Abstract
Widespread and repeated use of azoles has led to the rapid development of drug resistance in Candida albicans. Our previous study found Rta2p, a membrane protein with 7 transmembrane domains, was involved in calcineurin-mediated azole resistance and sphingoid long-chain base release in C. albicans. Conserved amino acids in the transmembrane domain of Rta2p were subjected to site-directed mutagenesis. The sensitivity of C. albicans to fluconazole in vitro was examined by minimum inhibitory concentration and killing assay, and the therapeutic efficacy of fluconazole in vivo was performed by systemic mice candidiasis model. Furthermore, dihydrosphingosine transport activity was detected by NBD labeled D-erythro-dihydrosphingosine uptake and release assay, and the sensitivity to sphingolipid biosynthesis inhibitors. We successfully constructed 14 mutant strains of Rta2p, screened them by minimum inhibitory concentration and found Ca2+ did not completely induce fluconazole resistance with G158E and G234S mutations. Furthermore, we confirmed that G234S mutant enhanced the therapeutic efficacy of fluconazole against systemic candidiasis and significantly increased the accumulation of dihydrosphingosine by decreasing its release. However, G158E mutant didn't affect drug therapeutic efficacy in vivo and dihydrosphingosine transport in C. albicans. G234 of Rta2p in C. albicans is crucial in calcineurin-mediated fluconazole resistance and dihydrosphingosine transport.
Keywords: Candida albicans, calcineurin pathway, drug resistance, dihydrosphingosine, Rta2p, functional sites
Introduction
Candida albicans is a commensal fungus colonizing the skin, mucosal surface and gastrointestinal tract in most healthy individuals. While in immunocompromised patients, C. albicans can cause oropharyngeal, esophageal or disseminated candidiasis.1,2 However, widespread and incorrect use of azoles in clinical have resulted in the fast development of multidrug resistance, which has been a tricky problem in the treatment of fungal infections.3,4 In recent years, a diverse range of drug resistance mechanisms have been identified, including multi-drug resistance efflux gene (MDR1) as well as CDR-type drug efflux pumps (CDR1 and CDR2),5-7 mutation of lanosterol 14-α demethylase Erg11p,8 and activation of calcineurin pathway.9,10 Previous studies indicated that some efflux pumps were found to be over-expressed in the fluconazole (FLC) resistance clinical C. albicans isolates from 12 hospitals in California.11
The protein information of C. albicans genome database indicates that RTA2 encodes a 50.9 kDa membrane protein, which is predicted to have 7 transmembrane domains. There are 4 Rta2p homologs (Rsb1p, Rta1p, Yer185w, and Ylr046c) in the S. cerevisiae genome database, with more than 40% similarity with RTA2.12 It was found in our previous study that disruption of RTA2 could increase the sensitivity to azoles in C. albicans, and the azole resistance was mediated in calcineurin by Rta2p and the function of Rta2p was to release sphingoid long chain base (LCB).13 Elevated RTA2 expression levels were also found in clinical isolates of azole-resistant C. albicans.12 As Rta2p is considered a potential target to combat antifungal resistance in clinical, the relationship between structure and function is important for designing antifungal drug targeting Rta2p. In this work, we developed and selected conserved amino acids in the transmembrane domains of Rta2p for site-directed mutagenesis, finding that G234 of RTA2 played a critical role in FLC resistance and the transport of dihydrosphingosine (DHS) in C. albicans.
Materials and Methods
Drugs
FLC was purchased from Changhai Hospital (Shanghai, China). Bovine serum albumin (BSA), fumonisin B1 and myriocin were from Sigma-Aldrich (St. Louis, MO, USA). C6 NBD labeled D-erythro-dihydrosphingosine (NBD-DHS) was obtained from Larodan Fine Chemical AB (Sweden).
C. albicans strains growth conditions
C. albicans strains used in this study were maintained on SDA agar plates (1% peptone, 4% dextrose, and 1.8% agar) and grown in YPD broth (1% yeast extract, 2% peptone, and 2% dextrose) at 30°C or SC medium lacking uridine.
Site-directed mutation and C. albicans transformation
Site-directed mutation was performed using the QuikChange® lightning site-directed mutagenesis kit from Stratagene (LaJolla, CA). The point mutation was introduced into the plasmid BES116 (ADE2-URA3-ADE2 AscI fragment in pBluescript II KS (+))14 according to the manufacturer's instructions (Table S1) and the desired nucleotide sequence alteration was confirmed by DNA sequencing. The original and mutant plasmids were linearized by NaeI and transformed to JXM101 using the Yeast Transformation System 2 from Clontech (Terra Bella, CA).
Determination of the minimum inhibitory concentration (MIC)
The drug MIC was determined by serial 2-fold dilutions in 96-well plates as described previously,15 based on the methods of the Clinical and Laboratory Standards Institute (M27-A2).16 The cells (100 μl; 5 × 103-1 × 104 cells/ml) were inoculated into a 96-well plate containing serial 2-fold dilutions of fluconazole (0.125–64 μg/ml) with or without 1 mM Ca2+ in RPMI 1640 medium. After 24 h incubation at 30°C, the optical density (OD) value was measured at 600 nm. The MIC80 value was determined as the lowest concentration of FLC inhibited growth by 80%.
C. albicans killing assay
C. albicans strains grown at 30°C were harvested and re-suspended in RPMI 1640 medium and prepared at the starting inoculum of 103 CFU/ml.17 The concentration of FLC was 0.125, 0.25, 0.5, 1, 2, 4, 8 and 16 μg/ml with or without 1 mM Ca2+. After 24 h, a 100 μl aliquot was serially diluted 10-fold in sterile water. A 100 μl aliquot from each dilution was spread on SDA plate. After 48 h incubation at 30°C, the number of colonies was counted and calculated. Three biological replicates were performed for the experiment.
Systemic mice candidiasis model. C
albicans strains grown at 30°C were harvested, washed and resuspended in sterile saline. ICR female mice (18–22 g) were from Shanghai SLAC Laboratory Animals and maintained in specific pathogen-free conditions at the Center for Experimental Medicine, Second Military Medical University. All mice were inoculated by injection of 0.2 ml C. albicans suspension into the lateral tail vein (1 × 106 c.f.u/mouse) to establish the systemic candidiasis model.18 The mice were initially treated with FLC (1.0 mg/kg) or sterile saline 2 h after infection by C. albicans, and then once a day intraperitoneally for 7 days. Mice were monitored daily for survival until Day 30. Duplicate independent experiments were conducted (n = 10 in each group). Kaplan-Meier analyses were used to estimate survival probabilities.
NBD-DHS uptake and release Assay
For uptake analysis, exponentially growing C. albicans cells were washed and resuspended in PBS buffer (OD600 = 0.45). The suspension was incubated with 5 mM NBD-DHS and 1 mg/ml BSA for 10, 30 and 60 min at 30°C. At each time cells were harvested and scanned by confocal microscopy (excitation wavelength, 488 nm; emission wavelength, 520–565 nm; Leica, Germany). In DHS release assay, cells were treated with 5 mM NBD-DHS for 1 h in PBS buffer and then incubated with 1 mg/ml BSA for the indicated time (0, 10 and 30 min) at 30°C. Cells were centrifugated and the concentrations of NBD-DHS in supernatant were measured by spectrophotometry using a standard curve for NBD-DHS in PBS buffer (excitation wavelength, 475 nm; emission wavelength, 525 nm; PolarStar, BMG).
Susceptibility testing
C. albicans cells (1 × 104 cells/ml) were inoculated into successive wells of a 96-well microtiter plate containing serial 2-fold dilutions of fumonisin B1 and myriocin (initial concentration at 25 μg/ml, respectively). After incubation at 30°C for 16 h, absorbance at 600 nm was measured in a microplate reader.
Molecular docking
The template protein d2qpea of the original or mutant Rta2p was predicted through remote homology modeling by CPHmodels-3.2 Server (http://www.cbs.dtu.dk/services/CPHmodels/).19 With appropriate parameters, D-erythro-dihydrosphingosine could dock with the predicted structure and bind into a protein by Discovery Studio™ software.20 The computer modeling results would indicate some core amino acids interacted directly between the nuclear protein structure and the chemical.
Ethics statement
The animal experiments were performed in compliance with the institutional guidelines and approved by the Animal Ethics Committee of Second Military Medical University, and all efforts were made to minimize suffering of the animals.
Statistical analysis
One-way analysis of variance (ANOVA) with Bonferroni post-tests was used to analyze multiple groups. Log-rank test was used to analyze the equality of survival curves by SPSS 17.0. P < 0.05 was considered statistically significant.
Results
Construction and expression of mutant RTA2 reintegration in C. albicans
Multiple sequence alignment of Rta2p horologes in S. cerevisiae genome (Rsb1p, Rta1p, Yer185w, and Ylr046c) showed 16 conserved amino acids in the transmembrane domain (Fig. 1). The transmembrane domain is the important topology determined domain of membrane protein,21 all conserved amino acids residues were selected for site-directed mutagenesis according to the length of side chain, with charge or not, and amino acid hydrophobicity. The whole alterations (C150W, E155A, G158E, L182F, P186R, F220L, D224A, Q231H, A232D, G234S, G235R, G258C, Q262H, D382G, F396L, and F402V) were introduced into the vector pBES116 and the mutant plasmids were transformed to competent cells. Positive mutant cells were further confirmed by DNA sequencing by extracting plasmid DNA (Fig. S1). Except D382G and F402V, the original RTA2, vector (pBES116) and other mutant plasmids were reintegrated to rta2Δ/Δ mutant strain JXM101, confirmed by PCR method (Fig. S2).
Figure 1.
The sequence alignment of Rta2p, Rsb1p, Rta1p, Yer185w and Ylr046c. The alignment was generated using the Clustal X2 programs. The black boxes indicate identical residues, and the gray boxes show amino acid similarity. The asterisks represent the conserved amino acids in transmembrane domains of homogenous proteins. TM1-TM7: indicated transmembrane domains predicted by C. albicans and S. cerevisiae genome database.11
Resistance to FLC of wild-type and mutant Rta2p in vitro
The susceptibilities of C. albicans to FLC were measured by determining the lowest drug concentrations that achieved MIC80. Our previous study found that RTA2 was involved in the resistance to FLC mediated by calcineurin pathway in C. albicans.22 It was found in this study that 1 mM CaCl2 was able to increase resistance to FLC in P-RTA2 dramatically, with MIC80 increasing from 1 to 64 μg/ml. However, the effect of Ca2+ on resistance to FLC completely disappeared without RTA2, with MIC80 remaining unchanged in P-B (Table 2). With the effect of Ca2+, the mutant strains of RTA2 increased resistance to FLC in different levels, while the MIC80 only increased from 0.25 to 1 μg/ml in M3 and remained unchanged in M10 (Table 2). The appearance of FLC resistance was further confirmed in C. albicans killing assay. Compared with P-RTA2, M10 was more sensitive to high concentrations of FLC, while M3 formed drug resistance in shaking state. The addition of 1 mM Ca2+ dramatically decreased the fungicidal effect of FLC from 0.125 to 64 μg/ml in P-RTA2 and M3 without affecting M10 and P-B (Table 3).
Table 1.
C. albicans strains used in this study
| Strain | Genotype | Parental strain | Reference |
|---|---|---|---|
| JXM101 | RM1000§rta2Δ::hisG/rta2Δ::hisG | RM1000 | Jia et al.12 |
| P-B | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3 | JXM101 | This study |
| P-RTA2 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2 | JXM101 | This study |
| M1 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2C150W | P-RTA2 | This study |
| M2 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2E155A | P-RTA2 | This study |
| M3 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2G158E | P-RTA2 | This study |
| M4 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2L182F | P-RTA2 | This study |
| M5 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2P186R | P-RTA2 | This study |
| M6 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2F220L | P-RTA2 | This study |
| M7 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2D224A | P-RTA2 | This study |
| M8 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2Q231H | P-RTA2 | This study |
| M9 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2A232D | P-RTA2 | This study |
| M10 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2G234S | P-RTA2 | This study |
| M11 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2G235R | P-RTA2 | This study |
| M12 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2G258C | P-RTA2 | This study |
| M13 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2Q262H | P-RTA2 | This study |
| M15 | RM1000§rta2Δ::hisG/rta2Δ::hisG ADE2/ADE2::URA3::RTA2F396L | P-RTA2 | This study |
Table 2.
The activity of fluconazole with or without CaCl2 (1 mM) against different Rta2p mutant strains in vitro (MIC80, μg/ml). Data are representative of 3 independent experiments
| Strains | TMD | Mutagenic Site | FLC only | FLC with CaCl2 | |
|---|---|---|---|---|---|
| 1 | P-RTA2 | RTA2 revertant | 1 | 64 | |
| 2 | P-B | rta2Δ/Δ mutant | 0.5 | 0.5 | |
| 3 | M1 | TMD2 | TGT→TGG(C150W) | 2 | 64 |
| 4 | M2 | TMD2 | GAG→GCG(E155A) | 1 | 32 |
| 5 | M3 | TMD2 | GGG→GAG(G158E) | 0.25 | 1 |
| 6 | M4 | TMD3 | TTA→TTC(L182F) | 0.5 | 64 |
| 7 | M5 | TMD3 | CCT→CGT(P186R) | 0.25 | 32 |
| 8 | M6 | TMD4 | TTC→TTG(F220L) | 4 | 64 |
| 9 | M7 | TMD4 | GAC→GCC(D224A) | 2 | 64 |
| 10 | M8 | TMD4 | CAA→CAC(Q231H) | 0.5 | 64 |
| 11 | M9 | TMD4 | GCT→GAT(A232D) | 4 | 64 |
| 12 | M10 | TMD4 | GGT→AGT(G234S) | 0.25 | 0.25 |
| 13 | M11 | TMD4 | GGA→CGA(G235R) | 2 | 64 |
| 14 | M12 | TMD5 | GGT→TGT(G258C) | 1 | 32 |
| 15 | M13 | TMD5 | CAA→CAC(Q262H) | 2 | 64 |
| 16 | M15 | TMD7 | TTC→TTG(F396L) | 4 | 64 |
Table 3.
The C. albicans killing assay (%) by FLC with or without Ca2+. Data represent mean (±SD) of triplicates of one representive experiment of 3
| FLC concentration (μg/ml) | P-RTA2 | P-B | M3 | M10 | ||||
|---|---|---|---|---|---|---|---|---|
| FLC | FLC + Ca2+ | FLC | FLC + Ca2+ | FLC | FLC + Ca2+ | FLC | FLC + Ca2+ | |
| 0.125 | 4.92 ± 0.11 | 2.86 ± 0.09 | 12.34 ± 0.04 | 9.68 ± 0.07 | 9.45 ± 0.10 | 4.28 ± 0.01 | 11.26 ± 0.01 | 8.49 ± 0.01 |
| 0.25 | 15.16 ± 0.09 | 3.74 ± 0.02 | 24.81 ± 0.02 | 15.83 ± 0.29 | 18.37 ± 0.08 | 6.77 ± 0.03 | 23.23 ± 0.02 | 11.72 ± 0.02 |
| 0.5 | 17.72 ± 0.08 | 4.41 ± 0.05 | 30.53 ± 0.05 | 21.67 ± 0.02 | 21.16 ± 0.09 | 9.36 ± 0.05 | 34.07 ± 0.01 | 14.89 ± 0.06 |
| 1 | 22.07 ± 0.13 | 4.98 ± 0.06 | 34.38 ± 0.03 | 25.89 ± 0.04 | 28.26 ± 0.08 | 11.45 ± 0.08 | 36.38 ± 0.03 | 20.63 ± 0.01 |
| 2 | 26.53 ± 0.17 | 7.84 ± 0.05 | 34.70 ± 0.02 | 35.99 ± 0.17 | 29.37 ± 0.05 | 15.82 ± 0.04 | 37.03 ± 0.02 | 24.42 ± 0.05 |
| 4 | 27.45 ± 0.10 | 9.03 ± 0.11 | 42.21 ± 0.01 | 41.07 ± 0.07 | 30.54 ± 0.02 | 16.31 ± 0.01 | 44.58 ± 0.03 | 34.01 ± 0.04 |
| 8 | 27.01 ± 0.04 | 10.11 ± 0.31 | 50.37 ± 0.04 | 44.37 ± 0.16 | 29.78 ± 0.05 | 18.53 ± 0.01 | 52.02 ± 0.02 | 40.57 ± 0.01 |
| 16 | 27.21 ± 0.19 | 13.09 ± 0.28 | 51.62 ± 0.02 | 53.15 ± 0.12 | 30.79 ± 0.03 | 19.01 ± 0.02 | 53.11 ± 0.05 | 49.31 ± 0.35 |
Rta2p mutation affected the therapeutic efficacy of FLC against candidemia in vivo
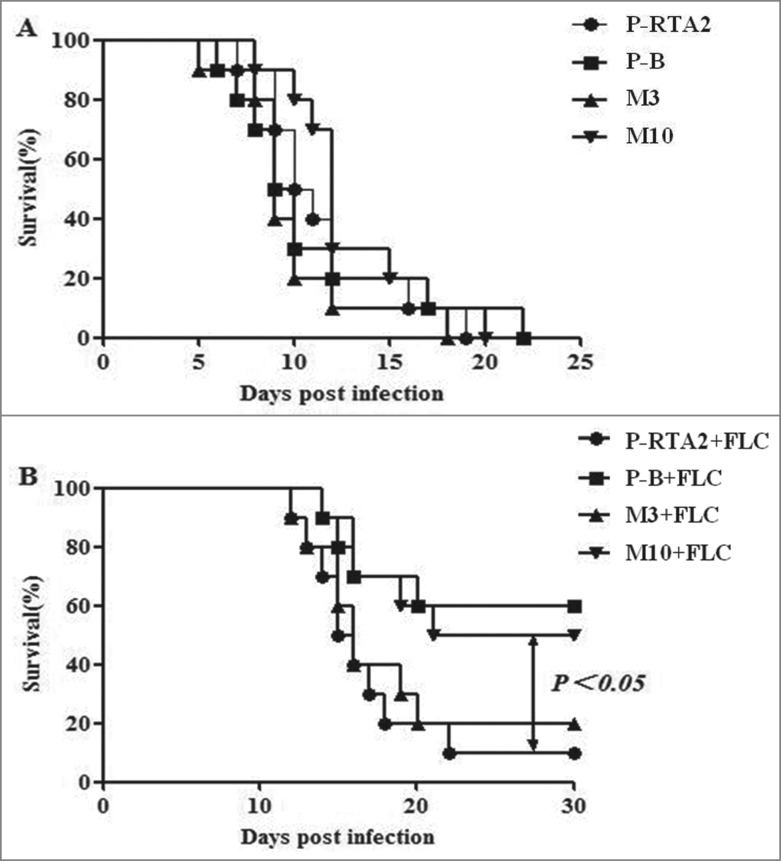
The Ca2+ mediated calcineurin pathway was reported to be crucial for the virulence and FLC resistance of C. albicans in the systemic infection model.23 Our previous study demonstrated that RTA2 was not involved in the virulence of C. albicans but could improve the survival rate of mice after FLC treatment.22 As the effect of Ca2+ failed to trigger FLC resistance completely in M10 in vitro, P-RTA2, P-B, M3 and M10 were selected to infect the mice. To examine the influences of mutant Rta2p on the efficacy of FLC against C. albicans in a murine model of systemic candidiasis, groups of 10 mice were intravenously injected with P-RTA2, P-B, M3 and M10, and the survival of mice was monitored for 30 days. As shown in Figure 2A, there was no significant difference in the survival rate between the groups of mice infected with P-RTA2, P-B, M3 and M10, while the mice infected by M10 after FLC treatment resulted in about 20% higher survival rate than the group infected by P-RTA2 in 30 days (P < 0.05; Fig. 2B). However, unlike the sensitivity to FLC with Ca2+ in settled state, the survival of M3 infected mice treated by FLC was similar with P-RTA2 infected mice (P > 0.05; Fig. 2B). These results suggested that G234, but not G158, was an important site of Rta2p affecting the therapeutic efficacy of FLC in the treatment of systemic candidiasis.
Figure 2.
Effects of FLC on survival curves of mice with systemic candidiasis. Groups of 10 mice were inoculated with 1 × 106 cells/ml of each strain via the lateral tail vein. Survival and weight were monitored daily. The survival time of mice infected with P-RTA2, P-B, M3, M10 treated with FLC (1.0 mg/kg) for 7 days. Groups without treatment were inoculated with NaCl. Data are representative of 3 independent experiments.
G234S mutation of Rta2p increased accumulation of DHS by reducing the export
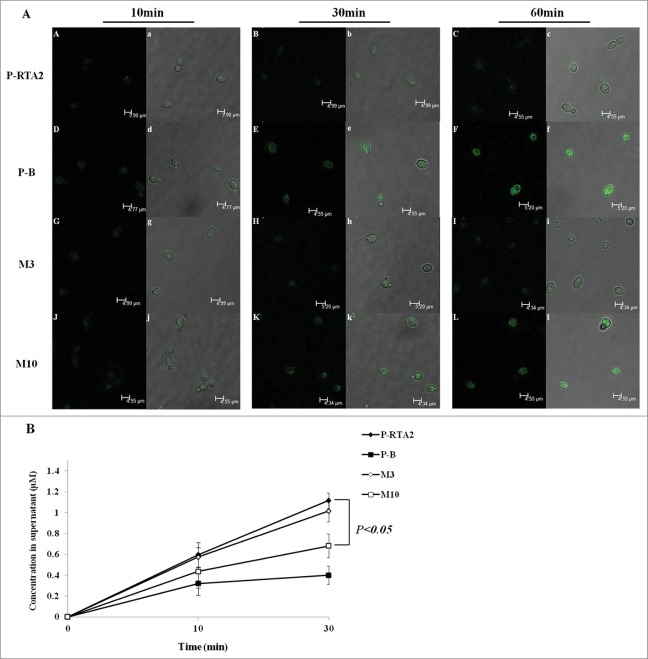
Like Rsb1p in S. cerevisiae, the function of Rta2p was to translocate DHS from the inner to the outer leaflet of the plasma membrane in C. albicans.13,24 To study the crucial site of Rta2p function, we examined whether the mutant strains could affect the intracellular accumulation and export of DHS. When cultured with NBD-DHS, P-B accumulated fluorescence-labeled DHS in the cells efficiently. The intracellular fluorescence density was increased dramatically at 30 and 60 min, and the similar appearance occurred in M10. However, P-RTA2 and M3 showed a much lower accumulation of NBD-DHS (Fig. 3A). On the other hand, we determined the NBD-DHS export in C. albicans cells in vivo. It was found that only 0.32 μM DHS of P-B was exported into the supernatant at 10 min and the level increased to 0.39 μM at 30 min (Fig. 3B, black squares). M10 exported a little more than P-B, from 0.43 μM to 0.68 μM during 10–30 min (Fig. 3B, white squares). In contrast, P-RTA2 exported 1.12 μM DHS at 30 min, which was approximately threefold higher than that in P-B (Fig. 3B, black diamonds). M3 exported 1.01 μM at 30 min, which was slightly lower than P-RTA2 (Fig. 3B, white diamonds). Together, M3 is equally capable of pumping out NBD-DHS as the P-RTA2, suggesting that the residue G158 is not involved in transport function, while G234 site appears to have a more direct effect on transport function.
Figure 3.
The uptake and release of dihydrosphingosine (DHS) by wild and mutant Rta2p. (A) For uptake experiments, C. albicans cells were incubated 5 mM NBD-DHS for 10, 30, 60 min. At each time point, the cells were washed and immobilized on slides; pictures were acquired with a Leica confocal microscope. (B) For release experiments, cells were pre-incubated with NBD-DHS for 1 h, resuspended in PBS containing 1 mg/ml BSA, and then incubated for 0, 10, 30 min. At each time point, cells were centrifuged and the supernatant was measured fluorimetrically. Data are shown as means ± SDs from 3 independent experiments.
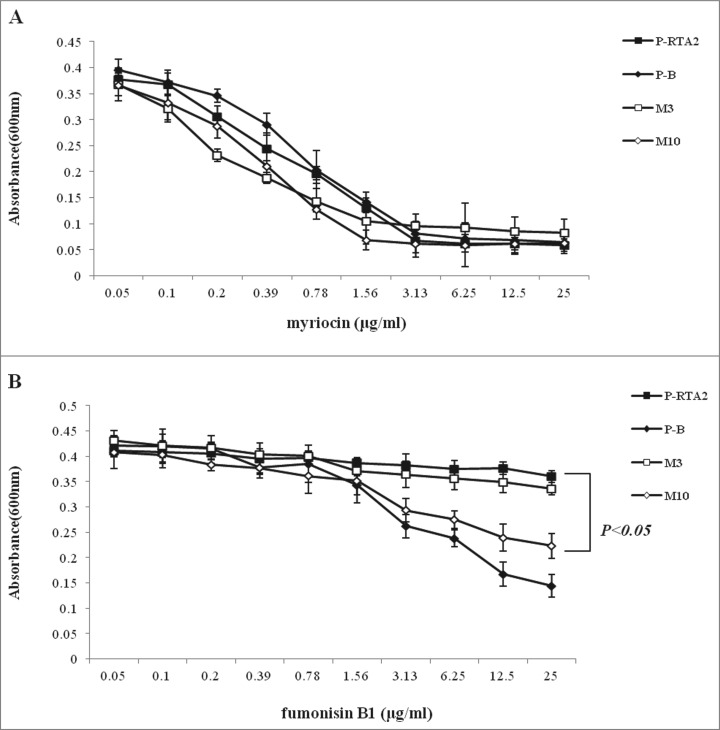
To get further verification, we tested the susceptibility of P-RTA2, P-B, M3 and M10 to myriocin and fumonisin B1, the upstream and downstream inhibitors of DHS in sphingolipid biosynthesis. As yeast cells could not survive in the presence of DHS over-accumulation,25 the decrease of myriocin and increase of fumonisin B1 could result in the accumulation of DHS in C. albicans. Microbroth dilution assay was used to determine the sensitivity of C. albicans to myriocin or fumonisin B1. All strains were equally sensitive to myriocin (Fig. 4A). Compared with P-RTA2 and M3, P-B and M10 were more sensitive to fumonisin B1 due to more DHS accumulation (Fig. 4B). Above all, G234S mutation, but not G158E of Rta2p, significantly increased the DHS accumulation in cells by decreasing the export.
Figure 4.
The sensitivity to myriocin (A) or fumonisinB1 (B) of C. albicans P-RTA2 (▪), P-B(⧫), M3(□) and M10 (⋄) cells by microbroth dilution assays. The absorbance at 600 nm was recorded after 24 h incubation at 30°C. Data are means ± SDs of triplicate wells and representative of 3 independent experiments.
Discussion
In the present study, we selected conserved amino acids in the transmembrane domain of Rta2p for site-directed mutagenesis and demonstrated that G234S, but not G158E of RTA2, could affect the FLC resistance in vitro and in vivo, also could increase the accumulation of DHS by reducing its export.
Site-directed mutagenesis has been widely applied to find key residues of MDR-associated proteins such as Cdr1p, Cdr2p in C. albicans and Pdr5p in S. cerevisiae.26-28 For example, mutation of C193, K901 and D327 of Cdr1p could change the ATPase activity of nucleotide-binding domains,29,30 and the transmembrane domains of Cdr1p could specifically transport and bind substrates of different chemicals such as azoles, phospholipids and sterols.31,32 The structure-functional relationship of Cdr1p determines the crucial amino acids and important domains of transport, which are considered as the base of designing suitable inhibitors or drugs targeting Crd1p of C. albicans. Like Cdr1p, Rta2p is also a membrane protein with several transmembrane domains, involved in azole resistance in clinical and sphingoid trasport in C. albicans.12,13 Above all, it's a new approach to study structure and functional relationship of Rta2p in C. albicans.
In this study, we used the site-directed method to find functional amino acids of the drug-resistance-related protein Rta2p. Among the conserved amino acids in transmembrane domains, we found that the resistance of Rta2p mutant strains to FLC was increased in varying degrees upon the effect of 1 mM Ca2+, with MIC80 of M3 (vector with RTA2-G158E) increasing from 0.25 to 1 μg/ml and that of M10 (vector with RTA2-G234S) remaining unchanged (Table 2). However, M3 formed FLC resistance with Ca2+ in shaking state in C. albicans killing assay (Table 3). We further found that the mutation of G234, but not G158, was more effective to treat systemic candidiasis by FLC in vivo (Fig. 2). With respect to Rta2p-mediated DHS translocation from inner to outer leaflet in C. albicans,13 we found that M10 could increase the accumulation of DHS by reducing the export, while M3 appeared not to have a more direct effect on transport function (Figs. 3 and 4). More importantly, the result of bioinformatics analysis suggested the interactions of key amino acids and D-erythro-dihydrosphingosine were completely different between the wild type and M10 (Fig. 5). The computer imitation results further suggested that G234 of Rta2p was an important amino acid involved in the binding between the nuclear structure and DHS.
Figure 5.
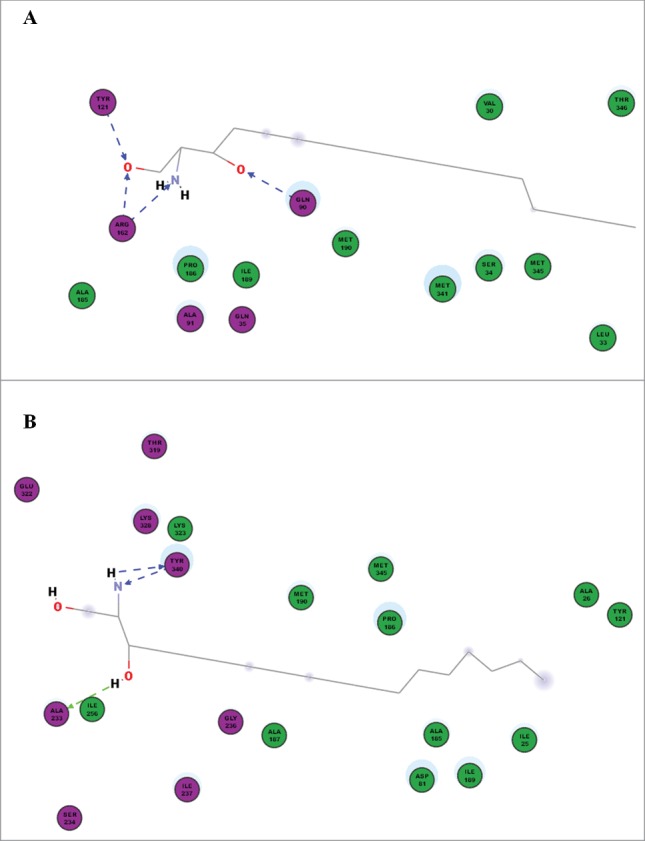
The computer modeling of amino acids and D-erythro-dihydrosphingosine between P-RTA2 (A) and M10 (B) by Discovery Studio™ software. Some key amino acids (violet) including Q90, Y121, R162 could directly link to DHS in the wild type strain, while the crucial amino acids were A233 and Y340 in the mutant strain.
n conclusion, we found that the mutant strain M10 was more sensitive to FLC in vitro and in vivo with Ca2+ than the other mutant strains as represented by decreased export of DHS. Our results suggest that the mutation in TMD4 (G234S) is only relevant in the presence of calcium when investigating FLC resistance in C. albicans. In addition, the concentration of 1 mM CaCl2 used in this study was almost equal to that of ionized calcium in mammalian serum (about 1–2 mM). Inhibition of calcium-mediated azole resistance has been proposed as a new therapeutic target of antifungal therapy.33 Sphingolipids, composed of dihydrosphingosine and phytosphingosine, are the primary lipids of plasma membrane in C. albicans. Our results also confirm that G234 is the functional site to affect DHS transport. Moreover, the susceptibility of C. albicans to FLC and transport of DHS are not merely dependent on the transmembrane domains. Further studies are needed to verify whether non-transmembrane domains and some amino acids residues of Rta2p have the function of lipid transport.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Xin-Ming Jia for providing C. albicans strain JXM101 and plasmid BES116.
Funding
This work was supported by the National Key Basic Research Program of China (No 2013CB531602), the National Science Foundation of China (No 81273556, 81330083), and Shanghai Science and Technology Major Project (11JC1415400).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol 2011; 12:112-22; PMID:22158429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams DW, Jordan RP, Wei XQ, Alves CT, Wise MP, Wilson MJ, Lewis MA. Interactions of Candida albicans with host epithelial cells. J Oral Microbiol 2013; 21:5:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perea S, Patterson TF. Antifungal Resistance in Pathogenic Fungi. Clin Infect Dis 2002; 35:1073-80; PMID:12384841; http://dx.doi.org/ 10.1086/344058 [DOI] [PubMed] [Google Scholar]
- 4.Chandra J, Kuhn DM, Mulherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol 2001; 183:5385-94; PMID:11514524; http://dx.doi.org/ 10.1128/JB.183.18.5385-5394.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White TC. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother 1997; 41:1482-87; PMID:9210670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prasad R, De Wergifosse P, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr Genet 1995; 27:320-9; PMID:7614555; http://dx.doi.org/ 10.1007/BF00352101 [DOI] [PubMed] [Google Scholar]
- 7.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 1997; 143:405-16; PMID:9043118; http://dx.doi.org/ 10.1099/00221287-143-2-405 [DOI] [PubMed] [Google Scholar]
- 8.Marichal P, Koymans L, Willemsens S, Bellens D, Verhasselt P, Luyten W, Borgers M, Ramaekers FC, Odds FC, Bossche HV. Contribution of mutations in the cytochrome P450 14alpha-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 1999; 145:2701-13; PMID:10537192 [DOI] [PubMed] [Google Scholar]
- 9.Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, Cardenas ME, Perfect JR, McCusker JH, Heitman J. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J 2002; 21:546-59; PMID:11847103; http://dx.doi.org/ 10.1093/emboj/21.4.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bader T, Schröppel K, Bentink S, Agabian N, Köhler G, Morschhäuser J. Role of calcineurin in stress resistance, morphogenesis, and virulence of a Candida albicans wild-type strain. Infect Immun 2006; 74:4366-9; PMID:16790813; http://dx.doi.org/ 10.1128/IAI.00142-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White TC, Holleman S, Dy F, Mirels LF, Stevens DA. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob Agents Chemother 2002; 46:1704-13; ; http://dx.doi.org/ 10.1128/AAC.46.6.1704-1713.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia XM, Ma ZP, Jia Y, Gao PH, Zhang JD, Wang Y, Xu YG, Wang L, Cao YY, Cao YB, et al.. RTA2, a novel gene involved in azole resistance in Candida albicans. Biochem Biophys Res Commun 2008; 373:631-6; PMID:18601908; http://dx.doi.org/ 10.1016/j.bbrc.2008.06.093 [DOI] [PubMed] [Google Scholar]
- 13.Jia XM, Wang Y, Jia Y, Gao PH, Xu YG, Wang L, Cao YY, Cao YB, Zhang LX, Jiang YY. RTA2 is involved in calcineurin-mediated azole resistance and sphingoid long-chain base release in Candida albicans. Cell Mol Life Sci 2009; 66:122-34; PMID:19002381; http://dx.doi.org/ 10.1007/s00018-008-8409-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Q, Summers E, Guo B, Fink G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol 1999; 181:6339-46; PMID:10515923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espinel-Ingroff A, Rodríguez-Tudela JL, Martínez-Suárez JV. Comparison of two alternative microdilution procedures with the National Committee for Clinical Laboratory Standards reference macrodilution method M27-P for in vitro testing of fluconazole-resistant and-susceptible isolates of Candida albicans. J Clin Microbiol 1995; 33:3154-8; PMID:8586692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamza OJ, van den Bout-van den Beukel CJ, Matee MI, Moshi MJ, Mikx FH, Selemani HO, Mbwambo ZH, Van der Ven AJ, Verweij PE. Antifungal activity of some Tanzanian plants used traditionally for the treatment of fungal infections. J Ethnopharmacol 2006; 108:124-32; PMID:16829001; http://dx.doi.org/ 10.1016/j.jep.2006.04.026 [DOI] [PubMed] [Google Scholar]
- 17.Klepser ME, Wolfe EJ, Jones RN, Nightingale CH, Pfaller MA. Antifungal pharmacodynamic characteristics of fluconazole and amphotericin B tested against Candida albicans. Antimicrob Agents Chemother 1997; 41:1392-5; PMID:9174207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Yan K, Zhang Y, Huang R, Bian J, Zheng C, Sun H, Chen Z, Sun N, An R, et al.. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc Natl Acad Sci USA 2007; 104:4606-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen M, Lundegaard C, Lund O, Petersen TN. CPHmodels-3.0-remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res 2010;38:W576-81; PMID:20542909; http://dx.doi.org/ 10.1093/nar/gkq535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Wang X, Li X, Zhang Y, Dai Y, Guo C, Zheng H. QSAR study and the hydrolysis activity prediction of three alkaline lipases from different lipase-producing microorganisms. Lipids Health Dis 2012; 11:124; PMID:23016923; http://dx.doi.org/ 10.1186/1476-511X-11-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raba M, Baumgartner T, Hilger D, Klempahn K, Härtel T, Jung K, Jung H. Function of transmembrane domain IX in the Na+/proline transporter PutP. J Mol Biol 2008; 382:884-93; PMID:18692508; http://dx.doi.org/ 10.1016/j.jmb.2008.07.070 [DOI] [PubMed] [Google Scholar]
- 22.Jia Y, Tang RJ, Wang L, Zhang X, Wang Y, Jia XM, Jiang YY. Calcium-Activated-Calcineurin Reduces the In Vitro and In Vivo Sensitivity of Fluconazole to Candida albicans via Rta2p. PLoS One 2012; 7:1-9; PMID:23118995; http://dx.doi.org/ 10.1371/annotation/7de63575-e5c9-4f1d-bb45-fc6420e92c71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blankenship JR, Wormley FL, Boyce MK, Schell WA, Filler SG, Perfect JR, Heitman J. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot Cell 2003; 2:422-30; PMID:12796287; http://dx.doi.org/ 10.1128/EC.2.3.422-430.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kihara A, Igarashi Y. Identification and Characterization of a Saccharomyces cerevisiae Gene, RSB1, Involved in Sphingoid Long-chain Base Release. J Biol Chem 2002; 277:30048-54; PMID:12034738; http://dx.doi.org/ 10.1074/jbc.M203385200 [DOI] [PubMed] [Google Scholar]
- 25.Mao C, Saba JD, Obeid LM. The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem J 1999; 342:667-75; PMID:10477278; http://dx.doi.org/ 10.1042/0264-6021:3420667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla S, Saini P, Smriti, Jha S, Ambudkar SV, Prasad R. Functional characterization of Candida albicans ABC transporter Cdr1p. Eukaryotic cell 2003; 2:1362-75; PMID:14665469; http://dx.doi.org/ 10.1128/EC.2.6.1361-1375.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Micheli M, Bille J, Schueller C, Sanglard D. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol Microbiol 2002; 43:1197-214; PMID:11918807; http://dx.doi.org/ 10.1046/j.1365-2958.2002.02814.x [DOI] [PubMed] [Google Scholar]
- 28.Egner R, Bauer BE, Kuchler K. The transmembrane domain 10 of the yeast Pdr5p ABC antifungal efflux pump determination both substrate specificity and inhibitor susceptibility. Mol Microbiol 2000; 35:1255-63; PMID:10712705; http://dx.doi.org/ 10.1046/j.1365-2958.2000.01798.x [DOI] [PubMed] [Google Scholar]
- 29.Jha S, Karnani N, Lynn AM, Prasad R. Covalent modification of cysteine 193 impairs ATPase function of nucleotide-binding domain of a Candida drug efflux pump. Biochem Biophys Res Commun 2003; 310:869-75; PMID:14550284; http://dx.doi.org/ 10.1016/j.bbrc.2003.09.094 [DOI] [PubMed] [Google Scholar]
- 30.Jha S, Dabas N, Karnani N, Saini P, Prasad R. ABC multidrug transporter Cdr1p of Candida albicans has divergent nucleotide-binding domains which display functional asymmetry. FEMS Yeast Res 2004; 5:63-72; PMID:15381123; http://dx.doi.org/ 10.1016/j.femsyr.2004.07.002 [DOI] [PubMed] [Google Scholar]
- 31.Krishnamurthy S, Gupta V, Snehlata P, Prasad P. Characterization of human steroid hormone transport mediated by Cdr1p, multidrug transporte of Candida albicans, belonging to the ATP binding cassette super family. FEMS Microbiol Lett 1998; 158:69-74; PMID:9453158; http://dx.doi.org/ 10.1111/j.1574-6968.1998.tb12802.x [DOI] [PubMed] [Google Scholar]
- 32.Dogra S, Krishnamurthy S, Gupta V, Dixit BL, Gupta CM, Sanglard D, Prasad R. Asymmetric distribution of phosphatidylethanolamine in C. albicans: possible mediation by CDR1, a multidrug transporter belonging to ATP binding cassette (ABC) superfamily. Yeast 1999; 15:111-21; PMID:10029989; http://dx.doi.org/ 10.1002/(SICI)1097-0061(19990130)15:2%3c111::AID-YEA350%3e3.0.CO;2-E [DOI] [PubMed] [Google Scholar]
- 33.Steinbach W, Reedy J, Cramer R, Perfect J, Heitman J. Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat Rev Microbiol 2007; 5:418-30; PMID:17505522; http://dx.doi.org/ 10.1038/nrmicro1680 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.