Abstract
Membrane lipid homeostasis is maintained by de novo synthesis, intracellular transport, remodeling, and degradation of lipid molecules. Glycerophospholipids, the most abundant structural component of eukaryotic membranes, are subject to acyl chain remodeling, which is defined as the post-synthetic process in which one or both acyl chains are exchanged. Here, we review studies addressing acyl chain remodeling of membrane glycerophospholipids in Saccharomyces cerevisiae, a model organism that has been successfully used to investigate lipid synthesis and its regulation. Experimental evidence for the occurrence of phospholipid acyl chain exchange in cardiolipin, phosphatidylcholine, phosphatidylinositol, and phosphatidylethanolamine is summarized, including methods and tools that have been used for detecting remodeling. Progress in the identification of the enzymes involved is reported, and putative functions of acyl chain remodeling in yeast are discussed.
Keywords: acyl chain exchange, membrane lipids, lipid homeostasis, phospholipases, acyltransferases, transacylases
Introduction
The proper functioning of biological membranes critically depends on the composition of their lipid matrix. In eukaryotes, the membrane lipid matrix is made up of glycerophospholipids, glycoglycerolipids (in chloroplasts), sterols, and sphingolipids, with the glycerophospholipids, also referred to as phospholipids, as the most abundant structural lipid.1 Phospholipids come in different classes defined by the nature of the phosphodiester at the sn-3 position of the glycerol backbone. Within each phospholipid class, we find multiple molecular species defined by the acyl chains esterified at the sn-1 and sn-2 positions of the glycerol backbone that vary in length and degree of unsaturation. Each phospholipid class has specific physicochemical properties modulated by the composition of the acyl chains. Acyl chain composition differs between phospholipid classes, eg, in yeast, with unsaturated acyl chains enriched in phosphatidylethanolamine (PE) and cardiolipin (CL) and saturated acyl chains in phosphatidylinositol (PI).2,3 The ensemble of the phospholipid class and acyl chain composition to a large extent determines the physical properties of a membrane, including membrane surface charge, membrane thickness, membrane fluidity, and membrane intrinsic curvature.4 Differences in lipid composition and physical properties between organellar membranes reflect the differences in the functions of organelles and require exquisite regulation of membrane lipid homeostasis.
Cellular membrane lipid homeostasis is maintained by de novo synthesis, intracellular transport, remodeling, and degradation of lipids. Our knowledge of these fundamental processes and their regulation has greatly benefited from research in the model eukaryote Saccharomyces cerevisiae. Whereas significant progress has been made in our understanding of the de novo synthesis of phospholipids and the underlying regulatory mechanisms in yeast,5 research into lipid transport has only recently gained momentum,6,7 while insight into lipid remodeling and degradation is lagging behind. This review focuses on acyl chain remodeling of phospholipids in yeast, ie, the process in which post-synthetically either one or both acyl chains of a phospholipid molecule are exchanged.
Lands was the first to report phospholipid acyl chain exchange.8 He observed that lysophosphatidylcholine (lyso-PC) formed by the action of phospholipase A (PLA) can be reacylated by an acyltransferase in an acyl-CoA-dependent manner. This two-step process known as the Lands’ cycle was proposed to adapt the phosphatidylcholine species profile to obtain the desired thickness and fluidity of cellular membranes.9 Alternative mechanisms of acyl chain exchange include an exchange of both acyl chains with glycerophosphodiesters as intermediates and acyl transfer independent of acyl-CoA or CoA in transacylation reactions in which a fatty acyl chain is transferred from a (lyso-) phospholipid donor to a lysophospholipid acceptor (reviewed by Yamashita et al).10 In mammalian systems, CoA-dependent transacylation reactions have also been reported, which comprise the reverse (ie, ATP-independent acyl-CoA synthesis) and forward reactions of acyl-CoA-dependent acyltransferases.10
In addition to maintaining phospholipid homeostasis, acyl chain remodeling serves specific functions, including the generation of disaturated glycerophospholipid species in pulmonary surfactants.11,12 Acyl chain remodeling also functions in the regulation of the free fatty acid content, in particular levels of fatty acids involved in lipid signaling, including arachidonic acid (C20:4).13 In the liver, the lyso-PC acyltransferase Lpcat3 preferentially uses C18:2–CoA and C20:4–CoA for acylating 1-acyl lyso-PC. Recently, the knockdown of Lpcat3 in mice was shown to increase hepatic inflammation, which is attributed to the accumulation of the lipid inflammatory mediators lyso-PC and C20:4.14 The incorporation of C20:4 into PC by Lpcat3 is also required for triacylglycerol secretion in the liver and intestine, presumably by conferring the optimal membrane fluidity required for the assembly of lipoprotein particles.15,16 In Caenorhabditis elegans, a reduction of C18:0 at the sn-1 position of PI due to defective remodeling interferes with the asymmetric division of stem-cell-like epithelial cells.17 This is proposed to be due to missorting of proteins resulting from a mislocalization of phosphoinositides that in turn is caused by the altered acyl chain composition of PI. Furthermore, acyl chain remodeling has been implicated in the replacement of oxidized acyl chains.18 In plants, newly synthesized fatty acids are primarily incorporated into glycerophospholipids through acyl chain exchange. While esterified to the glycerophospholipid, acyl chains are desaturated. Subsequently, they are released and used for the acylation of glycerol-3-phosphate in de novo glycerophospholipid synthesis.19,20
Here, we summarize the state-of-the-art knowledge on acyl chain remodeling of the major membrane lipids in yeast. The role of acyl chain remodeling in establishing the molecular species profiles of CL, PC, PI, and PE is addressed. Whereas the enzymes catalyzing the remodeling of the mitochondrial membrane lipid CL have been identified, the identity of the enzymes involved in the remodeling of PC, PI, and PE remains largely unknown. The phospholipid remodeling processes in yeast characterized so far have in common that their physiological functions, other than contributing to membrane lipid homeostasis, remain elusive.
Remodeling of CL
CL is a phospholipid containing four acyl chains that is synthesized and, for the most part, localized in the mitochondrial inner membrane.21 CL is important for the mitochondrial structure and function,22,23 eg, it supports the stability and activity of respiratory supercomplexes.24 However, CL is not confined to the inner membrane. It is also found in the mitochondrial outer membrane, where it is involved in the biogenesis of outer membrane proteins.25,26 CL has a characteristic fatty acyl chain composition, enriched in unsaturated fatty acids. In a human heart, 80% of all CL molecules contain four C18:2 acyl chains,27 while yeast CL contains almost exclusively C16:1 and C18:1.28 Remodeling by acyl chain exchange is crucial for obtaining these specific fatty acid compositions.
In mammalian cells, CL remodeling requires the acyl-CoA-independent transacylase tafazzin that shuttles acyl chains between CL and other glycerophospholipids.29 Mutations in the human tafazzin gene cause Barth syndrome, an X-linked disorder with symptoms such as cardiac and skeletal myopathy, neutropenia, and growth retardation.30 At the molecular level, Barth syndrome patients display severely decreased levels of mitochondrial CL, accumulation of monolysocardiolipin (MLCL), and a CL molecular species profile enriched in saturated acyl chains.31
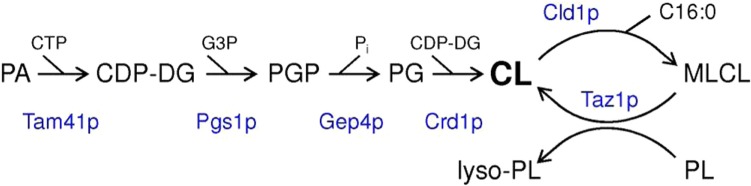
In yeast, CL remodeling depends on the release of C16:0 from CL catalyzed by the PLA Cld1p, localized at the matrix side of the mitochondrial inner membrane,32,33 followed by the reacylation of the resulting MLCL by Taz1p, the yeast homologue of tafazzin (Fig. 1).34 Taz1p has dual localization as a peripheral membrane protein of the mitochondrial inner and outer membranes facing the intermembrane space.35 Taz1p presumably does not display any acyl chain preference, similar to its tafazzin counterpart in Drosophila.36,37 Instead, the enrichment of unsaturated acyl chains in CL has been proposed to originate from the membrane environment where the enzyme is preferentially active, ie, in membrane zones of high negative curvature enriched in non-bilayer preferring lipids.38 The preferred phospholipid acyl chain donor of Taz1p (if any) and the metabolic fate of the lysophospholipid produced are unknown.
Figure 1.
Biosynthesis and acyl chain remodeling of CL in yeast mitochondria with the enzymes indicated in blue.
Abbreviations: PA, phosphatidic acid; CDP-DG, CDP-diacylglycerol; PGP, phosphatidylglycerolphosphate; PG, phosphatidylglycerol; MLCL, monolysocardiolipin; PL, phospholipid.
Recently, deletion of CLD1 was shown to completely restore the defective growth and respiration of a taz1 mutant, demonstrating that these are caused by the increased MLCL to CL ratio rather than by the increased saturation of CL.39,40 Moreover, the lack of a phenotype in cld1 and cld1taz1 mutants implies that acyl chain remodeling CL is not required under the conditions tested. This leaves us with the question of why CL remodeling had evolved in yeast. The upregulation of Cld1p expression during respiratory growth and the increased Cld1p activity in response to decreased mitochondrial membrane potential suggest that CL remodeling may serve to remove oxidatively damaged CL molecules.33,40
Remodeling of Phosphatidylcholine
In yeast, PC is synthesized via two biosynthetic pathways, the triple methylation of PE and the CDP-choline pathway.41 The CDP-choline pathway only contributes to net PC biosynthesis when choline is supplied exogenously. In the absence of exogenous choline, the production of PC relies on the methylation of PE, while the CDP-choline pathway serves to recycle choline from the PC turnover catalyzed by the phospholipase D Spo14p42 or by the combined action of the phospholipase B (PLB) Plb1p,43 or Nte1p44 with the phosphodiesterase Gde1p (Fig. 2).45,46
Figure 2.
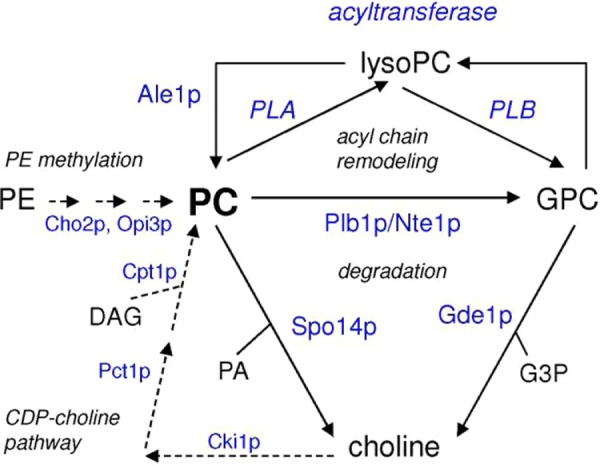
Metabolism of PC in yeast with the enzymes indicated in blue. Acyl chain remodeling of PC may proceed by deacylation to lyso-PC and/or GPC catalyzed by the PLBs Plb1p and Nte1p and/or by PLA and/or PLB that remain to be assigned. Subsequently, lyso-PC can be reacylated by the 1-acyl lyso-PC acyltransferase Ale1p. Acyl-CoA-dependent acyltransferases attaching an acyl chain at the sn-1 position remain to be identified. Alternatively, the de- and reacylation reactions could be catalyzed by transacylases (not shown). The biosynthesis routes of PC are indicated by the dashed arrows.
Yamada et al47 discovered acyl-CoA-dependent acyl-transferase activities in isolated yeast microsomes that acylated 1-acyl lyso-PC and 2-acyl lyso-PC. Given the relatively simple acyl chain composition of yeast membrane glycerophospholipids, Wagner and Paltauf48 wondered whether acyl chain remodeling of membrane lipids occurs in yeast. To address this question, they analyzed the lipid incorporation of radiolabeled oleic and palmitic acids supplied in the culture medium. Fast (within two minutes) and selective acyl chain incorporation was demonstrated at the sn-1 and sn-2 positions of the major glycerophospholipids, including PC, indicative of acyl chain exchange. The rate and extent of the exchange was dependent on the phospholipid class, the sn-1 or sn-2 position, and the nature of the supplied fatty acid (note that palmitic acid was rapidly desaturated into palmitoleic acid). After two minutes, the extent of labeling was the highest in PI and PC and relatively the modest in PE and PS. In PC, the radiolabel from palmitate was distributed in the ratio of 1:9 between sn-1 and sn-2 positions, while oleic acid was almost exclusively esterified at the sn-2 position.48 This study did not provide information with regard to the size of the lipid pools subject to acyl chain exchange.
Dynamic Lipidomic Approaches Addressing Synthesis and Remodeling of Phosphatidylcholine
Pulse-labeling with stable isotope-labeled lipid precursors and subsequent analysis by electrospray ionization tandem mass spectrometry (ESI-MS/MS) enable analysis of the substrate selectivity of lipid biosynthetic enzymes at the level of lipid molecular species. Moreover, time-dependent changes in the molecular species profile of lipid classes can be conveniently monitored in pulse-chase experiments. Applying these dynamic lipidomic approaches to PC metabolism in yeast, Boumann et al demonstrated the molecular species-selectivity of the two biosynthesis routes and the contribution of post-synthetic acyl chain exchange to the steady-state PC molecular species profile.49
The methylation of PE primarily generates di-unsaturated PC, while the CDP-choline route produces a more diverse PC species profile.49,50 Remodeling of PC by acyl chain exchange has been investigated in pct1 strains with an inactivated CDP-choline pathway (Fig. 2), to be able to distinguish acyl chain exchange from the contribution to the PC molecular species profile resulting from the recycling of choline through the CDP-choline pathway.49 So far, there is no evidence in yeast for the occurrence of molecular species-selective degradation of PC by phospholipases B, C, or D.51
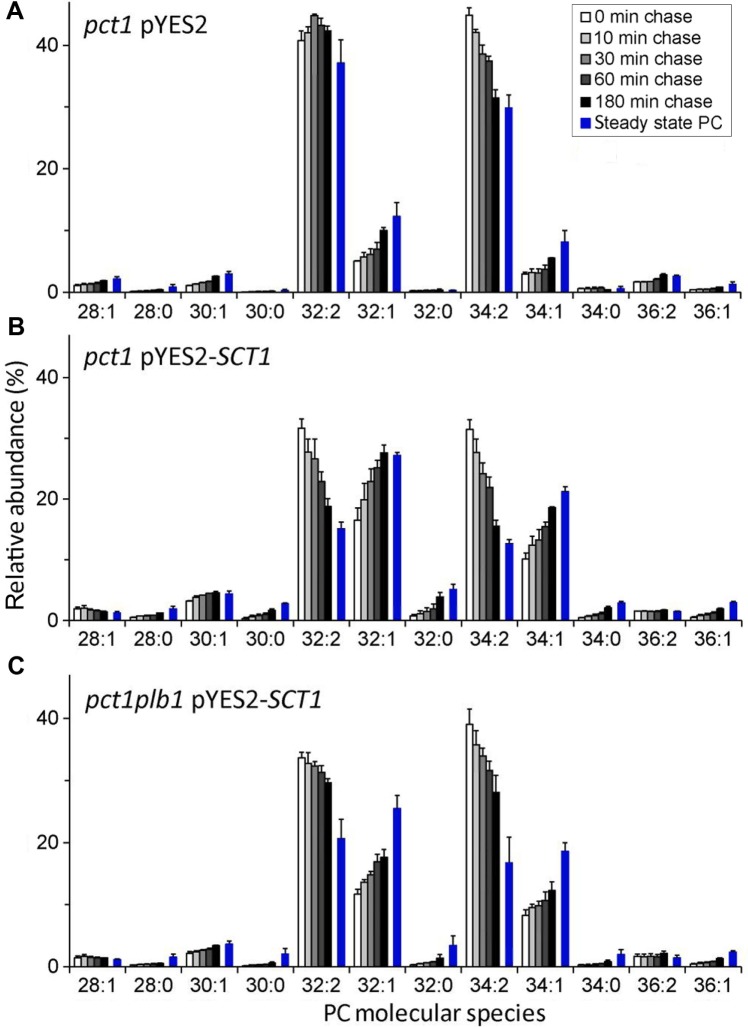
As shown in Figure 3A, labeling pct1 cells for 10 minutes with deuterium-labeled (methyl-D3)-methionine followed by ESI-MS/MS analysis of the newly synthesized PC pool (parent ion scanning for m/z 193) revealed a species profile dominated by 32:2 and 34:2 PC with relatively minor proportions of 32:1 and 34:1 PC. During the subsequent 3-hour chase with excess unlabeled methionine, the species profile of deuterated PC evolved into a profile resembling that of steady-state PC (blue bars in Fig. 3A). The initial proportions of 32:1 and 34:1 PC were almost doubled at the expense of the di-unsaturated species, indicative of the substitution of C16:1 and C18:1 by C16:0. Acyl chain exchange at the sn-1 position of the glycerol backbone was shown to be required in attaining the steady-state PC molecular species profile.51
Figure 3.
Time course of PC remodeling in live yeast cells at 30°C as recorded by ESI-MS/MS in pulse-chase experiments addressing PC synthesized by methylation of PE. Cells from pct1 pYES2 (empty vector control) (A), pct1 pYES2-SCT1 (B), and pct1plb1 pYES2-SCT1 (C) strains cultured in galactose-containing medium to induce overexpression of SCT1 from the GAL1 promoter (B and C), were pulsed for 10 minutes with (methyl-D3)-methionine. After rapid removal of the label, cells were chased in the presence of unlabeled methionine for the time intervals indicated. Molecular species profiles of (methyl-D3)3-PC and steady-state PC (blue bars) were analyzed by parent ion scanning of total lipid extracts in the positive ion mode at m/z 193 and 184, respectively (molecular species of PC are indicated by the total number of acyl carbon atoms:total number of double bonds). The overexpression of Sct1p slows down growth. The experimental conditions were such that the doubling time of the overexpressing strains was approximately 1.3× that of the empty vector control; deletion of PLB1 did not affect the growth rate. Data were taken from ref. 51.
The Ohta Lab has set up a different system for studying phospholipid acyl chain remodeling. Remodeling of (methyl-13C)3-labeled short-chain (C8) PC molecules exogenously supplied to a choline auxotrophic cho2opi3 strain was investigated.52 The di-C8:0 PC enabled growth of cho2opi3 cells in choline-free medium. ESI-MS/MS revealed the appearance of (methyl-13C)3-PC with C16 and C18 acyl chains within 5 minutes after administering (methyl-13C)3 di-C8:0 PC. Follow-up research, in which the CDP-choline pathway was inactivated by inserting a regulated promoter in front of the PCT1 gene, showed that degradation of di-C8:0 PC to choline or phosphocholine and incorporation into PC via the CDP-choline route contributed to the rapid appearance of the regular long chain PC molecular species.53 However, PC molecules in which only one short acyl chain was substituted by C16 were also detected, indicating that at least part of the observed PC remodeling originated from a deacylation–reacylation mechanism. The intermediates showed that C16:0 preferentially substitutes for C8:0 at the sn-1 position, whereas C16:1 substituted with similar efficiency at both positions of the glycerol backbone.52
Triple deletion of the PLB encoding genes PLB1, PLB2, and PLB3 in the cho2opi3 background did not affect the suppression of choline auxotrophy by di-C8:0 PC, suggesting a role for other phospholipases in remodeling di-C8:0 PC. Deletion of the ALE1 gene did cause partially impaired growth of cho2opi3 cells on di-C8:0 PC, consistent with a role for the acyltransferase Ale1p in PC remodeling. Ale1p is an acyl-CoA-dependent acyltransferase that catalyzes acyl transfer to the sn-2 position of a variety of lysophospholipids in vitro, with a preference for unsaturated acyl-CoA species.54–59 In support of a role for Ale1p in PC remodeling, ale1 cells accumulate more lyso-PC than wild type56 and more intracellular glycerophosphocholine (GPC) under conditions of an increased PC turnover.55 Deletion of ALE1 hardly affects the molecular species profiles of the major phospholipid classes.54
Overexpression of the Glycerol-3-Phosphate Acyltransferase Sct1p as a Tool for Studying PC Remodeling
A screen for identifying the phospholipases, acyltransferases, and/or transacylases catalyzing the acyl chain exchange of PC revealed that deletion of SCT1 reduced the extent of PC remodeling.60 Sct1p serves as one of the two glycerol-3-phosphate acyltransferases catalyzing the initial step in glycerolipid synthesis.61 Rather than being required for catalyzing acyl chain exchange, the activity of Sct1p was found to indirectly affect PC remodeling by its impact on cellular acyl chain composition. Deletion of the SCT1 gene was found to reduce the cellular content of C16:0 by 40%. Overexpression of Sct1p from the GAL1 promoter resulted in an almost threefold increase of the proportion of C16:0 at the expense of C16:1 and C18:1 that was reflected in the molecular species profiles of all lipid classes. The preferential incorporation of C16:0 into glycerolipids by Sct1p sequesters them from the yeast’s single desaturase Ole1p, thus increasing the cellular content of saturated acyl chains.60
Importantly, (methyl-D3)-methionine pulse-chase experiments showed that the increased cellular level of C16:0 induced by the overexpression of SCT1 strongly enhanced the incorporation of C16:0 during acyl chain remodeling of PC in pct1 cells (compare Fig. 3B to Fig. 3A),51 allowing for a quantitative analysis of the process. About 40% of the population of newly synthesized PC molecules is subject to detectable acyl chain exchange. Note that this number is an underestimate as only net changes in the total acyl chain length and number of double bonds of the two acyl chains are recorded by the MS/MS analysis. Acyl chain remodeling of PC proceeds with a half-time in the order of 30–60 minutes and occurs at both the sn-1 and sn-2 positions of the glycerol backbone.51 Since the overexpression of SCT1 did not affect the lyso-PC content of the cells or the overall kinetics of PC remodeling,51 it is considered as a reliable tool for monitoring PC remodeling in yeast. The picture emerging from the above studies reveals a continuous deacylation and reacylation of PC with the difference in acyl chain composition between the de novo-synthesized PC molecules and the composition of the pool of available acyl donors determining the extent of PC remodeling.
The Phospholipase B Plb1p Contributes to Remodeling of Phosphatidylcholine
The sensitive read-out of PC remodeling provided by overexpressing SCT1 was taken advantage of to identify the enzymes involved. Deletion of the PLB1 gene reduced the rate of PC remodeling (compare Fig. 3C to Fig. 3B) as well as the cellular lyso-PC content,51 implicating the PLB Plb1p in PC acyl chain exchange. Since Plb1p primarily hydrolyzes PC to GPC in vivo and in vitro,43,62 GPC is the proposed reaction intermediate. Consistent with a role in PC acyl chain remodeling, Plb1p was shown to localize to the ER in addition to its localization in the periplasmic space.63,64 Furthermore, the growth defect induced by overexpression of Plb1p is partially suppressed by an overexpression of Ale1p.56 Interestingly, the activity and expression of Plb1p are modulated by the kinase Ypk1p, a key player in the regulation of sphingolipid biosynthesis.65 Inhibition of sphingolipid biosynthesis upregulates the expression of PLB1, revealing a new link between glycerophospholipid and sphingolipid metabolism.63
Acyl-CoA synthetase (ACS)-deficient yeast mutants are incapable of recycling fatty acids produced by lipid deacylation and as a result secrete free fatty acids.66 Deletion of PLB1 in ACS-deficient cells (faa1faa2faa3faa4) was found not to affect the production of free fatty acids, arguing against the involvement of Plb1p in lipid remodeling or turnover.67 The apparent discrepancy with the above findings could be explained by the accumulation of free fatty acids and/or lysophospholipids in the mutant background that inhibit Plb1p activity by feedback regulation.
Remodeling of Other Membrane Lipids
As mentioned above, Wagner and Paltauf48 demonstrated rapid (within 2 minutes) labeling of PI, PE, and PS with radio-labeled palmitic or oleic acid. The proportion of the radiolabel associated with PI decreased over the two-hour labeling period, consistent with the high turnover of PI.68 The distribution of labeled acyl chain over the sn-1 and sn-2 positions within PI changed from 70% of the label at sn-2 after two minutes to 30% after two hours, indicative of the differential turnover of the acyl chains. Moreover, most of the radiolabeled stearic acid generated by elongation of C16:0 was recovered at the sn-1 position of PI. In contrast, the proportion of label associated with PE increased during the two-hour incubation with radiolabeled fatty acids, consistent with slow remodeling and/or a contribution by de novo biosynthesis. Oleic acid was preferentially incorporated at the sn-2 position, whereas the label from palmitic acid was recovered at both sn-1 and sn-2 positions.48 Since PS is an early metabolic intermediate in phospholipid biosynthesis, subject to rapid turnover,68 it is impossible to interpret its labeling data in terms of remodeling.
Based on sequence motifs, Cst26p was identified as a potential acyltransferase.69 Cst26p/Psi1p transfers C18:0 chains to 2-acyl-lysoPI in vitro and is the acyl-CoA-dependent acyltransferase required for the incorporation of C18:0 at the sn-1 position of PI in vivo.70 The corresponding deletion mutant has an overall reduced cellular C18:0 content, which has been attributed to the loss of a metabolic sink for C18:0 chains.60 This is compensated by a rise in the proportion of C16:0 that appears to be preferentially accommodated in PC,70 possibly via remodeling. The phospholipase or transacylase generating 2-acyl-lysoPI remains elusive. The post-synthetic incorporation of C18:0 at the sn-1 position of PI by Psi1p homologs (AGPAT8/ALCAT1) has been conserved in higher eukaryotes.17,71
Deng et al72 examined remodeling of short-chain di-C10:0 PE supplied in the culture medium, in a psd1psd2 strain with impaired PE synthesis. Remodeling of the exogenous PE by exchange of both acyl chains caused an evolution toward a wild-type PE profile. The study indicates that the established phospholipases B Plb1p, Plb2p, Plb3p, and Nte1p and the potential phospholipases Spo1p and Yor022cp were not required for remodeling of exogenous PE. The acyltransferases Ale1p and Slc1p were implicated in the incorporation of C16 and C18 acyl chains at the sn-2 position. Ale1p accounted for most of the observed incorporation of C16:1 and C18:1 at the sn-2 position, in agreement with its identification as the major lyso-PE acyltransferase.57
Candidate Enzymes Catalyzing Acyl Chain Remodeling
Except for Cld1p, Taz1p, Plb1p, Ale1p, and Cst26p/Psi1p alluded above, the phospholipases, acyltransferases, and transacylases catalyzing phospholipid acyl chain exchange in yeast are yet to be identified. Based on demonstrated enzyme activity, a short list of candidates is proposed.
The established phospholipases B could supply substrates for reacylation: Plb2p, although not involved in the remodeling of PC,51 degrades all phospholipid classes;73 Plb3p degrades PI to glycerophosphoinositol (GPI);73 and Nte1p hydrolyzes CDP-choline-derived PC.44 Similarly, the acyl-CoA-independent phospholipid:diacylglycerol acyltransferase Lro1p generates lyso-PE and lyso-PC in transferring acyl chains from PE and PC to diacylglycerol (DAG) yielding triacylglycerol.74–76 However, deletion of LRO1 did not affect PC remodeling assayed under conditions of SCT1-overexpression.51 The triacylglycerol lipase Tgl4p, in addition to its lipase motif, contains the conserved consensus sequence of a calcium-independent phospholipase A2. Purified Tgl4p indeed was found to hydrolyze PC to lyso-PC in vitro, and deletion of TLG4 specifically affects the PC molecular species profile, consistent with a role in PC remodeling.77 Tgl4p is truly multifunctional, as it also catalyzes the acyl-CoA-dependent acylation of 1-acyl lyso-PA to PA in vitro. Lpl1p contains a lipase motif and was identified as a broad specificity PLB that preferentially hydrolyzes at the sn-2 position.78
Yeast microsomal membranes contain an acyl-CoA-dependent GPC acyltransferase activity producing 1-acyl lyso-PC without acyl-CoA specificity in the yeast cell extract. The lyso-PC produced is converted to PC by Ale1p.79 The gene(s) encoding this enzyme activity, which is likely responsible for the incorporation of C16:0 at the sn-1 position during PC remodeling, is yet to be identified. Tgl3p is a dual function enzyme with TAG lipase and acyltransferase activity.80 In vitro Tgl3p acylates 1-acyl-lyso-PE with a preference for C18:1-CoA.
Conclusion and Perspective
Acyl chain remodeling adds an extra layer of complexity and versatility to membrane lipid homeostasis. Research into the post-synthetic modifications of membrane lipids in yeast is still in its infancy, as is the identification of the enzymes involved. This is to a large extent due to the lack of distinct phenotypes associated with defects in acyl chain remodeling, which is in agreement with the ability of yeast to tolerate huge variations in lipid composition.41 Yeast mutants deleted for genes known to encode enzymes catalyzing phospholipid acyl chain exchange (cld1, plb1, psi1, and ale1) do not exhibit a growth phenotype. Moreover, in plb1 and ale1 deletion strains, the molecular species profiles of the membrane lipids hardly differ from the wild type. These observations suggest the existence of additional enzymes with overlapping functions. In addition, there may be very strict coupling between reactions, eg, the absence of an acyltransferase would inhibit the upstream phospholipase to avoid accumulation of deleterious lysophospholipids. The identification of remodeling enzymes is further complicated by recent findings that lipid metabolism-related enzymes may serve multiple functions.77,80
On the other hand, the MS-based acyl chain remodeling assays now at hand provide a tool box to systematically test (combinations of) gene deletions for their role in acyl chain exchange. Candidate genes could be selected based on bioinformatics, on genetic interaction mapping,81 and on shotgun lipidomics screening of yeast mutant libraries.82 In such approaches, it is imperative to distinguish effects on acyl chain remodeling from effects on overall acyl chain composition (remember the Sct1p example stated above). Identification of the enzymes and their corresponding acyl chain exchange reactions will facilitate addressing their physiological functions.
Higher eukaryotes have evolved specific functions for at least a subset of lipid acyl chain remodeling reactions and products. In the absence of any such function assigned to acyl chain exchange in yeast yet, we can only conclude that acyl chain remodeling adds to the versatility of yeast to adapt its membrane lipid composition, eg, by replacing oxidatively damaged acyl chains (CL) and by accommodating (excess) saturated acyl chains (PI and PC). An additional function of acyl chain remodeling that could be envisioned is organelle-specific editing of membrane lipid acyl chain composition, which, eg, may account for the strong enrichment of saturated acyl chains in PS and PE in the yeast plasma membrane.2
Footnotes
ACADEMIC EDITOR: Tim Levine, Editor in Chief
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1563 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported by the Division of Chemical Sciences, with financial aid from The Netherlands Organization for Scientific Research and by the China Scholarship Council. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Jointly developed the structure and wrote the paper: MFR, XB, CHDS and AIdK. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.van Meer G, de Kroon AI. Lipid map of the mammalian cell. J Cell Sci. 2011;124(pt 1):5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- 2.Schneiter R, Brugger B, Sandhoff R, et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146(4):741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ejsing CS, Sampaio JL, Surendranath V, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A. 2009;106(7):2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Kroon AI, Rijken PJ, De Smet CH. Checks and balances in membrane phospholipid class and acyl chain homeostasis, the yeast perspective. Prog Lipid Res. 2013;52(4):374–394. doi: 10.1016/j.plipres.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190(2):317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang A, John Peter AT, Kornmann B. ER-mitochondria contact sites in yeast: beyond the myths of ERMES. Curr Opin Cell Biol. 2015;35:7–12. doi: 10.1016/j.ceb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Lahiri S, Toulmay A, Prinz WA. Membrane contact sites, gateways for lipid homeostasis. Curr Opin Cell Biol. 2015;33:82–87. doi: 10.1016/j.ceb.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lands WE. Metabolism of glycerolipids. 2. The enzymatic acylation of lysolecithin. J Biol Chem. 1960;235:2233–2237. [PubMed] [Google Scholar]
- 9.Lands WE. Lipid metabolism. Annu Rev Biochem. 1965;34:313–346. doi: 10.1146/annurev.bi.34.070165.001525. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita A, Hayashi Y, Nemoto-Sasaki Y, et al. Acyltransferases and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms. Prog Lipid Res. 2014;53:18–81. doi: 10.1016/j.plipres.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Hyatt BA, Mucenski ML, Mason RJ, Shannon JM. Identification and characterization of a lysophosphatidylcholine acyltransferase in alveolar type II cells. Proc Natl Acad Sci U S A. 2006;103(31):11724–11729. doi: 10.1073/pnas.0604946103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harayama T, Eto M, Shindou H, et al. Lysophospholipid acyltransferases mediate phosphatidylcholine diversification to achieve the physical properties required in vivo. Cell Metab. 2014;20(2):295–305. doi: 10.1016/j.cmet.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh M, Tucker DE, Burchett SA, Leslie CC. Properties of the group IV phospholipase A2 family. Prog Lipid Res. 2006;45(6):487–510. doi: 10.1016/j.plipres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Rong X, Albert CJ, Hong C, et al. LXRs regulate ER stress and inflammation through dynamic modulation of membrane phospholipid composition. Cell Metab. 2013;18(5):685–697. doi: 10.1016/j.cmet.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rong X, Wang B, Dunham MM, et al. Lpcat3-dependent production of arachidonoyl phospholipids is a key determinant of triglyceride secretion. Elife. 2015;4:e06557, 1–23. doi: 10.7554/eLife.06557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashidate-Yoshida T, Harayama T, Hishikawa D, et al. Fatty acid remodeling by LPCAT3 enriches arachidonate in phospholipid membranes and regulates triglyceride transport. Elife. 2015;4:e06328, 1–31. doi: 10.7554/eLife.06328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imae R, Inoue T, Kimura M, et al. Intracellular phospholipase A1 and acyl-transferase, which are involved in Caenorhabditis elegans stem cell divisions, determine the sn-1 fatty acyl chain of phosphatidylinositol. Mol Biol Cell. 2010;21(18):3114–3124. doi: 10.1091/mbc.E10-03-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cummings BS, McHowat J, Schnellmann RG. Phospholipase A(2)s in cell injury and death. J Pharmacol Exp Ther. 2000;294(3):793–799. [PubMed] [Google Scholar]
- 19.Bates PD, Ohlrogge JB, Pollard M. Incorporation of newly synthesized fatty acids into cytosolic glycerolipids in pea leaves occurs via acyl editing. J Biol Chem. 2007;282(43):31206–31216. doi: 10.1074/jbc.M705447200. [DOI] [PubMed] [Google Scholar]
- 20.Bates PD, Durrett TP, Ohlrogge JB, Pollard M. Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 2009;150(1):55–72. doi: 10.1104/pp.109.137737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baile MG, Lu YW, Claypool SM. The topology and regulation of cardiolipin biosynthesis and remodeling in yeast. Chem Phys Lipids. 2014;179:25–31. doi: 10.1016/j.chemphyslip.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi AS, Zhou J, Gohil VM, Chen S, Greenberg ML. Cellular functions of cardiolipin in yeast. Biochim Biophys Acta. 2009;1793(1):212–218. doi: 10.1016/j.bbamcr.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren M, Phoon CK, Schlame M. Metabolism and function of mitochondrial cardiolipin. Prog Lipid Res. 2014;55:1–16. doi: 10.1016/j.plipres.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta. 2009;1788(10):2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gebert N, Joshi AS, Kutik S, et al. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr Biol. 2009;19(24):2133–2139. doi: 10.1016/j.cub.2009.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauerwald J, Jores T, Eisenberg-Bord M, Chuartzman SG, Schuldiner M, Rapaport D. Genome-wide screens in Saccharomyces cerevisiae highlight a role for cardiolipin in biogenesis of mitochondrial outer membrane multispan proteins. Mol Cell Biol. 2015;35(18):3200–3211. doi: 10.1128/MCB.00107-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlame M, Shanske S, Doty S, et al. Microanalysis of cardiolipin in small biopsies including skeletal muscle from patients with mitochondrial disease. J Lipid Res. 1999;40(9):1585–1592. [PubMed] [Google Scholar]
- 28.Schlame M, Ren M, Xu Y, Greenberg ML, Haller I. Molecular symmetry in mitochondrial cardiolipins. Chem Phys Lipids. 2005;138(1–2):38–49. doi: 10.1016/j.chemphyslip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Kelley RI, Blanck TJ, Schlame M. Remodeling of cardiolipin by phospholipid transacylation. J Biol Chem. 2003;278(51):51380–51385. doi: 10.1074/jbc.M307382200. [DOI] [PubMed] [Google Scholar]
- 30.Bione S, D’Adamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat Genet. 1996;12(4):385–389. doi: 10.1038/ng0496-385. [DOI] [PubMed] [Google Scholar]
- 31.Schlame M, Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 2006;580(23):5450–5455. doi: 10.1016/j.febslet.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 32.Beranek A, Rechberger G, Knauer H, Wolinski H, Kohlwein SD, Leber R. Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast. J Biol Chem. 2009;284(17):11572–11578. doi: 10.1074/jbc.M805511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baile MG, Whited K, Claypool SM. Deacylation on the matrix side of the mitochondrial inner membrane regulates cardiolipin remodeling. Mol Biol Cell. 2013;24(12):2008–2020. doi: 10.1091/mbc.E13-03-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu Z, Valianpour F, Chen S, et al. Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Mol Microbiol. 2004;51(1):149–158. doi: 10.1046/j.1365-2958.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- 35.Claypool SM, McCaffery JM, Koehler CM. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J Cell Biol. 2006;174(3):379–390. doi: 10.1083/jcb.200605043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rijken PJ, Houtkooper RH, Akbari H, et al. Cardiolipin molecular species with shorter acyl chains accumulate in Saccharomyces cerevisiae mutants lacking the acyl coenzyme A-binding protein Acb1p: new insights into acyl chain remodeling of cardiolipin. J Biol Chem. 2009;284(40):27609–27619. doi: 10.1074/jbc.M109.016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malhotra A, Xu Y, Ren M, et al. Formation of molecular species of mitochondrial cardiolipin. 1. A novel transacylation mechanism to shuttle fatty acids between sn-1 and sn-2 positions of multiple phospholipid species. Biochim Biophys Acta. 2009;1791(4):314–320. doi: 10.1016/j.bbalip.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlame M, Acehan D, Berno B, et al. The physical state of lipid substrates provides transacylation specificity for tafazzin. Nat Chem Biol. 2012;8(10):862–869. doi: 10.1038/nchembio.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baile MG, Sathappa M, Lu YW, et al. Unremodeled and remodeled cardiolipin are functionally indistinguishable in yeast. J Biol Chem. 2014;289(3):1768–1778. doi: 10.1074/jbc.M113.525733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye C, Lou W, Li Y, et al. Deletion of the cardiolipin-specific phospholipase Cld1 rescues growth and life span defects in the tafazzin mutant: implications for Barth syndrome. J Biol Chem. 2014;289(6):3114–3125. doi: 10.1074/jbc.M113.529487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Kroon AI. Metabolism of phosphatidylcholine and its implications for lipid acyl chain composition in Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771(3):343–352. doi: 10.1016/j.bbalip.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 42.Sreenivas A, Patton-Vogt JL, Bruno V, Griac P, Henry SA. A role for phospholipase D (Pld1p) in growth, secretion, and regulation of membrane lipid synthesis in yeast. J Biol Chem. 1998;273(27):16635–16638. doi: 10.1074/jbc.273.27.16635. [DOI] [PubMed] [Google Scholar]
- 43.Lee KS, Patton JL, Fido M, et al. The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J Biol Chem. 1994;269(31):19725–19730. [PubMed] [Google Scholar]
- 44.Zaccheo O, Dinsdale D, Meacock PA, Glynn P. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J Biol Chem. 2004;279(23):24024–24033. doi: 10.1074/jbc.M400830200. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez-Murray JP, McMaster CR. Glycerophosphocholine catabolism as a new route for choline formation for phosphatidylcholine synthesis by the kennedy pathway. J Biol Chem. 2005;280(46):38290–38296. doi: 10.1074/jbc.M507700200. [DOI] [PubMed] [Google Scholar]
- 46.Fisher E, Almaguer C, Holic R, Griac P, Patton-Vogt J. Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae. J Biol Chem. 2005;280(43):36110–36117. doi: 10.1074/jbc.M507051200. [DOI] [PubMed] [Google Scholar]
- 47.Yamada K, Okuyama H, Endo Y, Ikezawa H. Acyltransferase systems involved in phospholipid metabolism in Saccharomyces cerevisiae. Arch Biochem Biophys. 1977;183(1):281–289. doi: 10.1016/0003-9861(77)90441-6. [DOI] [PubMed] [Google Scholar]
- 48.Wagner S, Paltauf F. Generation of glycerophospholipid molecular species in the yeast Saccharomyces cerevisiae. Fatty acid pattern of phospholipid classes and selective acyl turnover at sn-1 and sn-2 positions. Yeast. 1994;10(11):1429–1437. doi: 10.1002/yea.320101106. [DOI] [PubMed] [Google Scholar]
- 49.Boumann HA, Damen MJ, Versluis C, Heck AJ, de Kruijff B, de Kroon AI. The two biosynthetic routes leading to phosphatidylcholine in yeast produce different sets of molecular species. Evidence for lipid remodeling. Biochemistry. 2003;42(10):3054–3059. doi: 10.1021/bi026801r. [DOI] [PubMed] [Google Scholar]
- 50.Boumann HA, de Kruijff B, Heck AJ, de Kroon AI. The selective utilization of substrates in vivo by the phosphatidylethanolamine and phosphatidylcholine biosynthetic enzymes Ept1p and Cpt1p in yeast. FEBS Lett. 2004;569(1–3):173–177. doi: 10.1016/j.febslet.2004.05.043. [DOI] [PubMed] [Google Scholar]
- 51.De Smet CH, Cox R, Brouwers JF, de Kroon AI. Yeast cells accumulate excess endogenous palmitate in phosphatidylcholine by acyl chain remodeling involving the phospholipase B Plb1p. Biochim Biophys Acta. 2013;1831(6):1167–1176. doi: 10.1016/j.bbalip.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka K, Fukuda R, Ono Y, et al. Incorporation and remodeling of extracellular phosphatidylcholine with short acyl residues in Saccharomyces cerevisiae. Biochim Biophys Acta. 2008;1781(8):391–399. doi: 10.1016/j.bbalip.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 53.Kishino H, Eguchi H, Takagi K, et al. Acyl-chain remodeling of dioctanoyl-phosphatidylcholine in Saccharomyces cerevisiae mutant defective in de novo and salvage phosphatidylcholine synthesis. Biochem Biophys Res Commun. 2014;445(2):289–293. doi: 10.1016/j.bbrc.2014.01.136. [DOI] [PubMed] [Google Scholar]
- 54.Benghezal M, Roubaty C, Veepuri V, et al. SLC1 and SLC4 encode partially redundant acyl-coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast. J Biol Chem. 2007;282(42):30845–30855. doi: 10.1074/jbc.M702719200. [DOI] [PubMed] [Google Scholar]
- 55.Chen Q, Kazachkov M, Zheng Z, et al. The yeast acylglycerol acyltransferase LCA1 is a key component of lands cycle for phosphatidylcholine turnover. FEBS Lett. 2007;581(28):5511–5516. doi: 10.1016/j.febslet.2007.10.061. [DOI] [PubMed] [Google Scholar]
- 56.Tamaki H, Shimada A, Ito Y, et al. LPT1 encodes a membrane-bound O-acyltransferase involved in the acylation of lysophospholipids in the yeast Saccharomyces cerevisiae. J Biol Chem. 2007;282(47):34288–34298. doi: 10.1074/jbc.M704509200. [DOI] [PubMed] [Google Scholar]
- 57.Riekhof WR, Wu J, Jones JL, et al. Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2007;282(39):28344–28352. doi: 10.1074/jbc.M705256200. [DOI] [PubMed] [Google Scholar]
- 58.Riekhof WR, Wu J, Gijon MA, et al. Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: the role of P-type ATPases in transport and a broad specificity acyltransferase in acylation. J Biol Chem. 2007;282(51):36853–36861. doi: 10.1074/jbc.M706718200. [DOI] [PubMed] [Google Scholar]
- 59.Jain S, Stanford N, Bhagwat N, et al. Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2007;282(42):30562–30569. doi: 10.1074/jbc.M706326200. [DOI] [PubMed] [Google Scholar]
- 60.De Smet CH, Vittone E, Scherer M, et al. The yeast acyltransferase Sct1p regulates fatty acid desaturation by competing with the desaturase Ole1p. Mol Biol Cell. 2012;23(7):1146–1156. doi: 10.1091/mbc.E11-07-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng Z, Zou J. The initial step of the glycerolipid pathway: identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae. J Biol Chem. 2001;276(45):41710–41716. doi: 10.1074/jbc.M104749200. [DOI] [PubMed] [Google Scholar]
- 62.Merkel O, Oskolkova OV, Raab F, et al. Regulation of activity in vitro and in vivo of three phospholipases B from Saccharomyces cerevisiae. Biochem J. 2005;387(pt 2):489–496. doi: 10.1042/BJ20041272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Surlow BA, Cooley BM, Needham PG, Brodsky JL, Patton-Vogt J. Loss of Ypk1, the yeast homolog to the human serum- and glucocorticoid-induced protein kinase, accelerates phospholipase B1-mediated phosphatidylcholine deacylation. J Biol Chem. 2014;289(45):31591–31604. doi: 10.1074/jbc.M114.581157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Natter K, Leitner P, Faschinger A, et al. The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol Cell Proteomics. 2005;4(5):662–672. doi: 10.1074/mcp.M400123-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2011;108(48):19222–19227. doi: 10.1073/pnas.1116948108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scharnewski M, Pongdontri P, Mora G, Hoppert M, Fulda M. Mutants of Saccharomyces cerevisiae deficient in acyl-CoA synthetases secrete fatty acids due to interrupted fatty acid recycling. FEBS J. 2008;275(11):2765–2778. doi: 10.1111/j.1742-4658.2008.06417.x. [DOI] [PubMed] [Google Scholar]
- 67.Mora G, Scharnewski M, Fulda M. Neutral lipid metabolism influences phospholipid synthesis and deacylation in Saccharomyces cerevisiae. PLoS One. 2012;7(11):e49269. doi: 10.1371/journal.pone.0049269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patton-Vogt JL, Griac P, Sreenivas A, et al. Role of the yeast phosphatidylinositol/phosphatidylcholine transfer protein (Sec14p) in phosphatidylcholine turnover and INO1 regulation. J Biol Chem. 1997;272(33):20873–20883. doi: 10.1074/jbc.272.33.20873. [DOI] [PubMed] [Google Scholar]
- 69.Neuwald AF. Barth syndrome may be due to an acyltransferase deficiency. Curr Biol. 1997;7(8):R465–R466. doi: 10.1016/s0960-9822(06)00237-5. [DOI] [PubMed] [Google Scholar]
- 70.Le Guedard M, Bessoule JJ, Boyer V, et al. PSI1 is responsible for the stearic acid enrichment that is characteristic of phosphatidylinositol in yeast. FEBS J. 2009;276(21):6412–6424. doi: 10.1111/j.1742-4658.2009.07355.x. [DOI] [PubMed] [Google Scholar]
- 71.Imae R, Inoue T, Nakasaki Y, et al. LYCAT, a homologue of C. elegans acl-8, acl-9, and acl-10, determines the fatty acid composition of phosphatidylinositol in mice. J Lipid Res. 2012;53(3):335–347. doi: 10.1194/jlr.M018655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deng L, Fukuda R, Kakihara T, Narita K, Ohta A. Incorporation and remodeling of phosphatidylethanolamine containing short acyl residues in yeast. Biochim Biophys Acta. 2010;1801(6):635–645. doi: 10.1016/j.bbalip.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 73.Merkel O, Fido M, Mayr JA, et al. Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J Biol Chem. 1999;274(40):28121–28127. doi: 10.1074/jbc.274.40.28121. [DOI] [PubMed] [Google Scholar]
- 74.Oelkers P, Tinkelenberg A, Erdeniz N, Cromley D, Billheimer JT, Sturley SL. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J Biol Chem. 2000;275(21):15609–15612. doi: 10.1074/jbc.C000144200. [DOI] [PubMed] [Google Scholar]
- 75.Dahlqvist A, Stahl U, Lenman M, et al. Phospholipid: diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci U S A. 2000;97(12):6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Horvath SE, Wagner A, Steyrer E, Daum G. Metabolic link between phosphatidylethanolamine and triacylglycerol metabolism in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2011;1811(12):1030–1037. doi: 10.1016/j.bbalip.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rajakumari S, Daum G. Multiple functions as lipase, steryl ester hydrolase, phospholipase, and acyltransferase of Tgl4p from the yeast Saccharomyces cerevisiae. J Biol Chem. 2010;285(21):15769–15776. doi: 10.1074/jbc.M109.076331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Selvaraju K, Rajakumar S, Nachiappan V. Identification of a phospholipase B encoded by the LPL1 gene in Saccharomyces cerevisiae. Biochim Biophys Acta. 2014;1842(10):1383–1392. doi: 10.1016/j.bbalip.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 79.Stalberg K, Neal AC, Ronne H, Ståhl U. Identification of a novel GPCAT activity and a new pathway for phosphatidylcholine biosynthesis in S. cerevisiae. J Lipid Res. 2008;49(8):1794–1806. doi: 10.1194/jlr.M800129-JLR200. [DOI] [PubMed] [Google Scholar]
- 80.Rajakumari S, Daum G. Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol Biol Cell. 2010;21(4):501–510. doi: 10.1091/mbc.E09-09-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Surma MA, Klose C, Peng D, et al. A lipid E-MAP identifies Ubx2 as a critical regulator of lipid saturation and lipid bilayer stress. Mol Cell. 2013;51(4):519–530. doi: 10.1016/j.molcel.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tarasov K, Stefanko A, Casanovas A, et al. High-content screening of yeast mutant libraries by shotgun lipidomics. Mol Biosyst. 2014;10(6):1364–1376. doi: 10.1039/c3mb70599d. [DOI] [PubMed] [Google Scholar]