Abstract
The inherent simplicity of Caenorhabditis elegans and its extensive genetic toolkit make it ideal for studying complex biological processes. Recent developments further increase the usefulness of the worm, including new methods for: altering gene expression, altering physiology using optogenetics, manipulating large numbers of worms, automating laborious processes and processing high-resolution images. These developments both enhance the worm as a model for studying processes such as development and ageing and make it an attractive model in areas such as neurobiology and behaviour.
The nematode worm Caenorhabditis elegans has many features that make it an attractive model organism, including a defined, invariant cell lineage1–3 and extensive genetic tools. Research using C. elegans has provided fundamental advances in our knowledge of several biological processes, including development, apoptosis, neurobiology, gene regulation and ageing. As a simple organism, the worm is an ideal platform for implementing new technologies. It was the first multicellular organism to be sequenced4, and worm researchers were early adopters of genomic methods, such as microarrays. Despite its simplicity, studies in the worm also provide broader implications for fields such as biomedical research. For example, even though the worm genome is 30-fold smaller than the human genome, it contains almost as many genes4, and most of these have human homologues5.
In the past few years, several technological advances have allowed greater ease in manipulating the worm for existing applications, as well as providing new opportunities for using C. elegans to study a wider range of biological processes (TABLE 1). In this Review, we discuss selected examples of these new developments. We hope this Review serves not only to inform but also to encourage readers to incorporate some of these techniques into their own research and to take advantage of the worm research community’s culture of openness in the sharing of resources, protocols and reagents. Additionally, we hope that this Review inspires readers to reflect on areas that could be improved and to spend time and effort on developing new methodologies. Readers who want more background information should refer to well-written reviews that discuss basic worm techniques6 and genetic and genomic resources7,8.
Table 1.
The C. elegans toolkit: a summary of progress
| Type of approach | Method | Pros | Cons | Year | Refs |
|---|---|---|---|---|---|
| Loss-of-function | |||||
| Undirected mutagenesis | Chemical | Good for forward genetics | Many mutations in genome; long screening process | 1974 | 56 |
| Transposon | Good for forward genetics | Many mutations in genome; long screening process | 1985 | 57 | |
| Deficiencies | Good for forward genetics | Many genome rearrangements; long screening process | 1979 | 58 | |
| Targeted gene deletions | Mos transposon | Deletions are targeted; can delete or alter specific parts of genes rather than the entire gene | Need Mos site next to gene of interest | 2010 | 11 |
| ZFNs and TALENs | Can target any gene or part of the gene without the need for transposon sites | Expensive; complicated design considerations; potential off-target effects | 2011 | 16 | |
| RNAi | Whole-body RNAi | Fast; cheap; can perform knockdown in adults to avoid developmental defects | Potential off-target effects; affects all tissues | 1998 | 18 |
| Tissue-specific RNAi | Fast; cheap; can target specific tissues | Potential off-target effects; requires promoter specific to tissue of interest | 2007 | 20,21 | |
| Gain-of-function | |||||
| Generating transgenics | Microinjection | Fast; cheap | Can only generate high-copy extrachromosomal arrays | 1991 | 22 |
| Biolistic bombardment | Can generate integrated lines | Requires a large number of worms; slow; integration is random | 2006 | 23 | |
| Mos transposon | Can generate single-copy insertion into a defined locus | Long screening process | 2008 | 24 | |
| Site-specific recombination | Cre–loxP | Can control where and when a gene is expressed | Requires promoters that can drive proper expression | 2000 | 26 |
| FLP–FRT | Can control where and when a gene is expressed | Requires promoters that can drive proper expression | 2008 | 27,28 | |
| GAL4–UAS | Can control where and when a gene is expressed | Not yet available in C. elegans | |||
| Inducible systems | Heat shock | Can induce gene expression at a specific time | Induction is short and cannot be sustained robustly | 2007 | 59 |
| Drugs (for example, Tet and RU-486) | Can induce gene expression at a specific time | Not yet available in C. elegans | |||
| Other techniques | |||||
| Worm sorting | Flow cytometry | Can sort based on different criteria (such as size and fluorescence levels) | Expensive equipment required | 2006 | 60 |
| Microfluidics | Cheap; many applications | Requires knowledge of design and facility to make chips | 2004 | 61 | |
| Imaging | Lineage analysers | Expression data at the single-cell level | Requires specialized knowledge of the cell lineage; medium throughput | 2008 | 43,46 |
| Single mRNA in situ hybridization | Sensitive way to measure gene expression | Probes are expensive | 2008 | 47 | |
| Optogenetics | Can turn neurons on and off on a millisecond scale | Setup may involve expensive equipment | 2005 | 62 | |
FLP, flippase; FRT, flippase recognition target; TALEN, transcription activator-like effector nuclease; Tet, tetracycline; UAS, upstream activation sequence; ZFN, zinc finger nuclease.
Manipulation of gene expression
Understanding gene function is an important feature for any genetic model organism and requires the ability to alter gene expression easily and accurately. New techniques that precisely control where and when a gene is expressed are enabling C. elegans researchers to study the function of genes in ever-greater depth.
Loss-of-function
Traditionally, loss-of-function experiments have used random chemical-based mutagenesis or transposon-based insertional mutagenesis. These approaches have produced and continue to produce a large number of mutagenized strains, including those produced by two deletion consortiums: the C. elegans Gene Knockout Consortium and the National BioResource Project of Japan. With the advent of low-cost next-generation sequencing, generating mutants using random mutagenesis has become simpler than ever, as the previously cumbersome task of mutant identification is no longer required9,10. Still, techniques involving random mutagenesis are more appropriate for forward genetic screens (in which genes based on a phenotype of interest are screened for) than for reverse genetics (in which phenotypes associated with a particular gene of interest are screened for).
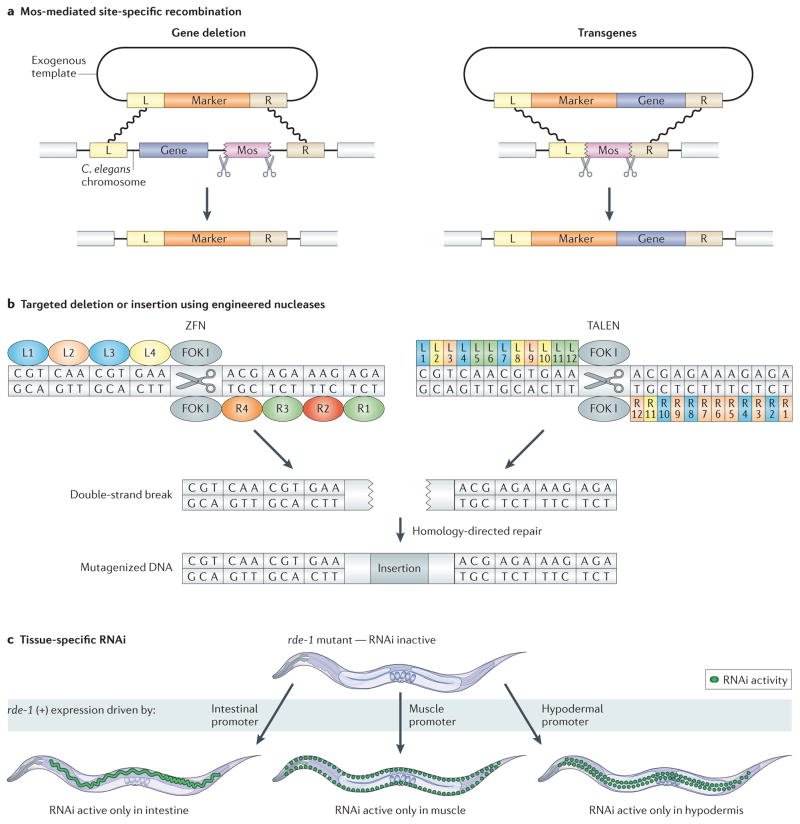
Recently developed technologies facilitate reverse genetics in the worm. One of these uses a Mos transposon from Drosophila melanogaster to make targeted gene deletions11. In contrast to previous work using C. elegans transposons, which are present in numerous copies throughout the genome, the D. melanogaster Mos transposon is not present in wild-type worms. Mos transposase is introduced into worms containing a Mos element near the target gene. As a result, Mos is excised, producing a double-strand DNA break, and a repair construct with homology arms that flank the target region serves as a template for homologous recombination. Positive selection markers are used to monitor for successful deletion (FIG. 1a). The authors of this method have successfully deleted regions of up to 25 kb with high efficiency. The NemaGENETAG consortium has generated a resource of Mos insertions in 14,000 known sites distributed throughout the C. elegans genome12. Importantly, 99.4% of all genes in C. elegans fall within 25 kb of at least one Mos element, so almost all worm genes can be targeted for deletion, and the resulting phenotypes can be studied.
Figure 1. New genetic methods in Caenorhabditis elegans.
a | Mos-mediated, site-specific recombination can be used for targeted gene deletions or for transgenesis. In both cases, excision of an integrated Mos transposon from a specific chromosomal site by Mos transposase results in a double-strand break, which is repaired by homologous recombination using an exogenous template. Left (L) and right (R) homology arms flank the region to be excised. b | Zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) can be designed to generate targeted mutations. For ZFNs, 3–6 zinc finger domains that each target three nucleotides are modularly fused to the nuclease domain of the FOK I restriction enzyme. For TALENs, a series of repeat units that each target one nucleotide is fused to the nuclease domain of FOK I. In both cases, FOK I endonuclease induces a double-strand break that is then repaired using error-prone non-homologous end-joining, which sometimes introduces small insertions (as illustrated) or deletions (not shown). For the ZFN, L1–4 and R1–4 specify left and right zinc finger domains, respectively. For the TALEN, L1–12 and R1–12 specify left and right repeat domains, respectively. c | Tissue-specific RNAi is accomplished by generating worms that are proficient for RNAi only in a tissue of interest. rde-1 mutants are RNAi-deficient. When wild-type rde-1 (+) is expressed under the control of a tissue-specific promoter, such as intestinal-, muscle- or hypodermal-specific promoters, worms become RNAi-proficient in the tissue where rde-1 (+) is expressed. When these worms are fed bacteria expressing double-stranded RNA against a gene of interest, knockdown will only occur in the tissue where rde-1 (+) is expressed.
Another method for making targeted mutations is to use site-specific nucleases. These are fusion proteins containing the nuclease domain of the FOK I restriction enzyme and either an array of zinc finger DNA-binding domains13 or the central repeat domain from the Xanthomonas spp. transcription activator-like effector (TALE) protein14,15. In both cases, the DNA-binding domains can be engineered in a modular series to recognize almost any sequence of interest and to generate a double-strand break at that site. The break is repaired by non-homologous end joining, an imprecise mechanism with a propensity to introduce insertions or deletions at the site of repair (FIG. 1b). Recently, both zinc finger nucleases (ZFNs) and TALE nucleases (TALENs) have been shown to be effective at creating targeted mutations in C. elegans with reasonable efficiency (1–5%) and without observable off-target effects16. The drawbacks to this technology include high cost17, complex design considerations and a lack of control over the types of mutations generated. However, ZFNs and TALENs have the potential to revolutionize reverse genetics, allowing researchers to generate mutations in any gene in wild-type animals without the need for random mutagenesis or nearby Mos sites.
To disrupt gene function without generating genetic mutants, RNAi can be performed, which causes potent and reproducible gene knockdown through use of double-stranded RNA (dsRNA)18. In worms, RNAi is generally administered by feeding with Escherichia coli expressing dsRNA against a gene of interest. An RNAi library containing E. coli strains expressing dsRNA against 86% of C. elegans genes is available, allowing researchers to conduct large-scale RNAi screens with relative ease19.
In wild-type worms, gene knockdown by RNAi occurs in the entire body, except for the nervous system, where knockdown efficiency is low. A complementary approach has been developed that allows RNAi knockdown to occur preferentially in neurons, but not in other tissues20. This is achieved by performing RNAi in transgenic worms expressing neuronal systemic RNA interference defective protein 1 (SID-1), a transmembrane protein required for cellular uptake of dsRNA that is normally excluded from neurons. In this system, efficiency of RNAi knockdown is markedly improved in neurons, but is decreased in other tissues. In non-neuronal tissues, tissue-specific RNAi knockdown can be performed by using a transgenic mutant strain that carries an rde-1 mutation (making them defective in RNAi) and then expressing wild-type rde-1 under the control of a tissue-specific promoter21. Worms can then be treated with dsRNA for a gene of interest, which will reduce activity of that gene only in the specific tissue in which rde-1 is expressed (FIG. 1c). Tissue-specific RNAi will make it easier for researchers to determine spatial requirements for gene function (genetic site of action) or to study genes that have pleitropic effects in different tissues.
Gain-of-function
Until recently, the generation of transgenic worms was carried out by one of two methods. DNA can be microinjected into the worm gonad, giving rise to extrachromosomal arrays that function as heritable mini-chromosomes22. An alternative method is to use biolistic bombardment23, which produces transgenic lines with a few copies of the transgene integrated into a random site in the genome. However, the high copy number associated with extrachromosomal arrays and the random site of integration associated with biolistic bombardment has placed limitations on the usefulness of these methods.
New approaches have overcome these limitations to open the door for studies in the worm that require predictable and stable levels of gene expression. One new method takes advantage of Mos transposition in order to generate transgenic worms with a single copy of a transgene inserted into a defined locus24 (FIG. 1a). Although the efficiency of insertion after transposase activation is fairly low, the use of selection markers ameliorates the identification of insertion events. The ability to insert only a single copy of a gene is particularly important for studies involving the worm germline, where highly expressed transgenes are generally silenced. Additionally, this approach will enable the study of gene expression noise in C. elegans, because it is possible to examine expression of single genes inserted into defined sites in single cells in individual worms.
An important consideration for gain-of-function experiments is the ability to control when and where a gene is expressed. Until recently, inducible approaches included placing a gene under the control of the heat-shock promoter or using the Cre–loxP system, in which the Cre recombinase, under the control of a cell-type specific promoter, is used to mediate recombination between loxP sites. The Cre–loxP system has been used both to activate25 and inactivate26 gene expression.
Recently, two groups independently configured the FLP–FRT system for inducible gene expression in C. elegans. This system uses the Saccharomyces cerevisiae recombination enzyme flippase (FLP) as another way of performing site-specific recombination in worms27,28. A gene of interest that is separated from its promoter by an inactivation cassette that is flanked by FLP recognition target (FRT) sites is introduced into the worm. Gene expression is inactive until the cassette is removed by activation of FLP recombinase, which is expressed from either a tissue-specific or a heat-shock promoter. The availability of both the Cre–loxP and FLP–FRT systems means that researchers can control gene expression with even greater specificity. For example, the FLP–FRT system was used to express a gene in a specific set of neurons (marked by two genetic markers) by expressing FLP under the control of one promoter and the gene of interest and inactivation cassette under a second promoter29. Expression of the gene only occurred in cells in which both promoters were active. If both Cre–loxP and FLP–FRT were used, a third level of control could be added, such as timing or further tissue specificity. Another application for these conditional systems could be to express RNAi hairpins that reduce the expression of a target gene28; this may be extremely useful for ageing studies, because E. coli-based RNAi methods are less effective in older worms.
Recent developments also improve the process of producing transgenic worms. One method simplifies the creation of DNA constructs for transgenesis by using DNA recombineering to modify C. elegans fosmid clones30. Each fosmid contains around 40 kb of C. elegans genomic DNA, enabling selection of specific fosmids that contain the full coding and regulatory regions for a gene of interest. Recombineering uses homologous recombination in E. coli to modify the inserted C. elegans sequences: for example, by adding a GFP tag to a gene of interest or adding a positive selection marker to the fosmid backbone. This technique has successfully been used by the modEncode group for high-throughput epitope tagging of a large number of transcription factors31 and should be useful for any researcher looking for an efficient way to generate and modify constructs for transgenesis experiments.
Another important development for creating transgenic worms is the use of antibiotics as selection markers for positive transgenesis events. Selection markers used in generating transgenic lines typically require visual screening and manual selection. Two groups have recently developed antibiotic selection systems for C. elegans using neomycin or puromycin for quickly and easily selecting transgenic worms32,33.
Worm manipulation
C. elegans worms are easy to grow, maintain and manipulate, making them excellent models for a variety of genetic and other screens. However, screening and manipulating large numbers of worms by hand is labour-intensive. New technologies use flow cytometry or microfluidics to automate the tedious processes that were previously involved in phenotype detection and drug screens.
Flow cytometry
Florescence-activated cell sorting (FACS) can now be applied to whole worms and may be particularly useful for isolating staged embryos. Obtaining large numbers of staged embryos is a difficult endeavour because embryos develop from 1 to 558 cells in under 7 hours3, and current methods for staging embryos suffer from insufficient temporal resolution; most synchronized populations still contain embryos in many developmental stages. Stoeckius et al.34 have developed a method to isolate large numbers of one-cell- stage embryos. The authors used a GFP reporter driven by the promoter for oma-1, a gene that is specifically expressed at this stage. They then used FACS to isolate GFP-expressing embryos, achieving sample purity of 98% fixed one-celled embryos. Using this sample, they were able to profile the changes in small RNA expression between one-cell-stage and 2–4-cell-stage embryos, providing new insights into the role of maternal input in early embryogenesis.
Whereas FACS is used to sort embryos, the COPAS Biosort instrument is used to perform size- and fluorescence- based flow cytometry on larval and adult worms and has been applied to tasks such as separating or identifying worms of different sizes35 and sorting live and dead worms36. Recent work reports that this instrument can also be used for genetic screens, as it is capable of isolating worm mutants with subtle changes in GFP expression37. To identify genes that affect the development of dopaminergic neurons, the authors of this study performed ethylmethane sulphonate (EMS)-based mutagenesis and screened for worms with modified expression of a GFP reporter that labels these cells. Using the worm sorter, the entire screening process was performed in 1.5 days. The most remarkable aspect of this technique is its ability to detect subtle changes in expression; there are only eight dopaminergic neurons, and the worm sorter was able to identify worms that have lost expression in just one of these.
Microfluidics
Microfluidic chambers and ‘lab-on-a-chip’ technologies have previously been used to streamline laboratory procedures. These technologies are now facilitating the semi-automation of previously labour-intensive processes involving manual worm manipulation (FIG. 2). In ageing studies, for example, lifespan analyses last from a few weeks to a few months and worms require semi-daily monitoring. To facilitate these assays, Hulme et al.38 have constructed a microfluidic system consisting of chambers that house individual worms from the end of development until death. The worms can be easily monitored in a semi-automated way for various phenotypes, such as growth, swimming frequency and survival. This approach could be used in conjunction with genetic biomarkers. For example, fluorescent biomarkers have been used in longitudinal studies to elucidate mechanisms underlying C. elegans ageing39. Combined with microfluidic chambers, such studies could be performed on the level of the individual, rather than the population, thereby providing a new approach for studying ageing in the worm.
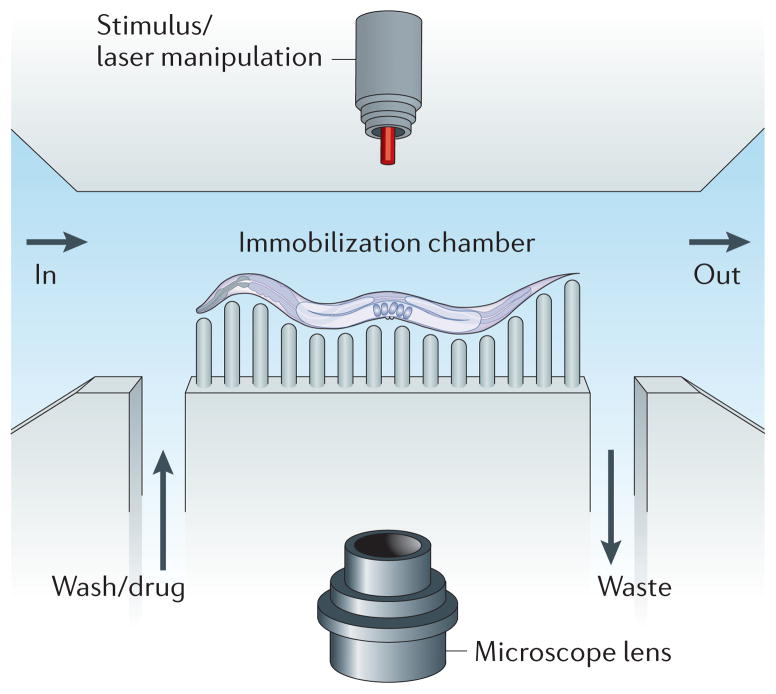
Figure 2. Caenorhabditis elegans microfluidics.
The basic layout for a worm microfluidic chamber consists of an inlet into the chamber (labelled ‘in’) and an outlet from the chamber (labelled ‘out’). A channel allows for input of media such as a wash solution or a drug. Another channel allows for the output of waste. Worms can be immobilized through many methods, including pressurization of a flexible polymer around the worm that restricts movement. A microscope can capture features of the worm for automated lifespan analysis or for longitudinal studies. Outside stimuli, such as laser manipulation, can also be added to the system.
Microfluidics also facilitates and semi-automates manual procedures that require great precision. Samara et al.40 devised a system that uses a microfluidic setup to perform neurosurgery. Their microfluidic device loads, isolates and immobilizes worms, laser-ablates the axon of an individual neuron and unloads the worms — all with minimal input from the user. The procedure is fast: the whole process takes an average of 20 seconds per worm. Additionally, software is capable of targeting the laser to the surgery site, meaning that precision is not required on the part of the user. The authors used this approach to study neurite regeneration, uncovering a potential role for protein kinase C activity in the regeneration of touch neurons.
Imaging
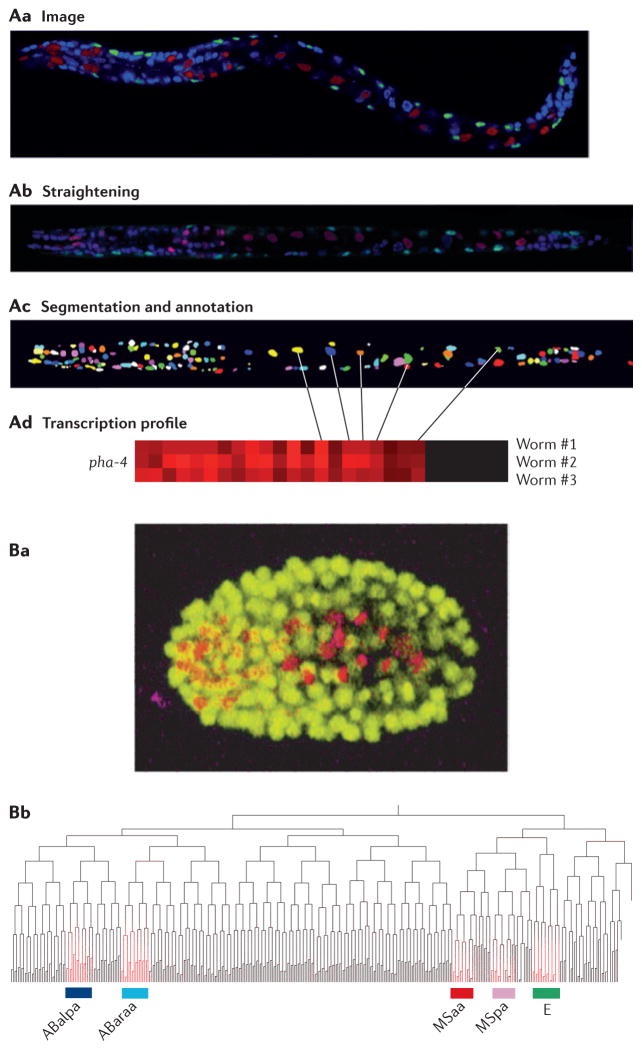
Advances in imaging have revolutionized biology by allowing researchers to study systems at ever-greater resolution. In C. elegans, new techniques in image processing allow for the digitization of high-resolution images and automated quantification of gene expression. To take advantage of the worm’s invariable cell lineage, Liu et al.41 built an automatic cell lineage analyser42–44, which identifies individual nuclei from a confocal image stack of a worm expressing a fluorescently tagged reporter. The first step is to straighten the three-dimensional confocal image so that worms have a uniform size and appearance. Then, the individual nuclei are identified and named according to their place in the cell lineage. Finally, the precise level of gene expression is measured in each nucleus. In a similar manner to microarray analysis, which turns images of spotted arrays into heat maps containing expression levels for each spot, the lineage analyser turns images of fluorescently spotted worms into a heat map containing expression levels for each cell (FIG. 3A). The authors defined a molecular signature for each cell that is based on the single-cell expression pattern of 93 genes44. They used this signature to address questions that are fundamental to developmental biology, such as whether gene expression is based on cell lineage or cell type.
Figure 3. Using imaging in Caenorhabditis elegans to study cell lineages.
A | Generating an expression profile from a three-dimensional confocal image stack. First, confocal images are generated from transgenic worms expressing cherry fluorescent protein from a promoter of interest (red) and GFP in muscle cells from the myo-3 promoter (green) and in which nuclei are stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue) (Aa). The images are then computationally straightened and DAPI-stained nuclei are identified (Ab). Finally, individual nuclei (pseudocoloured for visualization) are identified (Ac) and expression is quantified (Ad). B | Generating a lineage-specific expression profile from time-lapse images. Transgenic worms expressing both GFP from the histone promoter to mark all nuclei (yellow) and cherry fluorescent protein from a promoter of interest are imaged continuously throughout embryogenesis (Ba). The time and place of expression for a gene of interest can be mapped onto the defined C. elegans lineage tree (Bb). In this example, the gene of interest (highlighted in red) is expressed in the ABalpa, ABaraa, MSaa and MSpa developmental lineages, which comprise most of the cells in the pharynx, as well as the E lineage, which comprises intestinal cells. Part A is reproduced, with permission, from REF. 44 © (2009) Cell Press. Part B is reproduced, with permission, from REF. 46 © (2008) Macmillan Publishers Ltd. All rights reserved.
A similar technology uses time-lapse images of embryogenesis in order to follow gene expression through cell divisions45,46 (FIG. 3B). For example, previous studies of the C. elegans Neuro D orthologue, cnd-1, described its expression as beginning early in embryogenesis in descendents of the AB lineage. By mid-gastrulation, cnd-1 expression could be observed in numerous cells, but could not determine the identity of these cells. Using the lineage analyser, all cnd-1-expressing cells, which constitute a variety of different neurons, were identified46. These new imaging tools provide powerful ways to examine how gene expression drives developmental programs.
Imaging also offers a way to examine mRNA levels in individual cells, which allows for quantification of endogenous gene expression without needing to express an exogenous promoter. In Raj et al.47,48, a modified fluorescent in situ hybridization (FISH) technique is used to label individual mRNA molecules in worms. One application of this technology is for studying gene expression noise. The authors found that during intestine development, expression of erythroid-like transcription factor family (elt-2), an important determinant of intestinal fate, requires a threshold level of its upstream regulator endoderm-determining 1 (end-1) in a minimum of four cells in a specific developmental timeframe48. This result explains why mutants with decreased end-1 expression sometimes exhibit a complete loss of intestinal fates and, at other times, produce intestinal precursors. Examining endogenous gene expression at the single-cell and single-RNA level can help to address fundamental questions, such as how development is buffered from stochastic gene expression.
Altering physiology
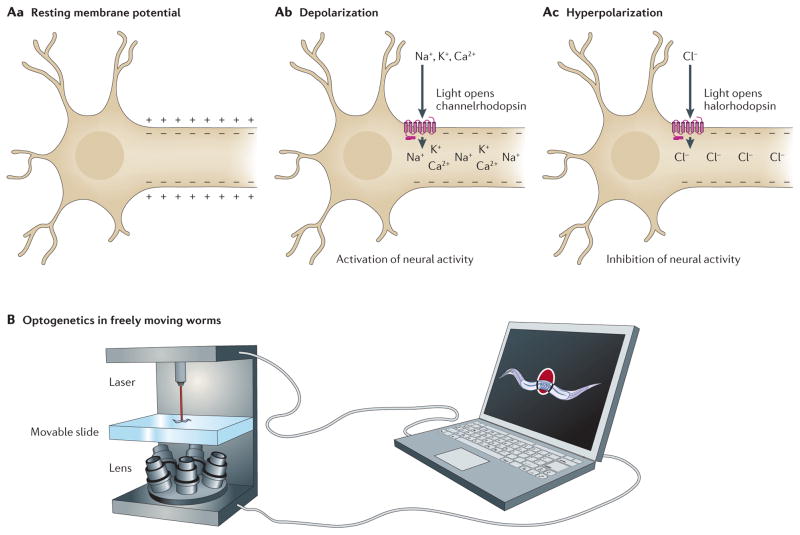
In the past decade, the field of neurobiology has been revolutionized by optogenetics49, a discipline based on the manipulation of light-sensitive proteins. Researchers introduce these proteins into cells of interest and can then activate or inhibit them using certain wavelengths of light, all on a millisecond timescale. For example, light-induced activation of channelrhodopsin causes membrane depolarization and activates neural activity. Conversely, light-induced activation of halorhodopsin causes membrane hyperpolarization and inhibits neural activity (FIG. 4A). Worms are an ideal platform for optogenetics owing to their transparent body and well-defined nervous system. Each worm contains only 302 neurons50, virtually all of which have distinct genetic markers51, enabling researchers in many cases to express their desired proteins in any neuron of interest. Additionally, a simple system such as C. elegans allows researchers to more easily tease apart the neural networks involved in behaviours such as movement, sensation and learning. Optogenetics has already contributed to several studies in the worm, including: establishing the functional relevance of neural connections52 and determining whether neurotransmission is a digital ‘all-or-none’ signal or an analogue graded signal53.
Figure 4. Using optogenetics in Caenorhabditis elegans.
A | Unstimulated neurons have a resting membrane potential (Aa), such that the inside of the cell contains more negative charge than the outside of the cell. Channelrhodopsin is a light-gated ion channel. On light-induced activation, channelrhodopsin transports cations into the cell, resulting in depolarization of the membrane potential and activation of neural activity (Ab). Halorhodopsin is a light-gated ion pump. On light-induced activation, halorhodopsin transports chloride ions into the cell, resulting in hyperpolarization of the membrane potential and inhibition of neural activity (Ac). B | By combining optogenetics with a tool for tracking freely moving worms, researchers can laser-activate channelrhodopsin or halorhodopsin in specific neurons, thereby activating or inactivating those neurons. They can then determine how altering the activity of those neurons affects behaviour.
Previous optogenetic studies in worms required immobilization, which precludes the examination of movement and other behaviours. To address this shortcoming, two groups have developed systems to perform optogenetics in freely moving worms54,55. To do this, a high-speed camera is used to move a computer-controlled microscope stage, which keeps a moving worm in the centre of view. Researchers can then illuminate the target cells in order to stimulate or suppress neural activity (FIG. 4B). As a proof of principle, Stirman et al. investigated the mechanosensory neurons involved in the touch response55. They created transgenic worms in which the mechanosensory neurons could be controlled by different intensities of light. Using this system, they determined how the presence of competing signals, such as stimulation of both anterior and posterior mechanosensory neurons, affected behaviour. Like genetic studies that determine how activation or inactivation of gene expression affects gene function, the optogenetic tools that enabled this work allow researchers to determine how activation or inactivation of defined neurons affect behaviour.
Conclusion
The simplicity of the worm has made it a popular multicellular model organism. Over the years, researchers have developed an extensive set of tools, including transgenesis methods and RNAi, which facilitate genetic studies in the worm. Of course, no model organism has every tool at its disposal. For example, the C. elegans toolkit lacks the arsenal of gene expression modifying techniques that has made bacteria a powerhouse for the fields of synthetic biology and bioengineering, nor does it have drug-inducible expression techniques, such as the tetracycline repressor (TetR) system, that are available in higher organisms. However, there are no theoretical reasons barring these technologies from working in C. elegans, so it is likely to be only a matter of time before they are developed for worms.
The C. elegans toolkit is constantly expanding, and the past few years have yielded a variety of new technologies that will open the door to further powerful studies that are ideally suited for the worm. For example, the cell lineage analyser only makes sense in an organism with an invariable and well-defined cell lineage and makes it possible to answer a multitude of questions on cell fate that have long puzzled developmental biologists. Even when a technique has been well-adapted in other organisms, as in the case of optogenetics, studying the worm provides many advantages. Neural circuits are incredibly complex, and the worm contains only 302 neurons and exhibits relatively simple behaviours. The addition of technologies that enable manipulation of neural activity in freely moving worms means that researchers can quickly decipher neural networks with-out perturbing those networks through immobilization. Now more than ever, these additions to the C. elegans genetic toolkit, coupled with the intrinsic simplicity of the worm, allow for rapid, simple and powerful ways to address important questions regarding development, behaviour and ageing.
Acknowledgments
We thank members of the worm research community and apologize for the numerous works we could not cite owing to space limitations. We thank D. Sagi, M. Schvarzstein and members of the Kim laboratory for helpful discussion. We thank A. Sánchez-Blanco, A. Friedland and the anonymous reviewers for their suggestions and improvements and for their critical reading of the manuscript. X.X. is supported by the Cancer Biology Training Grant at Stanford University, California, USA. Research in the laboratory of S.K.K. is supported by the US National Human Genome Research Institute, the US National Institute of General Medical Sciences, the US National Institute on Aging and the Glenn Foundation.
Glossary
- Biolistic bombardment
A technique for delivering nucleic acids into the cells, in which small metal particles coated with nucleic acid are fired into the target tissue under high pressure
- Recombineering
Molecular genetic engineering technique that uses homologous recombination to manipulate and/or alter DNA
- Fosmid
A low-copy vector for the construction of stable genomic libraries that uses the Escherichia coli F factor origin for replication. Each fosmid can store ~40 kb of library DNA, and these sequences are more stable in fosmids than in multi-copy vectors
- Florescence-activated cell sorting (FACS)
An experimental technology that can separate cells by their fluorescence and light-scattering properties
- Fluorescent in situ hybridization (FISH)
A microscopic technique that uses fluorescently tagged DNA probes to detect the cytological localization of specific RNA by in situ hybridization
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Stuart K. Kim’s homepage: http://cmgm.stanford.edu/~kimlab
C. elegans fosmid library: http://www.lifesciences.sourcebioscience.com/clone-products/genomic-dna-clones/c-elegans-fosmid-library-.aspx
C. elegans Gene Knockout Consortium: http://celeganskoconsortium.omrf.org
C. elegans RNAi library: http://www.gurdon.cam.ac.uk/~ahringerlab/pages/rnai.html
Caenorhabditis Genetics Center: http://www.cbs.umn.edu/CGC
Cell lineaging information from the Waterston laboratory: http://waterston.gs.washington.edu
Kim laboratory Automatic Cell Lineage Analyzer information: http://cmgm.stanford.edu/~kimlab/index_development.htm
National BioResource Project of Japan: http://www.shigen.nig.ac.jp/c.elegans/index.jsp
NemaGENETAG: http://elegans.gr/nemagenetag
Stanford Microfluidics Foundry: http://www.stanford.edu/group/foundry
WormBase: http://www.wormbase.org
WormBook: http://wormbook.org
Wormatlas: http://www.wormatlas.org
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- 2.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 3.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 4.C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 5.Lai CH, Chou CY, Ch’ang LY, Liu CS, Lin W. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 2000;10:703–713. doi: 10.1101/gr.10.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hulme SE, Whitesides GM. Chemistry and the worm: Caenorhabditis elegans as a platform for integrating chemical and biological research. Angew Chem Int Edn Engl. 2011;50:4774–4807. doi: 10.1002/anie.201005461. [DOI] [PubMed] [Google Scholar]
- 7.Antoshechkin I, Sternberg PW. The versatile worm: genetic and genomic resources for Caenorhabditis elegans research. Nature Rev Genet. 2007;8:518–532. doi: 10.1038/nrg2105. [DOI] [PubMed] [Google Scholar]
- 8.Van Nostrand EL, Kim SK. Seeing elegance in the gene regulatory networks of the worm. Curr Opin Genet Dev. 2011;21:1–10. doi: 10.1016/j.gde.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flibotte S, et al. Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics. 2010;185:431–441. doi: 10.1534/genetics.110.116616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarin S, et al. Analysis of multiple ethyl methanesulfonate-mutagenized Caenorhabditis elegans strains by whole-genome sequencing. Genetics. 2010;185:417–430. doi: 10.1534/genetics.110.116319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frokjaer-Jensen C, et al. Targeted gene deletions in C. elegans using transposon excision. Nature Methods. 2010;7:451–453. doi: 10.1038/nmeth.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bazopoulou D, Tavernarakis N. The NemaGENETAG initiative: large scale transposon insertion gene-tagging in Caenorhabditis elegans. Genetica. 2009;137:39–46. doi: 10.1007/s10709-009-9361-3. [DOI] [PubMed] [Google Scholar]
- 13.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 15.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 16.Wood AJ, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. Although there are still major design and cost considerations, ZFN- and TALEN-mediated mutagenesis have the potential to revolutionize the way mutants are generated. This paper describes the first time such methods have been employed in C. elegans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Francesco L. Move over ZFNs. Nature Biotech. 2011;29:681–684. doi: 10.1038/nbt.1935. [DOI] [PubMed] [Google Scholar]
- 18.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 19.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 20.Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nature Methods. 2010;7:554–559. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qadota H, et al. Establishment of a tissue-specific RNAi system in C. elegans. Gene. 2007;400:166–173. doi: 10.1016/j.gene.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. A classic, this paper describes how to introduce DNA into worms via microinjection. Microinjection is still the most common way of generating transgenic worms today and is also the basis for some of the newer techniques described in this Review, such as Mos transposition and mutagenesis mediated by TALENs and ZFNs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frokjaer-Jensen C, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nature Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macosko EZ, et al. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoier EF, Mohler WA, Kim SK, Hajnal A. The Caenorhabditis elegans APC-related gene apr-1 is required for epithelial cell migration and Hox gene expression. Genes Dev. 2000;14:874–886. [PMC free article] [PubMed] [Google Scholar]
- 27.Davis MW, Morton JJ, Carroll D, Jorgensen EM. Gene activation using FLP recombinase in C. elegans. PLoS Genet. 2008;4:e1000028. doi: 10.1371/journal.pgen.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voutev R, Hubbard EJ. A “FLP-Out” system for controlled gene expression in Caenorhabditis elegans. Genetics. 2008;180:103–119. doi: 10.1534/genetics.108.090274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ezcurra M, Tanizawa Y, Swoboda P, Schafer WR. Food sensitizes C. elegans avoidance behaviours through acute dopamine signalling. EMBO J. 2011;30:1110–1122. doi: 10.1038/emboj.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarov M, et al. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nature Methods. 2006;3:839–844. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- 31.Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giordano-Santini R, et al. An antibiotic selection marker for nematode transgenesis. Nature Methods. 2010;7:721–723. doi: 10.1038/nmeth.1494. [DOI] [PubMed] [Google Scholar]
- 33.Semple JI, Garcia-Verdugo R, Lehner B. Rapid selection of transgenic C. elegans using antibiotic resistance. Nature Methods. 2010;7:725–727. doi: 10.1038/nmeth.1495. [DOI] [PubMed] [Google Scholar]
- 34.Stoeckius M, et al. Large-scale sorting of C. elegans embryos reveals the dynamics of small RNA expression. Nature Methods. 2009;6:745–751. doi: 10.1038/nmeth.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moy TI, et al. High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem Biol. 2009;4:527–533. doi: 10.1021/cb900084v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doitsidou M, Flames N, Lee AC, Boyanov A, Hobert O. Automated screening for mutants affecting dopaminergic-neuron specification in C. elegans. Nature Methods. 2008;5:869–872. doi: 10.1038/nmeth.1250. Genetic screens are a time-consuming and laborious process. This paper describes a method that would not only automate such processes, but is exquisite in its ability to detect even minute changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hulme SE, et al. Lifespan-on-a-chip: microfluidic chambers for performing lifelong observation of C. elegans. Lab. Chip. 2010;10:589–597. doi: 10.1039/b919265d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez-Blanco A, Kim SK. Variable pathogenicity determines individual lifespan in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002047. doi: 10.1371/journal.pgen.1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samara C, et al. Large-scale in vivo femtosecond laser neurosurgery screen reveals small-molecule enhancer of regeneration. Proc Natl Acad Sci USA. 2010;107 doi: 10.1073/pnas.1005372107. Microfluidic technology has the potential to semi-automate many laborious processes in the laboratory. This paper presents one of the most elegant applications of microfluidics by converting the typically arduous process of neurosurgery into one that could be performed by a minimally trained user with ease, rapidity and precision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X, et al. Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell. 2009;139:623–633. doi: 10.1016/j.cell.2009.08.044. Although the invariant cell lineage of the worm has been defined for over 30 years, expression analysis still tends towards descriptive characterizations of large tissues rather than quantitative characterizations of single cells. The method described in this paper allows for a minimally trained user to measure gene expression at a level of detail that was not previously thought possible without having committed to knowledge the entire cell lineage of the worm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng H, Long F, Liu X, Kim SK, Myers EW. Straightening Caenorhabditis elegans images. Bioinformatics. 2008;24:234–242. doi: 10.1093/bioinformatics/btm569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long F, Peng H, Liu X, Kim SK, Myers G. Automatic recognition of cells (ARC) for 3D images of C. elegans. Lect Notes Comp Sci. 2008;4955:128–139. [Google Scholar]
- 44.Long F, Peng H, Liu X, Kim SK, Myers E. A 3D digital atlas of C. elegans and its application to single-cell analyses. Nature Methods. 2009;6:667–672. doi: 10.1038/nmeth.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aydin Z, Murray JI, Waterston RH, Noble WS. Using machine learning to speed up manual image annotation: application to a 3D imaging protocol for measuring single cell gene expression in the developing C. elegans embryo. BMC Bioinformatics. 2010;11:84. doi: 10.1186/1471-2105-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray JI, et al. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nature Methods. 2008;5:703–709. doi: 10.1038/nmeth.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deisseroth K. Optogenetics. Nature Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Trans R Soc Lond B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 51.Hobert O, Carrera I, Stefanakis N. The molecular and gene regulatory signature of a neuron. Trends Neurosci. 2010;33:435–445. doi: 10.1016/j.tins.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo ZV, Hart AC, Ramanathan S. Optical interrogation of neural circuits in Caenorhabditis elegans. Nature Methods. 2009;6:891–896. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Q, Hollopeter G, Jorgensen EM. Graded synaptic transmission at the Caenorhabditis elegans neuromuscular junction. Proc Natl Acad Sci USA. 2009;106:10823–10828. doi: 10.1073/pnas.0903570106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leifer AM, Fang-Yen C, Gershow M, Alkema MJ, Samuel AD. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nature Methods. 2011;8:147–152. doi: 10.1038/nmeth.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stirman JN, et al. Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans. Nature Methods. 2011;8:153–158. doi: 10.1038/nmeth.1555. Optogenetics has transformed the field of neurobiology by allowing researchers to study real-time behavioural phenotypes resulting from the activation or inactivation of specific neurons. This paper is exciting not only because it uses optogenetics to address important research questions, but also because it makes optogenetics accessible to most research laboratories by demonstrating that the required tools can be built from a standard three-colour liquid crystal display (LCD) projector. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greenwald I. lin-12, a nematode homeotic gene, is homologous to a set of mammalian proteins that includes epidermal growth factor. Cell. 1985;43:583–590. doi: 10.1016/0092-8674(85)90230-2. [DOI] [PubMed] [Google Scholar]
- 58.Sigurdson DC, Spanier GJ, Herman RK. Caenorhabditis elegans deficiency mapping. Genetics. 1984;108:331–345. doi: 10.1093/genetics/108.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bacaj T, Shaham S. Temporal control of cell-specific transgene expression in Caenorhabditis elegans. Genetics. 2007;176:2651–2655. doi: 10.1534/genetics.107.074369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pulak R. Techniques for analysis, sorting, and dispensing of C. elegans on the COPAS flow-sorting system. Methods Mol Biol. 2006;351:275–286. doi: 10.1385/1-59745-151-7:275. [DOI] [PubMed] [Google Scholar]
- 61.Gray JM, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 62.Nagel G, et al. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]