Summary
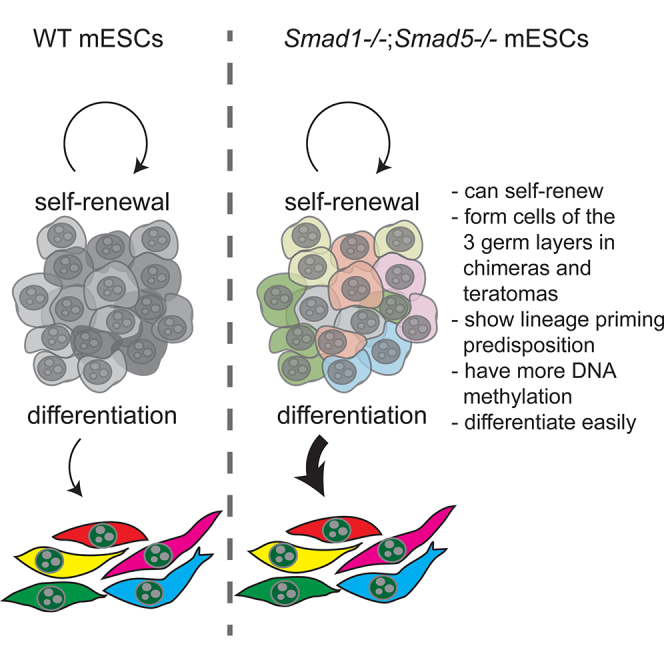
Naive mouse embryonic stem cells (mESCs) are in a metastable state and fluctuate between inner cell mass- and epiblast-like phenotypes. Here, we show transient activation of the BMP-SMAD signaling pathway in mESCs containing a BMP-SMAD responsive reporter transgene. Activation of the BMP-SMAD reporter transgene in naive mESCs correlated with lower levels of genomic DNA methylation, high expression of 5-methylcytosine hydroxylases Tet1/2 and low levels of DNA methyltransferases Dnmt3a/b. Moreover, naive mESCs, in which the BMP-SMAD reporter transgene was activated, showed higher resistance to differentiation. Using double Smad1;Smad5 knockout mESCs, we showed that BMP-SMAD signaling is dispensable for self-renewal in both naive and ground state. These mutant mESCs were still pluripotent, but they exhibited higher levels of DNA methylation than their wild-type counterparts and had a higher propensity to differentiate. We showed that BMP-SMAD signaling modulates lineage priming in mESCs, by transiently regulating the enzymatic machinery responsible for DNA methylation.
Graphical Abstract
Highlights
-
•
BMP-SMAD signaling in mESCs is more prominent in naive than ground state
-
•
BMP-SMAD signaling is dispensable for pluripotency in mESCs
-
•
BMP-SMAD signaling facilitates lineage priming in mESCs
-
•
BMP-SMAD signaling regulates Dnmt3b and hence levels of DNA methylation
In this article, Chuva de Sousa Lopes and colleagues show that the BMP-SMAD signaling is dispensable for the derivation, maintenance, and self-renewal of mESCs both in “serum” and/or “2i” pluripotency states. The BMP-SMAD signaling plays a role regulating the levels of DNA methylation (via Dnmt3a/b and Tet1/2) and hence lineage priming in pluripotent mESCs.
Introduction
Culture conditions affect features of mouse embryonic stem cells (mESCs), such as their proliferation, gene expression, epigenetic status, self-renewal, and capacity for multi-lineage differentiation (Marks et al., 2012, Tesar et al., 2007). In culture medium with fetal calf serum, naive mESCs grown on mouse embryonic fibroblast feeder cells (here abbreviated as “serum”) transit between inner cell mass (ICM)-like and epiblast-like pluripotency states (Sasai et al., 2013, Trott and Martinez Arias, 2013). However, when cultured in serum-free conditions with inhibitors of mitogen-activated protein kinase and glycogen synthase kinase 3 signaling, also called “2i” medium, mESCs become more homogeneous and adopt the more ICM-like or “ground” state (Marks et al., 2012, Nichols et al., 2009, Ying et al., 2003). The observation that naive mESCs interconvert between pluripotent states while remaining uncommitted has raised the suggestion that such heterogeneity may allow the cells to respond differently to environmental cues. In agreement, subpopulations of naive mESCs show different potentials to differentiate (Graf and Stadtfeld, 2008, Hanna et al., 2009, Hayashi et al., 2008). How the metastable transcriptional and epigenetic diversity of cultured mESCs is regulated and maintained has remained elusive.
The two notable characteristics of mESCs are their capacity to self-renew and differentiate into all embryonic lineages (Niwa et al., 1998). In mESCs, pluripotency is maintained by a core network of regulatory transcription factors, including Pou5f1, Sox2, and Nanog (Kashyap et al., 2009, Kim et al., 2008, Marson et al., 2008, Navarro et al., 2012); the balance between self-renewal and differentiation is regulated by protein-encoding genes that include Id1 and Dusp9, both downstream targets of the bone morphogenetic protein (BMP) signaling pathway (Li and Chen, 2013). Moreover, it has been shown that both the BMP and TGFβ (via NODAL) SMAD-mediated signaling pathways are involved in maintaining heterogeneity of NANOG in naive mESCs (Galvin-Burgess et al., 2013). Conversely, NANOG may attenuate BMP signaling via a feedback loop that involves titration of phosphorylated (P)SMAD1 by direct NANOG-SMAD1 interaction (Suzuki et al., 2006). However, the functional role of BMP-SMAD signaling in the metastable state of naive pluripotency has not been investigated.
Here, we report the derivation and characterization of transgenic mESCs that allow a real-time readout of SMAD-mediated BMP signaling activity. This transgenic BRE:gfp reporter mESC line expresses a well-characterized BMP responsive element (BRE) containing several PSMAD1/5 DNA-binding sites isolated from the Id1 promoter to drive GFP expression (Korchynskyi and ten Dijke, 2002, Monteiro et al., 2008). Activation of the BMP-SMAD reporter transgene was heterogeneous in serum mESCs (±50% GFP + cells) and 2i mESCs (±4% GFP + cells). By genetic abrogation of the core BMP pathway components SMAD1 and SMAD5, we demonstrated that BMP-SMAD signaling is dispensable for the maintenance and self-renewal of mESCs both in serum and 2i states, but that it regulates the levels of DNA methylation (via Dnmt3a/b and Tet1/2) and hence lineage priming in pluripotent mESCs.
Results
BMP-SMAD Signaling Is Activated during the Acquisition of Pluripotency
BMP signaling plays key roles in patterning of post-implantation mouse embryos (Kishigami and Mishina, 2005, Tam and Loebel, 2007). However, a role during pre-implantation development has been less evident because genetic ablation of single members of the BMP-SMAD pathway showed no evidence of a phenotype during the pre-implantation period (Goumans and Mummery, 2000, Graham et al., 2014, Reyes de Mochel et al., 2015, Zhao, 2003). We investigated whether the BMP-SMAD signaling pathway was active in pre-implantation embryos by examining BRE:gfp blastocysts at E3.5. We were unable to detect GFP at this stage (data not shown). As the BMP-SMAD pathway has been shown to play dual roles in self-renewal and differentiation of mESCs (Li and Chen, 2013), we monitored GFP during the derivation of mESCs from BRE:gfp blastocysts into the naive state (serum) and the ground state (2i). One day after plating (D1), GFP was still undetectable in blastocysts in either culture condition (Figure 1A); however, by D4, GFP+ cells were evident within the ICM-like cells of BRE:gfp blastocyst outgrowths in both serum and 2i (Figure 1A). This suggested that the BMP-SMAD pathway was activated during the acquisition of pluripotency in vitro.
Figure 1.
BMP-SMAD Signaling Activation in Serum and 2i Culture Conditions
(A) Derivation of BRE:gfp mESCs in serum and 2i conditions. D1, 1 day after blastocyst collection; D4, D1 plus 3 days after blastocyst plating, P3 mESCs, passage 3 of the derived mESCs. Scale bars represent 100 μm.
(B) Established serum and 2i BRE:gfp mESCs and their respective GFP expression profiles by FACS analysis. Scale bars represent 100 μm.
(C) Immunofluorescence of serum and 2i BRE:gfp mESCs for ID1, POU5F1, and NANOG. Scale bars represent 20 μm.
(D) Percentage (%) of NANOG-positive cells in the GFP+ and GFP− cells per colony BRE:gfp mESCs.
(E) Western blotting for PSMAD1/5/8, SMAD1/5/8, GFP and Tubulin in serum and 2i BRE:gfp and E14 mESCs as well as 2i E14 stimulated 1 hr with 25 ng/ml BMP4.
(F) Percentage (%) of GFP+ and GFP− cells in 2i BRE:gfp mESCs after 1 hr treatment with Activin A or BMP4. Bars represent the mean ± SD (N = 3).
(G) Percentage (%) of GFP+ and GFP− cells in 2i BRE:gfp mESCs switched to serum and serum BRE:gfp mESCs switched to 2i and cultured for four consecutive passages (P1–P4). See also Figure S1.
BMP-SMAD Signaling Activation in Serum and 2i mESCs
Once BRE:gfp mESCs lines had been established (Figures 1A and 1B) and karyotyped (Figure S1A), a striking difference was observed between the two conditions: serum BRE:gfp mESCs exhibited an heterogeneous pattern of GFP expression with about 50% of the cells being GFP+, whereas in 2i BRE:gfp mESCs less than 4% of cells were GFP+ (Figure 1B). In serum BRE:gfp mESCs, the GFP+ cells produced ID1 (Figure 1C), confirming that GFP expression corresponded to the activation of BMP-SMADs. The promoter of Id1 contains the PSMAD1/5 DNA-binding sites that were used to generate the BRE:gfp transgene (Figure S1B). Most 2i BRE:gfp mESCs showed no GFP and consequently no/low ID1 (Figure 1C). POU5F1 and NANOG were detected in both serum and 2i BRE:gfp mESCs. Quantification of NANOG suggested that it was more homogeneously expressed in GFP− cells per colony (Figure 1D) and this difference was statistically significant (n = 16; p < 0.05).
To measure BMP-SMAD signaling activation, we investigated the levels of PSMAD1/5/8, which were low in 2i medium in serum mESCs and high in 2i after 1 hr of stimulation with 25 ng/ml BMP4; in agreement, faint GFP was observed in 2i compared with serum BRE:gfp mESCs (Figure 1E). In addition, we examined the number of GFP+ cells present in 2i and showed that this increased in response to BMP4 but not to Activin A (which activates the NODAL pathway) (Figure 1F), and that BRE:gfp mESCs could be interconverted to adopt the GFP pattern associated with each culture medium within four cell passages (Figure 1G).
In Serum, GFP+ BRE:gfp mESCs Correlated with Low Levels of Dnmt3b and Lower DNA Methylation
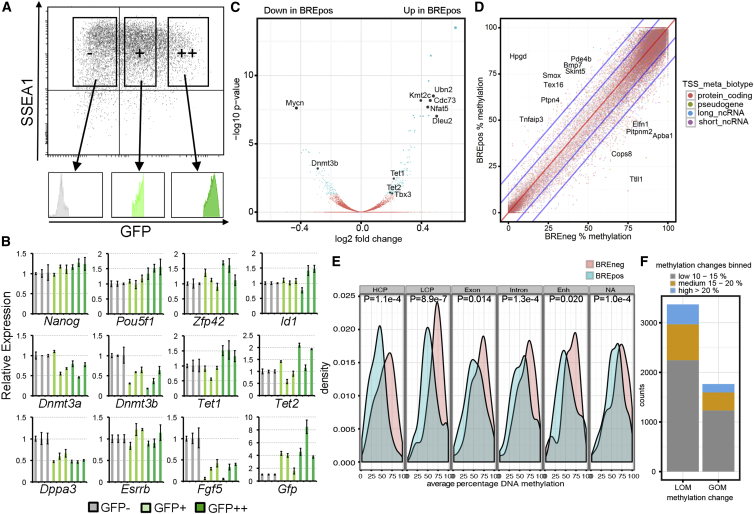
To further understand the role of BMP-SMAD signaling activation in pluripotency, fluorescence-activated cell sorting (FACS) sorted subpopulations of serum (GFP++, GFP+, GFP−) and 2i (GFP+, GFP−) BRE:gfp mESCs (Figures 2A and S2A) were analyzed by qPCR (Figures 2B and S2B). In serum, the sorted GFP++ mESCs (N = 3) exhibited lower levels of Dnmt3a/b, in particular Dnmt3b, and higher levels of Tet1/2, but similar high transcriptional levels of pluripotency genes (Figure 2B). A direct comparison between 2i and serum is provided in Figure S2B. Comparing whole transcriptome RNA sequencing (RNAseq) data of three independent serum GFP++ and GFP− mESC samples, we confirmed that Dnmt3b as well as Tet1/2 were among the few statistically significant differentially regulated genes observed (n = 315; p < 0.05), mostly protein-coding genes (Figures 2C, S2C, and S2D; Table S1). Next, using available single-cell RNAseq data (Sasagawa et al., 2013), we performed a hierarchical clustering of 38 individual cells from naive mESCs based on the expression of 30 selected genes. Interestingly, the cluster with the lowest transcriptional levels of Dnmt3b and high levels of Tet1 (Group 1) did not correlate with the cell clusters showing high transcriptional levels of Id1/Bmp4 (group 2/3) (Figure S2E). This is in agreement with our qPCR (Figure 2B) and RNAseq results (Id1 is not differentially expressed) (Figure 2C; Table S1) and suggests a clear discrepancy between the cells expressing ID1 protein (and GFP protein) and Id1 transcript. This discrepancy in the co-expression of proteins and transcripts is a well-known confounding but intrinsic property of cells, including mESCs (Torres-Padilla and Chambers, 2014).
Figure 2.
Transcriptome and Methylome in Subsets of Serum BRE:gfp mESCs
(A) Gatings used to FACS sort three subpopulations (GFP−, GFP+, GFP++) of serum BRE:gfp mESCs and the profile of the individual cell groups.
(B) Relative expression of several genes in the three subpopulations (GFP−, GFP+, GFP++) of serum BRE:gfp mESCs compared with the GFP− cells. Each bar represents the mean ± SD of technical triplicates and the three bars of the same color represent independent experiments (n = 9, N = 3).
(C) Volcano plot showing –log10 p values versus log2 fold transcriptional changes between GFP++ and GFP− fractions of serum BRE:gfp mESCs. Differentially expressed genes (DEGs) with p < 0.05 are blue, and genes with p > 0.05 are red; some highlighted DEGs are black.
(D) Scatterplot depicting a comparison of the percentage of DNA methylation in each 600-bp tile (dot) between GFP++ and GFP− fractions of serum BRE:gfp mESCs. Each tile was classified into a biotype category according to the nearest TSS. The red line represents no difference; the inner and outer blue lines represent borders for 10% and 20% change in methylation levels, respectively.
(E) Distribution of DNA methylation at specific genomic regions in GFP++ (in blue) and GFP− fractions of serum BRE:gfp (in red) mESCs. p Values were calculated with the two-sample Kolmogorov-Smirnov test. HCP, high CpG-content promoters; LCP, low CpG-content promoters; Enh, enhancers; NA, no annotation.
(F) Number of (600-bp tile) counts showing loss of methylation (LOM) or gain of methylation (GOM) in GFP++ compared with GFP− serum BRE:gfp mESC. See also Figure S2 and Tables S1 and S2.
We performed reduced-representation bisulfite sequencing (RRBS) of GFP++ and GFP− BRE:gfp mESCs and observed that DNA methylation levels were in general lower in mESCs with activation of the BMP-SMAD reporter transgene than in mESCs without reporter activity, as illustrated by the significant shifts toward lower DNA methylation at all genomic regions in GFP++ cells (Figures 2D–2F; Table S2). This is in agreement with the reduced levels of Dnmt3b expression in GFP++ cells.
BMP-SMAD Signaling Is Dispensable for Self-Renewal of mESCs
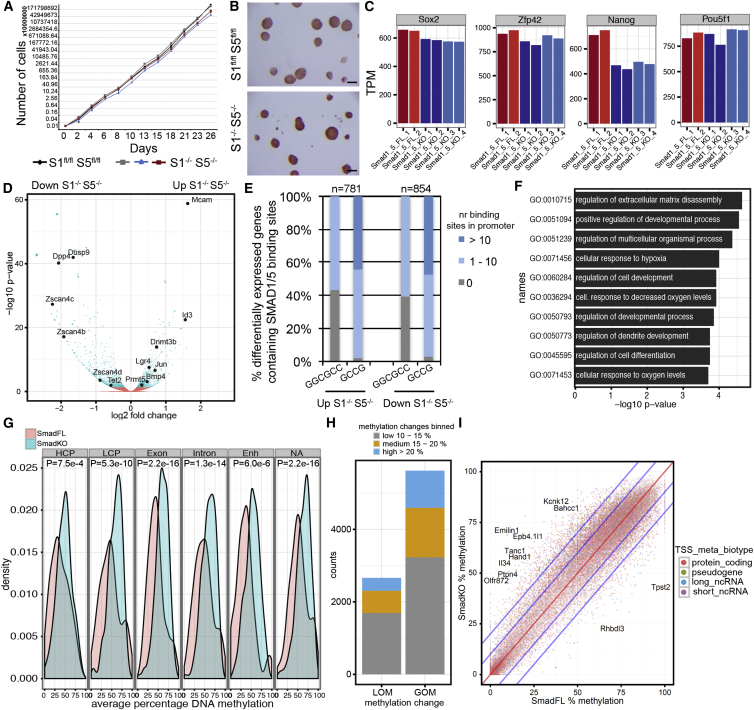
To clarify the role of BMP-SMAD signaling in the maintenance of the naive and ground state, we derived Smad1 and Smad5 double-knockout (S1−/−S5−/−) mESC lines in 2i from double homozygous floxed Smad1;Smad5 mESC lines (S1fl/flS5fl/fl) (Tremblay et al., 2001, Umans et al., 2003) that were hemizygous for the R26R Cre-reporter transgene (Soriano, 1999) using Cre recombinase (Figures S3A and S3B). We derived the S1−/−S5−/− mESC in 2i because BMP-SMAD signaling activation was less prominent in 2i and therefore the chance of deriving pluripotent S1−/−S5−/− mESCs was higher. The pluripotency of the S1−/−S5−/− mESCs was confirmed by showing its contribution to the three germ layers in S1−/−S5−/− <> wild-type chimeric embryos (Figure S3C), as well as in teratoma formation assays (Figure S3D) in independent lines with a normal karyotype (Figure S4A). Moreover, we showed that Smad8 was not upregulated in response to the deletion of Smad1 and Smad5, and that Id1 and Id2 were upregulated after stimulation with BMP4 only in the S1fl/flS5fl/fl parental line, as expected (Figure S4B). The 2i S1−/−S5−/− mESCs self-renewed at the same rate as the parental S1fl/flS5fl/fl mESCs (Figure 3A) and showed comparable alkaline phosphatase activity (Figure 3B). Unexpectedly, when S1−/−S5−/− mESCs were switched from 2i to serum, after an initial period of adaptation the cells continued to self-renew at similar rates as the parental S1fl/flS5fl/fl mESCs (Figure S4C) instead of differentiating. In general, the expression level of pluripotency genes remained high in the parental S1fl/flS5fl/fl and S1−/−S5−/− mESCs in 2i (Figure 3C) and serum (Figure S4D). Our results demonstrated that BMP-SMAD signaling is dispensable for self-renewal of mESCs.
Figure 3.
Transcriptome and Methylome in S1−/−S5−/− versus S1fl/flS5fl/fl mESCs
(A) Growth of S1fl/flS5fl/fl mESCs and three independent S1−/−S5−/− mESCs lines in 2i during 26 days. Means ± SD are depicted.
(B) Alkaline phosphatase activity in 2i S1fl/flS5fl/fl and S1−/−S5−/− mESC. Scale bars represent 100 μm.
(C) Expression of Sox2, Zfp42, Nanog, and Pou5f1 in transcripts per million (TPM) in 2i S1fl/flS5fl/fl (FL) and S1−/−S5−/− (KO) mESC.
(D) Volcano plot showing –log10 p values versus log2 fold transcriptional changes between S1fl/flS5fl/fl and S1−/−S5−/− mESCs in 2i. DEGs with p < 0.05 are blue, and genes with p > 0.05 are red; some highlighted DEGs are black.
(E) Percentage of DEGs (p < 0.01) (n = 781 upregulated in 2i S1−/−S5−/−; n = 854 downregulated in 2i S1−/−S5−/−) showing putative SMAD1/5 binding sites (GGCGCC/GCCG) in the promoter region.
(F) Top ten GO terms associated with biological processes (p < 0.05) in DEGs in 2i S1−/−S5−/− mESCs.
(G) Distribution of DNA methylation levels at specific genomic regions in 2i S1fl/flS5fl/fl (in red) and S1−/−S5−/− mESCs (in blue). p Values were calculated with two-sample Kolmogorov-Smirnov test. HCP, high CpG-content promoters; LCP, low CpG-content promoters; Enh, enhancers; NA, no annotation.
(H) Number of (600-bp tile) counts showing LOM or GOM in 2i S1fl/flS5fl/fl compared with S1−/−S5−/− mESCs.
(I) Scatterplot depicting a comparison of the percentage of DNA methylation in each 600-bp tile (dot) between 2i S1fl/flS5fl/fl and S1−/−S5−/− mESCs. Each tile was classified into a biotype category according to the nearest TSS. The red line represents no difference, and the inner and outer blue lines represent borders for 10% and 20% change in methylation levels, respectively. See also Figures S3, S4 and Tables S1, S2, and S3.
S1−/−S5−/− mESCs Have High Levels of Dnmt3b and High Levels of DNA Methylation
Next, we investigated the SMAD1/5-responsive genes using RNAseq (Figure 3D) and found that most differentially expressed genes (DEGs) between S1−/−S5−/− and S1fl/flS5fl/fl mESCs were protein-coding genes (Figure S4E). Interestingly, about half of the DEGs (including protein-coding, pseudogenes, and long non-coding RNAs) were upregulated (n = 781; p < 0.01) and half of the genes were downregulated (n = 854; p < 0.01) in S1−/−S5−/− mESCs (Figure 3E; Table S1).
To investigate whether the observed expression changes were consistent with direct transcriptional regulation, we integrated our RNAseq dataset with a list of direct SMAD1/5 targets (n = 562) identified by ChIP (Fei et al., 2010). Using gene set enrichment analysis, we found a significant enrichment of SMAD1/5 targets in genes that were downregulated in S1−/−S5−/− mESCs (p < 1 × 10−4) (Figure S4F).
Moreover, the great majority of the DEGs contained the sequence motifs GCCG and/or GGCGCC, well-characterized SMAD1/5 binding sites (Korchynskyi and ten Dijke, 2002), in their promoters, defined as ±2 kb from the transcriptional start site (TSS) (Figure 3E; Table S3). By contrast, genome-wide occurrence of GGCGCC and GCCG motifs at such promoters (including protein-coding, pseudogenes, and long non-coding RNAs) was not, or much less, enriched (Figure S4G), and significantly different from the enrichment observed at DEGs (p < 2.2 × 10−16). As an example, Dnmt3b was significantly upregulated in S1−/−S5−/− mESCs and contained 21x GCCG and 5x GGCGCC in the promoter region, suggesting direct (co-)regulation by DNA-binding BMP-SMADs. The DEGs were significantly enriched for gene ontology (GO) categories such as “regulation of developmental process,” “regulation of cell development,” and “regulation of cell differentiation” (Figure 3F), compatible with BMP-SMAD signaling not being involved in self-renewal of mESC, but rather predisposing mESCs to differentiate. The downregulation of Dnmt3b and enrichment in developmental genes in S1−/−S5−/− mESCs, led us to investigate the levels of DNA methylation by RRBS on several independent S1fl/flS5fl/fl and S1−/−S5−/− mESC lines (Table S2). S1−/−S5−/− mESCs displayed a significant shift toward higher levels of DNA methylation at all genomic regions analyzed when compared with S1fl/flS5fl/fl mESCs (Figures 3G–3I), suggesting that the enrichment in developmental genes is caused by the higher levels of DNA methylation.
mESCs Differentiated More Efficiently to Mesendoderm or Neurectoderm in the Absence of BMP-SMAD Signaling
Finally, we examined the differentiation capacity of S1−/−S5−/− mESCs in both serum and 2i and found that they formed endoderm (Sox17), mesoderm (T), and ectoderm (Pax6 and Sox1) more efficiently than the parental line (Figures 4A–4C) in the monolayer using differentiation protocols for either the mesendoderm (ME) or neuroectoderm (NE) lineages (Thomson et al., 2011). In addition, we investigated the capacity of the FACS-sorted subpopulations of serum BRE:gfp mESCs to differentiate to ME and NE and showed that GFP++ mESCs had lower levels of ME and NE early differentiation markers than GFP− mESCs (Figure 4D), demonstrating that GFP++ mESCs were less prone to differentiate. In agreement, GFP++ mESCs retained higher levels of pluripotency markers, at least after 4 days of differentiation to ME (Figure 4E). Our data showed that transient BMP-SMAD signaling activation tilted mESCs to a less differentiation-prone state, whereas in the absence of BMP-SMAD signaling the balance was shifted toward an increased predisposition to differentiate.
Figure 4.
BMP-SMAD Signaling during mESC Differentiation to Mesendoderm and Neurectoderm
(A) Schematic representation of the protocol to differentiate mESCs to mesendoderm (3 μM CHIR) or neurectoderm (500 nM retinoic acid [RA]).
(B) Relative expression of early lineage markers in differentiated serum and 2i S1−/−S5−/− and S1fl/flS5fl/fl mESCs.
(C) Immunofluorescence of differentiated serum S1fl/flS5fl/fl and S1−/−S5−/− mESCs for NANOG, SOX17, T, and SOX1. Scale bars represent 100 μm.
(D) Relative expression of early lineage markers in differentiated subpopulations (GFP−, GFP+, GFP++) of serum BRE:gfp mESCs compared with GFP− cells.
(E) Relative expression of pluripotency genes in differentiated subpopulations (GFP−, GFP+, GFP++) of serum BRE:gfp mESCs compared with GFP− cells.
Each bar represents the mean ± SD of technical triplicates and bars of the same color represent independent experiments (n = 9, N = 3) in (B) and independent experiments (n = 6, N = =2) in (D) and (E). Statistical analysis was performed on technical triplicates of independent experiments (n = 9, N = 3), ∗p ≤ 0.05, ∗∗p ≤ 0.01.
Discussion
A recent study reported the absence of Bmp4 and Id1 in (embryonic day) E3.5 ICMs and a high transient upregulation in E4.5 epiblasts, followed by downregulation of Bmp4 and Id3 expression during the next 6 days of the derivation of mESCs and their further maintenance in 2i (Boroviak et al., 2014). We now show this in real-time using BRE:gfp blastocysts to derive mESCs. Moreover, we demonstrated that BMP-SMAD signaling is not functionally implicated in self-renewal, in agreement with studies that have mapped genome-wide the genes that are directly regulated by SMAD1/5 (Chen et al., 2008, Fei et al., 2010). They showed that the genes regulated by SMAD1/5 were involved in fate determination, rather than self-renewal. Here, we provide functional evidence that SMAD1/5 are not necessary for mESC self-renewal in either naive (serum) or ground (2i) state.
Specific levels of DNA methylation and associated enzymes have been associated with the different pluripotency states (ground, naive, primed) (Habibi et al., 2013, Hackett et al., 2013, Smallwood et al., 2014), as well as with different levels of GFP in Nanog:gfp naive mESCs (Ficz et al., 2013). This reflects faithfully the rapid loss of genomic DNA methylation that the embryo undergoes in vivo during pre-implantation development, and the gain of DNA methylation during the transition between ICM and epiblast (Smith et al., 2012). Therefore, it is perhaps not surprising that the machinery to regulate rapid switches in genomic DNA methylation is present in pluripotent stem cells derived from ICM and epiblast. A role for BMP-SMAD signaling in LIF-dependent conversion between EpiSCs and ESCs has been reported (Onishi et al., 2014), but the association with changes in DNA methylation between EpiSCs and ESCs remains to be investigated.
Finally, it has been suggested that the epigenetic variation observed in pluripotent cells is stochastic and results in a diversity of predispositions to acquire specific cell fates when the cells are triggered to differentiate (Lee et al., 2014). Our data provide evidence that the cellular diversity of both serum and 2i mESCs regarding DNA methylation and associated enzymes is not a stochastic process as previously thought, but is in fact regulated by cell-cell signaling interactions involving the BMP-SMAD signaling pathway.
Experimental Procedures
mESCs Derivation and Culture
Derivation of BRE:gfp mESCs in 2i and serum and the conditional knockout mESCs for Smad1 and Smad5 (S1fl/flS5fl/fl) in 2i, as well as the Cre-recombination of S1fl/flS5fl/fl mESCs, are described in the Supplemental Experimental Procedures. Genotyping of the BRE:gfp mESCs was performed as described (Monteiro et al., 2008). E14 mESCs were cultured in either 2i or serum. Stimulation (1 hr) with BMP4 (R&D Systems) or Activin A (R&D Systems) was followed by FACS analysis or western blotting (see Supplemental Experimental Procedures). Details about generation of chimeric embryos, the teratoma formation assay, RNAseq, and RRBS are provided in the Supplemental Experimental Procedures.
mESCs Differentiation and Proliferation
mESCs were differentiated to ME or NE as described (Thomson et al., 2011). Briefly, mESCs (10,000 cells/cm2) were grown in N2B27 medium without supplements for 48 hr, after which either 3 μM CHIR99021 or 500 nM all-trans retinoic acid (RA) (Sigma-Aldrich) was added to the N2B27 medium for an additional 48 hr. Cells were then collected for immunofluorescence or qPCR (see Supplemental Experimental Procedures). For the proliferation assay, the total number of serum and 2i mESCs was monitored during each passage for 26 days of culture. Serum mESCs were pre-plated prior to counting.
Statistics
Quantification of NANOG-Positive Cells
Whole BRE:gfp mESC colonies (total n = 16) from three independent experiments (N = 3, 5–6 colonies per experiment) were manually counted three times and averaged. N refers to the number of independent experiments; n refers to total number or colonies counted. Statistical analysis was performed using the Student t-test (two-tailed, unequal variance), ∗p ≤ 0.05.
qPCR
In qPCR, each bar represents the average of technical triplicates. N refers to the number of independent experiments; n refers to total replicates. Statistical analysis was performed using the Student t-test (two-tailed, unequal variance), ∗p ≤ 0.05; ∗∗p ≤ 0.01.
RNAseq Expression Data
To determine significantly DEGs between GFP++ and GFP− or S1−/−S5−/− and S1fl/flS5fl/fl mESCs, we applied a cut-off of 0.01 and/or 0.05 on the p values adjusted for multiple testing hypothesis. N refers to the number of independent experiments; n refers to the number of genes.
RNAseq GO
Enrichment analysis for GO terms was done with the R package topGO based on DEGs (p < 0.05) and utilizing Fisher’s exact test.
RNAseq Motif Sequence Analysis
One-sided Fisher’s exact was used to determine significant over-representation of the analyzed motifs in promoter regions of DEGs relative to the genome-wide promoter regions. n refers to the number of genes.
SMAD1/5 ChIP-on-chip Data
To calculate the enrichment of SMAD1/5 targets identified p values were calculated by permuting genes. n refers to the number of genes.
RRBS Global Methylation Profile
To quantitatively assess global DNA methylation changes, we created histograms for tiles (methylation change >20%) and performed a one-sided two-sample Kolmogorov-Smirnov test to determine significant distribution differences between populations.
Author Contributions
M.G.F. designed and performed the experiments, analyzed the data, and wrote the manuscript. R.D., M.S.R., S.S., A.dM.B., R.P.D., R.R., K.S., E.M., L.U., D.S., V.A.E., D.E., W.V.C., and D.H. performed the experiments and/or analyzed the data. A.Z., C.M., and S.C.dS.L. designed the experiments, analyzed the data, and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
We acknowledge S. Kobayakawa and M. Bialecka for discussions, S. Mendes and C. Visseren for technical support, M. Bouma for technical help with teratomas, and Z. Zhang for generation of mouse chimeras using equipment provided by InfraMouse (KU Leuven-VIB) through a Hercules type 3 project (ZW09-03). This manuscript is dedicated to Cheryl Visseren, one of our most talented students, who carried out the initial experiments but tragically passed away on July 4, 2014. This work was supported by the Interuniversity Attraction Poles-Phase VII [IUAP/PAI P7/14] (to S.C.dL., C.M., A.Z., and D.H.) and individual grants by the Fundação para a Ciência e Tecnologia (FCT) [SFRH/BD/78689/2011 and SFRH/BD/94387/2013] to M.M.G.F. and A.dM.B., respectively, the Netherlands organization of Scientific Research (NWO) [ASPASIA 015.007.037] to S.M.C.dS.L., and the Bontius Stichting [PANCREAS] to M.S.R. The bio-informatics work in the D.H. team was also supported by Fund for Scientific Research-Flanders (FWO-V) GA.0941.11 and G.0782.14.
Published: December 17, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.11.012.
Accession Numbers
The GEO accession number for both the transcriptomics and methylomics data reported in this paper is GEO: GSE71556.
Supplemental Information
References
- Boroviak T., Loos R., Bertone P., Smith A., Nichols J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 2014;16:516–528. doi: 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Fei T., Xia K., Li Z., Zhou B., Zhu S., Chen H., Zhang J., Chen Z., Xiao H., Han J.D. Genome-wide mapping of SMAD target genes reveals the role of BMP signaling in embryonic stem cell fate determination. Genome Res. 2010;20:36–44. doi: 10.1101/gr.092114.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G., Hore T.A., Santos F., Lee H.J., Dean W., Arand J., Krueger F., Oxley D., Paul Y.L., Walter J. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin-Burgess K.E., Travis E.D., Pierson K.E., Vivian J.L. TGF-β-superfamily signaling regulates embryonic stem cell heterogeneity: self-renewal as a dynamic and regulated equilibrium. Stem Cells. 2013;31:48–58. doi: 10.1002/stem.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goumans M.J., Mummery C. Functional analysis of the TGFbeta receptor/Smad pathway through gene ablation in mice. Int. J. Dev. Biol. 2000;44:253–265. [PubMed] [Google Scholar]
- Graf T., Stadtfeld M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell. 2008;3:480–483. doi: 10.1016/j.stem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Graham S.J., Wicher K.B., Jedrusik A., Guo G., Herath W., Robson P., Zernicka-Goetz M. BMP signalling regulates the pre-implantation development of extra-embryonic cell lineages in the mouse embryo. Nat. Commun. 2014;5:5667. doi: 10.1038/ncomms6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E., Brinkman A.B., Arand J., Kroeze L.I., Kerstens H.H., Matarese F., Lepikhov K., Gut M., Brun-Heath I., Hubner N.C. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Dietmann S., Murakami K., Down T.A., Leitch H.G., Surani M.A. Synergistic mechanisms of DNA demethylation during transition to ground-state pluripotency. Stem Cell Reports. 2013;1:518–531. doi: 10.1016/j.stemcr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Markoulaki S., Mitalipova M., Cheng A.W., Cassady J.P., Staerk J., Carey B.W., Lengner C.J., Foreman R., Love J. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4:513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Lopes S.M.C.d.S., Tang F., Surani M.A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap V., Rezende N.C., Scotland K.B., Shaffer S.M., Persson J.L., Gudas L.J., Mongan N.P. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the Nanog, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J., Orkin S.H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S., Mishina Y. BMP signaling and early embryonic patterning. Cytokine Growth Factor Rev. 2005;16:265–278. doi: 10.1016/j.cytogfr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Korchynskyi O., ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Hore T.A., Reik W. Reprogramming the methylome: erasing memory and creating diversity. Cell Stem Cell. 2014;14:710–719. doi: 10.1016/j.stem.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Chen Y.-G. Functions of BMP signaling in embryonic stem cell fate determination. Exp. Cell Res. 2013;319:113–119. doi: 10.1016/j.yexcr.2012.09.016. [DOI] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Francis Stewart A., Smith A. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., Guenther M.G., Johnston W.K., Wernig M., Newman J. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro R.M., de Sousa Lopes S.M.C., Bialecka M., de Boer S., Zwijsen A., Mummery C.L. Real time monitoring of BMP Smads transcriptional activity during mouse development. Genesis. 2008;46:335–346. doi: 10.1002/dvg.20402. [DOI] [PubMed] [Google Scholar]
- Navarro P., Festuccia N., Colby D., Gagliardi A., Mullin N.P., Zhang W., Karwacki-Neisius V., Osorno R., Kelly D., Robertson M. OCT4/SOX2-independent Nanog autorepression modulates heterogeneous Nanog gene expression in mouse ES cells. EMBO J. 2012;31:4547–4562. doi: 10.1038/emboj.2012.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Burdon T., Chambers I., Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi K., Tonge P.D., Nagy A., Zandstra P.W. Local BMP-SMAD1 signaling increases LIF receptor-dependent STAT3 responsiveness and primed-to-naive mouse pluripotent stem cell conversion frequency. Stem Cell Reports. 2014;3:156–168. doi: 10.1016/j.stemcr.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes de Mochel N.S., Luong M., Chiang M., Javier A.L., Luu E., Toshihiko F., MacGregor G.R., Cinquin O., Cho K.W. BMP signaling is required for cell cleavage in preimplantation-mouse embryos. Dev. Biol. 2015;397:45–55. doi: 10.1016/j.ydbio.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa Y., Nikaido I., Hayashi T., Danno H., Uno K.D., Imai T., Ueda H.R. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013;14:R31. doi: 10.1186/gb-2013-14-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai M., Kawabata Y., Makishi K., Itoh K., Terada T.P. Time scales in epigenetic dynamics and phenotypic heterogeneity of embryonic stem cells. PLoS Comput. Biol. 2013;9:e1003380. doi: 10.1371/journal.pcbi.1003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood S.A., Lee H.J., Angermueller C., Krueger F., Saadeh H., Peat J., Andrews S.R., Stegle O., Reik W., Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z.D., Chan M.M., Mikkelsen T.S., Gu H., Gnirke A., Regev A., Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Suzuki A., Raya A., Kawakami Y., Morita M., Matsui T., Nakashima K., Gage F.H., Rodriguez-Esteban C., Izpisua Belmonte J.C. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2006;103:10294–10299. doi: 10.1073/pnas.0506945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P.P.L., Loebel D.A.F. Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D.G. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson M., Liu, Siyuan J., Zou L.-N., Smith Z., Meissner A., Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla M.E., Chambers I. Transcription factor heterogeneity in pluripotent stem cells: a stochastic advantage. Development. 2014;141:2173–2181. doi: 10.1242/dev.102624. [DOI] [PubMed] [Google Scholar]
- Tremblay K.D., Dunn N.R., Robertson E.J. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 2001;128:3609–3621. doi: 10.1242/dev.128.18.3609. [DOI] [PubMed] [Google Scholar]
- Trott J., Martinez Arias A. Single cell lineage analysis of mouse embryonic stem cells at the exit from pluripotency. Biol. Open. 2013;2:1049–1056. doi: 10.1242/bio.20135934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umans L., Vermeire L., Francis A., Chang H., Huylebroeck D., Zwijsen A. Generation of a floxed allele of Smad5 for cre-mediated conditional knockout in the mouse. Genesis. 2003;37:5–11. doi: 10.1002/gene.10219. [DOI] [PubMed] [Google Scholar]
- Ying Q.-L., Nichols J., Chambers I., Smith A. BMP induction of id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Zhao G.-Q. Consequences of knocking out BMP signaling in the mouse. Genesis. 2003;35:43–56. doi: 10.1002/gene.10167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.