Abstract
Background
Genome-wide studies reveal that genetic variants at chromosome 4q25 constitute the strongest locus associated with atrial fibrillation (AF), the most frequent arrhythmia. However, the mechanisms underlying this association are unknown. Our goal is to find and characterize left atrial expressed transcripts in the chromosome 4q25 AF risk locus that may play a role in AF pathogenesis.
Methods and Results
RNA sequencing performed on human left/right pairs identified an intergenic long noncoding RNA (lncRNA) adjacent to the PITX2 gene, which we have named PANCR (PITX2 adjacent noncoding RNA). In a human tissue screen, PANCR was expressed specifically in the left atria and eye, and in no other chambers of the heart. The levels of PANCR and PITX2c RNAs were highly correlated in 233 human left atrial appendage samples. PANCR levels were not associated with either atrial rhythm status or the genotypes of the chromosome 4q25 AF risk variants. Both PANCR and PITX2c RNAs were induced early during differentiation of human embryonic stem cells into cardiomyocytes. Since lncRNAs often control gene expression, we performed siRNA-mediated knockdown of PANCR; and, this treatment repressed PITX2c expression and mimicked the effects of PITX2c knockdown on global mRNA and miRNA expression. Cell fractionation studies demonstrate that PANCR is primarily localized in the cytoplasm.
Conclusions
PANCR and PITX2c are coordinately expressed early during cardiomyocyte differentiation from stem cells. PANCR knockdown decreased PITX2c expression in differentiated cardiomyocytes, altering the transcriptome in a manner similar to PITX2c knockdown.
Keywords: atrial fibrillation, gene expression, RNA sequencing, long intergenic noncoding RNA
Introduction
Atrial Fibrillation (AF), the most common sustained arrhythmia, is associated with a 2-fold increase in mortality and 4- to 5-fold increased risk for stroke worldwide.1, 2 Genome-wide association studies (GWAS) have consistently identified single nucleotide polymorphisms (SNPs) in the chromosome 4q25 locus as having the strongest association with AF.3-5 Within this locus, four independent linkage disequilibrium (LD) blocks contain multiple SNPs associated with AF risk.6, 7 These SNPs are located in an intergenic region from 26 to 217 kb distal to the closest gene on the chromosome, PITX2. Previously, we demonstrated that four strongest independent AF-associated SNPs at this locus are not associated with expression of PITX2c in a large cohort of human adult left atrial appendages.6 In a prior study, we performed RNA sequencing (RNAseq) on 4 pairs of left-right human atrial appendages and identified numerous transcripts with strong expression bias in the left or right atria. These included PITX2c, which is expressed only in the left atria.8 We also found many un-annotated novel spliced transcripts that were differentially expressed between the left and right atria which met our in silico criteria as long noncoding RNAs (lncRNAs).8 Here we report the characterization of PANCR, a PITX2 adjacent non coding RNA. Like PITX2c, PANCR is expressed specifically in the left atria. Intergenic lncRNAs are located between genes and typically have their own transcriptional machinery.9, 10 LncRNAs are characterized as transcripts >200 nucleotides in length that are 5’capped and 3’ polyadenylated, similar to mRNA; although, they do not code for a functional protein, they may contain short open reading frames.10 Many lncRNAs evolved recently and are conserved within lineages but not between lineages, with many lncRNAs conserved among primates that are not conserved with rodents.10-12 However, lncRNAs play important functional roles and they have been implicated in numerous biological processes such as epigenetic regulation, imprinting, cell-cycle control, cellular differentiation, splicing, nuclear/cytoplasmic trafficking, and transcription/ translation. Thus, lncRNAs may explain lineage differences in development.12-18 Recently, several lncRNAs associated with regulatory functions have been described in the heart and vascular system including Braveheart, Fendrr, CHRF, MALAT1, LIPCAR, and SENCR.13, 19-24 Since the AF-associated SNPs on chromosome 4q25 are intergenic, we hypothesized that they may alter AF susceptibility by regulating atrial gene expression. As we already found that PITX2c expression in human adult left atrial appendages is not associated with these AF risk SNPs6, we explored all transcripts in this region by RNAseq. We identified PANCR, a two exon lncRNA adjacent to PITX2 on chr 4q25. PANCR appears to be functional in cardiomyocytes differentiated from human embryonic stem cells, in that its knockdown has widespread effects on gene expression. However, similar to PITX2c, PANCR expression was not associated with the four independent AF-associated SNPs in the 4q25 locus. Therefore, the mechanisms by which the AF associated SNPs on chromosome 4q25 modify AF susceptibility remain enigmatic.
Materials and Methods
Human Left Atrial Tissue
Left atrial appendage tissues were obtained from a biorepository of human atrial tissues from patients who underwent cardiac surgery at the Cleveland Clinic and who consented to have discarded tissue used for research under a protocol approved by the Cleveland Clinic Institutional Review Board (IRB). Prior to 2008 verbal consent was obtained and documented in the medical records in a process approved by the IRB. From 2008 onward, patients provided IRB-approved written informed consent. AF history, type of AF, structural heart disease, demographics, and other clinical data were collected in a research database and a prospectively collected database of all cardiac surgeries. Subjects were categorized as “lone AF” if they had a history of AF and did not have coronary artery disease (CAD) or mitral valve disease (MVD). AF rhythm status was determined by review of electrocardiograms obtained prior to surgery. Samples were snap frozen in liquid nitrogen and kept at -80°C until RNA extraction.
RNA isolation and cDNA preparation
Left atrial appendage tissue was homogenized in TRIazol® (Invitrogen). RNA was isolated from the homogenate following the manufacturer’s protocol. 1 μg of RNA was reverse transcribed using Superscript® Vilo™ mastermix (Invitrogen).
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
Expanded methods for qRT-PCR are found in the Supplement. Delta C(t) values for PANCR and PITX2c expression levels were calculated relative to ACTC1 or PPIA expression, and the ΔΔCT method was used to compare expression among samples 25, 26, yielding log2 based expression values, which were converted to linear values when indicated by calculating 2-ΔΔCT. To calculate PANCR expression in human atrial samples, relative log2 gene expression levels were corrected for plate and batch effects using three standardized atrial RNA samples on each plate. Relative expression levels were fit to an additive linear model including age, gender, donor/surgical sample, atrial fibrillation history and pre-operative rhythm status, using the R statistical program. SNP genotyping and eQTL analyses were performed as previously described 6.
PANCR and PITX2c tissue panel expression assay
A Human Total RNA master tissue panel was purchased from Clontech (cat 636643). Additional ventricular samples from anonymous discarded tissue were obtained under an IRB approved protocol. IRB-exempt anonymous eye samples were obtained from the National Disease Research Interchange (Philadelphia, PA). The custom designed PANCR or PITX2c TaqMan assays (described above) were used with PPIA as the endogenous control. Reactions were run in triplicate and results were presented after conversion of log2 values into linear measures by applying a 2-ΔΔCT transformation.
Differentiation of H9 cells to cardiomyocytes
A monolayer protocol was used to differentiate H9 hES cells into cardiomyocytes as previously described27 with the following minor modifications. Cells were plated in 12-well dishes that were coated with diluted growth factor reduced matrigel (BD). The cells were cultured in mouse embryonic fibroblast media (R&D systems) for 3 days before growth factors were added as previously described 27. The cells were maintained in RPMI 1640 with B27 supplements (Invitrogen 17504-044) until experimentation. Beating foci of cardiomyocytes were first seen between days 11 and 14.
siRNA knockdown of PANCR and PITX2
100 pmol of a custom PANCR siRNA (Ambion silencer select, Supplemental Table 1), 33 pmol of each of 3 PITX2c siRNAs (Ambion silencer select cat#4392420, ids: S10557, S10558, and S10559), or 100 pmol of a control scramble siRNA (Ambion silencer select cat # 4390843) were transfected in differentiated H9 induced cardiomyocytes using the RNAiMax (Invitrogen) transfection reagent according to manufactures specifications. The siRNA complexes were incubated with cells for 48 hrs followed by RNA isolation, cDNA preparation, and qRT-PCR, as described above.
RNAseq and analysis
Expanded methods for RNAseq and analysis are found in the Supplement. Gene expression data for these studies is available at GEO accession number GSE67844. The search of miRNA target sites in PITX2 using 11 algorithms was performed using miRecords. 28
Statistical Methods
All data is shown as the mean ± SD, unless indicated otherwise. Patient membership in gender or disease status classes were subjected to chi-square tests. Patient BMI and age were not normally distributed and were analyzed by Krusakal-Wallis nonparametric ANOVA. Human atrial PITX2c and PANCR RNA levels were adjusted for by the factors listed in the table or figure legends, used as linear covariates. The p-values for the association of SNPs with PANCR expression was performed by linear regression using the R statistical software, with or without correction using sex, age, BMI, hypertension, CAD, MVD, and AF history/rhythm as linear covariates. Left-right atrial PANCR gene expression was compared using a paired t-test. Gene expression data from the H9 cell line was assumed to be normally distributed, thus parametric T-test and ANOVA analyses were used, along with posttests indicated in figure legends. All column-based and correlation analyses were performed using Prism 6.0 software (GraphPad). RNAseq statistical analyses and false discovery rate p-value adjustments are described in the Supplement.
Results
Discovery of a lncRNA adjacent to the PITX2 gene in human left atria
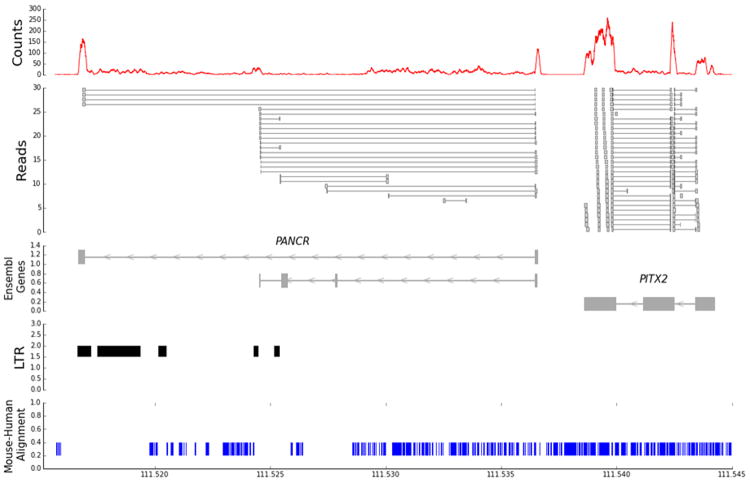
RNAseq was previously performed in four pairs of human left-right atrial samples 8. We detected a left atrial expressed RNA adjacent to PITX2, with a 1937 bp intergenic region, which corresponds to Ensembl transcript RP11-380D23.2-002 (ENST00000513690, release 74), which is classified as a long intergenic noncoding RNA (Figure 1). This transcript will be referred to as PANCR (PITX2 Adjacent Non-Coding RNA). The most common transcript isoform was 446 nucleotides long, encoded by two-exons derived from a gene of ~ 20 kb (ENSG00000250103). We excluded the possibility that PANCR could be due to read through transcription of the PITX2c gene; in the four left atrial samples we found 94 RNAseq reads spanning the two exons of PANCR and 0 reads spanning the last exon of PITX2c and the first exon of PANCR. Our analysis confirmed the lncRNA annotation, with the longest open reading frame (ORF) encoding only 35 amino acid residues, typical for lncRNAs. 29 Other minor splice junctions were observed and confirmed upon sequencing additional left atrial appendages, yielding a second 4-exon transcript that corresponds to Ensembl transcript ENST00000503456.1 (RP11-380D23.2); but, the read coverage for this alternate transcript was low (Figure 1). Our exon-spanning reads also detected a novel alternatively spliced version of the 4-exon ENST00000503456.1 transcript that contained only the first and last exons. Overall, 90.8% of the junctional reads spanned the two major exons in PANCR, with the first exon and second exons comprising 152 and 294 bp, respectively. The entire second exon of PANCR is composed of a long terminal repeat (LTR) in the ERVL-MaLR retrotransposon family. This ORF and the entire PANCR transcript sequence are well conserved in chimpanzee with 100% and 98.9% identity, respectively. However, the mouse shares only a small 55 bp region of identity in exon 1 (54/55 nucleotides conserved) located in the orthologous region on mouse chromosome 3 between the Enpep and Pitx2 genes, with only the first 19 nucleotides of the ORF conserved perfectly. We performed RNAseq of mouse left atria and did not find any detectable expression overlapping the 55 bp region of identity with human PANCR.
Figure 1.
RNAseq identification of PANCR. The top panel shows aggregate read counts from a human left atrial appendage in a portion of the chr.4q25 region. The second panel shows a non-representational set of the individual reads that span exon junctions (horizontal lines). The third panel shows the Ensembl annotated transcripts, the three-exon PITX2c transcript, the major two-exon PANCR transcript, and the minor four-exon transcript. The fourth panel shows the locations of long terminal repeats (LTRs) in this region, which overlaps with the second exon of PANCR. The bottom panel shows the mouse-human alignment (in blue), with only a 55 bp fragment of PANCR exon 1 conserved.
Left atrial PANCR expression is not associated with AF rhythm status
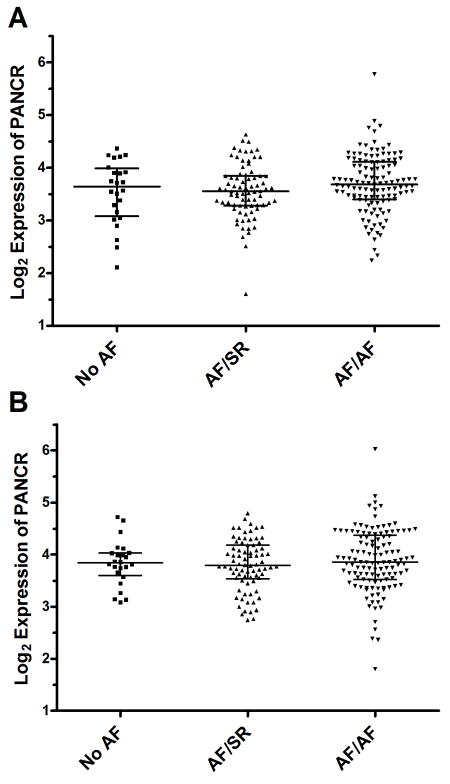
We obtained left atrial appendage specimens from 223 cardiac surgery patients of European ancestry. Samples were subdivided into three groups based on their history of AF and their preoperative rhythm status: no history of AF (No AF, N=24); history of AF in sinus rhythm (SR) at time of sample collection (AF/SR, N=78); and history of AF in AF rhythm at time of sample collection (AF/AF, N=121) (Table 1). AF/rhythm status was examined for association with sex, age, BMI, and history of hypertension, CAD, and MVD. The only significant characteristic associated with AF rhythm status was age, with the No AF group being the oldest and the AF/SR group being the youngest (p-value = 0.0035). There were also trends for association with BMI, history of hypertension, and history of mitral valve disease. Among the 199 AF subjects, there were 35 subjects with lone AF equally represented in the AF/SR and AF/AF groups. PANCR RNA levels in the left atrial appendages, normalized to ACTC1 were measured by qPCR. There was no association of PANCR levels with AF rhythm status, either before or after adjusting for age, sex, BMI, hypertension, CAD, and MVD (Figure 2).
Table 1.
Left atrial appendage surgical patient characteristics
| Patient Characteristics | Total N=223 | No History of AF n=24, 11% | History of AF/SR n=78, 35% | History of AF/AF n=121, 54% | P-Value |
|---|---|---|---|---|---|
| Sex, Female, % | 22% | 27% | 25% | 19% | 0.45† |
| Age (years)* | 62 (55, 69) | 68 (62,75) | 59 (52, 65) | 64 (55, 70) | 0.0035‡ |
| BMI (kg/m2)* | 27.8 (24.7, 31.6) | 26.5 (23.4,28.3) | 27.0 (24.0, 31.4) | 28.2 (25.2, 32.2) | 0.064§ |
| Hypertension, % | 51% | 60% | 41% | 56% | 0.085† |
| CAD, % | 34% | 50% | 29% | 34% | 0.17† |
| MVD, % | 49% | 70% | 53% | 43% | 0.076† |
| Lone AF, % | 16% | 0% | 17% | 18% | 0.78§ |
Median (interquartile range)
p-value by chi-square test
p-value by Kruskal Wallis nonparametric ANOVA
p-value by chi-square comparing only AF/SR and AF/AF groups
Figure 2.
PANCR expression not associated with AF. Unadjusted (A) and sex and age adjusted (B) PANCR expression levels, normalized to ACTC1 expression are not significantly associated with AF history and rhythm status in 223 human left atrial appendages. Sex and age were used as linear covariates for adjusting PANCR expression.
PANCR and PITX2c expression in human tissues
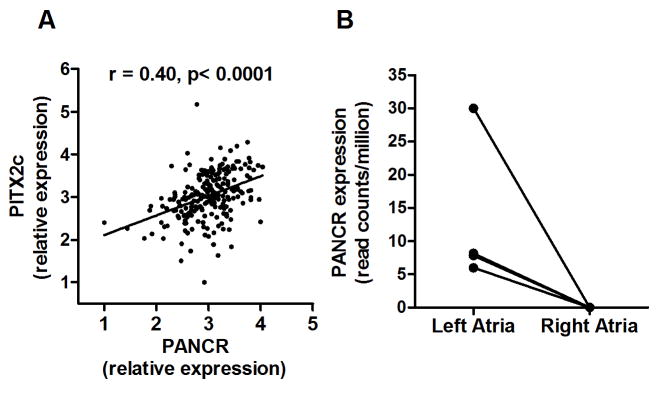
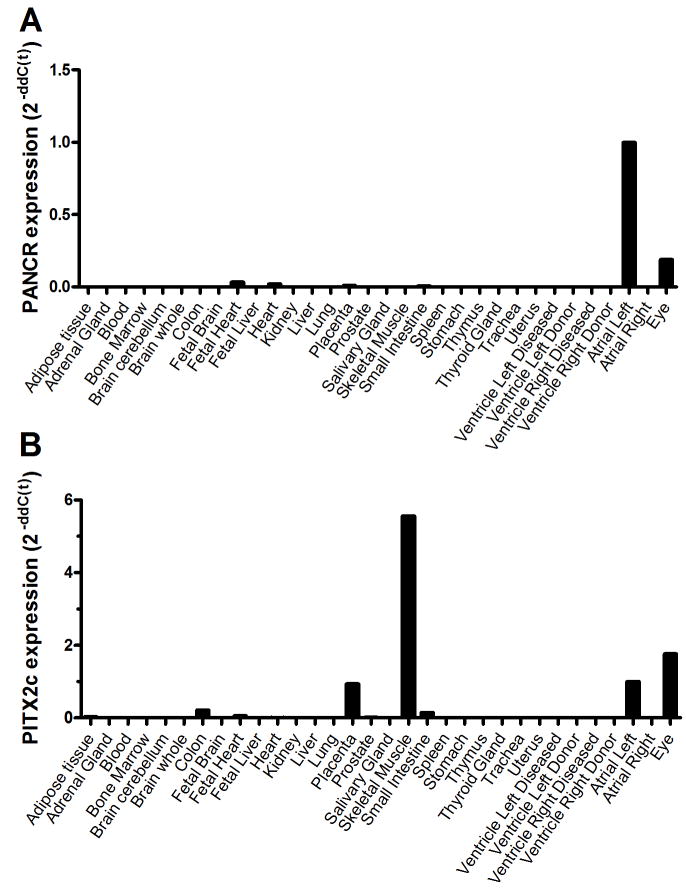
Using RNA from the same 223 human left atrial appendage subjects described above, we also measured the levels of PITX2c mRNA, as PITX2c is the only isoform of the adjacent gene expressed in left atria. Overall, there was a robust and significant positive correlation between PANCR and PITX2c RNA levels (r= 0.40, p<0.0001, Figure 3A). Analysis of our left/right atrial RNAseq data demonstrated that PANCR expression was left atrial specific with an average of 13.0 reads per million mapped reads while no reads above background mapped to this transcript in the right atria (p-value = 0.05, Figure 3B). To determine the tissue distribution of the PANCR expression, qPCR was performed on 33 human tissue samples, including the left atrium as a positive control. Expression was highest in the left atrium, with ~ 10-fold lower expression in the eye and lower levels detected in fetal heart, total heart, placenta, and small intestine (Figure 4A). PANCR was not detected in adult right atrium, left or right ventricle, or any other tissue examined. We also evaluated PITX2c expression in these same tissue samples. PITX2c was most highly expressed in skeletal muscle (~ 5.6-fold greater than left atrium), followed by eye (~1.8-fold vs. left atrium) and left atrium (Figure 4B). PITX2c expression was also detected in placenta ≫ colon, small intestines, fetal and adult total heart, prostate, and adipose tissue. In agreement with our prior studies, PITX2c expression was much higher in the left vs. right atrial appendage 6, 8.
Figure 3.
PANCR expression correlated with PITX2c expression in human left atrial appendages. A. PANCR and PITX2c levels, measured by qPCR normalized to ACTC1, are significantly and positively correlated in human adult left atrial appendages from 233 subjects of European ancestry (r= 0.40; p < 0.0001). B. RNAseq demonstrated that PANCR was expressed in the left atria but not in the right atria in 4 left/right atrial pairs (paired t-test p=0.05).
Figure 4.
Tissue specific expression of PANCR and PITX2c. RNA from 33 different human tissue samples was used to determine specific tissue expression of PANCR and PITX2c by qPCR normalized to PPIA expression. A. PANCR was most highly expressed in the left atria and in the eye. B. PITX2c was most highly expressed in skeletal muscle, left atria and in the eye.
Lack of Identification of common cis variants associated with PANCR expression in left atria
Previously, we determined that common SNPs in the 4q25 region are not associated with PITX2c expression in human adult left atrial appendages.6 To determine if common genetic variants in this locus were associated with PANCR expression, we performed a cis-expression quantitative trait (cis-eQTL) analysis by calculating the association of PANCR expression with SNP genotypes obtained from microarrays (+/- 500 kb from PANCR). First we examined the four independent AF associated SNPs on chromosome 4q25 for association with PANCR expression. These four SNPs (Table 2) were identified from the Cleveland Clinic Lone AF GWAS 6; and, the independent association of these SNPS with AF, or proxies in perfect LD (HapMap 22), was recently confirmed in a large meta-analysis.7 None of these SNPs was associated with PANCR expression, normalized to ACTC1, before or after adjustment for sex, age, body mass index, hypertension, coronary artery disease, mitral valve disease, and AF history/rhythm (p≥0.25, Table 2). A simulation using our log2 PANCR expression data, with a coefficient of variation of 14.32% in 223 subjects, and a minor allele frequency of 0.10 (the lowest frequency of these four AF associated SNPs) demonstrates that we were powered to observe a 7% allele effect on PANCR expression at p<0.05. Thus, we were well powered to observe cis-effects on gene expression. Extending this analysis to 169 genotyped SNPs in this region, none were associated with the expression of PANCR at a Bonferroni corrected p-value threshold of 0.05. Thus, the AF associated SNPs do not appear to regulate PANCR expression in adult human left atrial appendages.
Table 2.
Chr. 4 AF risk SNPs not associated with PANCR expression
| SNP | location | PANCR p-value uncorrected | PANCR p-value phenotype corrected* |
|---|---|---|---|
| rs2200733 | 111929618 | 0.87 | 0.80 |
| rs3853445 | 111980936 | 0.60 | 0.68 |
| rs1448818 | 111789672 | 0.26 | 0.25 |
| rs10033464 | 111940210 | 0.35 | 0.31 |
corrected for sex, age, BMI, hypertension, CAD, MVD, and AF history/rhythm as linear covariates.
PANCR and PITX2c are coordinately expressed during cardiomyocyte differentiation of human H9 embryonic stem cells
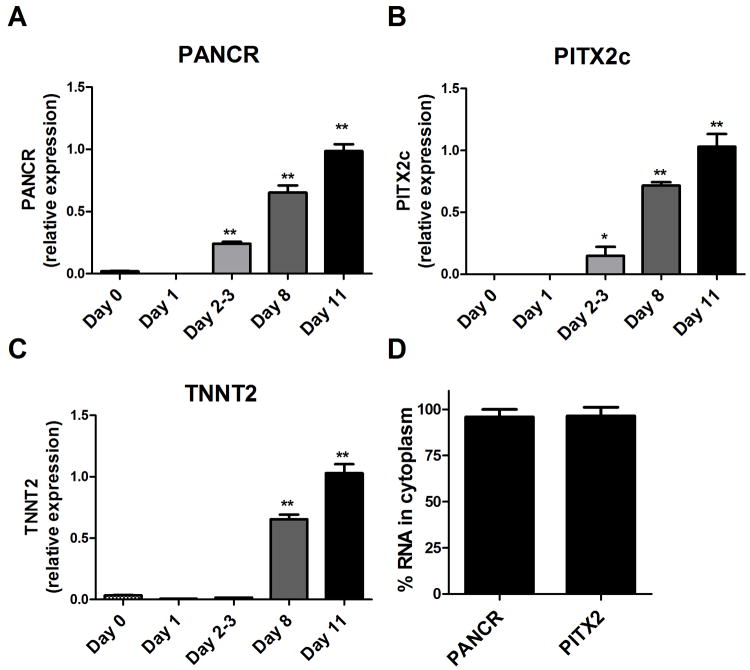
H9 embryonic stem cells were differentiated into cardiomyocytes as described in the Methods section. Cells were harvested prior to the addition of each new factor to determine when PANCR, PITX2c, and the cardiomyocyte specific marker Troponin T2 were induced during differentiation. Expression levels were measured by qPCR, normalized to PPIA. PANCR and PITX2c were both induced at day 2-3, while cardiac troponin was not induced until day 8 (Figure 5A-C), indicating that PANCR and PITX2c were coordinately induced during differentiation prior to the expression of this cardiomyocyte structural protein. We fractionated these differentiated cardiomyocytes into nuclear and cytoplasmic fractions and determined that >95% of both PANCR RNA and PITX2c mRNA were located in the cytoplasm (Figure 5D).
Figure 5.
PANCR and PITX2c coordinately induced during cardiomyocyte differentiation. A-C. During differentiation of H9 ES cells to cardiomyocytes, PANCR and PITX2c were coordinately induced at day 2-3, preceding the induction of cardiac troponin, a cardiomyocyte specific structural protein (N=3, mean ± SD; *, p<0.05; **, p<0.001 compared to Day 0 by ANOVA with Dunnett posttest). D. Nuclear and cytoplasmic RNAs were prepared and qRT-PCR demonstrated that both PANCR and PITX2c RNAs were predominantly cytoplasmic (N=2, mean ± S.D)
PANCR and PITX2c knockdowns in H9 derived cardiomyocytes
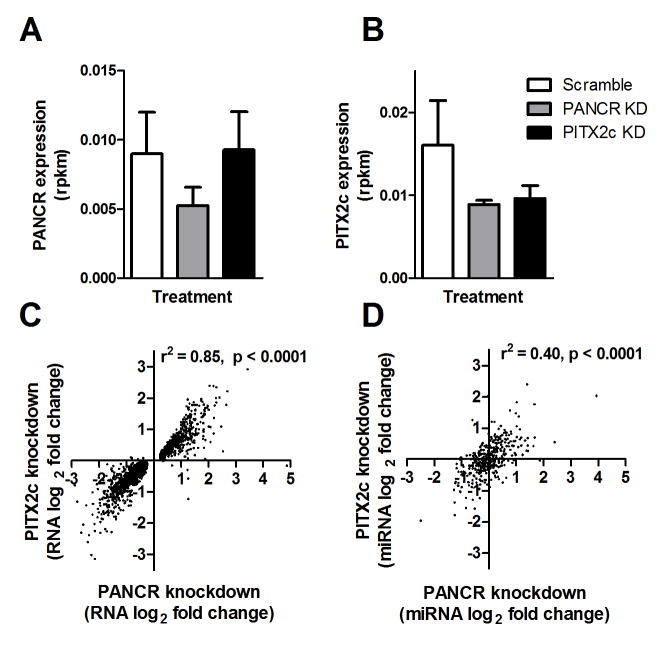
We demonstrated above that the expression of PANCR and PITX2c were positively correlated in human adult left atrial appendages. We further hypothesized that PANCR and the adjacent transcription factor, PITX2c, might regulate each other’s expression, and both might also regulate expression of other genes in trans. PANCR and PITX2c were knocked down in H9 differentiated cardiomyocytes (3 biological replicates) using siRNA transfection, and global gene expression was ascertained by RNAseq. We first evaluated regulation of the expression of the two adjacent genes (Figure 6A,B). PITX2c knock down reduced its own expression by 40%, but expression of PANCR was not affected. In contrast, PANCR knockdown decreased expression of both itself and PITX2c (42% and 44%, respectively). These effects of PITX2c and PANCR knockdown were confirmed and found significant in an independent experiment where the expression of PITX2c and PANCR were measured using qPCR (Supplemental Figure 1). There is no sequence similarity between PITX2c and PANCR RNAs, as determined using blast2n, thus the siRNA to PANCR cannot directly interact with PITX2c mRNA.
Figure 6.
siRNA knockdown of PANCR and PITX2c in differentiated cardiomyocytes. After differentiation H9 hESC cells to cardiomyocytes, siRNA was used to knockdown PANCR and PITX2c RNAs and changes in gene expression were determined by RNAseq. A. PANCR expression was significantly decreased only by PANCR siRNA. B. PITX2c expression was significantly decreased when either PITX2c or PANCR siRNA (expression calculated in reads per million per kb transcript size, rpkm; N=3, ± SD). C. Fold changes in RNA expression were significantly correlated after PANCR vs. PITX2c knockdown. Only RNAs whose expression was significantly altered by PANCR knockdown are plotted. D. Fold changes in miRNA expression were significantly correlated after PANCR vs. PITX2c knockdown.
An analysis of the expressed genes detected by RNAseq showed that PANCR knockdown resulted in changes in gene expression that were remarkably similar to those mediated by PITX2c knockdown. There were 2029 genes regulated by PANCR knockdown and 760 gene regulated by PITX2c knockdown at p<0.05, but this difference in number is simply due to slightly higher variability in the PITX2c knockdown. Thus, instead of looking at overlap of p<0.05 threshold genes in both sets that would arbitrarily exclude many potentially co-regulated genes, we examined the 2029 genes that were regulated by PANCR knockdown, and found that their direction and fold change effects of PANCR or PITX2c knockdown were highly conserved (Figure 6C, r2 = 0.85). Thus, we conclude that the major activity of PANCR is to positively regulate PITX2c mRNA. Gene ontology searches for the set of genes regulated by PANCR or PITX2c knockdown also showed agreement and revealed that cell adhesion was the top biological process, extracellular region was the top cellular compartment, and calcium ion binding was the top molecular function, for both knockdowns (all < 0.03 FDR). The top 20 genes (by fold change) altered by each knockdown are shown in Supplemental Tables 2 and 3.
We also performed small RNAseq on the H9 differentiated cardiomyocytes after PANCR or PITX2c knockdown to assess their effects on the miRNA transcriptome. Many miRNAs were significantly up- or down-regulated after PANCR or PITX2c knockdown. The top 20 miRNAs (by p-value) altered by each knockdown are shown in Supplemental Tables 4 and 5. We compared the fold change effects of the 386 expressed miRNAs in the PANCR knockdown dataset with the fold change effects after PITX2c knockdown and again found good conservation in the direction and effect size (Figure 6D, r2 = 0.40). Similar to the RNAseq analysis, this miRNAseq analysis shows that the effects of PANCR knockdown on miRNA expression may be largely mediated by its effect on PITX2c expression.
Discussion
Multiple genome wide association studies have identified the chromosome 4q25 region as the strongest AF susceptibility locus.3, 5 Our earlier RNAseq study of four left/right atria pairs revealed an uncharacterized lncRNA, which we have now named PANCR, located just 2 kb 3’ of the PITX2 gene near the chromosome 4q25 AF susceptibility region.8 Here we report that PANCR is most highly expressed in human left atria and eye; while, PITX2c is also highly expressed in the left atria and eye, its highest expression level is in skeletal muscle. The left atria-eye connection has been previously noted in Axenfeld–Rieger syndrome, which is attributed to mutations in the PITX2 gene. These patients are characterized by abnormal development of the anterior eye and of the heart, among other malformations.30, 31 Complete knockout of the PITX2 gene in mice leads to developmental lethality with malformation of several organs, while hemizygous mice have eye abnormalities and are susceptible to pacing induced atrial arrhythmia.31-35
We previously found that AF susceptibility SNPs in the 4q25 region were not associated with PITX2c expression in human adult left atrial appendages from 239 subjects.6 Thus, we were excited by the possibility that these AF susceptibility SNPs might be associated with PANCR expressions. However, we report here that these SNPs in the 4q25 region were not associated with expression of PANCR. Another group recently looked for association of chr4q25 SNPs with PITX2 expression in 122 right and 12 left atrial appendage patient samples; and, the only significant association found was for the PITX2a isoform in right atrial appendages, where the AF risk alleles were associated with higher PITX2a expression.36 Thus, there is still no evidence in human adult atrial appendages that expression of the PITX2c isoform is associated the AF risk alleles at 4q25. However, this region of 4q25 does contain histone marks (H3K4me1) in human fetal heart that are associated with enhancer activity.37 One small region near the AF risk SNPs was identified to have weak enhancer activity, albeit not tissue specific to atrial like cells or to the heart, tested in reporter gene transfection studies and transgenic mice, respectively.37 We still suspect that these SNPs may regulate PITX2c and/or PANCR expression, but perhaps at an earlier time in development, and/or in a different region of the heart. In fact, a lacZ PITX2c knockin is expressed highest during development and in the early postnatal period, compared to older mice.34 In addition, isoform specific knockout of PITX2c in mice alters the pulmonary vein connections to the left atrium; and, the atrial appendage tissue samples we studied have a different embryological origin from the secondary heart field where PITX2c is expressed during early development38, and thus gene regulation may be different in these regions.
LncRNAs are still an enigma. In contrast to miRNAs that function by binding directly to target mRNAs to decrease their translation and/or stability, lncRNAs do not have a single mechanism of action.9 Some lncRNAs reside in the nucleus, others are in the cytoplasm, and still others are found in both compartments. Among the many reported functions of lncRNAs, they may modulate gene expression via recruitment of histone modifying proteins to alter epigenetic regulation, recruitment of transcription activators or repressors, alteration of nuclear structure, acting as antisense transcripts to overlapping or partially complementary mRNAs, acting as sponges to bind to RNA binding proteins or miRNAs. Some lncRNAs are actually mis-identified and are actually mRNAs that code for small proteins detected by mass spectrometry.39 In addition, many lncRNAs are not well conserved between mice and humans; however, even some of these non-conserved lncRNAs have been shown to have significant functions.40, 41 In our human left atrial samples, we found a striking correlation between the levels of PANCR lncRNA and PITX2x mRNA. Additionally, during hES cell cardiomyocyte differentiation, PANCR and PITX2c were coordinately induced prior to cardiac troponin, suggesting that these genes are expressed before the cells are fully differentiated into cardiomyocytes. This finding, combined with mouse data showing reduced, but present, PITX2c levels in adult vs. postnatal heart 34, suggests that PANCR and PITX2c may play a larger role during development and in childhood than in adult heart.
Our co-expression data suggest that PANCR may regulate PITX2c expression, or alternatively that PITX2 may regulate the expression of PANCR. To test these hypotheses we separately performed siRNA knockdown of PANCR and PITX2 in cardiomyocytes differentiated from H9 human ES cells. We found that while PANCR knockdown reduced PITX2c levels, PITX2c knockdown did not reduce PANCR levels. This suggests that PANCR may act as a positive regulator of PITX2c. However, cell fractionation studies found that PANCR is localized in the cytoplasm of induced cardiomyocytes; thus, PANCR is less likely to increase PITX2c transcription via either epigenetic modifier or transcription factor recruitment in the nucleus. The transcription of a lncRNA in itself could act to promote transcription of an adjacent gene.9 Since PANCR siRNA knockdown would not be expected to alter PANCR transcription in the nucleus but its turnover in the cytoplasm, it seems likely that just the transcription of PANCR is not sufficient to positively regulate the expression of PITX2c in-cis. Rather, we speculate that it is more likely that PANCR stabilizes PITX2c mRNA in the cytoplasm. However, this mechanism would need to work despite our RNAseq finding that PANCR is expressed at lower levels than PITX2c in our ES cell derived cardiomyocytes.
Our RNAseq study with the induced cardiomyocytes revealed that PANCR knockdown yielded similar changes in mRNA levels as PITX2c knockdown, indicating that the other effects of PANCR knockdown were probably mediated indirectly, via modulation of PITX2c mRNA levels. Likewise, the PANCR and PITX2c knockdowns had similar overall effects on the miRNA transcriptome; however, changes in miR-143 and miR-501 both caught our attention. We had previously noted that miR-143 was the most abundant miRNA in human atria, accounting for ~30% of all mapped small RNA reads.8 Here, we also observed that miR-143 was the most abundant miRNA in the H9 differentiated cardiomyocytes. Knockdown of either PANCR or PITX2c led to significant ~ 2-fold upregulation of miR-143 (Supplemental Tables 4, 5). The Miranda and RNAhybrid miRNA target algorithms predict that PITX2c contain a miR-143 binding site, although several other algorithms fail to detect this site. In zebrafish studies, miR-143 has been shown to alter cardiac chamber morphogenesis and play a role in retinoic acid signaling and cardiac outflow tract development.42, 43 In addition, knockout of the miR143-145 gene cluster in mice leads to impaired vascular function.44 miR-501 was induced 3.2-fold after PANCR knockdown and 1.6-fold after PITX2c knockdown (Supplemental Tables 4, 5). miR-501 is predicted to bind to PITX2c by the Miranda, miRTarget, Pita, and RNAhybrid algorithms. Thus, one potential mechanism for the cytoplasmic PANCR lncRNA to regulate PITX2c mRNA is by acting as a miRNA sponge and protecting the binding of specific miRNAs to PITX2c mRNA that would otherwise increase its degradation.
Supplementary Material
WHAT IS KNOWN
Genome wide association studies have identified the chromosome 4q25 locus as the strongest locus associated with atrial fibrillation.
The PITX2c gene resides in this locus, but its expression in human adult left atrial appendages is not associated with the chromosome 4q25 genetic variants discovered in the atrial fibrillation genome wide association study.
WHAT THE STUDY ADDS
RNA sequencing of human left atrial appendages identified a gene for a long noncoding RNA adjacent to the PITX2c gene, named PANCR.
PANCR is expressed highest in the left atrium and eye, tissues that also express PITX2c highly.
PANCR and PITX2c were coordinately up regulated during cardiomyocyte differentiation from stem cells, and PANCR knockdown down regulated PITX2c mRNA levels in these cardiomyocytes, illuminating a novel mechanism to control PITX2c expression and its downstream targets.
Acknowledgments
We wish to thank Dr. Bela Anand-Apte for procuring eye tissue for this study.
Funding Sources: This work was supported by the NIH grants R01 HL111314 (MKC, JB, DRVW, JDS), supplement to HL111314 (SRGP), F31 HL103088 (SRGP), and T32 GM088088 (JH)
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Benjamin EJ, Chen PS, Bild DE, Mascette AM, Albert CM, Alonso A, Calkins H, Connolly SJ, Curtis AB, Darbar D, Ellinor PT, Go AS, Goldschlager NF, Heckbert SR, Jalife J, Kerr CR, Levy D, Lloyd-Jones DM, Massie BM, Nattel S, Olgin JE, Packer DL, Po SS, Tsang TS, Van Wagoner DR, Waldo AL, Wyse DG. Prevention of atrial fibrillation: Report from a national heart, lung, and blood institute workshop. Circulation. 2009;119:606–618. doi: 10.1161/CIRCULATIONAHA.108.825380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lloyd-Jones DM, Wang TJ, Leip EP, Larson MG, Levy D, Vasan RS, D’Agostino RB, Massaro JM, Beiser A, Wolf PA, Benjamin EJ. Lifetime risk for development of atrial fibrillation: The framingham heart study. Circulation. 2004;110:1042–1046. doi: 10.1161/01.CIR.0000140263.20897.42. [DOI] [PubMed] [Google Scholar]
- 3.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Muller-Nurasyid M, Krijthe BP, Lubitz SA, Bis JC, Chung MK, Dorr M, Ozaki K, Roberts JD, Smith JG, Pfeufer A, Sinner MF, Lohman K, Ding J, Smith NL, Smith JD, Rienstra M, Rice KM, Van Wagoner DR, Magnani JW, Wakili R, Clauss S, Rotter JI, Steinbeck G, Launer LJ, Davies RW, Borkovich M, Harris TB, Lin H, Volker U, Volzke H, Milan DJ, Hofman A, Boerwinkle E, Chen LY, Soliman EZ, Voight BF, Li G, Chakravarti A, Kubo M, Tedrow UB, Rose LM, Ridker PM, Conen D, Tsunoda T, Furukawa T, Sotoodehnia N, Xu S, Kamatani N, Levy D, Nakamura Y, Parvez B, Mahida S, Furie KL, Rosand J, Muhammad R, Psaty BM, Meitinger T, Perz S, Wichmann HE, Witteman JC, Kao WH, Kathiresan S, Roden DM, Uitterlinden AG, Rivadeneira F, McKnight B, Sjogren M, Newman AB, Liu Y, Gollob MH, Melander O, Tanaka T, Stricker BH, Felix SB, Alonso A, Darbar D, Barnard J, Chasman DI, Heckbert SR, Benjamin EJ, Gudnason V, Kaab S. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, Sinner MF, de Bakker PI, Mueller M, Lubitz SA, Fox E, Darbar D, Smith NL, Smith JD, Schnabel RB, Soliman EZ, Rice KM, Van Wagoner DR, Beckmann BM, van Noord C, Wang K, Ehret GB, Rotter JI, Hazen SL, Steinbeck G, Smith AV, Launer LJ, Harris TB, Makino S, Nelis M, Milan DJ, Perz S, Esko T, Kottgen A, Moebus S, Newton-Cheh C, Li M, Mohlenkamp S, Wang TJ, Kao WH, Vasan RS, Nothen MM, MacRae CA, Stricker BH, Hofman A, Uitterlinden AG, Levy D, Boerwinkle E, Metspalu A, Topol EJ, Chakravarti A, Gudnason V, Psaty BM, Roden DM, Meitinger T, Wichmann HE, Witteman JC, Barnard J, Arking DE, Benjamin EJ, Heckbert SR, Kaab S. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, MacRae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 6.Gore-Panter SR, Hsu J, Hanna P, Gillinov AM, Pettersson G, Newton DW, Moravec CS, Van Wagoner DR, Chung MK, Barnard J. Atrial fibrillation associated chromosome 4q25 variants are not associated with PITX2c expression in human adult left atrial appendages. PLoS One. 2014;9:e86245. doi: 10.1371/journal.pone.0086245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lubitz SA, Lunetta KL, Lin H, Arking DE, Trompet S, Li G, Krijthe BP, Chasman DI, Barnard J, Kleber ME. Novel genetic markers associate with atrial fibrillation risk in europeans and japanese. J Am Coll Cardiol. 2014;63:1200–1210. doi: 10.1016/j.jacc.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu J, Hanna P, Van Wagoner DR, Barnard J, Serre D, Chung MK, Smith JD. Whole genome expression differences in human left and right atria ascertained by RNA SequencingClinical perspective. Circ Cardiovasc Genet. 2012;5:327–335. doi: 10.1161/CIRCGENETICS.111.961631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell Guttman IA, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 13.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schonrock N, Harvey RP, Mattick JS. Long noncoding RNAs in cardiac development and pathophysiology. Circ Res. 2012;111:1349–1362. doi: 10.1161/CIRCRESAHA.112.268953. [DOI] [PubMed] [Google Scholar]
- 15.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai M, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Morales DR, Thomas K, Presser A, Bernstein BE, Van Oudenaarden A. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thum T, Kumarswamy R. The smooth long noncoding RNA SENCR. Arterioscler Thromb Vasc Biol. 2014;34:1124–1125. doi: 10.1161/ATVBAHA.114.303504. [DOI] [PubMed] [Google Scholar]
- 20.Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M. The tissue-specific lncRNA fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, Liu CY, Sun T, Zhang XJ, Li PF. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014;114:1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- 22.Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 23.Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F, Thum T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114:1569–1575. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 24.Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, Miano JM. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol. 2014;34:1249–1259. doi: 10.1161/ATVBAHA.114.303240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 26.Reich DE, Goldstein DB. Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol. 2001;20:4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q, Jiang J, Han P, Yuan Q, Zhang J, Zhang X, Xu Y, Cao H, Meng Q, Chen L. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–587. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: An integrated resource for microRNA-target interactions. Nuc Acids Res. 2009;37:D105–10. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eddy SR. Computational genomics of noncoding RNA genes. Cell. 2002;109:137–140. doi: 10.1016/s0092-8674(02)00727-4. [DOI] [PubMed] [Google Scholar]
- 30.Semina EV, Reiter R, Leysens NJ, Alward WLM, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in rieger syndrome. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 31.Franco D, Christoffels VM, Campione M. Homeobox transcription factor Pitx2: The rise of an asymmetry gene in cardiogenesis and arrhythmogenesis. Trends Cardiovasc Med. 2014;24:23–31. doi: 10.1016/j.tcm.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- 33.Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, Izpisúa-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmentry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–281. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XHT, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci USA. 2010;107:9753. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld HH, Rotering H, Fortmueller L, Laakmann S, Verheule S. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene ExpressionClinical perspective. Circ Cardiovasc Genet. 2011;4:123–133. doi: 10.1161/CIRCGENETICS.110.958058. [DOI] [PubMed] [Google Scholar]
- 36.Martin RI, Babaei MS, Choy M, Owens WA, Chico TJ, Keenan D, Yonan N, Koref MS, Keavney BD. Genetic variants associated with risk of atrial fibrillation regulate expression of PITX2, CAV1, MYOZ1, C9orf3 and FANCC. J Mol Cell Cardiol. 2015;85:207–214. doi: 10.1016/j.yjmcc.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Aguirre LA, Alonso ME, Badía-Careaga C, Rollán I, Arias C, Fernández-Miñán A, López-Jiménez E, Aránega A, Gómez-Skarmeta JL, Franco D. Long-range regulatory interactions at the 4q25 atrial fibrillation risk locus involve PITX2c and ENPEP. BMC Biol. 2015;13:26. doi: 10.1186/s12915-015-0138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C, Liu W, Palie J, Lu MF, Brown NA, Martin JF. Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development. 2002;129:5081–5091. doi: 10.1242/dev.129.21.5081. [DOI] [PubMed] [Google Scholar]
- 39.Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, Krajewski K, Strahl BD, Chen X, Bickel P, Giddings MC, Brown JB, Lipovich L. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paralkar VR, Mishra T, Luan J, Yao Y, Kossenkov AV, Anderson SM, Dunagin M, Pimkin M, Gore M, Sun D, Konuthula N, Raj A, An X, Mohandas N, Bodine DM, Hardison RC, Weiss MJ. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123:1927–1937. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deacon DC, Nevis KR, Cashman TJ, Zhou Y, Zhao L, Washko D, Guner-Ataman B, Burns CG, Burns CE. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis. Development. 2010;137:1887–1896. doi: 10.1242/dev.050526. [DOI] [PubMed] [Google Scholar]
- 43.Miyasaka KY, Kida YS, Banjo T, Ueki Y, Nagayama K, Matsumoto T, Sato M, Ogura T. Heartbeat regulates cardiogenesis by suppressing retinoic acid signaling via expression of miR-143. Mech Dev. 2011;128:18–28. doi: 10.1016/j.mod.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Norata GD, Pinna C, Zappella F, Elia L, Sala A, Condorelli G, Catapano AL. MicroRNA 143-145 deficiency impairs vascular function. Int J Immunopathol Pharmacol. 2012;25:467–474. doi: 10.1177/039463201202500216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.