and general functions of PML-NBs and their components with respect to invading viral genomes, more detailed studies of other DNA viruses are needed.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Iggo and T. Dobner for the antibodies, D. Pickets for the EGFP-ATRX plasmid, and J. Dechanet for HFFs. We thank T. Sternsdorf for antibody and helpful discussions. We thank the Plateforme de Vectorologie (SFR Transbiomed) for preparation of the lentiviral vectors. The microscopy was done in the Bordeaux Imaging Center, a service unit of the CNRS-INSERM and Bordeaux University, a member of National Infrastructure France BioImaging. The help of Christel Poujol is acknowledged.
This work was supported through ANR grant (ANR 14 IFEC 0003-04) Infect-ERA, project eDEVILLI (H.W.), a BIS-Japan travel grant from the Excellence Initiative (IdEX) of the Bordeaux University (T.K.), and a grant-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (K.N.). H.W. is an INSERM fellow.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02545-15.
REFERENCES
- 1.Lallemand-Breitenbach V, de Thé H. 2010. PML nuclear bodies. Cold Spring Harb Perspect Biol 2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ching RW, Dellaire G, Eskiw CH, Bazett-Jones DP. 2005. PML bodies: a meeting place for genomic loci? J Cell Sci 118:847–854. doi: 10.1242/jcs.01700. [DOI] [PubMed] [Google Scholar]
- 3.Schreiner S, Wodrich H. 2013. Virion factors that target Daxx to overcome intrinsic immunity. J Virol 87:10412–10422. doi: 10.1128/JVI.00425-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, Hoischen C, Maul G, Dittrich P, Hemmerich P. 2008. Dynamics of component exchange at PML nuclear bodies. J Cell Sci 121:2731–2743. doi: 10.1242/jcs.031922. [DOI] [PubMed] [Google Scholar]
- 5.Ishov AM, Maul GG. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol 134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sourvinos G, Tavalai N, Berndt A, Spandidos DA, Stamminger T. 2007. Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells. J Virol 81:10123–10136. doi: 10.1128/JVI.01009-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell P, Lieberman PM, Maul GG. 2000. Lytic but not latent replication of Epstein-Barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J Virol 74:11800–11810. doi: 10.1128/JVI.74.24.11800-11810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day PM, Baker CC, Lowy DR, Schiller JT. 2004. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc Natl Acad Sci U S A 101:14252–14257. doi: 10.1073/pnas.0404229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett RD. 2013. The spatial organization of DNA virus genomes in the nucleus. PLoS Pathog 9:e1003386. doi: 10.1371/journal.ppat.1003386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett RD. 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell Microbiol 8:365–374. [DOI] [PubMed] [Google Scholar]
- 11.Boutell C, Sadis S, Everett RD. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J Virol 76:841–850. doi: 10.1128/JVI.76.2.841-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett RD, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol 72:6581–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett RD, Murray J. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J Virol 79:5078–5089. doi: 10.1128/JVI.79.8.5078-5089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukashchuk V, Everett RD. 2010. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J Virol 84:4026–4040. doi: 10.1128/JVI.02597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol 80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett RD, Parada C, Gripon P, Sirma H, Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol 82:2661–2672. doi: 10.1128/JVI.02308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiner S, Wimmer P, Sirma H, Everett RD, Blanchette P, Groitl P, Dobner T. 2010. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J Virol 84:7029–7038. doi: 10.1128/JVI.00074-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berscheminski J, Wimmer P, Brun J, Ip WH, Groitl P, Horlacher T, Jaffray E, Hay RT, Dobner T, Schreiner S. 2014. Sp100 isoform-specific regulation of human adenovirus 5 gene expression. J Virol 88:6076–6092. doi: 10.1128/JVI.00469-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tavalai N, Papior P, Rechter S, Stamminger T. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J Virol 82:126–137. doi: 10.1128/JVI.01685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol 80:8006–8018. doi: 10.1128/JVI.00743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell AM, Hirsch ML, Li C, Samulski RJ. 2014. Promyelocytic leukemia protein is a cell-intrinsic factor inhibiting parvovirus DNA replication. J Virol 88:925–936. doi: 10.1128/JVI.02922-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erickson KD, Bouchet-Marquis C, Heiser K, Szomolanyi-Tsuda E, Mishra R, Lamothe B, Hoenger A, Garcea RL. 2012. Virion assembly factories in the nucleus of polyomavirus-infected cells. PLoS Pathog 8:e1002630. doi: 10.1371/journal.ppat.1002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stepp WH, Meyers JM, McBride AA. 2013. Sp100 provides intrinsic immunity against human papillomavirus infection. mBio 4:e00845-13. doi: 10.1128/mBio.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giberson AN, Davidson AR, Parks RJ. 2012. Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res 40:2369–2376. doi: 10.1093/nar/gkr1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Berna AJ, Marion S, Chichon FJ, Fernandez JJ, Winkler DC, Carrascosa JL, Steven AC, Siber A, San Martin C. 2015. Distribution of DNA-condensing protein complexes in the adenovirus core. Nucleic Acids Res 43:4274–4283. doi: 10.1093/nar/gkv187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komatsu T, Haruki H, Nagata K. 2011. Cellular and viral chromatin proteins are positive factors in the regulation of adenovirus gene expression. Nucleic Acids Res 39:889–901. doi: 10.1093/nar/gkq783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haruki H, Gyurcsik B, Okuwaki M, Nagata K. 2003. Ternary complex formation between DNA-adenovirus core protein VII and TAF-Iβ/SET, an acidic molecular chaperone. FEBS Lett 555:521–527. doi: 10.1016/S0014-5793(03)01336-X. [DOI] [PubMed] [Google Scholar]
- 28.Puntener D, Engelke MF, Ruzsics Z, Strunze S, Wilhelm C, Greber UF. 2011. Stepwise loss of fluorescent core protein V from human adenovirus during entry into cells. J Virol 85:481–496. doi: 10.1128/JVI.01571-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komatsu T, Dacheux D, Kreppel F, Nagata K, Wodrich H. 2015. A method for visualization of incoming adenovirus chromatin complexes in fixed and living cells. PLoS One 10:e0137102. doi: 10.1371/journal.pone.0137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pombo A, Ferreira J, Bridge E, Carmo-Fonseca M. 1994. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J 13:5075–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dekker J, Kanellopoulos PN, Loonstra AK, van Oosterhout JAWM, Leonard K, Tucker PA, van der Vliet PC. 1997. Multimerization of the adenovirus DNA-binding protein is the driving force for ATP-independent DNA unwinding during strand displacement synthesis. EMBO J 16:1455–1463. doi: 10.1093/emboj/16.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ching W, Koyuncu E, Singh S, Arbelo-Roman C, Hartl B, Kremmer E, Speiseder T, Meier C, Dobner T. 2013. A ubiquitin-specific protease possesses a decisive role for adenovirus replication and oncogene-mediated transformation. PLoS Pathog 9:e1003273. doi: 10.1371/journal.ppat.1003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wodrich H, Henaff D, Jammart B, Segura-Morales C, Seelmeir S, Coux O, Ruzsics Z, Wiethoff CM, Kremer EJ. 2010. A capsid-encoded PPxY-motif facilitates adenovirus entry. PLoS Pathog 6:e1000808. doi: 10.1371/journal.ppat.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schreiner S, Martinez R, Groitl P, Rayne F, Vaillant R, Wimmer P, Bossis G, Sternsdorf T, Marcinowski L, Ruzsics Z, Dobner T, Wodrich H. 2012. Transcriptional activation of the adenoviral genome is mediated by capsid protein VI. PLoS Pathog 8:e1002549. doi: 10.1371/journal.ppat.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komatsu T, Nagata K. 2012. Replication-uncoupled histone deposition during adenovirus DNA replication. J Virol 86:6701–6711. doi: 10.1128/JVI.00380-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bérubé NG, Healy J, Medina CF, Wu S, Hodgson T, Jagla M, Picketts DJ. 2008. Patient mutations alter ATRX targeting to PML nuclear bodies. Eur J Hum Genet 16:192–201. doi: 10.1038/sj.ejhg.5201943. [DOI] [PubMed] [Google Scholar]
- 37.Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JHJ, Sung PA, Jasin M, Rosenbluh J, Zwang Y, Weir BA, Hatton C, Ivanova E, Macconaill L, Hanna M, Hahn WC, Lue NF, Reddel RR, Jiao Y, Kinzler K, Vogelstein B, Papadopoulos N, Meeker AK. 2012. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet 8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiner S, Bürck C, Glass M, Groitl P, Wimmer P, Kinkley S, Mund A, Everett RD, Dobner T. 2013. Control of human adenovirus type 5 gene expression by cellular Daxx/ATRX chromatin-associated complexes. Nucleic Acids Res 41:3532–3550. doi: 10.1093/nar/gkt064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez R, Huang W, Finnen R, Bragg C, Flint SJ. 2006. Adenovirus E1B 55-kilodalton protein is required for both regulation of mRNA export and efficient entry into the late phase of infection in normal human fibroblasts. J Virol 80:964–974. doi: 10.1128/JVI.80.2.964-974.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bishop CL, Ramalho M, Nadkarni N, May Kong W, Higgins CF, Krauzewicz N. 2006. Role for centromeric heterochromatin and PML nuclear bodies in the cellular response to foreign DNA. Mol Cell Biol 26:2583–2594. doi: 10.1128/MCB.26.7.2583-2594.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haruki H, Okuwaki M, Miyagishi M, Taira K, Nagata K. 2006. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J Virol 80:794–801. doi: 10.1128/JVI.80.2.794-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang M, Imperiale MJ. 2012. Design stars: how small DNA viruses remodel the host nucleus. Future Virol 7:445–459. doi: 10.2217/fvl.12.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmid M, Speiseder T, Dobner T, Gonzalez RA. 2014. DNA virus replication compartments. J Virol 88:1404–1420. doi: 10.1128/JVI.02046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Everett RD. 14 October 2015. The dynamic response of IFI16 and PML nuclear body components to HSV-1 infection. J Virol doi: 10.1128/JVI.02249-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.