Abstract
In the title compound, C14H12BrN, the tricyclic ring system is essentially planar (r.m.s. deviation 0.026 Å). The carbon atoms of the ethyl group deviate from the mean plane by 0.148 (9) (CH2) and 1.59 (1) Å (CH3). In the crystal, H⋯π contacts [2.698–2.898 Å] shorter than the van der Waals contact distance of 3.70 Å are observed. A scalable to gram quantities selective synthesis of mono-bromine-substituted carbazole derivatives was developed.
Keywords: crystal structure, carbazole, C—H⋯π interactions
Related literature
N-substituted carbazole derivatives are important for anti-cancer research (Caulfield et al., 2002 ▸) and as materials for opto-electronic devices (Niu et al., 2011 ▸; Miyazaki et al., 2014 ▸; Grigalevicius et al., 2002 ▸). The crystal structure of 1,3,6,8-tetrabromo-9-ethyl-9H-carbazole was reported by Bezuglyi et al. (2015 ▸).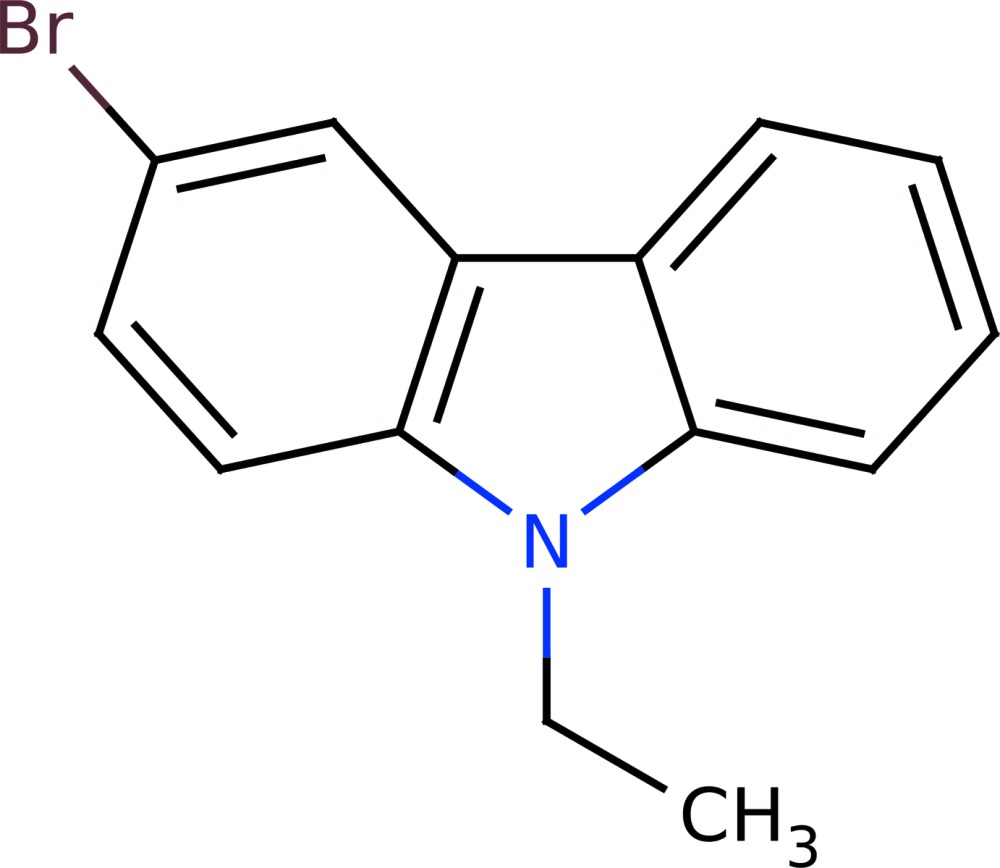
Experimental
Crystal data
C14H12BrN
M r = 274.16
Orthorhombic,

a = 15.263 (16) Å
b = 7.745 (8) Å
c = 20.41 (2) Å
V = 2413 (5) Å3
Z = 8
Mo Kα radiation
μ = 3.39 mm−1
T = 293 K
0.40 × 0.09 × 0.08 mm
Data collection
Rigaku XtaLAB mini diffractometer
Absorption correction: multi-scan (REQAB; Rigaku, 1998 ▸) T min = 0.450, T max = 0.763
8316 measured reflections
2721 independent reflections
1383 reflections with F 2 > 2.0σ(F 2)
R int = 0.056
Refinement
R[F 2 > 2σ(F 2)] = 0.078
wR(F 2) = 0.236
S = 1.05
2721 reflections
145 parameters
H-atom parameters constrained
Δρmax = 1.37 e Å−3
Δρmin = −0.46 e Å−3
Data collection: CrystalClear-SM Expert (Rigaku, 2011 ▸); cell refinement: CrystalClear-SM Expert; data reduction: CrystalClear-SM Expert; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▸); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▸); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▸); software used to prepare material for publication: CrystalStructure (Rigaku, 2010 ▸).
Supplementary Material
Crystal structure: contains datablock(s) General, I. DOI: 10.1107/S2056989015023907/nk2233sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015023907/nk2233Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015023907/nk2233Isup3.cml
. DOI: 10.1107/S2056989015023907/nk2233fig1.tif
The molecular structure of the title molecule with displacement ellipsoids drawn at the 50% probability level.
CCDC reference: 1442215
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 are the centroids of the N1/C1/C6/C7/C12 and C1–C6 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C8—H8⋯Cg1i | 0.93 | 2.81 | 3.637 (7) | 149 |
| C11—H11⋯Cg2ii | 0.93 | 3.01 | 3.922 (8) | 167 |
Symmetry code: (i) 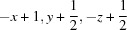 ; (ii)
; (ii)  .
.
Acknowledgments
This research was supported by FP7 REGPOT-2012–2013-1 ICT project CEOSeR under grant agreement No 316010. The authors are grateful to Dr Vasyl Kinzhybalo from the Institute of Low Temperature and Structure Research, Polish Academy of Sciences, for valuable recommendations.
supplementary crystallographic information
S1. Synthesis and crystallization
9-ethyl-carbazole (1.00 g, 5.12 mmol) was added to a solution of N-bromosuccinimide (0.911 g, 5.12 mmol) in 10 ml of DMF. The reaction mixture was refluxed at room temperature for 24 h. When the reaction was completed (monitored via TLC) the solution was poured into a large amount of water with ice and extracted with ethyl acetate. The organic layer was dried over anhydrous sodium sulfate followed by solvent evaporation in rotary evaporator. The product was crystallized from methanol to afford a white needle-like crystals. Yield: 0.88 g (62%), melting point 58–60°C. 1H NMR (700 MHz, CDCl3) δ 8.10 (d, J = 7.7 Hz, 1H), 7.63 (d, J = 2.5 Hz, 1H), 7.49 (ddd, J = 8.2, 7.1, 1.1 Hz, 1H), 7.42 (d, J = 8.2 Hz, 1H), 7.35 (d, J = 8.7 Hz, 1H), 7.25 – 7.22 (m, 1H), 7.16 (dd, J = 8.8, 2.5 Hz, 1H), 4.37 (q, J = 7.3 Hz, 2H), 1.45 (t, J = 7.3 Hz, 4H).
S2. Refinement
All hydrogen atoms were placed in geometrically idealized positions and constrained to ride on their parent atoms, with C—H = 0.930 Å for aromatic C—H, with 0.969 Å for methylene C—H, 0.957 Å for methyl distances and Uiso(H) = 1.2 Ueq.
Figures
Fig. 1.

The molecular structure of the title molecule with displacement ellipsoids drawn at the 50% probability level.
Crystal data
| C14H12BrN | F(000) = 1104.00 |
| Mr = 274.16 | Dx = 1.509 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71075 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 3894 reflections |
| a = 15.263 (16) Å | θ = 3.1–27.5° |
| b = 7.745 (8) Å | µ = 3.39 mm−1 |
| c = 20.41 (2) Å | T = 273 K |
| V = 2413 (5) Å3 | Chip, colorless |
| Z = 8 | 0.40 × 0.09 × 0.08 mm |
Data collection
| Rigaku XtaLAB mini diffractometer | 1383 reflections with F2 > 2.0σ(F2) |
| Detector resolution: 13.653 pixels mm-1 | Rint = 0.056 |
| ω scans | θmax = 27.5° |
| Absorption correction: multi-scan (REQAB; Rigaku, 1998) | h = −18→18 |
| Tmin = 0.450, Tmax = 0.763 | k = −10→7 |
| 8316 measured reflections | l = −26→21 |
| 2721 independent reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.078 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.236 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0926P)2 + 4.1576P] where P = (Fo2 + 2Fc2)/3 |
| 2721 reflections | (Δ/σ)max < 0.001 |
| 145 parameters | Δρmax = 1.37 e Å−3 |
| 0 restraints | Δρmin = −0.46 e Å−3 |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. ENTER SPECIAL DETAILS OF THE MOLECULAR GEOMETRY |
| Refinement. Refinement was performed using all reflections. The weighted R-factor (wR) and goodness of fit (S) are based on F2. R-factor (gt) are based on F. The threshold expression of F2 > 2.0 σ(F2) is used only for calculating R-factor (gt). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.28798 (6) | 0.61687 (12) | 0.45898 (4) | 0.0841 (5) | |
| N1 | 0.6116 (4) | 0.2371 (7) | 0.3684 (3) | 0.0635 (15) | |
| C1 | 0.5364 (5) | 0.3073 (8) | 0.3969 (4) | 0.0570 (17) | |
| C2 | 0.4935 (6) | 0.2633 (10) | 0.4542 (4) | 0.0680 (19) | |
| C3 | 0.4189 (6) | 0.3557 (10) | 0.4719 (4) | 0.070 (2) | |
| C4 | 0.3891 (5) | 0.4915 (9) | 0.4327 (4) | 0.0629 (18) | |
| C5 | 0.4317 (5) | 0.5360 (9) | 0.3759 (4) | 0.0559 (16) | |
| C6 | 0.5065 (5) | 0.4456 (8) | 0.3573 (4) | 0.0527 (16) | |
| C7 | 0.5643 (4) | 0.4553 (8) | 0.3016 (4) | 0.0519 (16) | |
| C8 | 0.5686 (5) | 0.5583 (9) | 0.2455 (4) | 0.0608 (18) | |
| C9 | 0.6333 (5) | 0.5278 (10) | 0.1987 (4) | 0.071 (2) | |
| C10 | 0.6935 (5) | 0.3978 (11) | 0.2092 (4) | 0.072 (3) | |
| C11 | 0.6939 (5) | 0.2950 (10) | 0.2632 (5) | 0.067 (2) | |
| C12 | 0.6283 (5) | 0.3243 (9) | 0.3100 (4) | 0.0577 (17) | |
| C13 | 0.6556 (6) | 0.0790 (9) | 0.3911 (4) | 0.074 (3) | |
| C14 | 0.6145 (6) | −0.0826 (9) | 0.3648 (5) | 0.083 (3) | |
| H3 | 0.4107 | 0.6258 | 0.3500 | 0.0671* | |
| H8 | 0.7363 | 0.3795 | 0.1775 | 0.0866* | |
| H9 | 0.6358 | 0.5943 | 0.1608 | 0.0853* | |
| H10 | 0.5139 | 0.1737 | 0.4803 | 0.0816* | |
| H13 | 0.3888 | 0.3269 | 0.5100 | 0.0840* | |
| H14 | 0.5283 | 0.6470 | 0.2394 | 0.0730* | |
| H15 | 0.7358 | 0.2091 | 0.2688 | 0.0804* | |
| H17A | 0.6458 | −0.1813 | 0.3809 | 0.0995* | |
| H17B | 0.6167 | −0.0812 | 0.3178 | 0.0995* | |
| H17C | 0.5545 | −0.0887 | 0.3789 | 0.0995* | |
| H18A | 0.6540 | 0.0757 | 0.4386 | 0.0894* | |
| H18B | 0.7166 | 0.0827 | 0.3778 | 0.0894* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0778 (7) | 0.0894 (7) | 0.0852 (7) | 0.0084 (5) | 0.0147 (5) | −0.0107 (5) |
| N1 | 0.067 (4) | 0.053 (3) | 0.071 (4) | 0.017 (3) | −0.015 (3) | −0.007 (3) |
| C1 | 0.071 (5) | 0.043 (4) | 0.056 (4) | −0.003 (4) | −0.017 (4) | −0.005 (3) |
| C2 | 0.080 (5) | 0.059 (4) | 0.065 (5) | 0.004 (4) | −0.012 (4) | 0.004 (4) |
| C3 | 0.091 (6) | 0.063 (5) | 0.056 (5) | −0.006 (5) | 0.001 (4) | 0.001 (4) |
| C4 | 0.062 (5) | 0.053 (4) | 0.074 (5) | 0.000 (4) | 0.002 (4) | −0.008 (4) |
| C5 | 0.061 (4) | 0.045 (4) | 0.061 (4) | −0.003 (4) | −0.010 (4) | −0.001 (3) |
| C6 | 0.055 (4) | 0.045 (4) | 0.058 (4) | −0.003 (3) | −0.007 (4) | −0.009 (3) |
| C7 | 0.049 (4) | 0.043 (4) | 0.063 (4) | −0.000 (3) | −0.017 (3) | −0.007 (3) |
| C8 | 0.062 (4) | 0.056 (4) | 0.065 (5) | −0.001 (4) | −0.011 (4) | 0.003 (4) |
| C9 | 0.069 (5) | 0.069 (5) | 0.075 (5) | −0.011 (5) | −0.001 (4) | −0.005 (4) |
| C10 | 0.064 (5) | 0.083 (6) | 0.070 (5) | −0.006 (5) | −0.002 (4) | −0.021 (5) |
| C11 | 0.054 (4) | 0.060 (5) | 0.087 (6) | 0.007 (4) | −0.014 (4) | −0.019 (4) |
| C12 | 0.054 (4) | 0.050 (4) | 0.068 (5) | 0.001 (4) | −0.007 (4) | −0.004 (4) |
| C13 | 0.080 (5) | 0.064 (5) | 0.080 (5) | 0.020 (4) | −0.023 (5) | 0.007 (4) |
| C14 | 0.104 (7) | 0.051 (4) | 0.094 (6) | 0.010 (5) | 0.001 (5) | 0.003 (4) |
Geometric parameters (Å, º)
| Br1—C4 | 1.901 (7) | C10—C11 | 1.358 (12) |
| N1—C1 | 1.396 (9) | C11—C12 | 1.403 (11) |
| N1—C12 | 1.393 (10) | C13—C14 | 1.499 (11) |
| N1—C13 | 1.471 (10) | C2—H10 | 0.930 |
| C1—C2 | 1.382 (10) | C3—H13 | 0.930 |
| C1—C6 | 1.418 (9) | C5—H3 | 0.930 |
| C2—C3 | 1.392 (12) | C8—H14 | 0.930 |
| C3—C4 | 1.397 (11) | C9—H9 | 0.930 |
| C4—C5 | 1.373 (10) | C10—H8 | 0.930 |
| C5—C6 | 1.391 (9) | C11—H15 | 0.930 |
| C6—C7 | 1.442 (10) | C13—H18A | 0.970 |
| C7—C8 | 1.397 (10) | C13—H18B | 0.970 |
| C7—C12 | 1.419 (9) | C14—H17A | 0.960 |
| C8—C9 | 1.394 (11) | C14—H17B | 0.960 |
| C9—C10 | 1.381 (11) | C14—H17C | 0.960 |
| N1···C5 | 3.594 (9) | C10···H3vi | 2.9041 |
| N1···C8 | 3.594 (10) | C10···H15ix | 2.9077 |
| C1···C4 | 2.761 (10) | C11···H3vi | 3.1000 |
| C1···C14 | 3.311 (11) | C11···H14vi | 3.5811 |
| C2···C5 | 2.811 (10) | C11···H15ix | 3.3834 |
| C2···C13 | 3.133 (12) | C11···H17Aix | 3.4343 |
| C3···C6 | 2.781 (11) | C11···H17Bix | 3.2429 |
| C5···C8 | 3.388 (10) | C11···H18Bix | 3.5072 |
| C7···C10 | 2.764 (11) | C12···H14vi | 2.9363 |
| C8···C11 | 2.819 (10) | C12···H18Bix | 3.3937 |
| C9···C12 | 2.767 (11) | C13···H17Aix | 3.5602 |
| C11···C13 | 3.156 (12) | C14···H13vii | 3.1805 |
| C12···C14 | 3.351 (11) | C14···H14x | 3.5604 |
| C2···C3i | 3.574 (11) | C14···H14vi | 3.5260 |
| C2···C4i | 3.486 (11) | C14···H15xi | 3.4162 |
| C3···C2i | 3.574 (11) | H3···C9ii | 3.3372 |
| C3···C3i | 3.527 (12) | H3···C10ii | 2.9041 |
| C4···C2i | 3.486 (11) | H3···C11ii | 3.1000 |
| Br1···H3 | 2.9084 | H3···H8ii | 3.0356 |
| Br1···H13 | 2.9144 | H3···H8iii | 3.3234 |
| N1···H10 | 2.7717 | H3···H15ii | 3.3615 |
| N1···H15 | 2.7889 | H3···H17Cviii | 3.1708 |
| N1···H17A | 3.2924 | H8···Br1vi | 3.4695 |
| N1···H17B | 2.6741 | H8···Br1xii | 3.4304 |
| N1···H17C | 2.6783 | H8···C4xii | 3.3551 |
| C1···H3 | 3.2683 | H8···C5xii | 3.3993 |
| C1···H13 | 3.2280 | H8···C9xi | 3.4012 |
| C1···H17C | 3.1012 | H8···H3vi | 3.0356 |
| C1···H18A | 2.6759 | H8···H3xii | 3.3234 |
| C1···H18B | 3.2769 | H8···H9xi | 2.9669 |
| C2···H17C | 3.2655 | H8···H15ix | 3.1884 |
| C2···H18A | 2.8668 | H9···Br1xii | 3.3766 |
| C3···H3 | 3.2516 | H9···C1ii | 3.3199 |
| C4···H10 | 3.2612 | H9···C2ii | 3.3337 |
| C5···H13 | 3.2457 | H9···C3ii | 3.4827 |
| C5···H14 | 3.2684 | H9···C6ii | 3.5009 |
| C6···H10 | 3.2790 | H9···H8ix | 2.9669 |
| C6···H14 | 2.8883 | H9···H15ix | 3.0795 |
| C7···H3 | 2.8662 | H9···H17Cii | 3.3331 |
| C7···H9 | 3.2568 | H10···C4i | 3.4733 |
| C7···H15 | 3.3072 | H10···H10vii | 2.8396 |
| C8···H3 | 3.2607 | H10···H17Cvii | 3.1274 |
| C8···H8 | 3.2241 | H13···Br1xiii | 3.3182 |
| C9···H15 | 3.2540 | H13···C1i | 3.5975 |
| C10···H14 | 3.2341 | H13···C14vii | 3.1805 |
| C11···H9 | 3.2440 | H13···H17Avii | 2.5518 |
| C11···H17B | 3.3353 | H13···H17Cvii | 3.0494 |
| C11···H18B | 2.8803 | H13···H18Avii | 3.3550 |
| C12···H8 | 3.1954 | H13···H18Bv | 3.5574 |
| C12···H14 | 3.2644 | H14···N1ii | 3.1441 |
| C12···H17B | 3.1500 | H14···C1ii | 3.2025 |
| C12···H18A | 3.2783 | H14···C6ii | 3.0863 |
| C12···H18B | 2.6882 | H14···C7ii | 2.8977 |
| C13···H10 | 2.9205 | H14···C8ii | 3.5256 |
| C13···H15 | 2.9579 | H14···C11ii | 3.5811 |
| C14···H10 | 3.4427 | H14···C12ii | 2.9363 |
| C14···H15 | 3.5181 | H14···C14viii | 3.5604 |
| H3···H14 | 2.8901 | H14···C14ii | 3.5260 |
| H8···H9 | 2.2887 | H14···H17Bviii | 2.9697 |
| H8···H15 | 2.2820 | H14···H17Bii | 3.0636 |
| H9···H14 | 2.3299 | H14···H17Cviii | 3.5302 |
| H10···H13 | 2.3284 | H14···H17Cii | 3.2796 |
| H10···H17C | 2.9668 | H15···C8xi | 3.2416 |
| H10···H18A | 2.4234 | H15···C9xi | 2.8304 |
| H15···H17B | 3.0595 | H15···C10xi | 2.9077 |
| H15···H18B | 2.4483 | H15···C11xi | 3.3834 |
| H17A···H18A | 2.3159 | H15···C14ix | 3.4162 |
| H17A···H18B | 2.3126 | H15···H3vi | 3.3615 |
| H17B···H18A | 2.8071 | H15···H8xi | 3.1884 |
| H17B···H18B | 2.3301 | H15···H9xi | 3.0795 |
| H17C···H18A | 2.3268 | H15···H17Aix | 3.0376 |
| H17C···H18B | 2.8068 | H15···H17Bix | 2.9508 |
| Br1···H8ii | 3.4695 | H17A···C3vii | 3.4391 |
| Br1···H8iii | 3.4304 | H17A···C7x | 3.4783 |
| Br1···H9iii | 3.3766 | H17A···C11xi | 3.4343 |
| Br1···H13iv | 3.3182 | H17A···C13xi | 3.5602 |
| Br1···H18Ai | 3.2895 | H17A···H13vii | 2.5518 |
| Br1···H18Av | 3.2825 | H17A···H15xi | 3.0376 |
| N1···H14vi | 3.1441 | H17A···H18Bxi | 2.7853 |
| C1···H9vi | 3.3199 | H17B···C8x | 3.2433 |
| C1···H13i | 3.5975 | H17B···C8vi | 3.2915 |
| C1···H14vi | 3.2025 | H17B···C11xi | 3.2429 |
| C2···H9vi | 3.3337 | H17B···H14x | 2.9697 |
| C3···H9vi | 3.4827 | H17B···H14vi | 3.0636 |
| C3···H17Avii | 3.4391 | H17B···H15xi | 2.9508 |
| C4···H8iii | 3.3551 | H17C···C5x | 3.4596 |
| C4···H10i | 3.4733 | H17C···C8vi | 3.3569 |
| C5···H8iii | 3.3993 | H17C···C9vi | 3.3968 |
| C5···H17Cviii | 3.4596 | H17C···H3x | 3.1708 |
| C6···H9vi | 3.5009 | H17C···H9vi | 3.3331 |
| C6···H14vi | 3.0863 | H17C···H10vii | 3.1274 |
| C7···H14vi | 2.8977 | H17C···H13vii | 3.0494 |
| C7···H17Aviii | 3.4783 | H17C···H14x | 3.5302 |
| C8···H14vi | 3.5256 | H17C···H14vi | 3.2796 |
| C8···H15ix | 3.2416 | H18A···Br1i | 3.2895 |
| C8···H17Bviii | 3.2433 | H18A···Br1xiv | 3.2825 |
| C8···H17Bii | 3.2915 | H18A···H13vii | 3.3550 |
| C8···H17Cii | 3.3569 | H18B···C11xi | 3.5072 |
| C9···H3vi | 3.3372 | H18B···C12xi | 3.3937 |
| C9···H8ix | 3.4012 | H18B···H13xiv | 3.5574 |
| C9···H15ix | 2.8304 | H18B···H17Aix | 2.7853 |
| C9···H17Cii | 3.3968 | ||
| C1—N1—C12 | 108.6 (6) | C1—C2—H10 | 120.636 |
| C1—N1—C13 | 124.6 (6) | C3—C2—H10 | 120.638 |
| C12—N1—C13 | 126.1 (6) | C2—C3—H13 | 119.841 |
| N1—C1—C2 | 130.3 (7) | C4—C3—H13 | 119.848 |
| N1—C1—C6 | 108.7 (6) | C4—C5—H3 | 120.246 |
| C2—C1—C6 | 121.0 (7) | C6—C5—H3 | 120.252 |
| C1—C2—C3 | 118.7 (7) | C7—C8—H14 | 120.069 |
| C2—C3—C4 | 120.3 (7) | C9—C8—H14 | 120.061 |
| Br1—C4—C3 | 119.2 (6) | C8—C9—H9 | 120.379 |
| Br1—C4—C5 | 119.7 (6) | C10—C9—H9 | 120.368 |
| C3—C4—C5 | 121.2 (7) | C9—C10—H8 | 118.092 |
| C4—C5—C6 | 119.5 (6) | C11—C10—H8 | 118.084 |
| C1—C6—C5 | 119.3 (6) | C10—C11—H15 | 121.478 |
| C1—C6—C7 | 107.0 (6) | C12—C11—H15 | 121.484 |
| C5—C6—C7 | 133.7 (6) | N1—C13—H18A | 108.980 |
| C6—C7—C8 | 134.8 (6) | N1—C13—H18B | 108.983 |
| C6—C7—C12 | 106.8 (6) | C14—C13—H18A | 108.979 |
| C8—C7—C12 | 118.4 (6) | C14—C13—H18B | 108.984 |
| C7—C8—C9 | 119.9 (7) | H18A—C13—H18B | 107.770 |
| C8—C9—C10 | 119.3 (8) | C13—C14—H17A | 109.478 |
| C9—C10—C11 | 123.8 (8) | C13—C14—H17B | 109.463 |
| C10—C11—C12 | 117.0 (7) | C13—C14—H17C | 109.470 |
| N1—C12—C7 | 108.9 (6) | H17A—C14—H17B | 109.470 |
| N1—C12—C11 | 129.5 (7) | H17A—C14—H17C | 109.473 |
| C7—C12—C11 | 121.6 (7) | H17B—C14—H17C | 109.472 |
| N1—C13—C14 | 113.0 (7) | ||
| C1—N1—C12—C7 | 1.4 (7) | Br1—C4—C5—C6 | 179.0 (4) |
| C1—N1—C12—C11 | −176.9 (6) | C3—C4—C5—C6 | −0.8 (10) |
| C12—N1—C1—C2 | 179.1 (6) | C4—C5—C6—C1 | 1.0 (9) |
| C12—N1—C1—C6 | −2.1 (7) | C4—C5—C6—C7 | 178.2 (6) |
| C1—N1—C13—C14 | 83.0 (8) | C1—C6—C7—C8 | 178.2 (6) |
| C13—N1—C1—C2 | 8.6 (11) | C1—C6—C7—C12 | −1.1 (7) |
| C13—N1—C1—C6 | −172.6 (6) | C5—C6—C7—C8 | 0.7 (13) |
| C12—N1—C13—C14 | −85.8 (8) | C5—C6—C7—C12 | −178.6 (7) |
| C13—N1—C12—C7 | 171.7 (6) | C6—C7—C8—C9 | −177.5 (6) |
| C13—N1—C12—C11 | −6.5 (11) | C6—C7—C12—N1 | −0.1 (7) |
| N1—C1—C2—C3 | 179.7 (6) | C6—C7—C12—C11 | 178.3 (5) |
| N1—C1—C6—C5 | 179.9 (5) | C8—C7—C12—N1 | −179.6 (6) |
| N1—C1—C6—C7 | 2.0 (7) | C8—C7—C12—C11 | −1.2 (9) |
| C2—C1—C6—C5 | −1.2 (10) | C12—C7—C8—C9 | 1.7 (9) |
| C2—C1—C6—C7 | −179.1 (6) | C7—C8—C9—C10 | −1.3 (10) |
| C6—C1—C2—C3 | 1.1 (10) | C8—C9—C10—C11 | 0.1 (12) |
| C1—C2—C3—C4 | −0.8 (11) | C9—C10—C11—C12 | 0.4 (12) |
| C2—C3—C4—Br1 | −179.2 (6) | C10—C11—C12—N1 | 178.1 (7) |
| C2—C3—C4—C5 | 0.6 (11) | C10—C11—C12—C7 | 0.1 (10) |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+1, y+1/2, −z+1/2; (iii) x−1/2, y, −z+1/2; (iv) −x+1/2, y+1/2, z; (v) x−1/2, −y+1/2, −z+1; (vi) −x+1, y−1/2, −z+1/2; (vii) −x+1, −y, −z+1; (viii) x, y+1, z; (ix) −x+3/2, y+1/2, z; (x) x, y−1, z; (xi) −x+3/2, y−1/2, z; (xii) x+1/2, y, −z+1/2; (xiii) −x+1/2, y−1/2, z; (xiv) x+1/2, −y+1/2, −z+1.
Hydrogen-bond geometry (Å, º)
Cg1 are the centroids of the N1/C1/C6/C7/C12 and C1–C6 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C8—H8···Cg1ii | 0.93 | 2.81 | 3.637 (7) | 149 |
| C11—H11···Cg2ix | 0.93 | 3.01 | 3.922 (8) | 167 |
Symmetry codes: (ii) −x+1, y+1/2, −z+1/2; (ix) −x+3/2, y+1/2, z.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: NK2233).
References
- Bezuglyi, M., Grybauskaite, G., Bagdziunas, G. & Grazulevicius, J. V. (2015). Acta Cryst. E71, o373. [DOI] [PMC free article] [PubMed]
- Caulfield, T., Cherrier, M. P., Combeau, C. & Mailliet, P. (2002). Eur. Patent No. 1 253 141.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Grigalevicius, S., Ostrauskaite, J., Grazulevicius, J. V., Gaidelis, V., Jankauskas, V. & Sidaravicius, J. (2002). Mater. Chem. Phys. 77, 281–284.
- Miyazaki, T., Shibahara, M., Fujishige, J., Watanabe, M., Goto, K. & Shinmyozu, T. (2014). J. Org. Chem. 79, 11440–11453. [DOI] [PubMed]
- Niu, F., Niu, H., Liu, Y., Lian, J. & Zeng, P. (2011). RSC Adv. 1, 415–423.
- Rigaku (1998). REQAB. Rigaku Corporation, Tokyo, Japan.
- Rigaku (2010). CrystalStructure. Rigaku Corporation, Tokyo, Japan.
- Rigaku (2011). CrystalClear-SM Expert. Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) General, I. DOI: 10.1107/S2056989015023907/nk2233sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015023907/nk2233Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015023907/nk2233Isup3.cml
. DOI: 10.1107/S2056989015023907/nk2233fig1.tif
The molecular structure of the title molecule with displacement ellipsoids drawn at the 50% probability level.
CCDC reference: 1442215
Additional supporting information: crystallographic information; 3D view; checkCIF report


