Abstract
Understanding the structure and function of the centrosome will require identification of its constituent components and a detailed characterization of the interactions among these components. Here, we describe the application of proximity-dependent biotin identification (BioID) to identify spatial and temporal relationships among centrosome proteins. The BioID method relies on protein fusions to a promiscuous mutant of the Escherichia coli biotin ligase BirA, which biotinylates proteins that are in a ~10 nm labeling radius of the enzyme. The biotinylated proteins are captured by affinity and are identified by mass spectrometry. Proteins identified in this way are referred to as “proximity interactors.” Application of BioID to a set of centrosome proteins demonstrated the utility of this approach in overcoming inherent limitations in probing centrosome structure. These studies also demonstrated the potential of BioID for building large-scale proximity interaction maps among centrosome proteins. In this chapter, we describe the work flow for identification of proximity interactions of centrosome proteins, including materials and methods for the generation and characterization of a BirA*-fusion protein expression plasmid, expression of BirA*-fusion proteins in cells, and purification and identification of proximity partners by mass spectrometry.
INTRODUCTION
Understanding the structure and function of the centrosome will require the identification of its constituent components and a detailed characterization of the interactions among these components. A combination of proteomic, bioinformatics, and comparative genomic studies has identified many, likely most, of the components of mammalian centrosomes (Alves-Cruzeiro, Nogales-Cadenas, & Pascual-Montano, 2014; Andersen et al., 2003; Firat-Karalar, Sante, Elliott, & Stearns, 2014; Hoh, Stowe, Turk, & Stearns, 2012; Jakobsen et al., 2011; Li et al., 2004). However, our understanding of the interactions between these components has been limited because the centrosome presents two challenges for such studies. First, there is only one centrosome in most nondividing cells, limiting the amount of material for biochemical studies; second, the centrosome is an insoluble structure, making the relevant interactions inaccessible to standard techniques such as co-immunoprecipitation. Efforts to overcome these limitations have included salt extraction of centrosome proteins from isolated centrosomes (Moritz et al., 1995; Schnackenberg, Khodjakov, Rieder, & Palazzo, 1998; Schnackenberg & Palazzo, 1999), and use of Xenopus egg extracts, which lack centrosomes but contain all of the components, as the starting material for immunoprecipitation (Hatch, Kulukian, Holland, Cleveland, & Stearns, 2010).
Recently, superresolution microscopy techniques have been applied to a subset of centrosome proteins, yielding new information on the spatial organization of centrosome proteins within subdomains of the organelle (Fu & Glover, 2012; Lawo, Hasegan, Gupta, & Pelletier, 2012; Mennella et al., 2012; Sonnen, Schermelleh, Leonhardt, & Nigg, 2012). Other approaches that have some promise to address this problem include fluorescence resonance energy transfer (Lukinavicius et al., 2013; Muller et al., 2005) and chemical cross-linking (Lukinavicius et al., 2013). Here, we describe the application of proximity-dependent biotin identification (BioID) (Roux, Kim, & Burke, 2013; Roux, Kim, Raida, & Burke, 2012) to the study of the mammalian centrosome. BioID has unique advantages in overcoming the limitations of studying interactions in the centrosome and has the potential for building large-scale interaction maps among centrosome proteins.
In the BioID approach, the protein of interest is tagged with a mutant form of E. coli biotin ligase BirA (R118G) (hereafter BirA*) (Roux et al., 2012) (Figure 1(A)). Wild-type BirA normally only transfers a biotin to a substrate bearing a specific recognition sequence. BirA* is promiscuous in that it activates biotin for transfer in the absence of a substrate peptide, and the activated biotinoyl-5′-AMP is free to diffuse away from the enzyme and covalently modify primary amines of nearby proteins. Since biotinoyl-5′-AMP is highly reactive and short-lived, the zone of modification in BioID is thought to extend only about 10 nm in radius around the BirA*-tagged protein. We will refer to biotin-labeled proteins in a BioID experiment as proximity partners. Biotinylated proximity partner proteins are purified by streptavidin-binding and identified by mass spectrometry (MS). It is important to note that the BioID biotinylation is a mark of potential proximity and not an evidence for physical interactions. In this respect, BioID is a screen for proteins that are proximal to the protein of interest at some point during labeling; a positive result might reflect direct or indirect physical interaction, or simple proximity without physical contact. Thus, the proximity interactions identified by BioID should be tested by subsequent experiments to, for example, determine which reflect physical interactions, and whether the identified proteins colocalize within the cell. Comparison of results from BioID and immunoprecipitation experiments for a number of proteins demonstrated that the two approaches yield an overlapping set of proteins (Couzens et al., 2013; Lambert, Tucholska, Go, Knight, & Gingras, 2014).
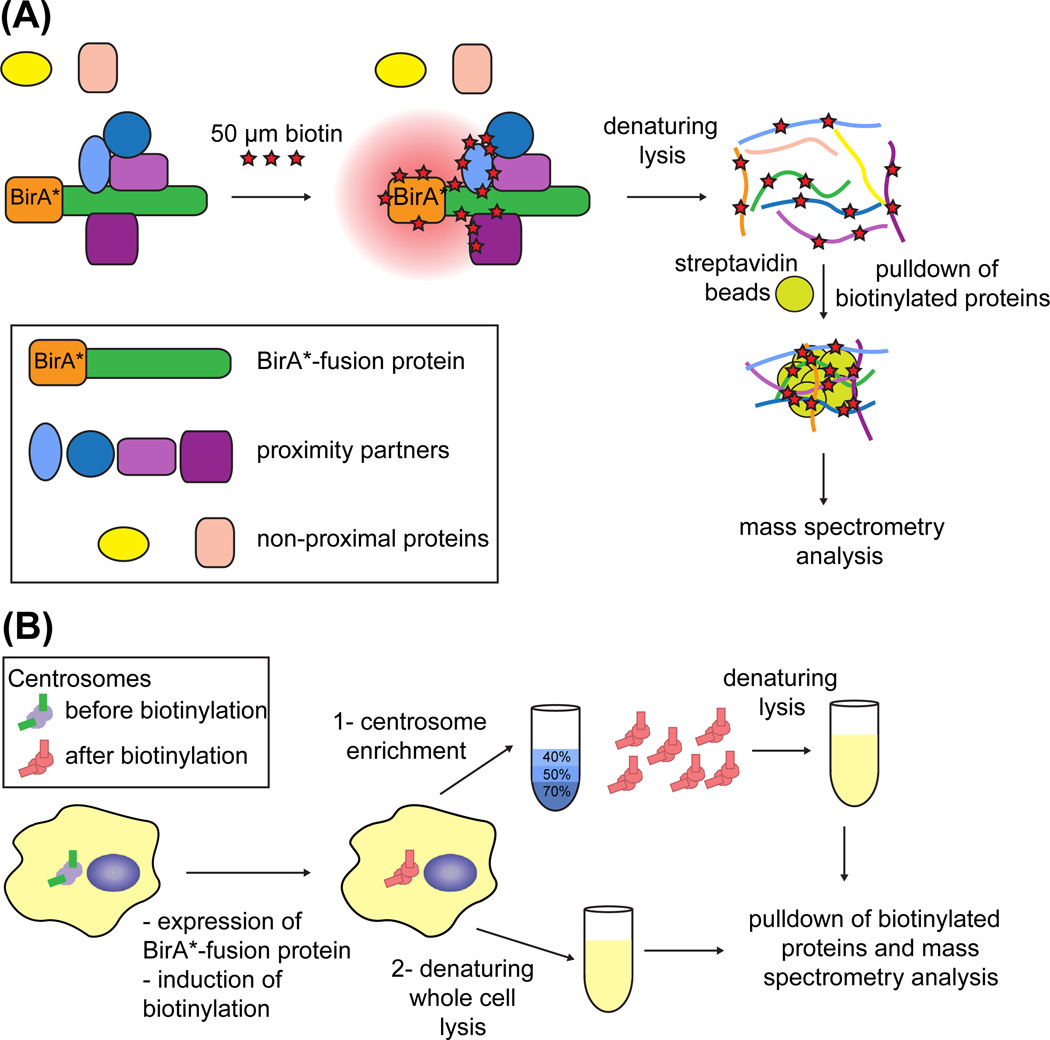
FIGURE 1. Work flow for the application of the BioID approach to the centrosome.
(A) In the BioID approach, the protein of interest is tagged with a mutant form of E. coli biotin ligase BirA (R118G), denoted as BirA*. Following incubation of cells with 50 µM of biotin for 18–24 h, BirA* biotinylates proteins that are in the proximity of the BirA*-fusion protein and we refer to such proteins as “proximity interactors.” The labeling radius of BirA* is estimated to be around 10 nm. The proteins outside the proximity radius are denoted as “nonproximal proteins.” Following incubation of cells with biotin, proteins are solubilized and biotinylated proteins are precipitated by streptavidin beads for subsequent mass spectrometry analysis. (B) Cell line of choice is transfected with the expression plasmid for the BirA*-fusion protein. Cells transiently or stably expressing the BirA*-fusion protein are incubated with excess biotin to induce biotinylation of the proximity interactors. These cells are then processed for identification of the biotinylated proteins by the following two methods: (1) Centrosome-enriched fractions are prepared from these cells by sucrose gradient fractionation, and centrosomes are solubilized under denaturing conditions. (2) Cells are lysed under denaturing conditions. Following lysis, the biotinylated proteins are affinity captured and are identified by mass spectrometry. (See color plate)
BioID has several advantages over traditional approaches for studying interactions at the centrosome, or other “solid-state” cell structures. First, due to the strong affinity of the biotin–streptavidin interaction, purification of biotinylated proteins can take place under denaturing conditions. This allows one to solubilize the centrosome while preserving the proximity relationship information, which is encoded in covalent protein modification. Second, proteins that are associated with the target protein only transiently can be more robustly identified by BioID, since such proteins are marked by biotinylation before cell lysis. These advantages of BioID have been successfully exploited in screens for proximity interactions in the mammalian centrosome (Comartin et al., 2013; Firat-Karalar, Rauniyar, Yates, & Stearns, 2014), nuclear lamina and envelope (Kim et al., 2014; Roux et al., 2012), chromatin (Lambert et al., 2014), bilobe in Trypanosoma bruce (Morriswood et al., 2013) and Hippo signaling pathway (Couzens et al., 2013).
In this chapter, we describe the application of BioID to identify spatial and temporal proximity relationships among centrosome proteins. The basis for this description is our application of BioID to a set of core centrosome duplication and maturation proteins (Firat-Karalar, Rauniyar, et al., 2014). This work identified most of the known interactions between these proteins, and also identified previously uncharacterized relationships and proteins. We will describe the work flow for identification of proximity interactions of centrosome proteins, including materials and methods for the generation and characterization of a BirA*-fusion protein expression plasmid, expression of BirA*-fusion proteins in cells, and purification and identification of proximity partners by MS (Figure 1). In addition, we describe methods for combining BioID labeling with standard techniques for centrosome purification to enrich for centrosomal interactions (Figure 1(B)).
1. MATERIALS
Expression plasmid for generating BirA*-fusion proteins
Cells of choice for validation experiments and growth media for those cells
Biotin (2000× stock): 10 mM
Fixative (i.e., methanol, paraformaldehyde)
Antibodies specific to the epitope tag of the BirA* (i.e., myc, FLAG)
Antibodies specific to biotinylated proteins (Alexa Fluor-streptavidin, horseradish peroxidase (HRP)-streptavidin)
Antibodies that mark the centrosome (i.e., gamma-tubulin antibody)
Secondary antibodies that detect the chosen primary antibodies
DNA dyes (i.e., DAPI)
Streptavidin agarose resin
Standard immunofluorescence and immunoblotting reagents.
1.1 EQUIPMENT
Cell culture incubator, humidified, set at 37 °C, 5% CO2
Refrigerated clinical centrifuge
Microcentrifuge
Ultracentrifuge
Sonicator with pulse capacity (i.e., Branson Digital Sonifier)
Rotator for microcentrifuge tubes
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transfer unit.
1.2 RECIPES
1.2.1 Biotin stock (2000X): 10 mM
Dissolve 122 mg biotin (Sigma #B4501) in 50 mL of dimethyl sulfoxide (DMSO). Aliquot to the desired volume and store at −20 °C.
1.2.2 Polyethylenimine (1 mg/mL)
Dissolve 50 mg polyethylenimine (PEI) 25 kD (Polysciences # 23966-2) in 50 mL purified water and adjust the pH to 7.0. Filter and sterilize the solution through 0.22 µm membrane, then aliquot to the desired volume, and store the PEI at −80 °C.
1.2.3 HB buffer
HEPES: 20 mM, pH 7.8
K-acetate: 5 mM
MgCl2: 0.5 mM
DTT: 0.5 mM
Protease inhibitors.
1.2.4 BioID lysis buffer
Tris: 50 mM, pH 7.4
NaCl: 500 mM
SDS: 0.4%
Ethylene diamine tetraacetic acid (EDTA): 5 mM
DTT: 1 mM
Triton X-100: 2%
Protease inhibitors (10 µg/mL each of aprotinin, leupeptin, pepstatin, and 1 mM phenyl-methylsulfonyl fluoride).
1.2.5 Wash buffer 1
SDS (2%) in dH2O.
1.2.6 Wash buffer 2
Deoxycholate: 0.2%, Triton X-100: 1%
NaCl: 500 mM
EDTA: 1 mM
HEPES: 50 mM, pH 7.5.
1.2.7 Wash buffer 3
Tris: 10 mM, pH 8.1
LiCl: 250 mM, 0.5% NP-40
Deoxycholate: 0.5%
Triton X-100: 1%
NaCl: 500 mM
EDTA: 1 mM.
1.2.8 Wash buffer 4
Tris: 50 mM, pH 7.4
NaCl: 50 mM.
1.2.9 SDS-PAGE sample buffer
Tris: 50 mM, pH 6.8
SDS: 2%
Glycerol: 10%
β-mercaptoethanol: 1%
Bromophenol-blue 0.005%.
2. METHODS
2.1 GENERATION OF BirA*-FUSION PROTEIN EXPRESSION PLASMID
The first step in the BioID approach is constructing an expression plasmid in which the gene of interest is fused with BirA*. To increase the likelihood that the BirA*-fusion protein will localize and function correctly, several points must be considered in the cloning strategy. BirA* is a 33.5 kDa protein, about 5 kDa larger than green fluorescent protein (GFP), and for GFP, the BirA*-fusion is usually made at either the N or C terminus of the target protein. Ideally, this fusion would not impair localization, stability, or function of the target protein, but this must be empirically determined for each protein. Our experience is that most proteins that are functional when tagged with GFP are also functional when tagged in the same site with BirA*; thus, prior evidence on successful GFP-fusion proteins should be used in deciding on BirA*-fusion orientation. We routinely make fusions to both termini for proteins about which we have no prior knowledge of fusion behavior. Second, if available, the protein structure or the spatial organization of the protein can also provide useful information; for example, superresolution imaging studies demonstrated that CEP152 has an elongated structure within the pericentriolar material, such that its N and C termini are in different spatial domains (Sonnen et al., 2012). Accordingly, analysis of both N- and C-terminal CEP152 fusions with BirA* yielded subprotein-level spatial information (Firat-Karalar, Rauniyar, et al., 2014).
In addition to choosing the orientation of the BirA*-fusion, one must also consider other desired properties of the construct. For example, most BirA*-fusion constructs also include an epitope tag (i.e., myc, FLAG) that allows both microscopic and biochemical detection of the fusion protein. One should also consider the promoter for the fusion protein, the nature of the vector, and the means for introducing that vector into the desired cells. Since these are all standard considerations for any mammalian cell expression experiment, we will not consider in depth here, except as they relate to the specifics of the experiments described.
2.2 CHOICE OF CELL LINE AND EXPRESSION METHOD
The centrosome is a small, single-copy organelle in most cultured cells. Therefore, limiting material is an ever-present challenge in studying centrosome protein interactions in situ. One can maximize the amount of relevant material by choosing cell lines that have small surface area-to-volume ratio. One such cell line, HEK293T, is highly transfectable, and has been successfully used for large-scale BioID pulldown of centrosome proteins. As is often the case with mammalian cells, there are tradeoffs associated with the choice of cell line. Although HEK293T cells are easy to manipulate, they typically do not make a primary cilium, which limits their utility for some centrosome-related studies, for example. In contrast, RPE1 cells make a primary cilium at a high frequency, but are more difficult to grow to high density, and have a higher area-to-volume ratio than HEK293T cells. Thus, the cell line should be chosen based on the specific experimental requirements and constraints.
Once the cell line is chosen, the means of introducing and expressing the BirA*-fusion protein must also be chosen, based on the experimental objectives and the cell line. The BirA*-fusion proteins can be expressed either transiently or stably in cells using various means of introducing the vector (i.e., viral infection, lipid- or polymer-based transfection). Transient expression, which is usually accomplished by transfection of a plasmid vector, results in heterogeneous expression of the BirA*-fusion protein; depending on the vector used, this can range from lower to much higher than the endogenous protein is expressed. This is acceptable in some cases, but in others, particularly for the proteins that mislocalize upon overexpression, it would be preferable to have the BirA*-fusion protein expressed more homogeneously, and at a level more similar to the endogenous protein level. This can be accomplished by making stable cell lines and identifying those with the appropriate expression level. There are many methods for generating stable cell lines, including random integration by transient transfection of plasmid vectors or infection with integrating viral vectors (i.e., lentivirus), or integration at a specific genome location (i.e., Flip-in or homologous recombination). In the case for proteins that are toxic when stably expressed, inducible expression can be employed. Perhaps most ideally, the ability to target genomic loci by CRISPR/Cas9 endonuclease-mediated genome editing allows the creation of knock-in alleles in which BirA* is integrated in frame at the genomic locus of the gene of interest.
2.3 VALIDATION OF THE BirA*-FUSION PROTEIN IN CELLS
As the goal of BioID is to identify proximity interactions in the normal context of a target protein, the expression, localization, function, and biotinylation activity of the BirA*-fusion protein should be validated. Localization of the fusion protein can be compared to the endogenous protein by immunofluorescence microscopy (note that the BirA*-fusion vectors typically contain an epitope tag) (Figure 2(A)). Expression of a protein with the correct molecular weight can be determined by immunoblotting (Figure 2(B)). Function can be tested by complementation of a loss-of-function phenotype, or by presence of an overexpression phenotype that is the same as that observed with untagged protein (Figure 2(A)). However, this is not possible in many cases, as it requires that the function of the protein be known, and that there be an assay for that function. Lastly, the biotinylation activity of the BirA*-fusion protein, which is essential to the success of the method, can be assessed in cells by immunofluorescence with fluorescent-streptavidin and on blots with streptavidin-HRP. Although we have not characterized this in detail, most fusion proteins that localize to the structure of interest yield detectable biotinylation at that structure, either due to self-biotinylation of the BirA*-fusion protein, biotinylation of proximal proteins, or both. The experimental outline for validation experiments in a typical transfection-based experiment is described in this section.
- For each experimental condition, place a single glass coverslip in one well of a standard six-well plate and plate cells to the wells the day before transfection. For maximum transfection efficiency, cells should be 80–90% confluent at the time of transfection.To monitor the induction of biotinylation, we recommend the following two experimental conditions: (1) transfected cells incubated with biotin (i.e., plus biotin condition) and (2) transfected cells incubated with vehicle (DMSO) (i.e., minus biotin condition).
- Express the BirA*-fusion protein in cells using the choice of the transfection reagent. To induce biotinylation, change the medium 6–8 h after transfection to medium supplemented with 50 µm biotin. For the minus biotin condition, change the medium to medium supplemented with DMSO (vehicle).Many transfection reagents are available and the reagent must be chosen depending on the cell line. The ideal method should have high transfection efficiency, low toxicity, and minimal effects on normal physiology of the cells. For most cell lines, Lipofectamine LTX (Life Technologies) works well with the manufacturer’s suggested protocol.
- After 12–24 h biotin incubation, process the cells for immunofluorescence and immunoblotting experiments.Incubation time with biotin can be adjusted depending on the protein of interest and the objective of the experiment. We note that levels of biotinylated proteins increase in parallel with biotin exposure time, reaching saturation within 12–24 h.
- For immunofluorescence experiments, fix the coverslips with the choice of the fixation solution, block and stain the cells with the appropriate antibodies. After mounting the coverslips, image the cells by fluorescence microscopy.We recommend the following primary antibodies for staining: (1) antibody against the epitope tag antibody to confirm the expression of the fusion protein, (2) fluorescent-streptavidin antibody to confirm biotinylation of proximity partners, and (3) antibody to mark the centrosome (i.e., gamma-tubulin antibody). The signal form the staining of the epitope tag and the biotinylated peptides are expected to overlap given that the biotinylation radius of BirA* was estimated to be 10 nm.
For immunoblotting experiments, wash the cells twice with PBS. Add 1 mL of PBS +2 mM EDTA to the cells and incubate for 5 min at 37° to detach the cells from the wells. Transfer the cells to a microcentrifuge tube and pellet the cells at 15,000 rpm for 1 min. Following the centrifugation, resuspend the pellet in a volume of SDS-PAGE sample buffer appropriate for the number of cells to be lysed and boil the lysate at 95 °C for 5 min. Run the lysate on an SDS-PAGE gel, block and blot the membrane with primary antibodies against the epitope tag of the BirA*-fusion protein. Once the expression of the fusion protein is confirmed, strip the blot using standard methods, block and blot with streptavidin-HRP antibody to confirm biotinylation of the proximity partners of the BirA*-fusion protein.
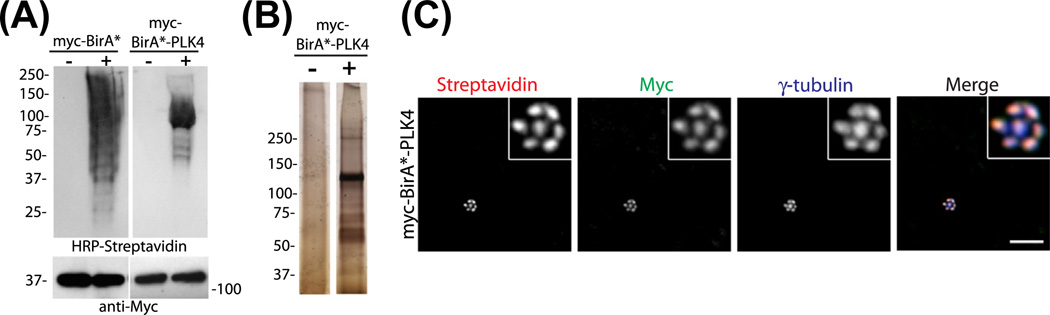
FIGURE 2. Localization and activity of myc-BirA*-PLK4 in mammalian cells.
(A) HEK293T cells were transfected with Myc-BirA* or Myc-BirA*-PLK4. After 18 h incubation with 50 µM biotin or the vehicle control DMSO, cells were lysed. Cell lysates were ran on an SDS-PAGE gel and processed for Western blotting using the following antibodies: (1) anti-Myc antibody to detect expression of the BirA*-fusion protein, (2) HRP-streptavidin to detect biotinylation of the proximity interactors of the BirA*-fusion protein. (B) HEK293T cells expressing Myc-BirA*-PLK4 were incubated with 50 µM biotin or DMSO for 18 h, lysed, ran on an SDS-PAGE gel, and stained by silver-staining. (C) U2OS cells were transfected with Myc-BirA*-PLK4. After 18 h incubation with biotin, cells were fixed, and stained for Myc-BirA*-PLK4 with anti-Myc antibody, for biotinylated proteins with fluorescent streptavidin and centrosomes with anti-γ-tubulin antibody. Scale bars = 10 µm, all insets show 4× enlarged centrosomes.
2.3.1 Expression of BirA*-fusion protein in cells for large-scale pulldowns
Both transient and stable expressions of the BirA*-fusion protein in cells have been successfully used to identify proximity partners of centrosome proteins (Comartin et al., 2013; Firat-Karalar, Rauniyar, et al., 2014). The choice of transient or stable transfection depends on the properties of the protein of interest and objective of the experiment.
2.3.2 Transient expression (PEI)
For the transient transfection approach, it is important to use a cell line that has high transfection efficiency. Since untransfected cells are a source of contaminating background proteins, low transfection efficiencies will decrease the sensitivity of identifying proteins specific to the BirA*-fusion protein. Given their high transfection efficiency and availability of cost-effective transfection reagents, HEK293T cells are ideal for such experiments and they were successfully used for identifying proximity interactions at the centrosome (Firat-Karalar, Rauniyar, et al., 2014).
One day before transfection, plate the cells to five 15 cm dishes so that they are 70–90% at the time of transfection next day.
- Transfect the cells using standard PEI-based transfection approach. For each 15 cm plate, mix 30 µg expression plasmid and the optimized PEI amount (3:1 dilution of 1 mg/mL PEI stock) in serum-free medium, incubate the transfection mixture for 30 min at room temperature, and add it dropwise to the cells. Then 4–6 h after transfection, change the medium with medium supplemented with 50 µm biotin.Other transfection reagents of choice can be used for transient expression of the BirA*-fusion protein for large-scale experiments. We chose PEI-based transfection approach given its high transfection efficiency and the low cost for HEK293T cells.We found 3:1 ratio of PEI to DNA (w/w) to be optimal for most genes we expressed in HEK293T cells. However, this ratio should be screened for each gene tested. Ratios between 1:1 and 5:1 can be used for screening.
As described in Section 3, 24 h after transfection, process the transfected cells for large-scale BioID pulldown experiments.
2.3.3 Stable cell lines
Stable cell lines can be generated using random integration or integration at a specific genome location. Inducible Flip-In T-Rex system (Life Technologies) was successfully used to generate isogenic stable mammalian cell lines that express BirA*-fusion protein from a specific genomic location under a tetracycline-inducible promoter (Comartin et al., 2013; Lambert et al., 2014). Once the correct localization and biotinylation activity of the BirA*-fusion protein is validated by the experiments described in Section 2.3, these cell lines could be used for large-scale BioID pulldown experiments.
3. LARGE-SCALE BioID PULLDOWN OF BirA*-FUSION PROTEINS
Experimental design
Appropriate controls are essential to the success of a BioID experiment. The chosen controls might differ based on the nature of the experiment, but at the least should include cells transfected with BirA* itself expressed in a manner similar to the BirA*-fusion protein, and a cell population that is not expressing any BirA* protein, that is, mock-transfected cells for a transient expression experiment, or the parental cell line for a stable cell line experiment. These control samples should be processed in parallel to the experimental samples for both validation (above) and large-scale experiments. Finally, at least three biological replicates for each experimental condition are required for the statistical analysis of the MS data, as described in Section 4.
3.1 CENTROSOME ENRICHMENT BEFORE STREPTAVIDIN AFFINITY PURIFICATION
Grow five 15 cm dishes of cells transiently or stably expressing BirA*-fusion protein at 37 °C, 5% CO2. Once they are about 70–80% confluent, replace the medium with medium containing 50 µm biotin and incubate the cells further for 18–14 h to induce biotinylation.
- Replace the medium with complete medium containing 5 µg/mL nocodazole and 5 µg/mL cytochalasin B and incubate the cells at 37 °C, 5% CO2 for 1 h.During this incubation time, prepare discontinuous sucrose gradient by progressively layering 5 mL of 70%, 3 mL of 50%, and 3 mL of 40% sucrose solutions upon one another in a Thinwall, Ultra-Clear Beckman tube (max volume 38.5 mL).
Wash the cells twice with ice-cold HB buffer and incubate with 20 mL HB buffer at 4 °C for 10 min.
Scrape the cells from the plate gently with a cell scraper, and transfer the cells to a 15 mL dounce homogenizer and homogenize the cells by applying 10 strokes of the tight-fitting pestle of the dounce homogenizer.
Transfer the lysate to a 15 mL conical tube and centrifuge at 3000 rpm at 4 °C for 5 min to pellet nuclei.
Transfer the supernatant to a 50 mL conical tube and wash the pellet with 10 mL ice-cold HB buffer to remove centrosomes that pelleted with nuclei. Centrifuge the lysate at 3000 rpm at 4 °C for 5 min to pellet nuclei.
Collect the supernatant and combine with the first supernatant in the 50 mL conical tube. Add 0.1% Triton X-100 and centrifuge the lysate again at 3000 rpm at 4 °C for 5 min.
Collect the supernatant and load onto the sucrose gradient in Ultra-Clear Beckman tubes and centrifuge at 26,000 rpm in SW32Ti at 4 °C for 1 h.
- Collect 0.5 mL fractions either by fractionating the gradient from the bottom of the tube by poking a hole or from the top using an automated fraction collector.The centrosome peak should be approximately in fractions 9–11, starting from the bottom of the tube. To determine the peak centrosome fractions, collect 10 µL of each fraction in the 40–60% sucrose range, run on an SDS-PAGE gel, and process for immunoblotting using antibodies against gamma-tubulin (marker for the centrosomes).
- Pool the centrosome-containing fractions in a 15 mL conical tube and resuspend in lysis buffer (1.5 mL resuspension volume per 0.5 mL fraction pooled from the sucrose gradient).Given that 0.5 mL of each fraction will be resuspended in a final volume of 1.5 mL lysis buffer, add 1.5X lysis buffer for a final resuspension concentration of 1X lysis buffer.
Process the samples for streptavidin affinity purification experiments, as described in Section 3.3.
3.2 WHOLE-CELL LYSIS BEFORE THE STREPTAVIDIN AFFINITY PURIFICATION
Grow five 15 cm dishes of cells transiently or stably expressing BirA*-fusion protein at 37 °C, 5% CO2. Once they are about 70–80% confluent, replace the medium with medium containing 50 µm biotin and incubate the cells further for 18–14 h to induce biotinylation.
Wash the cells twice with 15 mL of room temperature 1X PBS.
- Add 1.5 mL of room temperature lysis buffer to each plate and scrape the cells gently with a cell scraper.High concentration of detergents and salt in the lysis buffer are required to denature and solubilize the centrosome proteins.High concentration of detergents and salt in the lysis buffer cause it to precipitate if the lysis buffer is stored at 4 °C.
Transfer the cell lysate to a 15 mL conical tube and process for streptavidin affinity purification experiments, as described in Section 3.3.
3.3 STREPTAVIDIN AFFINITY PURIFICATION EXPERIMENTS
- Sonicate the sample in a Branson Digital Sonifier using the following parameters: (1) 30% amplitude for 1 min in pulses (15 s on, 45 s off), then wait for 2 min to allow sample to chill, and repeat the entire sequence two times; then (2) 40% amplitude for 30 s in pulses (15 s on, 45 s off), wait for 2 min to allow sample to chill, and repeat the entire sequence two times.Efficiency of lysis can be monitored by change in the viscosity of the sample or more quantitatively by the Bradford Colorimetric Assay for protein concentration. On different sonication machines, adjust the sonication regime for optimal lysis.
- Add equal volume of ice-cold 50 mM of Tris, pH 7.4 to the lysate and mix well.Dilution of the lysis buffer twofold at this step is important for making the conditions more favorable for streptavidin–biotin binding.
- Aliquot the lysate to microcentrifuge tubes and pellet the insoluble material by spinning the lysate at 15,000 rpm for 10 min at 4 °C.During the centrifugation step, wash streptavidin agarose resin (60 µL per plate) twice with 0.5X lysis buffer (lysis buffer diluted in 50 mM of Tris, pH 7.4) in a microcentrifuge tube, centrifuging between washes at 1000 rpm for 1 min.
- Transfer the supernatant to a 15-mL conical tube on ice and add the bead suspension to the supernatant. Rotate the bead-lysate mixture at 4 °C overnight.If time is constrained during the experiment, the affinity capture time can be shortened. Due to the strong binding affinity between streptavidin and biotin, the binding will be saturated within several hours of incubation at 4 °C.
Pellet the beads at 1000 rpm for 1 min and carefully remove the supernatant by pipetting without disturbing the beads.
Resuspend the beads in 2 mL wash buffer one and transfer the beads to a microcentrifuge tube. Rotate the tube at room temperature for 10 min and pellet the beads as described in Step 5.
Wash the beads once with 2 mL wash buffer 2 as described in Step 6.
Wash the beads once with 2 mL wash buffer 3 as described in Step 6.
Wash the beads once with 2 mL wash buffer 4 as described in Step 6.
- Pellet the beads at 1000 rpm for 1 min and resuspend the beads in 100 µL of 50 mM ammonium bicarbonate.Save 10 µL of beads (10% total) for immunoblot analysis. Prior to sending samples to MS, it is important to confirm successful pulldown of biotinylated proteins. Immunoblot analysis of the final beads using HRP-streptavidin antibody will detect biotinylated proteins and using an antibody against the epitope tag fused to BirA* will detect BirA*-fusion protein.
3.4 PREPARATION OF SAMPLE FOR MS
The protocol for releasing biotinylated peptides from the streptavidin beads should be carefully considered. The biotin–streptavidin interaction is one of the strongest noncovalent interactions known, and therefore, it is difficult to release biotinylated proteins from the streptavidin beads without losing significant material or without interfering with subsequent MS analysis. To circumvent this problem, we recommend releasing the peptides for analysis by MS using on-bead tryptic digestion. This method also prevents removal of the streptavidin itself from the beads that might interfere with MS analysis. If, instead, elution of the proteins followed by tryptic digest is the method of choice, elution conditions must be optimized to ensure efficient release of biotinylated proteins from the beads. Note that the method by which proteins are released from the streptavidin beads will affect the expected results: digestion of proteins from the beads will leave the biotinylated peptide on the beads, whereas release by elution will include the biotinylated peptide in the sample, and thus might allow identification of the site of biotinylation on peptides.
4. DATA ANALYSIS
In most cases, the identification of proteins in the BioID precipitates will be determined by MS followed by filtering of the data to generate a list of significant “hits.” The MS methodology itself is not the subject of this article, and is subject to change as methods improve. However, various aspects of the data analysis are important and will be covered here.
Standard liquid chromotography-MS/MS analysis of the BioID samples will generate data on peptide masses, which are then compared to a database of virtually digested predicted proteins from the species in use. This will ultimately result in a list of proteins with associated information for each protein, including the number and identity of peptides, as well as the spectral counts, a measure of the abundance of that protein in the sample. We recommend that only proteins that were identified in at least two of the three recommended replicates, and that have a spectral count greater than four, be considered for further analysis.
Once the MS data from the experimental replicates and control samples are obtained, the next step in analysis is the normalization of data across experimental runs and among different samples. There are several normalization methods developed to quantitatively analyze label-free MS data (Neilson et al., 2011); we have used normalized spectral abundance factor (NSAF) analysis (Zybailov et al., 2006). The normalization in the NSAF approach takes into account the number of residues in each protein, as well as their spectral counts. As larger proteins are expected to generate more spectral counts than smaller proteins present in the same molar amount, the number of spectral counts for each protein is divided by number of residues, defining the spectral abundance factor (SAF). To account for variability between independent experimental runs, individual SAF values are normalized against the sum of all SAF values for a particular run, yielding the NSAF, which is a unitless, arbitrary value that can be used to compare the relative abundance of proteins across samples and experiments. The NSAF values are used to rank the mass spec hits by relative abundance.
To distinguish specific proximity interactions from nonspecific interactions, the ratio of the NSAFs of each protein in control and experimental datasets is calculated. A threshold for this NSAF ratio must be determined empirically based on the experimental objectives and knowledge of the system. For BioID analysis of centriole duplication proteins, a protein was considered a specific proximity interactor if the NSAF ratio was higher than 2.5 (Firat-Karalar, Rauniyar, et al., 2014). Information about specific proteins that are common contaminants in mass spec experiments can, if available, also be used to filter the data. For example, for some human and mouse cell lines, there is a publicly available repository of such contaminants maintained for this purpose (Mellacheruvu et al., 2013). In the case of proteins with multiple isoforms, the largest isoform should be chosen to assign spectral count for these proteins. It should be noted that the abundance, or lack thereof, does not necessarily correlate with biological relevance, and subsequent interaction and functional experiments are required to identify proximity interactors specific to the protein of interest.
CONCLUSION
Here we described the work flow of the application of BioID to the centrosome. So far, BioID has only been used to identify proximity interactions of a set of centrosome duplication and maturation proteins (Comartin et al., 2013; Firat-Karalar, Rauniyar, et al., 2014) and the results of these studies demonstrated the utility of the BioID approach in identifying relevant interactions at the centrosome. Future work to expand this approach to other centrosome proteins will be invaluable to ultimately build a proximity interaction map for the centrosome. It is important to note that the proximity information derived from BioID experiments is different from the physical information derived from biochemical approaches typically used to map protein–protein interactions. Therefore, BioID should be used complementary to such approaches. Together these studies can ultimately lead to the generation of an interactome for the centrosome that is essential for our understanding of the structure and function of the centrosome.
Proximity-labeling techniques have advanced significantly in recent years. In addition to the BioID approach, a new proximity-labeling approach that is capable of much higher temporal resolution has been developed recently (Hung et al., 2014; Lam et al., 2014; Rhee et al., 2013). This approach makes use of an engineered ascorbate peroxidase (APEX) and its substrate biotin-phenol to biotinylate proteins proximal to the fusion protein. Biotinylation in this case is achieved after only 1 min induction as opposed to 18–24 h induction in the BioID approach. Interactions among centrosome proteins are dynamic during the cell cycle and currently this is not a good approach available to identify temporal interactions among centrosome proteins. Given its short-labeling time, APEX-based proximity labeling has strong potential for identifying such interactions. This approach so far has only been used to identify the proteome of mitochondria (Hung et al., 2014; Rhee et al., 2013) and has not been tested for its feasibility in identifying temporal proximity interactions among proteins. Thus, the application of this approach in the context of the centrosome should be investigated by future studies.
Acknowledgments
This work was supported by NRSA grant 5 F32 GM106620 to ENF and NIH grant R01 GM52022 to TS.
REFERENCES
- Alves-Cruzeiro JM, Nogales-Cadenas R, Pascual-Montano AD. CentrosomeDB: a new generation of the centrosomal proteins database for Human and Drosophila melanogaster. Nucleic Acids Research. 2014;42(Database issue):D430–D436. doi: 10.1093/nar/gkt1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426(6966):570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Comartin D, Gupta GD, Fussner E, Coyaud E, Hasegan M, Archinti M, et al. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Current Biology. 2013;23(14):1360–1366. doi: 10.1016/j.cub.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Couzens AL, Knight JD, Kean MJ, Teo G, Weiss A, Dunham WH, et al. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Science Signaling. 2013;6(302):rs15. doi: 10.1126/scisignal.2004712. [DOI] [PubMed] [Google Scholar]
- Firat-Karalar EN, Rauniyar N, Yates JR, 3rd, Stearns T. Proximity interactions among centrosome components identify regulators of centriole duplication. Current Biology. 2014;24(6):664–670. doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Sante J, Elliott S, Stearns T. Proteomic analysis of mammalian sperm cells identifies new components of the centrosome. Journal of Cell Science. 2014;127:4128–4133. doi: 10.1242/jcs.157008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Glover DM. Structured illumination of the interface between centriole and peri-centriolar material. Open Biology. 2012;2(8):120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. The Journal of Cell Biology. 2010;191(4):721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh RA, Stowe TR, Turk E, Stearns T. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS One. 2012;7(12):e52166. doi: 10.1371/journal.pone.0052166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V, Zou P, Rhee HW, Udeshi ND, Cracan V, Svinkina T, et al. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Molecular Cell. 2014;55(2):332–341. doi: 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO Journal. 2011;30(8):1520–1535. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proceedings of the National Academy of Sciences of the USA. 2014;111(24):E2453–E2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JP, Tucholska M, Go C, Knight JD, Gingras AC. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. Journal of Proteomics. 2014;118:81–94. doi: 10.1016/j.jprot.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nature Methods. 2014;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nature Cell Biology. 2012;14(11):1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117(4):541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Lukinavicius G, Lavogina D, Orpinell M, Umezawa K, Reymond L, Garin N, et al. Selective chemical crosslinking reveals a Cep57-Cep63-Cep152 centrosomal complex. Current Biology. 2013;23(3):265–270. doi: 10.1016/j.cub.2012.12.030. [DOI] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nature Methods. 2013;10(8):730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, et al. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nature Cell Biology. 2012;14(11):1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Fung JC, Sedat JW, Alberts BM, Agard DA. Three-dimensional structural characterization of centrosomes from early Drosophila embryos. The Journal of Cell Biology. 1995;130(5):1149–1159. doi: 10.1083/jcb.130.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriswood B, Havlicek K, Demmel L, Yavuz S, Sealey-Cardona M, Vidilaseris K, et al. Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryotic Cell. 2013;12(2):356–367. doi: 10.1128/EC.00326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller EG, Snydsman BE, Novik I, Hailey DW, Gestaut DR, Niemann CA, et al. The organization of the core proteins of the yeast spindle pole body. Molecular Biology of the Cell. 2005;16(7):3341–3352. doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, et al. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11(4):535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339(6125):1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Burke B. BioID: a screen for protein–protein interactions. Current Protocols in Protein Science. 2013;74(Unit 19):23. doi: 10.1002/0471140864.ps1923s74. [DOI] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. The Journal of Cell Biology. 2012;196(6):801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackenberg BJ, Khodjakov A, Rieder CL, Palazzo RE. The disassembly and reassembly of functional centrosomes in vitro. Proceedings of the National Academy of Sciences of the USA. 1998;95(16):9295–9300. doi: 10.1073/pnas.95.16.9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackenberg BJ, Palazzo RE. Identification and function of the centrosome centromatrix. Biology of the Cell. 1999;91(6):429–438. [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biology Open. 2012;1(10):965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. Journal of Proteome Research. 2006;5(9):2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]