Abstract
Campylobacter coli are one of the most common bacteria in bacterial gastroenteritis and acute enterocolitis in humans. However, relatively little is known regarding the mechanisms of pathogenesis and host response to C. coli infections. To investigate the influence of genetic changes, we first used PCR to demonstrate the presence of the known virulence genes cadF, virB11, cdtB, cdtC and ceuE in the clinical isolate C. coli 26536, which was isolated from the liver of infected BALB/c mice. Sequence analyses of the cadF, virB11, cdtB and ceuE genes in C. coli 26536 confirmed the stability in these virulence genes during their transmission through the host. We further investigated C. coli infection for the bacterial clearance from the liver and spleen of infected mice, and for their immune response. C. coli persisted well in both organs, with better survival in the liver. We also determined the levels of several pro-inflammatory cytokines (i.e., interleukin [IL]-6, IL-12, interferon-γ, tumor necrosis factor-α) and the anti-inflammatory cytokine IL-10 in plasma and in liver homogenates from the infected mice, using enzyme-linked immunosorbent assays. The lowest levels among these cytokines were for tumor necrosis factor-α in the plasma and IL-6 in the liver on days 1, 3 and 8 post-infection. The most pronounced production was for IL-10, in both plasma (days 1 and 8 post-infection) and liver (day 8 post-infection), which suggests that it has a role in healing of the organ inflammation. Our findings showed dynamic relationships between pro- and anti-inflammatory cytokines and thus contribute toward clarification of the healing processes involved in the resolution of C. coli infections.
Keywords: Campylobacter coli, cytokine profile, mouse model, virulence factors
Abbreviations
- CDT
cytolethal distending toxin
- CFU
colony forming units
- IFN
interferon
- IL
interleukin
- PCR
polymerase chain reaction
- TNF
tumor necrosis factor.
Introduction
Foodborne illnesses continue to present a public health challenge, and Campylobacter spp. represent the most common gastrointestinal bacterial pathogens in the European Union, where infection rates continue to increase.1 Human infections with Campylobacter spp most often arise as a direct result of consumption of chicken products that have been contaminated during processing2 and have not been cooked well enough. Intestinal campylobacteriosis manifests as enteritis of the terminal part of the ileum and colon. Patients can have mild to severe symptoms, which commonly include watery, and sometimes bloody, diarrhea, and intense abdominal pain, fever, headache and nausea. Infection is usually self-limiting and lasts only a few days.3
Although intestinal infection is rarely associated with a systemic illness, extra-intestinal manifestations or post-infection complications can occur, such as Guillain-Barré syndrome, glomerulonephritis, and reactive arthritis, especially in immunocompromised subjects.4 Both bacterial and host factors contribute to the differences in the clinical manifestations of such diseases in human.5 Campylobacter spp. have been reported to have different virulence factors, such as for motility, bacterial adherence to and invasion of epithelial cells, and production of toxins.5-8 Several pathogenic genes are responsible for these virulence properties, which include: cadF,9 which is responsible for adherence and colony formation; virB11,10 which is responsible for adherence and invasion; cdtB and cdtC,11-14 which are responsible for toxin production and have additional roles in adherence and invasion; and ceuE, which is responsible for hemolytic activity.15 However, it is well known that Campylobacter spp are naturally competitive, and combined with their high rate of recombination, this aspect can contribute to their genetic diversity.16 Although high prevalence of some virulence and toxin genes among Campylobacter spp. isolates has been noted, several isolates also show wide variations in the presence of these pathogenic genes.17,18 The mechanisms that induce this genetic diversity in Campylobacter spp still remain poorly understood, and thus the detection, and a better understanding of, these factors will allow the definition of the virulence factors associated with disease.18
The interactions of Campylobacter spp. with intestinal epithelial cells result in the production of cytokines, which are regulators of host responses to infection, and which have important roles in the pathogenesis of infectious diseases.19-21 Cytokines are produced by immune cells, and according to their function, they are divided into the pro-inflammatory cytokines, such as interleukin (IL)-1, IL-2, IL-6, IL-8, IL-12, IL-17 and IL-18, tumor necrosis factor (TNF)-α, and interferon (IFN)-γ, and the anti-inflammatory cytokines, such as IL-4, IL-10 and tumor growth factor-β.2,22 Both pro-inflammatory and anti-inflammatory cytokines are induced during Campylobacter spp infection, and it is important to determine whether the immune system is successful in providing protection against these specific pathogens.5 Some cytokines can be produced in excess, and thus these might contribute strongly to disease pathology.19 Indeed, it has been reported that macrophages produce IL-1α, IL-1β, IL-6, IL-8 and TNF-α when infected with Campylobacter spp..23 However, the pathogenesis of C. coli infection is still poorly understood, which is mainly a consequence of the lack of a suitable animal model that can be used to mimic the course of infection in human.24
We thus monitored the stability of the virulence and toxin genes (i.e., cadF, virB11, cdtB, cdtC, ceuE) by comparing the sequence data of C. coli 26536 before and after transmission through the host, and their colonisation and cytokine induction in a murine model. In previous studies, we showed that C. jejuni can successfully invade epithelial cells in vitro8,40 and produce systemic infection in vivo.25 Our earlier data obtained in vitro showed that C. jejuni can survive both in phagocytes (for 72 h) and in intestinal epithelial cells (for 96 h). This is in agreement with the conclusions of Day et al. (2000),41 who considered C. jejuni to be a facultative intracellular pathogen, and not a typical extracellular bacterium.
In the present study, we focused on the clearance from the liver and spleen of BALB/c mice infected with the C. coli 26536 clinical isolate, and the association between systemic and local cytokine responses following C. coli dissemination. The levels of several pro-inflammatory cytokines (i.e., IL-6, IL-12, TNF-α, IFN-γ) and one anti-inflammatory cytokine (i.e., IL-10) in plasma and liver homogenates were determined on days 1, 3 and 8 post-infection, using enzyme-linked immunosorbent assays. As the cytokine network is one of the main controlling elements in the inflammation process, we wanted to determine any correlation between cytokine production and course of infection.
Results
Presence of virulence genes in C. coli
The presence of the cadF, virB11, cdtB, cdtC and ceuE virulence genes in C. coli 26536 was analyzed using polymerase chain reaction (PCR) (Table 1). Of these, the cdtC gene was not detected in the C. coli 26536 strain (Table 2). Furthermore, we analyzed the stability of these genes by sequencing of the virulence genes cadF, virB11, cdtB and ceuE in C. coli 26536 after transmission through the mice. These examinations of the DNA sequence data of each of these individual genes allowed comparisons of the analyzed gene sequences in C. coli 26536 obtained after 48 h of cultivation on Columbia selective agar in a microaerobic atmosphere at 42°C, and in C. coli 26536 isolated from the spleen of infected BALB/c mice. We confirmed the stability of the tested cadF, virB11, cdtB and ceuE genes in C. coli 26536 (Table 2).
Table 1.
PCR primer pairs and conditions32
| Target | Primer pair | n-mer | Sequence (5′–3′) | Amplication size (bp) | PCR conditions | Reference |
|---|---|---|---|---|---|---|
| cadF | F2B | 20 | TGG AGG GTA ATT TAG ATA TG | 400 | 94°C, 1 min | Konkel et al., 1999 |
| R1B | 20 | CTA ATA CCT AAA GTT GAA AC | 45°C, 1 min (30 cycles) | |||
| 72°C, 3 min | ||||||
| virB11 | virB11 | 23 | GAA CAG GAA GTG GAA AAA CTA GC | 708 | 95°C, 2 min | Bacon et al., 2000 |
| virBR | 21 | TTC CGC ATT GGG CTA TAT G | 95°C, 30 s | |||
| 52°C, 30 s (35 cycles) | ||||||
| 72°C, 2 min | ||||||
| cdtB | VAT2 | 24 | GTT AAA ATC CCC TGC TAT CAA CCA | 495 | 94°C, 1 min | Pickett et al., 1996 |
| WMI-R | 24 | GTT GGC ACT TGG AAT TTG CAA GGC | 42°C, 2 min (30 cycles) | |||
| 72°C, 3 min | ||||||
| cdtC | WMI-F | 24 | TGG ATG ATA GCA GGG GAT TTT AAC | 555 | 94°C, 1 min | Pickett et al., 1996 |
| LPF-X | 24 | GTT GGC ACT TGG AAT TTG CAA GGC | 42°C, 2 min (30 cycles) | |||
| 72°C, 3 min | ||||||
| ceuE | COL1 | 24 | ATG AAA AAA TAT TTA GTT TTT GCA | 894 | 95°C, 30 s | Gonzales et al., 1997 |
| COL2 | 21 | ATT TTA TTA TTT GTA GCA GCG | 57°C, 30 s (30 cycles) | |||
| 72°C, 1 min |
Table 2.
Target genes and GenBank accession numbers of the C. coli 23536 isolate
|
C. coli 26536 |
|||||
|---|---|---|---|---|---|
| Virulence properties | Target gene | Isolate from −80°C | Isolate from infected mouse liver | GenBank accession No. | |
| Adherence, colonisation | cadF | X | X | KJ875966 | |
| Adherence, invasion | virB11 | X | X | KJ875960 | |
| Adherence, invasion, CDT toxin | cdtB | X | X | KJ875957 | |
| Adherence, invasion, CDT toxin | cdtC | ND | ND | / | |
| Hemolytic activity | ceuE | X | X | KJ875961 | |
X, gene present; ND, gene not detected.
Survival of C. coli 26536 in the livers and spleens of infected mice
BALB/c mice were infected intravenously with C. coli 26536 and the course of infection was monitored over 8 days, in terms of the bacterial colony forming units (CFU) in the livers and spleens of the infected mice. Infected mice did not show signs of diarrhea, but they were lethargic with bristled fur, and they appeared not to be in good health (e.g., also showing lack of appetite and shaking). On days 1, 3 and 8 post-infection, the mice were sacrificed and the culturability of the bacteria from their livers and spleens was determined.
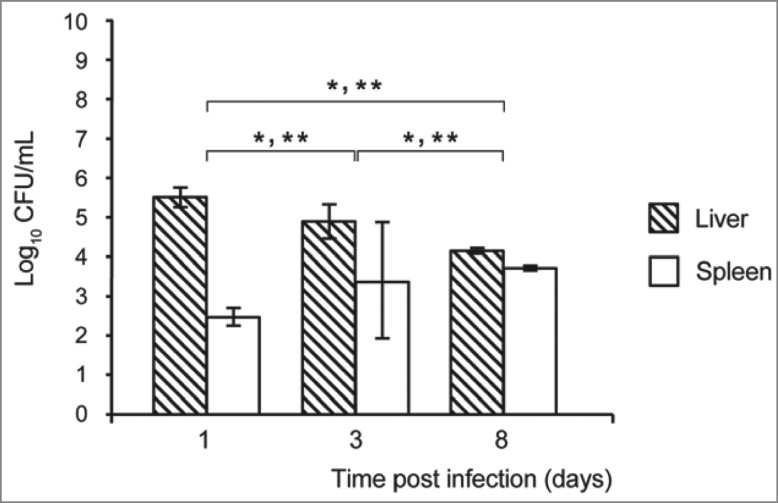
On day 3 post-infection, the livers of infected mice were enlarged and yellowish nodes were noted (Fig. 1). Microscopic examination of haematoxylin and eosin stained sections showed local tissue necroses and inflammatory infiltration, with neutrophils and rare plasma cells in the surrounding tissue, although bacteria could not be seen. These macroscopic liver changes were more prominent than those of the spleen. However, C. coli 26536 was detected in both livers and spleens of the infected mice through the entire experimental period (Fig. 2), although the growth patterns differed. At each time point, the CFU of C. coli 26536 isolated from the spleen was significantly lower than the CFU of C. coli 26536 recovered from the liver (P < 0.005). C. coli 26536 reached their maximal CFU in the liver on day 1 post-infection, which then slowly, but significantly, decreased through day 3 to day 8 post-infection (P < 0.05). In contrast, C. coli 26536 CFU in the spleen increased significantly through day 3 to day 8 post-infection (P < 0.05), which was the last monitored day of infection in the present study. Kupffer cells in the liver and macrophages, and other phagocytic cells in the spleen, appeared to provide a reservoir where C. coli survived for some time, and consequently were isolated from these organs up to day 8 post-infection.
Figure 1.
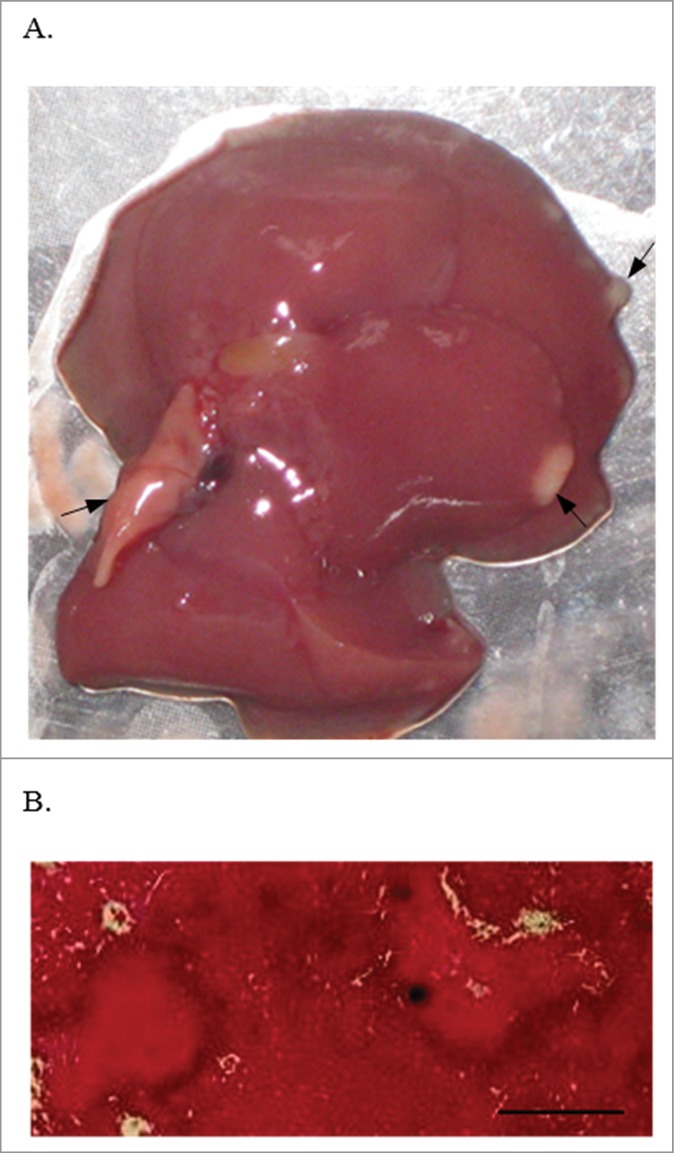
Liver from a BALB/c mouse on day 3 post intravenous infection with 0.5–1.0 × 109 CFU C. coli 26536. Macroscopic (A) and microscopic examination of haematoxylin and eosin stained liver section (B). Arrows, local tissue necrosis. Bar, 500 µm.
Figure 2.
Survival of C. coli 26536 in the liver and spleen (as indicated) of mice infected intravenously with 0.5–1.0 × 109 CFU C. coli 26536. At each indicated time, the mice were sacrificed, and the number of bacteria per organ was determined using the CFU assay. Each point represents the log10 of the median value ± standard deviation, as CFU/organ (* P ≤ 0.05).
Levels of different cytokines in the plasma and liver during C. coli 26536 infection of mice
The impact of C. coli 26536 on systemic cytokine production and in the liver of infected mice was studied, in comparison with the cytokine production patterns in the plasma and liver of non-infected mice. We determined the concentrations of the pro-inflammatory IL-6, IL-12, TNF-α and IFN-γ, and the anti-inflammatory IL-10, in the plasma and in liver homogenates on days 1, 3 and 8 post-infection (Fig. 3).
Figure 3.
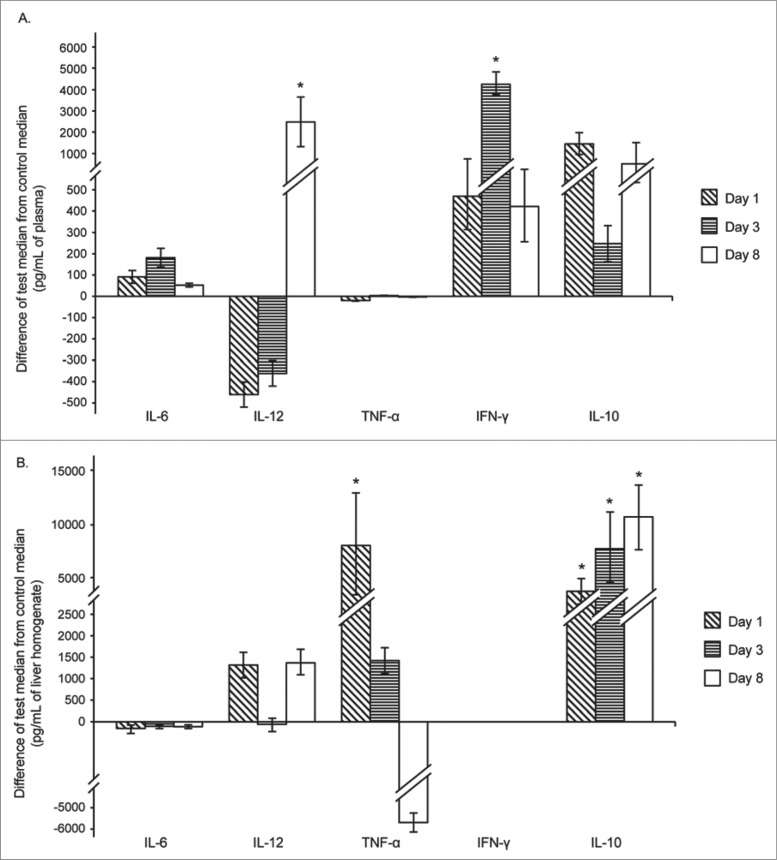
Differences from the control mice of the median cytokine levels (as indicated) in the plasma (A) and liver (B) of mice infected with C. coli 26536. Median control levels represent zero on the Y axis. Each bar represents the difference between the values for the infected mice minus those for the control mice, at each time point (as indicated) (* P < 0.05). Error bars represent ranges (maximum or minimum test value minus median control value). Cytokine levels in control mice were as follows: (A) plasma: IL-6, 12.83 pg/mL; IL-12, 1529.5 pg/mL; TNF-α, 27.61 pg/mL; IFN-γ, 42.84 pg/mL; IL-10, 37 pg/mL. (B) liver: IL-6, 853 pg/mL; IL-12, 1659.5 pg/mL; TNF-α, 14445 pg/mL; IFN-γ, 3300 pg/mL; IL-10, 1637.8 pg/mL.
When the mice were infected with C. coli 26536, the levels and the dynamics of the production of these investigated cytokines differed. The differences in the concentrations of IL-6 and TNF-α in the plasma between the infected and control mice were minimal throughout the experimental period. TNF-α in the plasma remained at about 22 pg/mL and IL-6 at about 100 pg/mL. On days 1 and 3 post-infection, the levels of IL-12 in the plasma tended to be lower in the infected mice than in the control mice, but by day 8 post-infection, IL-12 in the plasma was higher in the infected mice than in the control mice. However, for the plasma levels of the cytokines, on day 3 post-infection there were also significant differences between the mice infected with C. coli 26536 and the control mice, with IFN-γ significantly higher in the infected mice (Fig. 3A). At the same time, although the concentrations of IL-10 in the plasma of the infected mice were higher than in the control mice, these differences did not reach significance.
Different cytokine production patterns were revealed in the liver of the mice infected with C. coli 26536, as can be seen in Figure 3B. Here, when IL-10 was analyzed, there were marked differences between the infected and control mice through the entire period of infection. The concentrations of IL-10 in the liver showed an increasing trend, and reached the highest values on day 8 post-infection. In addition, there was a pronounced difference between the infected mice and the control mice for TNF-α production in the liver that was already apparent on day 1 of infection. By the last day of infection, day 8, this TNF-α concentration had dropped, and it was slightly lower in the infected mice compared to the control mice, although this difference was not significant. The differences in IL-6 production in the liver between the infected and control mice were minimal throughout the entire infection period, at around 50 pg/mL to 100 pg/mL in the infected and control mice, and these differences did not reach significance. Furthermore, no significant differences were seen between the infected mice and the control mice for IFN-γ in the liver. These results clearly show different cytokine patterns between systemic and local cytokine production during infection, probably linked to the repair of damage to the affected organs.
Discussion
The prevalence of intestinal campylobacteriosis is very high in developed countries, and this represents a great burden for public healthcare systems.1 Despite this, the pathogenesis of campylobacteriosis is still not completely understood for infections with both C. jejuni and, in particular, C. coli, although infections with C. coli are less frequent than those with C. jejuni. Most cases of campylobacteriosis are associated with eating raw or undercooked contaminated poultry or pork meat, or from cross-contamination of other foods by these items.28 Comparisons of pulsed-field gel electrophoresis and virulence profiles have shown high genetic diversity of the different strains examined, which leads us to believe that there can be different degrees of pathogenicity within any given Campylobacter spp population.29
More specifically considering the virulence mechanisms for the pathogenesis of Campylobacter spp. during human infections, these might contribute to their flagella-mediated motility, adherence to the intestinal mucosa, invasive capability, and production of toxins.6,7 Indeed, several potential virulence factors have been identified11,13-15,23 that show different prevalence according to the human, animal or environmental sources of the Campylobacter spp strain.18,30-32 One of these is the cadF gene, which encodes a protein belonging to the group of outer-membrane proteins that facilitates the binding of Campylobacter spp. in the host cells,33 as was confirmed by Campylobacter spp colonisation using a chicken model.34 Another virulence gene linked with Campylobacter spp. invasiveness is the invasion-associated marker virB11 gene, which is associated preferentially with both bacterial adherence and invasion.10 Campylobacter spp can also secrete a toxin known as cytolethal distending toxin (CDT), which is a complex of the ctd gene products CdtA, CdtB and CdtC. These three subunits act together to block cell division through cell-cycle arrest, and all 3 are necessary for the cytotoxin activity that is known to be lethal for host enterocytes.11,12 The ceuE gene encodes a lipoprotein component of a binding-protein-dependent transport system for the siderophore enterochelin.35 Iron acquisition is a crucial aspect of bacterial infectivity, and it has been suggested that this system has an important role in bacterial virulence.
In the present study, we initially investigated the presence of the cadF, virB11, cdtB, cdtC and ceuE genes in the C. coli 26536 clinical isolate using PCR. In this way, we confirmed the presence of the cadF, virB11, cdtB and ceuE genes, but not of the cdtC gene, in this C. coli 26536 isolate. Analysis of the cadF gene has revealed its high prevalence (i.e., 100%) among Campylobacter spp. isolates derived from poultry carcasses, broiler faeces, bovine faeces, retail raw meat, pig and cattle,18,30-32,36 and also among isolates of human origin.30,31,37 The cadF gene is probably conserved among Campylobacter spp isolates,18 regardless of their origin or species, as is the case also in the C. coli 26536 isolate used here. In contrast, several studies have shown wide variations for the presence of virB11, which was not found in C. jejuni and C. coli isolates from poultry faeces,18 although it was identified in 4% to 15% of isolates tested from poultry meat, pig and cattle,31,32,36 and in 10% to 17% of human clinical samples.31,38 However, the prevalence of the plasmid pVir appears to be similar among poultry and human isolates.17,31,38 The production of the pVir plasmid genes in the C. coli 26536 clinical isolate used in the present study thus indicates the possibility of severe invasive infection.
In recent studies, the distribution and prevalence of the CDT gene cluster of the cdtA, cdtB and cdtC genes in C. jejuni and C. coli isolates was investigated, and these genes were found in different ratios.30,32 The genes encoding the cdt gene cluster are widespread among poultry and human Campylobacter spp. isolates, with the cdtC gene showing higher prevalence among C. jejuni strains (93%–100%) than for C. coli (66%–100%), and with similar cdtB gene prevalence for both C. jejuni and C. coli (91%–100%).18,32,36,37 The absence of the PCR product of the cdtC gene in the clinical C. coli isolate in the present study is similar to data from previous studies, where C. jejuni can produce much higher levels of CDT in comparison to C. coli. The functional toxin is a tripartite complex in which CdtB is the active subunit that has an intrinsic DNase-I–like activity, while the CdtA and CdtC subunits are required for the delivery of CdtB into the host cell.7,11-13 Furthermore, the interactions of CdtA, CdtB and CdtC are necessary to form an active tripartite holotoxin that has full cellular toxicity.12 Hence, the absence of the cdtC gene indicates that C. coli 26536 cannot produce a fully active CDT toxin. The ceuE gene encodes a lipoprotein, and this has been detected in all Campylobacter spp isolates,32 and was also present in the clinical C. coli 26536 isolate in the present study, thus confirming the high prevalence of this virulence factor.
As determined above, studies of the prevalence of these virulence genes in isolates from various sources are essential to provide more information on the hazards of the Campylobacter spp. that might be circulating. As different determinants can influence the characteristics of these virulence determinants,39 we also followed the stability of the cadF, virB11, cdtB and ceuE genes that we detected in C. coli 26536 after transmission through a host. Here, we infected BALB/c mice with C. coli 26536 and then we collected the isolate from the spleen of the infected mice from day 3 post-infection. Each of the cadF, virB11, cdtB and ceuE genes was detected by PCR in isolates after storage conditions at −80°C, and again in isolates from the livers of these infected mice. Interestingly, all of these virulence genes were maintained in the C. coli isolates after the imitation food chain and after the transmission through the host mouse model. We further followed the stability in the cadF, virB11, cdtB and ceuE genes by comparing the sequence data for the isolates obtained through the imitation transition route using transmission through the host model. These sequence analyses showed that the sequence data of the virulence and toxin genes cadF, virB11, cdtB and ceuE remained the same in both of these isolates, and thus these genes were conserved through the transmission through the host, which supports their involvement in the virulence process.
It is well known that innate immunity is the first line of defense against microbial infection, and that this is required for the removal of the pathogen from the host.42,43 For colonisation and cytokine induction, we used the experimental model of murine campylobacteriosis, which has been adopted for the study of systemic infection and dissemination of Campylobacter spp, and the pathogenesis of Campylobacter spp. infection.25-27 We thus followed the systemic spread and infection for the internal organs (i.e., liver, spleen) after intravenous infection of BALB/c mice with this human clinical isolate of C. coli. We followed the course of infection and cytokine production.
After intravenous inoculation, C. coli 26536 established both liver and spleen infections, with better survival in the liver than the spleen of these infected mice. The CFU of C. coli isolated from the liver of the infected mice on day 8 post-infection was significantly lower in comparison to days 1 and 3 post-infection. This falling pattern of bacterial numbers isolated from the liver has also been seen previously in the liver of mice infected with C. jejuni.44 In contrast, the CFU of C. coli isolated from the spleen of the infected mice in the present study was increasing toward the last day post-infection (day 8). This might be explained by the potentially lower number of macrophages in the liver. Kupffer cells in the liver and macrophages and other cells in the spleen (i.e., neutrophils, eosinophils, monocytes) also have phagocytic functions.45 It has been shown that any prolongation of bacterial survival in macrophages increases the probability of their transmission within the host organism, and this has further implications in the pathogenesis of campylobacteriosis.46,47 Thus these cells might represent a reservoir, whereby the Campylobacter spp can survive for some time after intravenous inoculation in an experimental animal, which might be why C. coli was still isolated from the selected organs at day 8 post-infection.
During a systemic bacterial infection, numerous pro-inflammatory cytokines are produced, and their blood levels are elevated, although this greatly depends on the microbial characteristics and structure.43 Furthermore, the immune functions of cytokines result from the coordinated action of T cells, macrophages and dendritic cells, and these largely depend on the recruitment of these calls to the sites of infection and inflammation, and to other pathological lesions.48 Previously, Hu et al. (2006)49 demonstrated that C. jejuni infection can trigger innate inflammatory responses, and induce human dendritic cell maturation and production of pro-inflammatory cytokines, including IL-6, IL-8, IL-12 and TNF-α. In this way, cytokines can induce Campylobacter-specific Th1 effector-cell responses, and contribute to the pathogenesis and clinical symptoms of Campylobacter spp. infection. However, the molecular mechanisms underlying the dendritic-cell-mediated induction of adaptive immune responses to C. jejuni are not known.50 Interestingly, in the present study, the numbers of bacteria infecting the liver and spleen did not completely correlate with the cytokine profiles in the plasma and liver. Pro-inflammatory cytokines persisted during the whole infection period, both in the plasma and the liver, although C. coli 26536 did not provoke the production of IFN-γ in the liver, and there was only weak production of IL-12. In addition, high TNF-α production on day 1 post-infection was accompanied by the lowest production of the anti-inflammatory IL-10 in the liver of the infected mice, although not in their plasma.
The spectrum of cytokines in the plasma did not completely correlate with the cytokine profile in the liver. In plasma, for example, the levels of TNF-α were similar to those in the control, non-infected, mice during the whole period of infection, but in the liver, TNF-α peaked on day 1 post-infection and then dropped to even below the level of TNF-α in the control mice. Previous studies have demonstrated central roles for IFN-γ, IL-1 and TNF-α in inflammatory reactions, which can lead to the eradication of obligate and facultative intracellular pathogens.51,52 TNF-α has been shown to have a role in limiting the severity of bacterial infections through various mechanisms, which include the selective killing of cells that harbour bacteria, activation of monocytes and granulocytes, and stimulation of specific immune responses. Particularly in infections with facultative intracellular bacteria, anti-TNF-α antibody treatment promoted massive proliferation of the infecting organisms. These data suggest that the ability to inhibit TNF-α is advantageous to the pathogen.53 Unexpectedly, in the present study, C. coli did not show this effect: in spite of the extremely low concentrations of TNF-α in the liver of infected mice on day 8 post-infection, the number of C. coli isolated also dropped.
Overall, following the infections with C. coli, the levels of IL-6 for both the plasma and the liver of the infected mice were almost the same as in the control mice during the entire course of the infection. Infection of the mice with C. coli 26536 provoked the greatest production of IFN-γ in the plasma on day 3 post-infection. IFN-γ is known to enhance major histocompatibility complex class I and II expression on nucleated cells and to stimulate many effector functions of mononuclear phagocytes. In addition, IFN-γ induces the expression of many adhesion proteins on the vascular endothelium, to potentiate the function of TNF-α and to promote T-lymphocyte infiltration at sites of infection. For this reason, these high levels of TNF-α in the liver at the beginning of the infection and IFN-γ in the plasma might indicate the activation of macrophages and Th1 cells.5 Our data show time-dependent secretion of IL-12 and an increase in IL-12 production toward day 8 post-infection in the plasma of the infected mice. Secreted IL-12 can enhance CD4+ T-cell proliferation and IFN-γ production, although this was not seen here. In addition, recent studies have shown that IL-12 mediates dendritic cell activation of natural killer cells, and in this way contributes to initial resistance to bacterial pathogens, and also controls reinfection of C. jejuni in mice.54-56 High levels of production of IL-12 initiate a Th1 polarized adaptive immune response.49
Interleukin-10 functions as an anti-inflammatory cytokine, and it is known to suppress production of pro-inflammatory cytokines such as IL-12, TNF-α and IFN-γ. In addition, its relationship with some pro-inflammatory cytokines is important for the outcome of infection and sepsis.48 In the present study, the first cytokine that began to rise in the liver of the infected mice was IL-10. C. coli 26536 induced the strongest anti-inflammatory IL-10 response in the liver by the last day of infection (day 8). As indicated above, there was an early rise in TNF-α in the liver, which is as expected, as TNF-α is a prerequisite for the production of other pro-inflammatory cytokines. On the other hand, an early increase in IL-10 is hard to explain, especially as it occurred simultaneously with the rise in the pro-inflammatory cytokines, as seen in the liver of the infected mice. This will need further investigation for a systematic explanation. However, this simultaneous production of both pro-inflammatory and anti-inflammatory cytokines has also been demonstrated in an in-vitro cell culture model of the INT407 human intestinal cell line.57
In conclusion, along with C. jejuni, C. coli are one of the most common bacteria that cause bacterial gastroenteritis and acute enterocolitis in human in developed countries. Despite its obvious global importance, relatively little is known regarding the mechanisms of pathogenesis and host response to C. coli infection. In the present study, we have confirmed the presence of the cadF, virB11, cdtB and ceuE virulence genes in this C. coli 26536 isolate, and we monitored the stable virulence and toxin genes following C. coli transmission through a host, as such differences might influence Campylobacter spp colonisation and cytokine induction in an in-vivo model. Furthermore, we used our previously established experimental model of systemic campylobacteriosis in mice. In this way, we followed the course of infection and cytokine production after intravenous challenge of BALB/c mice with this human clinical C. coli 26536 isolate. The levels of several pro-inflammatory cytokines (IL-6, IL-12, TNF-α, IFN-γ) and one anti-inflammatory cytokine (IL-10) in plasma and liver homogenate were determined on days 1, 3 and 8 post-infection using enzyme-linked immunosorbent assays. The C. coli produced systemic infection in these immunocompetent BALB/c mice, but did not induce production of the pro-inflammatory cytokine IL-6, either in the plasma or in the liver of the infected mice. In contrast, in these infected mice, the anti-inflammatory cytokine IL-10 showed markedly greater production in the liver, with dynamics that correlated with the declining number of C. coli in the liver. It is well known that the cytokine network is one of the main controlling elements in the inflammation and immune reactions that occur during bacterial infection. Dynamic relationships between pro- and anti-inflammatory cytokines, their rates of production, and their levels in different tissues are among the variables in the control of healing processes and the resolution of infections. Thus, it is important to determine any correlations between cytokine production and the course of infection. Here, our investigations have contributed further toward the clarification of these relationships.
Materials and Methods
Bacterial isolate
The C. coli 26536 clinical isolate was provided by the Laboratory for Diagnostics of Enteric Infections of the Teaching Institute of Public Health of the Primorsko-Goranska county, Croatia, and was previously characterized.58 The isolate was then stored at −80°C in brain–heart infusion broth supplemented with blood (5%, Oxoid, SR0048) and glycerol (10%, Kemika, 0709704) until use, and cultured using microaerobic growth (5% O2, 10% CO2, 85% N2) in Preston broth (Oxoid, CM0689) at 42°C.59
Virulence genes and sequence analysis
The cadF gene encodes adhesin, which is responsible for certain steps in cell invasion, while the virB11 gene is a marker for the pVir plasmid, the cdtB and cdtC genes are involved in CDT production, and the ceuE gene encodes a lipoprotein that is associated with haemolysins. These five genes were assessed using PCR. For the present analysis using PCR and for the sequence analysis, we used C. coli 26536 revitalised on Columbia selective agar (Oxoid, CM0331), after storage at −80°C. This isolate was further used to infect BALB/c mice, and then recovered from the mouse spleen on day 3 post-infection.
The primer sequences of the virulence and toxin genes, the expected sizes of their products, and the cycling conditions for the determination of the presence of these 5 putative virulence factors cadF, virB11, cdtB, cdtC and ceuE of C. coli are listed in Table 1. Bacterial DNA was extracted using the PrepMan Ultra sample preparation reagent (Life Technologies, 4318930), following the manufacturer recommendations, and the extracted DNA preparations were stored at −20°C. PCR amplifications were performed in 50 µL reaction volumes that contained 10× RED Taq PCR buffer (Sigma-Aldrich GmbH, B5926), 20 mM dNTP (Life Technologies, N8080007), 300 nM forward primer, 300 nM reverse primer (Table 1), 1 U/µL RED Taq polymerase (Sigma-Aldrich GmbH, D4309), and 5 µL DNA lysate. The PCR was performed in a thermal cycler PCR system (2400 GeneAmp; Perkin Elmer). The cycling conditions varied according to the specific gene determination (Table 1). The PCR products were electrophoresed on 2% agarose gels. The virulence genes cdtB, virB11, cadF and ceuE of C. coli were further investigated by sequence analysis (Macrogen), to study the genetic differences of this C. coli isolate following the storage conditions, after sub-cultivation in the food model, and after isolation from the infected mice. The sequences of cdtB, cdtC, cadF and ceuE have been deposited in GenBank, with their accession numbers given in Table 2.
C. coli 26536 infection in mice
Eight to 12-week-old BALB/c (H-2d) mice were obtained from the Central Animal Facility of the Medical Faculty, University of Rijeka, and the experiments were conducted according to the Guidelines of the International Guiding Principles for Biomedical Research Involving Animals (NCR, 2004). The Ethical Committee of the University of Rijeka approved all of the animal experiments described here.
The mice were given standard laboratory rodent food (Mucedola, 4RF21GLP) and water ad libitum. They were infected intravenously via the lateral tail vein with a single dose (200 µL) of 0.5–1.0 × 109 CFU C. coli 26536 cells, as determined by the turbidity of the bacterial suspension and confirmed retrospectively by plating the inoculum on blood agar and incubating them microaerobically for 48 h at 42°C. On days 1, 3 and 8 post-infection, the mice were anaesthetised with sodium pentobarbital and sacrificed, and their livers and spleens were removed aseptically. The C. coli 26536 CFU in these organs were determined as previously described.25 At least 6 mice per time point were infected. For organ burden determination, at least 3 mice were sacrificed per time point, and at least another 3 for cytokine analyses. All of the experiments were repeated 3 times and the data from all of the replicate experiments were pooled and are presented as means ± standard deviation.
Cytokine analysis
For the determination of the cytokine concentrations, mice were anaesthetised with sodium pentobarbital and sacrificed, with their blood collected in sterile vials (Eppendorf AG, 00298–00) that contained ethylendiamine tetra-acetic acid. The tubes were centrifuged, and the plasma was separated and stored at −20°C until assayed. The livers of these mice were removed aseptically, dissected from the surrounding tissue, and collected on ice in preweighed sterile vials (Eppendorf AG). The weights of the livers were recorded using an electronic balance (PB602-S). The livers were then frozen in liquid nitrogen, and stored at −80°C.
The livers were homogenized and the organ supernatants collected as previously described.44 Briefly, immediately before analysis, the livers were thawed on ice and homogenized in ice-cold phosphate-buffered saline (5 mL per 1 g tissue, wet weight) using a hand-held tissue homogenizer. Following homogenization, the liver samples were centrifuged (14,000 × g, 10 min, 4°C) to precipitate the cell debris as a pellet. These supernatants were stored at −20°C until they were assayed for cytokine levels. The levels of IL-6, IL-10, IL-12, TNF-α and IFN-γ were determined in duplicate, using mouse cytokine enzyme-linked immunosorbent assay kits (Thermo Scientific, EM2IL6, EM2IL10, EMIL12TOT2, EMTNFA5, ESS0020), and the data are expressed in pg/mL plasma or tissue homogenate, as described previously.44 The sensitivity levels were: IL-6, 7 pg/mL; IL-10, 12 pg/mL; TNF-α, 9 pg/mL; and IFN-γ, 10 pg/mL. Control mice of the same age and sex were injected with sterile saline. All of the experiments were independently repeated 3 times, and the data are presented as means ± standard deviation.
Statistical analysis
The differences for the CFU in the organs and the cytokine levels in the plasma and liver homogenates were calculated using Kruskal Wallis tests. Mann Whitney tests were used to verify differences between pairs of groups. Differences were considered significant at the P level of 0.05. Statistical analysis was performed using SPSS 15.0 for Windows (Statsoft Inc.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by the Ministry of Higher Education, Science and Technology of the Republic of Slovenia, by the Z1–2190 post-doctoral project of AK, by research program P4–0116, and through the research project Molecular Mechanisms of Bacterial Pathogenesis and Stress Response, supported by the University of Rijeka, Croatia.
References
- 1.European Food Safety Authority The European Union report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA J 2013; 11:3129 [Google Scholar]
- 2.Young KT, Davis LM, DiRita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol 2007; 5(9):665-79; PMID:17703225; http://dx.doi.org/ 10.1038/nrmicro1718 [DOI] [PubMed] [Google Scholar]
- 3.Haddad N, Marce C, Magras C, Cappelier JM. An overview of methods used to clarify pathogenesis mechanisms of Campylobacter jejuni. J Food Protect 2010; 73(4):786-802; PMID:20377972 [DOI] [PubMed] [Google Scholar]
- 4.Zilbauer M, Dorrell N, Wrenb BW, Bajaj-Elliott M. Campylobacter jejuni-mediated disease pathogenesis: an update. Trans R Soc Trop Med Hyg 2008; 102(2):123-9; PMID:18023831; http://dx.doi.org/ 10.1016/j.trstmh.2007.09.019 [DOI] [PubMed] [Google Scholar]
- 5.Al-Banna N, Raghupathy R, Albert MJ. Correlation of proinflammatory and anti-inflammatory cytokine levels with histopathological changes in an adult mouse lung model of Campylobacter jejuni infection. Clin Vacc Immunol 2008; 15(12):1780-7; PMID:18827187; http://dx.doi.org/ 10.1128/CVI.00193-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ketley JM. Pathogenesis of enteric infection by Campylobacter. Microbiology 1997; 143:5-21; PMID:9025274; http://dx.doi.org/ 10.1099/00221287-143-1-5 [DOI] [PubMed] [Google Scholar]
- 7.Wassenaar TM. Toxin production by Campylobacter spp.. Clin Microbiol Rev 1997; 10(3):466-76; PMID:9227862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubeša Mihaljević R, Šikić Pogačar M, Klančnik A, Brumini G, Smole Možina S, Abram M. Environmental stress factors affecting survival and virulence of Campylobacter jejuni. Microb Pathogen 2007; 2/3:120-5; PMID:17512161; http://dx.doi.org/ 10.1016/j.micpath.2007.03.004 [DOI] [PubMed] [Google Scholar]
- 9.Ziprin RL, Young CR, Byrd JA, Stanker LH, Hume ME, Gray SA, Kim BJ, Konkel ME. Role of Campylobacter jejuni potential virulence genes in cecal colonization. Avian Dis 2001; 45(3):549-57; PMID:11569726; http://dx.doi.org/ 10.2307/1592894 [DOI] [PubMed] [Google Scholar]
- 10.Bacon DJ, Alm RA, Burr DH, Hu L, Kopecko DJ, Ewing CP, Trust TJ, Guerry P. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect Immun 2000; 68(8):4384-90; PMID:10899834; http://dx.doi.org/ 10.1128/IAI.68.8.4384-4390.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickett CL, Pesci EC, Cottle DL, Russell G, Erdem AN, Zeytin H. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB gene. Infect Immun 1996; 64(6):2070-8; PMID:8675309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lara-Tejero M, Galán JE. CdtA, CdtB, and CdtC form a tripartite complex that is required for cytolethal distending toxin activity. Infect Immun 2001; 69(7):4358-65; PMID:11401974; http://dx.doi.org/ 10.1128/IAI.69.7.4358-4365.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konkel ME, Monteville MR, Rivera-Amill V, Joens LA. The pathogenesis of Campylobacter jejuni-mediated enteritis. Curr Iss Intest Microbiol 2001; 2(2):55-71; PMID:11721281 [PubMed] [Google Scholar]
- 14.Jain D, Prasad KN, Sinha S, Husain N. Differences in virulence attributes between cytolethal distending toxin positive and negative Campylobacter jejuni strains. J Med Microbiol 2008; 57:267-72; PMID:18287287; http://dx.doi.org/ 10.1099/jmm.0.47317-0 [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez I, Grant KA, Richardson PT, Park SF, Collins MD. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli using a PCR test based on the ceuE gene encoding a putative virulence determinant. J Clin Microbiol 1997; 35(3):759-63; PMID:9041429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dingle KE, Colles FM, Wareing DR, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJ, Urwin R, Maiden MC. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol 2001; 39(1):14-23; PMID:11136741; http://dx.doi.org/ 10.1128/JCM.39.1.14-23.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Deun K, Haesebrouck F, Heyndrickx M, Favoreel H, Dewulf J, Ceelen L, Dumez L, Messens W, Leleu S, Van Immerseel F, et al.. Virulence properties of Campylobacter jejuni isolates of poultry and human origin. J Med Microbiol 2007; 56:1284-9; PMID:17893162; http://dx.doi.org/ 10.1099/jmm.0.47342-0 [DOI] [PubMed] [Google Scholar]
- 18.Khoshbakht R, Tabatabaei M, Hosseinzadeh S, Shekarforoush SS, Aski SH. Distribution of nine virulence-associated genes in Campylobacter jejuni and C. coli isolated from broiler feces in Shiraz, southern Iran. Foodborne Pathog Dis 2013; 10(9):764-70; PMID:23789768; http://dx.doi.org/ 10.1089/fpd.2013.1489 [DOI] [PubMed] [Google Scholar]
- 19.Smith CK, Kaiser P, Rothwell L, Humphrey T, Barrow PA, Jones MA. Campylobacter jejuni - induced cytokine responses in avian cells. Infect Immun 2005; 73(4):2094-2100; PMID:15784550; http://dx.doi.org/ 10.1128/IAI.73.4.2094-2100.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janssen R, Krogfelt KA, Cawthraw SA, van Pelt W, Wagenaar JA, Owen RJ. Host-pathogen interactions in Campylobacter infections: the host perspective. Clin Microbiol Rev 2008; 21(3):505-18; PMID:18625685; http://dx.doi.org/ 10.1128/CMR.00055-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinarello CA. Proinflammatory cytokines. Chest J 2000; 118(2):503-8; http://dx.doi.org/ 10.1378/chest.118.2.503 [DOI] [PubMed] [Google Scholar]
- 22.Bashashati M, Rezaei N, Andrews CN, Chen CQ, Daryani NE, Sharkey KA, Storr MA. Cytokines and irritable bowel syndrome: where do we stand? Cytokine 2012; 57:201-9; PMID:22178716; http://dx.doi.org/ 10.1016/j.cyto.2011.11.019 [DOI] [PubMed] [Google Scholar]
- 23.Hu L, Kopecko DJ. Cell biology of human host cell entry by Campylobacter jejuni In: Campylobacter. Nachamkin I, Syzmanski CM, Blaser MJ ed; Washington, DC: ASM Press; 2008. pp:297-315 [Google Scholar]
- 24.Lane JA, Mehra RK, Carrington SD, Hickey RM. The food glycome: a source of protection against pathogen colonization in the gastrointestinal tract. Int J Food Microbiol 2010; 142:1-13; PMID:20580113; http://dx.doi.org/ 10.1016/j.ijfoodmicro.2010.05.027 [DOI] [PubMed] [Google Scholar]
- 25.Vučković D, Abram M, Dorić M. Primary Campylobacter jejuni infection in different mice strains. Microb Pathog 1998; 24:263-8; PMID:9533898; http://dx.doi.org/ 10.1006/mpat.1997.0194 [DOI] [PubMed] [Google Scholar]
- 26.Vučković D, Abram M, Bubonja M, Wraber B, Dorić M. Host resistance to primary and secondary Campylobacter jejuni infections in C57Bl/6 mice. Microb Pathog 2006; 40(1):35-9; PMID:16324818; http://dx.doi.org/ 10.1016/j.micpath.2005.10.004 [DOI] [PubMed] [Google Scholar]
- 27.Trevijano-Contador N, Zaragoza O. Expanding the use of alternative models to investigate novel aspects of immunity to microbial pathogens. Virulence 2014; 5(4):454-6; PMID:24717215; http://dx.doi.org/ 10.4161/viru.28775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smole Možina S, Kurinčič M, Klančnik A, Mavri A. Campylobacter and its multi-resistance in the food chain. Trends Food Sci Technol 2011; 22:91-8; http://dx.doi.org/ 10.1016/j.tifs.2010.09.003 [DOI] [Google Scholar]
- 29.Giannatale E, Serafino G, Zilli K, Alessiani A, Sacchini L, Garofolo G, Aprea G, Marotta F. Characterization of antimicrobial resistance patterns and detection of virulence genes in Campylobacter isolates in Italy. Sensors 2014; 14(2):3308-22; PMID:24556669; http://dx.doi.org/ 10.3390/s140203308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozynek E, Dzierzanowska-Fangrat K, Jozwiak P, Popowski J, Korsak D, Dzierzanowska D. Prevalence of potential virulence markers in Polish Campylobacter jejuni and Campylobacter coli isolates obtained from hospitalized children and from chicken carcasses. J Med Microbiol 2005; 54:615-9; PMID:15947425; http://dx.doi.org/ 10.1099/jmm.0.45988-0 [DOI] [PubMed] [Google Scholar]
- 31.Datta S, Niwa H, Itoh K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J Med Microbiol 2003; 52:345-8; PMID:12676874; http://dx.doi.org/ 10.1099/jmm.0.05056-0 [DOI] [PubMed] [Google Scholar]
- 32.Bang DD, Nielsen EM, Scheutz F, Pedersen K, Handberg K, Madsen M. PCR detection of seven virulence and toxin genes of Campylobacter jejuni and Campylobacter coli isolates from Danish pigs and cattle and cytolethal distending toxin production of the isolates. J Appl Microbiol 2003; 94:1003-14; PMID:12752808; http://dx.doi.org/ 10.1046/j.1365-2672.2003.01926.x [DOI] [PubMed] [Google Scholar]
- 33.Konkel ME, Kim BJ, Rivera-Amill V, Garvis SG. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mammalian cells. Mol Microbiol 1999; 32(4):691-701; PMID:10361274; http://dx.doi.org/ 10.1046/j.1365-2958.1999.01376.x [DOI] [PubMed] [Google Scholar]
- 34.Ziprin RL, Young CR, Stanker LH, Hume ME, Konkel ME. The absence of cecal colonization of chicks by a mutant of Campylobacter jejuni not expressing bacterial fibronectin-binding protein. Avian Dis 1999; 43(3):586-9; PMID:10494431; http://dx.doi.org/ 10.2307/1592660 [DOI] [PubMed] [Google Scholar]
- 35.Park SF, Richardson PT. Molecular characterization of Campylobacter jejuni lipoprotein with homology to periplasmic siderophore-binding proteins. J Bacteriol 1995; 177(9):2259-64; PMID:7730251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieczorek K, Szewczyk R, Osek J. Prevalence, antimicrobial resistance, and molecular characterization of Campylobacter jejuni and C. coli isolated from retail raw meat. Poland Veterinarni Medicina 2012; 57(6):293-9 [Google Scholar]
- 37.Rizal A, Kumar A, Vidyarthi AS. Prevalence of pathogenic genes in Campylobacter jejuni isolated from poultry and human. Int J Food Saf 2010; 12:29-34 [Google Scholar]
- 38.Tracz DM, Keelan M, Ahmed-Bentley J, Gibreel A, Kowalewska-Grochowska K, Taylor DE. pVir and bloody diarrhea in Campylobacter jejuni enteritis. Emerg Infect Dis 2005; 11:838-43; PMID:15963277; http://dx.doi.org/ 10.3201/eid1106.041052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ganan M, Campos G, Muñoz R, Carrascosa AV, de Pascual-Teresa S, Martinez-Rodriguez AJ. Effects of growth phase on the adherence to and invasion of Caco-2 epithelial cells by Campylobacter. Int J Food Microbiol 2010; 140(1):14-8; PMID:20223546; http://dx.doi.org/ 10.1016/j.ijfoodmicro.2010.02.021 [DOI] [PubMed] [Google Scholar]
- 40.Šikić Pogačar M, Klančnik A, Smole Možina S, Cencič A. Attachment, invasion and translocation of Campylobacter jejuni in pig small intestinal epithelial cells. Foodborne Pathog Dis 2010; 7:589-95; PMID:20039793; http://dx.doi.org/ 10.1089/fpd.2009.0301 [DOI] [PubMed] [Google Scholar]
- 41.Day WA Jr, Sajecki JL, Pitts TM, Joens LA. Role of catalase in Campylobacter jejuni intracellular survival. Infect Immun 2000; 68(11):6337-45; http://dx.doi.org/ 10.1128/IAI.68.11.6337-6345.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aderem A. Phagocytosis and the inflammatory response. J Infect Dis 2003; 187(2):S340-345; http://dx.doi.org/ 10.1086/374747 [DOI] [PubMed] [Google Scholar]
- 43.Akira S, Uematsu A, Takeuch O. Pathogen recognition and innate immunity. Cell 2006; 124:783-801; http://dx.doi.org/ 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- 44.Klančnik A, Vučković D, Plankl M, Abram M, Smole Možina S. In-vivo modulation of Campylobacter jejuni virulence in response to environmental stress. Foodborne Pathog Dis 2013; 10(6):566-72.; http://dx.doi.org/ 10.1089/fpd.2012.1298 [DOI] [PubMed] [Google Scholar]
- 45.Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nature Immunol 2013; 14:986-95; http://dx.doi.org/ 10.1038/ni.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lastovica AJ. Emerging Campylobacter spp: the tip of the iceberg. Clin Microbiol Newsletter 2006; 28(7):49-56; http://dx.doi.org/ 10.1016/j.clinmicnews.2006.03.004 [DOI] [Google Scholar]
- 47.Wassenaar TM, Engelskirchen M, Park S, Lastovica AJ. Differential uptake and killing potential of Campylobacter jejuni by human peripheral monocytes/macrophages. Med Microbiol Immun 1997; 186(2-3):139-44; http://dx.doi.org/ 10.1007/s004300050056 [DOI] [PubMed] [Google Scholar]
- 48.Zídek Z, Anzenbacher P, Kmoníčková E. Current status and challenges of cytokine pharmacology. Br J Clin Pharmacol 2009; 157(3):342-61; http://dx.doi.org/ 10.1111/j.1476-5381.2009.00206.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu L, Bray MD, Osorio M, Kopecko DJ. Campylobacter jejuni induces maturation and cytokine production in human dendritic cells. Infect Immun 2006; 74(5):2697-705; http://dx.doi.org/ 10.1128/IAI.74.5.2697-2705.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu L, Bray MD, Geng Y, Kopecko DJ. Campylobacter jejuni - mediated induction of CC and CXC chemokines and chemokine receptors in human dendritic cells. Infect Immun 2012; 80(8):2929-39; http://dx.doi.org/ 10.1128/IAI.00129-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mielke MEA, Ehlers S, Hahn H. The role of cytokines in experimental listeriosis. Immunobiol 1993; 189(3-4):285-315; http://dx.doi.org/ 10.1016/S0171-2985(11)80363-3 [DOI] [PubMed] [Google Scholar]
- 52.Havell EA. Role of TNF-α in resistance to bacteria. Immunol Ser 1992; 56:341-63 [PubMed] [Google Scholar]
- 53.Beuscher HU, Rodel F, Forsberg A, Rollinghoff M. Bacterial evasion of host immune defense: Yersinia enterocolitica encodes a suppressor for tumor necrosis factor alpha expression. Infect Immun 1995; 63(4):1270-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mori S, Jewett A, Cavalcanti M, Murakami-Mori K, Nakamura S, Bonavida B. Differential regulation of human NK cell-associated gene expression following activation by IL-2, IFN-alpha and PMA/ionomycin. Int J Oncol 1998; 12(5):1165-70 [DOI] [PubMed] [Google Scholar]
- 55.Hafsi N, Voland P, Schwendy S, Rad R, Reindl W, Gerhard M, Prinz C. Human dendritic cells respond to Helicobacter pylori, promoting NK cell and Th1-effector responses in vitro. J Immunol 2004; 173(2):1249-57; http://dx.doi.org/ 10.4049/jimmunol.173.2.1249 [DOI] [PubMed] [Google Scholar]
- 56.Rathinam VAK, Hoag KA, Mansfield LS. Dendritic cells from C57BL/6 mice undergo activation and induce Th1-effector cell responses against Campylobacter jejuni. Microb Infect 2008; 10(12-13):1316-24; http://dx.doi.org/ 10.1016/j.micinf.2008.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al-Salloom FS, Al Mahmeed A, Ismaeel A, Botta GA, Bakhiet M. Campylobacter-stimulated INT407 cells produce dissociated cytokine profiles. J Infect 2003; 47(3):217-24; http://dx.doi.org/ 10.1016/S0163-4453(03)00076-8 [DOI] [PubMed] [Google Scholar]
- 58.Zorman T, Smole Možina S. Classical and molecular identification of thermotolerant campylobacters from poultry meat. Food Technol Biotechnol 2002; 40:177-83 [Google Scholar]
- 59.Klančnik A, Botteldoorn N, Herman L, Smole Možina S. Survival and stress-induced expression of groEL and rpoD of Campylobacter jejuni from different growth phases. Int J Food Microbiol 2006; 112(3):200-7; http://dx.doi.org/ 10.1016/j.ijfoodmicro.2006.03.015 [DOI] [PubMed] [Google Scholar]