Abstract
Streptococcal histidine triad proteins HTPs are widely distributed within the Streptococcus genus. Based on the phylogenetic relationship and domain composition, HTPs are classified into type I and type II subfamilies. Previous studies revealed that several pathogenic streptococci contain more than one htp gene. We found that the highly virulent strain of Streptococcus suis 2 (S. suis 2), 05ZYH33 encodes 3 HTPs, designated HtpsA (previously described as HtpS), HtpsB, and HtpsC. Among them, HtpsC is the only member that contains leucine-rich repeat (LRR) domains at the C-terminal. In this study, we demonstrated that the recombinant HtpsC could bind to 2 different components of human ECM complex laminin and fibronectin in vitro, suggesting that it is a novel adhesin of S. suis 2. Having constructed an htpsC mutant, we evaluated its role in the pathogenesis of the highly virulent S. suis 2 strain 05ZYH33. Our data showed that inactivation of htpsC significantly affected adherence of S. suis 2 to Hep-2 cells and shortened the survival of the bacteria in whole blood. Furthermore, deletion of htpsC significantly attenuated the virulence of S. suis 2 in mice. These results demonstrated that htpsC was involved in the pathogenesis of the highly virulent S. suis 2 strain 05ZYH33. In line with the observation, immunization with HtpsC significantly prolonged mice's survival after S. suis 05ZYH33 challenge, indicating its potential use in the vaccine development against S. suis.
Keywords: histidine triad protein, Streptococcus suis serotype 2, virulence
Introduction
During the infection process, Streptococcus often expresses many surface-exposed proteins to facilitate host infection.1,2 These proteins play various roles in the physiology, invasion, and immune escape of the pathogen via different mechanisms. For example, the zinc binding protein AdcA is essential for zinc homeostasis in S. pneumoniae;3 Laminin-binding protein (Lmb), glyceraldehyde-3-phosphate dehydrogenase (GADPH), and enolase are involved in adherence of several Streptococci to host tissues;4-7 Pneumococcal surface protein A (PspA) and histidine triad protein (HTP) help Streptococcus evade the host immune system.8,9 These factors cooperatively enable streptococci to colonialize successfully within their host. Of note, some surface-exposed proteins associated with bacterial virulence such as enolase, Lmb, and HTP, are conserved across pathogenic streptococci such as S. pneumoniae, S. pyogenes, S. agalactiae and S. suis.5, 6, 10-12 Exploring the roles of these proteins in pathogenicity will provide a general understanding of streptococcal infection. Furthermore, conservation of these proteins among different strains and various species suggests that they are potential candidates for the vaccine development against Streptococcus. In fact, many of these proteins were shown to have excellent immunogenic properties, and some have already been used for vaccine development.5,6, 13-15
HTPs are a group of surface-exposed proteins widely distributed in many streptococcal species.10 Based on the phylogenetic relationship and domain composition, HTPs in streptococcal species were divided into 2 subfamilies: HTP I and HTP II.10 Both subfamily proteins play important roles in streptococcal physiology and pathogenicity.16 S. pneumoniae contains 4 HTPs (PhtA, PhtB, PhtD and PhtE,) and all of them belong to the HTP I subfamily.10,17 Experimental tests revealed that the 4 proteins play important but redundant roles in the virulence of S. pneumoniae.9 S. pyogenes contains one HTP I protein (HtpA) and one HTP II protein (Slr).10,18, 19 Independent studies suggested that the 2 proteins were involved in the pathogenesis of S. pyogenes.18,19 Although the HTPs are involved in streptococcal pathogenicity, the underlining mechanisms have been poorly understood, especially for the HTPII subfamily. Many studies have demonstrated that the HTP I proteins can bind zinc, and are thus involved in zinc homeostasis in bacterial infections.19,20 Work by Ogunniyi and colleagues suggested that HTP I proteins were required for inhibition of complement deposition on the pneumococcal surface through recruitment of complement factor H.9 Compared with HTP I protein, most of the HTPII proteins encode additional leucine-rich repeat (LRR) domains at their C-terminals.10 Most LRR-containing proteins are involved in protein-protein interactions for signal transduction and cell adhesion,21 indicating that HTP II protein may function as adhesin in streptococcal infections.
Streptococcus suis serotype 2 (S. suis 2) is an important zoonotic pathogen that infects swine and humans, giving rise to many serious diseases. Previously, we identified 3 htp genes in the S. suis 2 Chinese isolate 05ZYH33.14 One (ORF number SSU0332) was downstream of the lmb gene5 and identified as a type I htp gene (previously referred to as HtpS14). To distinguish it from the other 2 htp genes in S. suis, we named it hereafter HtpsA, and the other 2 genes (SSU1267 and SSU1577) were referred to as HtpsB and HtpsC, respectively. Among the 3 HTPs in 05ZYH33, HtpsC is the only one that contains an LRR domain. This study sought to elucidate whether this HTP II protein functions as an adhesin in S. suis 2 05ZYH33 and its potential role in S. suis pathogenicity.
Results
Sequence analysis of the htpsC gene
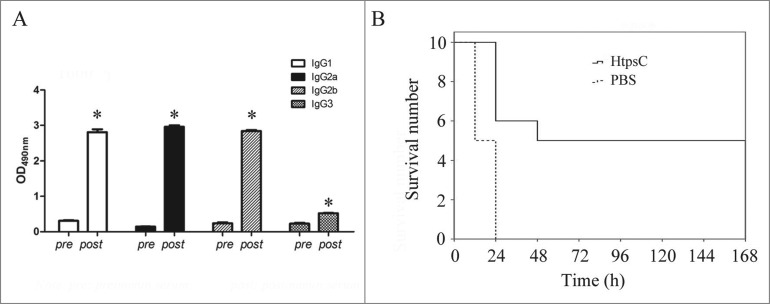
Our previous studies revealed that the genome of the S. suis 2 Chinese strain 05ZYH33 encoded 3 htp genes belonging to 2 separate subfamilies.10,14 However, the annotated open reading frame (ORF) of htpsC in the genome release of 05ZYH3322 was shorter than its orthologous gene in the S. suis 2 strain P1/7. Re-sequencing and re-annotate of this region in this study updated the ORF of htpsC (genbank accession number: KJ149804) in 05ZYH33 and revealed that the sequence is actually identical to its ortholog in strain P1/7. According to the sequence result, the re-annotated HtpsC in 05ZYH33 contains 858 amino acids. The deduced protein sequence of HtpsC is comprised of 4 HTP motifs at the N-terminal region and 13 successively appearing LRR repeats at the C-terminal (Fig. 1). By aligning HtpsC with its orthologs Slr (Genbank accession number: HQ908654.1) and Blr (GenBank accession number: DQ242614.1), we found the amino acid sequences to be considerably conserved among the 3 species, especially at the C-terminal half occupied by the LRR domain.
Figure 1.
Sequence analysis of S. suis HtpsC. (A) Schema of S. suis HtpsC protein. HTP motifs and LRR repeats are indicated with black and red boxes, respectively. (B) Multiple sequence alignments of S. suis HtpsC with related homologous proteins at the amino acid level. HtpsC orthologous proteins (Blr and Slr) are from S. agalactiae (Genbank accession: DQ242614.1) and S. pyogenes (GenBank accession: HQ908654.1). HTP motifs and LRR repeats are indicated with underlines and arrowed lines, respectively.
Construction and characterization of an htpsC mutant
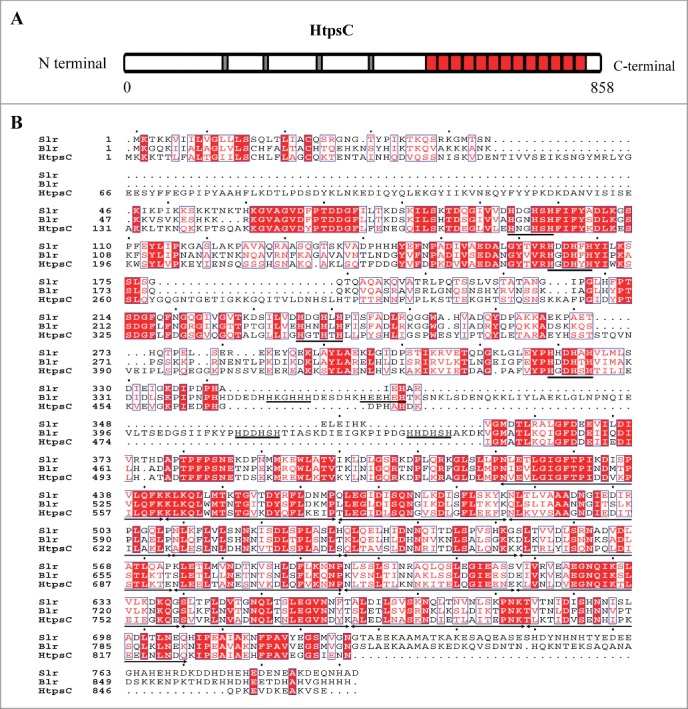
To investigate the role of htpsC in S. suis strain 05ZYH33, an isogenic knockout mutant of htpsC was constructed. The 5′ fragment of the htpsC ORF was replaced by a Spcr gene based on homologous recombination as described in Materials and Methods (Fig. 2A). The insertion of Spcr gene also resulted in frame-shift for the remaining 3′ region of htpsC. The obtained ΔhtpsC strain was confirmed by combined PCR analysis with different primer sets (Fig. 2B) and RT-PCR (data not shown).
Figure 2.
The effect of htpsC mutation on the morphology of S. suis. (A) Cartoon description for knockout of htpsC from the S. suis 2 chromosome. pUC-HtpsC is the recombinant vector constructed to specifically inactivate htpsC. LA and RA separately indicate the left and right border of the target gene. Two pairs of specific primers (Out1 & Out2, Check1 & Check2) were used to detect htpsC in S. suis 2 genome with PCR. The former is located in up and down adjacent regions of the htpsC gene, whereas the latter one is located at the 5′ and 3′ region of the htpsC gene. WT, wild type S. suis 2; ΔhtpsC, an isogenic mutant of the htpsC gene. (B) Multiple-PCR analysis of ΔhtpsC. PCR products were separated by electrophoresis on a 1.0% agarose gel stained with ethidium bromide (EB). (C) Growth characteristics of ΔhtpsC, CΔhtpsC and WT strains measured spectrometrically at 600 nm. (D) Separate aliquots of the bacterial suspensions were serially diluted and plated to determine CFU numbers per milliliter.
Prior to evaluating the effect of htpsC inactivation on the virulence of S. suis 2 05ZYH33, we first examined in vitro phenotype characteristics of the htpsC mutant. Measuring growth curves of the bacterial strains under normal growth conditions without antibiotics revealed that the ΔhtpsC grew slightly slower than both the wild type (WT) and the complementary strains (Fig. 2C). However, when we determined the growth rates of the 3 strains cultures by counting the CFU, no obvious differences in the CFU counts were observed during the initial 6 to 7 hours of growth. At this time, the WT strain had already reached the post-exponential-growth phase, but the ΔhtpsC strain continued to reproduce nearly one more generation than the WT strain (Fig. 2D). Therefore, the growth rate of ΔhtpsC was not slower than the WT strain in terms of living cells. The observed lower OD600 value of the ΔhtpsC culture than that of the WT culture indicated that the deletion of htpsC might affected the light-scattering properties of S. suis 2.
S. suis HtpsC is a surface-expressed adhesin
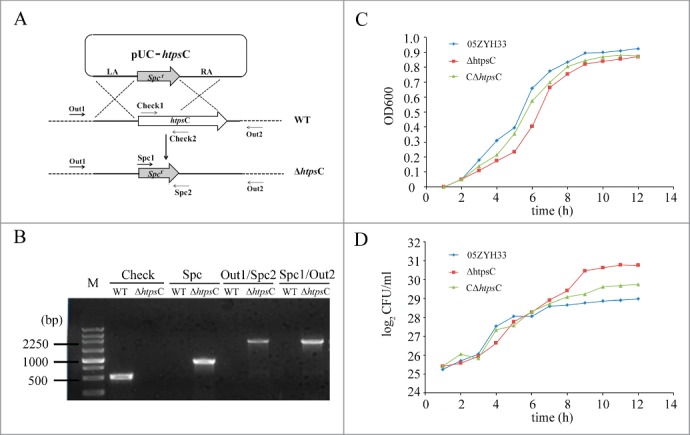
To determine whether HtpsC also functions as an adhesin of S. suis, we examined its surface expression and performed in vitro adherence analyses. rHtpsC was expressed and purified from the E. coli expression system as described in Materials and Methods. Purified rHtpsC protein was then used for preparing polyclonal antibodies, which were used for subsequent analysis. The cell surface exposure of HtpsC was determined by FCM. As shown in Figure 3A, the mean fluorescence intensity (MFI) of both WT strain and CΔhtpsC strain incubated with rabbit anti-HtpsC serum was higher than that of the ΔhtpsC strain, implying that HtpsC is expressed on the cell surface of S. suis 2. We then used ELISA to test whether HtpsC could bind human extracellular matrix (ECM) in vitro. As shown in Figure 3B, HtpsC strongly adhered to human ECM in a concentration dependent manner, indicating its function as an adhesion in S. suis 2.
Figure 3.
HtpsC is a surface-expressed adhesin of S. suis. (A) Cell surface localization of HtpsC on S. suis 2 cells was measured by flow cytometry. WT (yellow) and CΔhtpsC (blue) had greater MFI than that of the ΔhtpsC strain (red) after being incubated with anti-HtpsC antibodies. (B) ELISA for binding of different concentrations of HtpsC to immobilized human ECM. HtpsC strongly adhered to immobilized human ECM in a concentration-dependent manner. (C) Inactivation of htpsC significantly decreased the adherence of S. suis 2 to Hep2 cells. The normalized mean fluorescence intensities (NMFI) of Hep-2 cells after incubation with the bacteria are shown as columns with standard errors. (* indicates P<0.05, Student's t-test).
To explore the role of HtpsC in S. suis 2 adherence to human cells, we compared adherence of ΔhtpsC and WT S. suis 2 strains to Hep-2 cells. As expected, ΔhtpsC had decreased adherence to Hep-2 cells compared with the WT strain. As shown in Figure 3C, the binding of ΔhtpsC to Hep-2 cells is significantly weaker (P < 0.01, Student's t-test) than that of the WT strain. The adhesion ability was recovered to the normal level in the complementary strain CΔhtpsC. This observation confirmed that HtpsC plays an important role in the adherence of S. suis 2 to human epithelial cells.
Both laminin and fibronectin are host receptors of HtpsC
Collagens, fibronectin (Fn) and laminin (Lm), 3 components of human ECM, are also expressed on the surface of Hep-2 cells,23 indicating they might be common host receptors of HtpsC. Furthermore, a previous study reported that S. pyogenes Slr protein, the orthologous of HtpsC, could bind type I collagen of human.24 To elucidate whether some of these ECM components are receptors of HtpsC, adhesion of rHtpsC to type I collagen, Fn and Lm was tested. As shown in Figure 4A, rHtpsC could bind to immobilized Fn and Lm with similar affinity in a dose-dependent manner, but did not bind to type I collagen. The observation suggested that rHtpsC binds 2 different components of ECM, both Fn and Lm.
Figure 4.
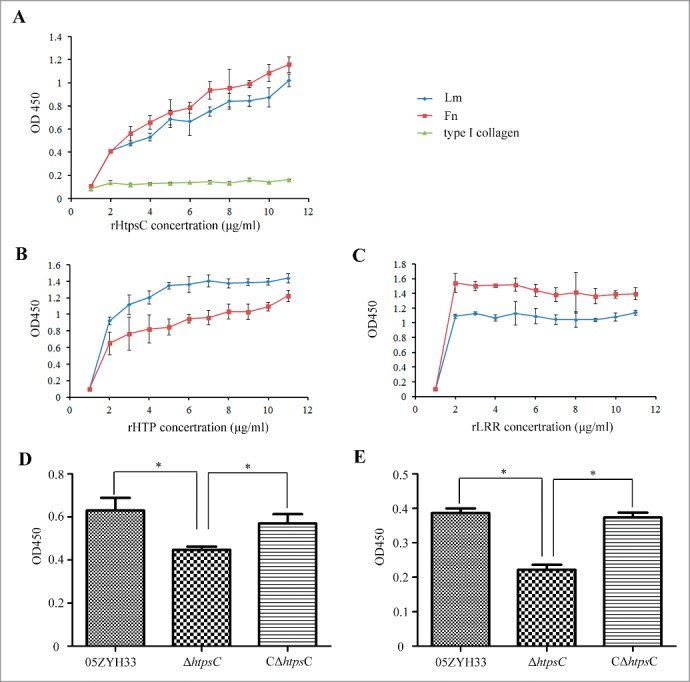
Determine the host receptors of HtpsC. (A) The adherence of rHtpsC to 3 different components of human ECM was tested to reveal that both Fn and Lm are receptors of HtpsC. (B) The adherence of HTP domain to Fn and Lm were determined. (C) The adherence of LRR domain to Fn and Lm were determined. (D) Inactivation of htpsC decreased 05ZYH33 adherence to Lm. E. Inactivation of htpsC decreased 05ZYH33 adherence to Fn. (* indicates P < 0.05, Student's t-test).
To further test whether HtpsC is involved in the binding of whole S. suis 2 cells to Fn and Lm, the adherence to these 2 ECM components were compared among △htpsC, 05ZYH33 and C△htpsC. It was found that both the WT strain adhered well to Fn and Lm (Fig. 4D and E). In contrast, the adherence of △htpsC to human Fn and Lm were both decreased significantly compared to either the WT strain or C△htpsC strain. These results suggested that the adherence of S. suis 2 to host ECM components Fn and Lm was mediated by HtpsC.
HTP domain and LRR domain independently contribute to HtpsC adherence to Fn and Lm
The observed binding of rHtpsC to 2 different components of ECM prompted us to explore whether the adherence ability of HtpsC was contributed independently by the 2 domains. Therefore, the N-terminal HTP domain (amino acids 28 to 403) and the C-terminal LRR domain (amino acids 496 to 821) of HtpsC were in vitro expressed separately. The resulted recombinant proteins rHTP and rLRR were then subjected to test their adherence to Fn and Lm. As shown in Figure 4B and C, both rHTP and rLRR could adhere strongly to human Fn and Lm in a dose dependent fashion. In addition, rLRR showed higher affinity to Fn than to Lm, whereas rHTP protein showed an opposite pattern. These results demonstrated that HTP domain and LRR domain independently contributed to the adherence ability of HtpsC with different affinities toward Fn and Lm.
HtpsC is required for the full virulence of S. suis 2
To determine the role of HtpsC in S. suis 2 infection, we measured survival of the △htpsC in human whole blood. In a whole-blood killing test, we observed that △htpsC strain survival is significantly decreased (P < 0.05, Student's t-test) compared with the WT strain (Fig. 5A), whereas the survival of the C△htpsC in whole blood is similar to the WT strain. This suggested that HtpsC may confer bacterial resistance to whole-blood killing during the infection process.
Figure 5.
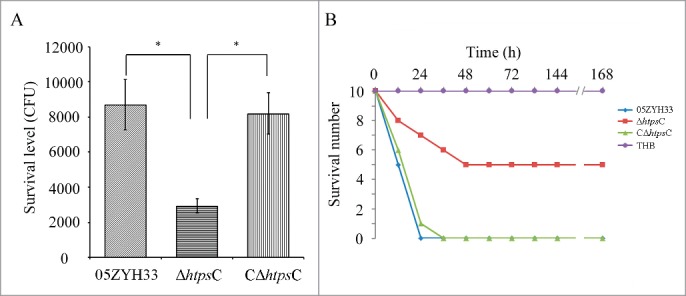
Deletion of htpsC attenuated the virulence of S. suis 2 strain 05ZYH33. (A) Resistance to whole blood killing of the △htpsC strain. △htpsC strain survival was significantly decreased compared with C△htpsC and the WT strain (P < 0.05, Student's t-test). (B) Comparative analysis of bacterial virulence of the htpsC mutant with the WT strain using a mouse model. Mice challenged with △htpsC strain had greater survival than those challenged with the WT strain or C△htpsC strain (P<0.01, Kaplan-Meier survival analysis), indicating that inactivation of the htpsC gene significantly decreased the virulence of S. suis 2.
We then compared the ability of WT and mutant strains to cause mouse mortality. After challenging with the WT 05ZYH33 or C△htpsC strain, 5 and 4 mice died within 12 h, but only 2 of 10 mice challenged with the △htpsC strain died during the same period. Subsequently, all of the mice challenged by 05ZYH33 or C△htpsC strain died during the first 36 h. In contrast, only 4 mice challenged by △htpsC strain died in the same period and 5 of 10 mice survived the entire experimental period. All mice injected with THB survived. Data summarized in Figure 5B revealed that the rate of mortality was significantly reduced in mice injected with the mutant strain (P < 0.01, Kaplan-Meier survival analysis). Thus, we concluded that htpsC is required for full virulence of S. suis 2 under the conditions tested.
HtpsC protects mice against S. suis 2 infection
To evaluate whether rHtpsC could confer protection against S. suis 2 infection, mice were immunized with rHtpsC and challenged with S. suis 2. When mice of the negative control group were challenged with 05ZYH33, 5 mice died within 12 h post-inoculation and another 5 mice died within 24 h post-inoculation. In contrast, no mice immunized with rHtpsC died during the first 12 h, and only 4 died within 24 h post-inoculation. Finally, 5 mice in the immunized group survived during the experimental period (Fig. 6B). Our results indicate that rHtpS confers protection in mice.
Figure 6.
HtpsC confers protection against S. suis 2 infection in mice. (A) IgG subclasses elicited in response to HtpsC immunization. IgG1, IgG2a, IgG2b and IgG3 were significantly increased (P < 0.05, Student's t-test) in the sera of mice immunized with HtpsC. (B) Kaplan-Meier survival analysis showed greater survival for mice immunized with HtpsC compared to those immunized with PBS (p < 0.01).
To gain further insights into the immune reaction induced by rHtpsC in mice, IgG subclasses elicited in response to HtpsC immunization were measured by ELISA with class-specific anti-sera. Figure 6A indicates that IgG1, IgG2a, IgG2b and IgG3 are significantly increased in the sera of mouse immunized by HtpsC. The proportion of IgG1, IgG2a and IgG2b induced by HtpsC are similar to each other, whereas the amount of IgG3 is relatively small.
Discussion
The type II HTP is a novel class of adhesin in Streptococcus
Streptococcus species have shown to inherit 2 subfamilies of HTPs from their common ancestor.10 While type I HTPs are widely distributed across different streptococci, type II HTPs were retained only in the pyogenic group species and S. suis.10 Different from HTP I proteins that only contain HTP motif in their sequences, most HTP II proteins possess LRR domain at the C-terminal.10 Binding to host cells through surface-expressed adhesion is a first step of pathogenic bacteria in the colonization of host mucosal surfaces and host cell invasion.5 Many studies have revealed that the LRR domain is adopted to achieve protein-protein interactions across different life domains.21 Several bacteria also encode LRR-containing proteins to facilitate their adhesion and invasion into host cells.25 A previous study suggested that S. suis 2 strains could bind different components of human ECM, including Fn, Lm and collagen proteins.26 Recently, several proteins involved in S. suis 2 adherence to host proteins, such as GADPH, enolase and Lmb were identified.5,6,27 The LRR-containing structure of HtpsC implied that it might be a new adhesin in S. suis. Therefore, we expressed HtpsC and evaluated its binding to human ECM complex in vitro. As expected, HtpsC strongly adhered to human ECM complex. Furthermore, we demonstrated that deletion of this gene significantly decreased the adhesion ability of S. suis 2 to Hep2 cell, which further supported its role in adhesion in vivo. Human ECM contains 3 major components collagen, Fn and Lm, which are also presented at the surface of Hep-2 cells.23 This indicates that some of these proteins might be the receptors of HtpsC protein, and mediate the adherence of HtpsC to Hep-2 cells. As expected, both in vitro and in vivo experiments in our study supported that Fn and Lm are receptors of HrpsC. Since both Fn and Lm are detected at the surface of Hep-2 cells, HtpsC at least partially contributed to the adherence of S. suis 2 to Hep-2 cells.
A recent study revealed that S. agalactiae Slr, the ortholog of HtpsC, could bind to human type I collagen.24 However, very weak adhesion was observed between HtpsC and human type I collagen, indicating that recognition specificity among HTP II proteins may be different. Actually, the receptor specificity of LRR domain is very sensitive. Even a few amino acid substitutions would alter its specificity.28 Furthermore, positive selection has also been observed at some LRR unites among HTP II proteins.10 Additionally, our results also revealed that the HTP domain alone could adherence to both Fn and Lm. This expanded our understanding of the function of HTP family. Although have been evidenced to bind different host components, both Slr and HtpsC are involved in adherence of streptococci to their host. Considering their conserved domain composition, the type II HTP may represent a novel class of adhesin in streptococcus.
HtpsC is involved in the full virulence of S. suis 2
During the infection process, S. suis express a diverse range of pathogenic factors and these virulent proteins have varied roles for host cell adhesion, invasion, and immune evasion, cooperatively contributing to the full virulence of S. suis.29 In this study, by constructing a knockout strain, we demonstrated that htpsC is a requirement for the full virulence of S. suis in a mice model.
HTP protein family was first observed in S. pneumoniae, and subsequently found in S. pyogenes and S. suis.14,17,19 A recent comprehensive analysis suggested that HTPs could be classified into 2 subfamilies10 and both proteins are involved in the pathogenicity of different streptococcal species.9,19 During evolution, HTP genes expanded to different extents among streptococcal species.10 The S. pneumoniae genome has the maximal 4 htp genes among all sequenced streptococci, and all belong to type I subfamily.10,17 In a previous study, deletion of each of the 4 paralogs did not abolish the pathogenicity of S. pneumoniae.9 However, when 2 or more genes were deleted simultaneously, the pathogenicity of S. pneumoniae was weakened or even abolished, which indicated that the 4 paralogs might play redundant roles.9 In S. pyogenes, 2 htp genes were identified and assigned to different subfamilies. Different than S. pneumoniae, the 2 genes appeared not to complement each other, because knockout of each gene eliminated the pathogenicity of S. pyogenes.18,19 Interestingly, S. suis is not the same with respect to S. pyogenes or S. pneumoniae. The S. suis genome contains 3 htp genes,10,14 which is more than S. pyogenes but less than S. pneumoniae. Two of the S. suis htp genes belong to the HTP II subfamily, but only one (htpsC) encodes the LRR domain. Therefore, we investigated whether this gene could independently affect the virulence of S. suis. Herein, our data revealed that inactivation of htpsC is sufficient to attenuate the pathogenicity of S. suis.
Pathogens adhesion to host tissue is a critical early step in the process of infection.1 Our data revealed that the htpsC mutant had significantly deceased adhesion ability, which may contributed to its weakened virulence. Deletion of htpsC also decreased the survival of S. suis in whole blood, suggesting its role in resistance to host immunity system clearance. This kind of dual-function protein has been described in several bacteria. For example, Group B Streptococci ScpB protein mediates the adherence of GBS to epithelial cells by binding to Fn and involved in bacteria evasion of the host immune system through cleavage of C5a on the bacterial surface, which inhibits the activation of host complement system.30 The HTP I subfamily members are also dual function proteins that are involved in bacteria adherence to host cells and are capable of blocking the host complement pathway by recruiting complement factor H on bacteria surface.9,31 As HtpsC is also belonging to HTP family, a similar mechanism may adopted by the protein to help S. suis evasion from (or resistance to) host immunity clearance. This still needs further studies to test. Nevertheless, the deletion of HtpsC indeed attenuated the virulence of S. suis. These results together suggest that HtpsC is involved in the pathogenicity of S. suis.
HtpsC confers protection against infection of S. suis 2
In the past, many studies have been devoted to exploring and developing effective vaccines for preventing S. suis infection.5,6,14,32 Surface-exposed immunogenic proteins were believed to be good candidates for vaccine development, and several proteins in this regard have been identified recently by different groups.5,6,14,32 HTPs are surface exposed proteins14,19,33 and have been proved to be immunogenic antigens.20,25, 38 Some of them, including PhtD, HtpA and HtpS (termed as HtpsA in this study) confer significant protection to the host against the infection of streptococcus.13,14,19 Our work revealed that HtpsC is located on the surface of S. suis and was expressed during infection. Immunization with HtpsC elicited an immune response in mouse. Of note, a proportion of mice immunized with HtpsC had long survival times or survived after a challenge of S. suis. These results suggest that HtpsC could confer protection against infection from the highly virulent S. suis 2 strain.
IgG subclasses induced by antigen reflect the pattern of immune responses in the host.34-36 By analyzing the IgG composition of antibodies induced by HtpsC in mice, we found that IgG1, IgG2a, IgG2b and IgG3 are significantly increased in the sera of mouse immunized with HtpsC. In the mouse, IgG1 was associated with a Th2-like immune response (humoral immunity), whereas IgG2a and IgG2b were associated with a Th1-like immune response (cellular immunity).34,37 The latter type response is particularly effective at mediating bacterial opsonophagocytosis.32 In our work, the proportion of IgG1, IgG2a and IgG2b induced by HtpsC were similar, indicating both Th1- and Th2-like immune responses might have been induced by HtpsC. The amount of IgG2a+IgG2b exceeds IgG1, suggesting that Th1-like immune responses dominated the process. Notably, our previous study have revealed that S. suis protein that induces both Th1- and Th2-like immune responses is more efficient to confer protection than those only inducing humoral immunity,36 which indicated that HtpsC is a good candidate for the vaccine development against S. suis 2.
Materials and Methods
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are list in Table 1. E. coli DH5α and BL21 (DE3) were grown in Luria Broth (LB) liquid medium or plated on LB agar at 37°C. S. suis 2 strains were maintained in Todd-Hewitt broth (THB; Difco Laboratories, Detroit, MI) liquid and/or agar medium. The two cloning vectors, pMD18-T and pUC18 were used for either direct DNA sequencing or gene cloning. The pET32a vector was used for protein expression. When necessary, appropriate antibiotics were supplemented as follows: spectinomycin (Spc, Sigma) for htpsC mutant of S. suis 05ZYH33, 100 μg/ml and ampicillin (Amp, Sigma) for E. coli strains, 100 μg/ml.
Table 1.
Bacterial strains and plasmids used in this study
| Strains or plasmids | Characteristics and/or functiona | Source |
|---|---|---|
| S. suis 05ZYH33 | Virulent strain isolated from a patient with STSS | Lab collection |
| S. suis △htpsC | Isogenic △htpsC deletion mutant of strain 05ZYH33; SpcR | This study |
| E. coli DH5α | Cloning host for recombinant plasmid | Transgene |
| E. coli BL21 | Expression host for recombinant plasmid | Transgene |
| pMD18-T | Cloning vector; AmpR | Takara |
| pET32a | Expression vector; AmpR | Novagen |
| pET32a-htpsC | A recombinant expression vector containing htpsC; AmpR | This study |
| pUC18 | E. coli cloning vector, lacZ, AmpR | Takara |
| pUC18-htpsC | A recombinant vector with the background of pUC18, designed for knock-out of htpsC; AmpR, SpcR | This study |
| pSET2 | E. coli-S. suis shuttle vector; SpcR | Takamatsu et al. 39 |
| pEAZY-T5 | E. coli cloning vector, lacZ, AmpR | Transgene |
| pVA838 | E. coli-S. suis shuttle vector; EryR, CmR | Lab collection |
| pVA838-htpsC | pVA838 containing the intact htpsC gene and its upstream promoter; EryR, CmR | This study |
Ampr, ampicillin-resistant; Spcr, spectinomycin-resistant.
Sequence analysis of htpsC
To verify the sequence of htpsC in the previous genome release of 05ZYH33,22 the genomic region encompassing the ORF 1577 was resequenced based on PCR amplification with primers A1 and A2 (Table 2). The coding sequence of htpsC was reannotated by using ortholog gene of S. suis 2 strain P1/7 in the PATRIC database (http://patric.vbi.vt.edu/) as a template. Multiple sequence alignments were conducted using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html), and ESPript 2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) was used to process the output.
Table 2.
Primers used for PCR amplification and detection
| Primer | Sequences (5′–3′)a | Restriction enzyme | Function |
|---|---|---|---|
| A1 | GTCAATGATCGTAGGGATGGA | Resequence htpsC | |
| A2 | CAGTCATTCCTTGCTTATCGA | Resequence htpsC | |
| E1 | GGATCCCAAGCGTCTAACTATAGAG | BamHI | Up primer for rHtpsC expression |
| E2 | CTCGAGTTAAGGAAATATAAAATCCTCA | XhoI | Down primer for rHtpsC expression |
| LA1 | CGGAATTCTGAAGCGCAGATAAATC | EcoRI | Left arm of htpsC |
| LA2 | CGGGATCCAGTCCAAGGTCAAA | BamHI | Left arm of htpsC |
| RA1 | CGTCGACTTTTGTCCTCCTAAGCTC | SalI | Right arm of htpsC |
| RA2 | CGGTCGACAGGTAGCAGAGATAGTAC | SphI | Right arm of htpsC |
| Spc1 | CCGGATCCGTTCGTGAATACAT | BamHI | SpcR gene |
| Spc2 | CGGTCGACGTTTTCTAAAATCTGA | SalI | SpcR gene |
| Check1 | GCCATTCGCATCTTCAGCCA | Internal region of htpsC | |
| Check2 | TTGAAGGTCCGATCCCGTATGC | Internal region of htpsC | |
| Out1 | CATTAGCTGTCAGTTCTTCTAAATTCTCTGT | For combined PCR detection | |
| Out2 | GACGGAAATATCGTATGTAGTCAGCTG | For combined PCR detection | |
| C1 | GGATCCCTCGCTTACCTTTGCT | BamHI | Amplification of htpsC gene and its upstream promoter |
| C2 | CGTCGACTGCACTTTAACAAGGAT | SalI | Amplification of htpsC gene and its upstream promoter |
| F1 | GGATCCATGACCCTTCCAGATTCT | BamHI | Up primer for rHTP expression |
| F2 | CTCGAGCTATCCAACTTCAACTTTCTC | XhoI | Down primer for rHTP expression |
| F3 | GGATCCATGCTAAAGAGAGCCG | BamHI | Up primer for rLRR expression |
| F4 | CTCGAGTTATGCTTCCTTATCAACTTC | XhoI | Down primer for rLRR expression |
The underlined sequences are the restriction sites
Construction of the htpsC mutant and complementary strains
A recombination based method was used to construct the htpsC gene mutant strain of S. suis 05ZYH33 as previously described.38 Briefly, the spectinomycin resistance (Spcr) gene was amplified from pSET2 (provided by Doctor Daisuke Takamatsu,39) and then inserted into a pUC18 vector (Takara) to create the recombinant plasmid pUC18-Spcr. Two DNA fragments (LA and RA) flanking the htpsC gene were amplified from 05ZYH33 genomic DNA and cloned into pUC18-Spc to generate the knockout plasmid pUC-htpsC. The pUC-htpsC plasmid was then used for transformation of 05ZYH33 competent cells. The acquired transformants were confirmed with multiplex-PCR assays with a series of specific primers (Table 2), followed by direct DNA sequencing. Reverse transcription-PCR (RT-PCR) detection was also performed to confirm the successful inactivation of the htpsC gene in the deletion mutant, designated △htpsC.
For complementation assay, a DNA fragment containing the entire htpsC gene and its upstream promoter was amplified using primers C1 and C2, which introduced BamH I and Sal I sites, respectively. After digestion with the appropriate restriction enzymes, the resulting fragment was cloned into the E. coli-S. suis shuttle vector pVA838, to generate the htpsC-complementing plasmid pVA838-htpsC. This vector was electrotransformed directly into the htpsC mutant to obtain the complemented strain C△htpsC by using our previously reported method.38
Evaluation of bactericidal activity in human whole blood
Whole-blood bactericidal activity was conducted to measure possible bacterial virulence attenuation in blood. Bacterial strains △htpsC, C△htpsC and WT S .suis 2 were cultured to a mid-logarithmic growth phase (˜108CFU/ml), and harvested by centrifugation for 2 min at 8,000 × g. After washing 3 times with PBS, the bacteria were adjusted to 8×105CFU/ml with PBS. Then, 10 μl of △htpsC, C△htpsC or WT S .suis 2 05ZYH33 was added into 350 μl of heparinized whole blood. The mixtures were then incubated at 37°C for 8 h. Subsequently, 50 μl of sample of each group was diluted, plated on THB agar plates and cultured at 37°C for 48 h before the colonies on each plate were counted. Experiment for each sample was performed in triple, and repeated 3 times.
Assays for cell adhesion
Adhesion ability to Hep-2 cells of △htpsC, C△htpsC and WT strains was tested according to our previous publication.40 In brief, mid-logarithmic growth phase bacteria were harvested, washed, and resuspended in PBS. Carboxyfluorescein diacetate succinimidyl ester (CFDA-SE) solution was added to the bacterial suspensions to a final concentration of 10 μM. Bacterial cells were then incubated with gentle rotation at 37°C for 20 min. Human laryngeal epithelial cell line Hep-2 (CCTCC GDC004) cells were first cultured to monolayer at 37°C with 5% CO2 in RPMI 1640 medium (Gibco), and then detached with 0.05% trypsin. The cells were washed twice, and resuspended in PBS. CFDA-SE Labeled bacteria (△htpsC, C△htpsC or 05ZYH33) were added into Hep-2 cells in a ratio of 100:1. The mixtures were incubated at 37°C with gentle agitation for 2 h, and then fixed with 4% (wt/vol) paraformaldehyde for flow cytometry (BD) analysis. The mean fluorescent intensity (MFI) values of each group were used to measure adherent bacteria. Experiment for each sample was performed in triple, and repeated 3 times.
Evaluation of the pathogenicity of △htpsC in a mouse model
To compare the pathogenicity of △htpsC, C△htpsC and WT strains, a bacterial challenge experiment was performed in a previously constructed mouse model.41 BALB/c mice (female, 4-weeks-old) were divided into 4 groups (N=10 ). Bacterial cultures of △htpsC, C△htpsC and 05ZYH33) were harvested, washed 3 times by THB and then adjusted to 1×108 CFU/ ml in THB to inoculate of BALB/c mice by intraperitoneal injection (1 ml per mouse). The THB without any bacteria were inoculated into the control group (1 ml per mouse). Mice were monitored for one week and deaths were recorded. A Kaplan-Meier survival analysis was performed using the SPSS package to test the significant difference among different groups. All animal experiments were approved by the Research Institute for Medicine of Nanjing Command ethics committee, PLA.
Expression of recombinant proteins and preparation of antiserum
The full length HtpsC protein and the 2 truncated fragments that encompass either HTP domain or LRR domain were expressed as previously described5,14 to obtain the recombinant protein rHtpsC, rHTP and rLRR. Briefly, the coding sequences were amplified from S. suis 2 Chinese strain 05ZYH33 genomic DNA with 3 pairs of primers (Table 2), and subcloned into the pET32a vector. The recombinant vectors were then transformed into E. coli BL21 for over-expression of the 3 recombinant proteins. Recombinant proteins were purified using Ni–NTA columns (Novagen).
Antiserum against rHtpsC, rHTP and rLRR were prepared respectively as previously described.14 Pre-immune serum was collected from New Zealand White rabbits (female, 1.5 kg) and then animals were injected subcutaneously at multiple sites with approximately 1 mg/kg of purified recombinant protein emulsified with Freund's complete adjuvant (1:1). After two weeks, each rabbit received the first booster injection with the same antigen concentration emulsified with Freund's incomplete adjuvant (1:1). After one week, each rabbit received the second booster injection with the same antigen concentration emulsified with Freund's incomplete adjuvant (1:1). Then, sera was collected when the booster was administered 7 d later and assayed for biological activity.
Flow cytometry detect the surface expression of HtpsC
To determine the surface localization of HtpsC on S. suis 2, flow cytometry (FCM) was performed as previously described6,14 with some modifications. Briefly, Mid-logarithmic growth phase (∼108CFU/ml) cultures of WT, △htpsC and complementary strain (C△htpsC) were incubated with rabbit-anti-HtpsC serum for 1 hour at 4°C. After incubation, cells were then incubated with FITC-conjugated goat anti-rabbit IgG (Boster, China) for 1 h at 4°C. Cells were then washed 3 times and fixed with 2% paraformaldehyde for 30 min, and analyzed by FCM (BD). The experiment was performed in triplicate and repeated 3 times.
Analyzing binding of recombinant proteins to human ECM, Fn, Lm and type I collagen
An Enzyme-Linked ImmunoSorbent Assay (ELISA) was used to evaluate binding of HtpsC, rHTP and rLRR to immobilized human ECM complex (composed of collagens, fibronectin and laminin, as well as some heparin sulfate proteoglycan) or different ECM components. Human ECM (BD), Fn (Sigma), Lm (Sigma) and type I collagen (BD) were diluted to 25 μg/ml with 0.1 M carbonate coating buffer (pH 9.6) and added to a 96-well ELISA plate (Greiner Bio-One) overnight at 4°C. After washing 3 times with PBS containing 0.05% (v/v) Tween 20 (PBST, pH 7.3) and blocking with 1% BSA (in PBST), recombinant proteins (1–10 μg/ml) were added to wells following 3 times washes with PBST and incubated for 2 h at room temperature. Rabbit-anti-recombinant protein antiserum and horseradish peroxidase labeled goat-anti-rabbit antibodies (BIOSS) were then added to the wells successively with each following 3 times of PBST washes. After incubation for 1 h at room temperature and 3 times washes, the substrate TMB (Beyotime) was added for visualization of binding. The plate was analyzed using a 550 microplate reader (Bio-Rad) at 450 nm. The experiment was performed in triplicate and repeated 3 times.
Adherence of bacteria to Fn and Lm
Adherence of bacteria to Fn and Lm was conducted to analyze the interactions between bacteria and host as previously described with some modification.42 05ZYH33, △htpsC and C△htpsC strains were used in the binding assays. The microplates were coated with Fn and Lm (10 μg/ml) respectively overnight at 4°C, washed, and subsequently blocked with 3% BSA (in PBST) for 2h at 37°C. Bacterial cells (05ZYH33, △htpsC and C△htpsC) were grown in THB to OD600 = 0 .6, then cultures were harvested, washed with PBS. Bacteria were suspended in PBS to give 5×108 bacteria per ml in 1% BSA, and added to the wells. Background values were obtained from wells containing no bacteria culture. Following incubated for 2h at 37°C, the plates were washed and reacted with the rabbit-anti-05ZYH33 antiserum. Bindings were detected with secondary HRP-conjugated antibody as described above. Each assay was performed in triplicate and repeated 3 times.
Protection assay
An assay of protection of HtpsC was performed as previously described36 with some modifications. Briefly, 4-week-old, specific pathogen free (SPF) grade (SLAC, China) female BALB/c mice were divided into 2 groups (N = 10 mice/group). Mice in both groups were immunized subcutaneously with purified rHtpsC protein (25μg) or PBS that were emulsified with Freund's complete adjuvant (sigma). After 14 and 21 days, the mice were booster immunized with the same amount of rHtpsC or PBS emulsified with Freund's incomplete adjuvant, respectively. Then 3 d after the last booster, mouse blood samples were drawn via the vena caudalis and a specific antibody titer and IgG types were measured with indirect ELISA as described in a previous study.36
A week after the last injection, 1×108 CFU of highly pathogenic S. suis 2 strain 05ZYH33 suspended in sterile Todd-Hewitt broth was injected intraperitoneally into the mice. After the challenge, mice were monitored for 7 d A Kaplan-Meier survival analysis was used to test the significant difference between the 2 groups. All animal experiments were approved by the Research Institute for Medicine of Nanjing Command ethics committee, PLA.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Doctor Daisuke Takamatsu at National Institute of Animal Health of Japan for kindly providing the pSET2 E. coli-S. suis shuttle vector.
Funding
This work was supported by the National Natural Science Foundation of China (81172794, 81071317, 31170124, 81471920 and 81171527), Medical science and technology innovation project of Nanjing Military Command, China (ZX39) and Natural Science Foundation of Jiangsu Province (BK2011098, BK2011097, BK2012080 and BK20140096). Dr. Feng is a recipient of the “Young 1000 Talents” Award.
References
- 1.Singh B, Fleury C, Jalalvand F, Riesbeck K. Human pathogens utilize host extracellular matrix proteins laminin and collagen for adhesion and invasion of the host. FEMS Microbiol Rev 2012; 36:1122-80; PMID:22537156; http://dx.doi.org/ 10.1111/j.1574-6976.2012.00340.x. [DOI] [PubMed] [Google Scholar]
- 2.Baums CG, Valentin-Weigand P. Surface-associated and secreted factors of Streptococcus suis in epidemiology, pathogenesis and vaccine development. Anim Health Res Rev 2009; 10:65-83; PMID:19558750; http://dx.doi.org/ 10.1017/S146625230999003X [DOI] [PubMed] [Google Scholar]
- 3.Bayle L, Chimalapati S, Schoehn G, Brown J, Vernet T, Durmort C. Zinc uptake by Streptococcus pneumoniae depends on both AdcA and AdcAII and is essential for normal bacterial morphology and virulence. Mol Microbiol 2011; 82:904-16; PMID:22023106; http://dx.doi.org/ 10.1111/j.1365-2958.2011.07862.x [DOI] [PubMed] [Google Scholar]
- 4.Fu Q, Wei Z, Liu X, Xiao P, Lu Z, Chen Y. Glyceraldehyde-3-phosphate dehydrogenase, an immunogenic Streptococcus equi ssp. zooepidemicus adhesion protein and protective antigen. J Microbiol Biotechnol 2013; 23:579-85; PMID:23568215; http://dx.doi.org/ 10.4014/jmb.1209.09037 [DOI] [PubMed] [Google Scholar]
- 5.Zhang YM, Shao ZQ, Wang J, Wang L, Li X, Wang C, Tang J, Pan X. Prevalent distribution and conservation of Streptococcus suis Lmb protein and its protective capacity against the Chinese highly virulent strain infection. Microbiol Res 2014; 169(5–6):395-401. [DOI] [PubMed] [Google Scholar]
- 6.Feng Y, Pan X, Sun W, Wang C, Zhang H, Li X, Ma Y, Shao Z, Ge J, Zheng F, et al.. Streptococcus suis enolase functions as a protective antigen displayed on the bacterial cell surface. J Infect Dis 2009; 200:1583-92; PMID:19848587; http://dx.doi.org/ 10.1086/644602 [DOI] [PubMed] [Google Scholar]
- 7.Moschioni M, Pansegrau W, Barocchi MA. Adhesion determinants of the Streptococcus species. Microb Biotechnol 2010; 3:370-88; PMID:21255337; http://dx.doi.org/ 10.1111/j.1751-7915.2009.00138.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tu AH, Fulgham RL, McCrory MA, Briles DE, Szalai AJ. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect Immun 1999; 67:4720-4; PMID:10456922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogunniyi AD, Grabowicz M, Mahdi LK, Cook J, Gordon DL, Sadlon TA, Paton JC. Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J 2009; 23:731-8; PMID:18971260; http://dx.doi.org/ 10.1096/fj.08-119537 [DOI] [PubMed] [Google Scholar]
- 10.Shao ZQ, Zhang YM, Pan XZ, Wang B, Chen JQ. Insight into the Evolution of the Histidine Triad Protein (HTP) Family in Streptococcus. PLoS One 2013; 8:e60116; PMID:23527301; http://dx.doi.org/ 10.1371/journal.pone.0060116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cork AJ, Jergic S, Hammerschmidt S, Kobe B, Pancholi V, Benesch JL, Robinson CV, Dixon NE, Aquilina JA, Walker MJ. Defining the structural basis of human plasminogen binding by streptococcal surface enolase. J Biol Chem 2009; 284:17129-37; PMID:19363026; http://dx.doi.org/ 10.1074/jbc.M109.004317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pancholi V, Fischetti VA. α-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem 1998; 273:14503-15; PMID:9603964; http://dx.doi.org/ 10.1074/jbc.273.23.14503 [DOI] [PubMed] [Google Scholar]
- 13.Plumptre CD, Ogunniyi AD, Paton JC. Vaccination against Streptococcus pneumoniae Using Truncated Derivatives of Polyhistidine Triad Protein D. PLoS One 2013; 8:e78916; PMID:24205351; http://dx.doi.org/ 10.1371/journal.pone.0078916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao Z, Pan X, Li X, Liu W, Han M, Wang C, Wang J, Zheng F, Cao M, Tang J. HtpS, a novel immunogenic cell surface-exposed protein of Streptococcus suis, confers protection in mice. FEMS Microbiol Lett 2011; 314:174-82; PMID:21133988; http://dx.doi.org/ 10.1111/j.1574-6968.2010.02162.x [DOI] [PubMed] [Google Scholar]
- 15.Seiberling M, Bologa M, Brookes R, Ochs M, Go K, Neveu D, Kamtchoua T, Lashley P, Yuan T, Gurunathan S. Safety and immunogenicity of a pneumococcal histidine triad protein D vaccine candidate in adults. Vaccine 2012; 30:7455-60; PMID:23131206; http://dx.doi.org/ 10.1016/j.vaccine.2012.10.080 [DOI] [PubMed] [Google Scholar]
- 16.Plumptre CD, Ogunniyi AD, Paton JC. Polyhistidine triad proteins of pathogenic streptococci. Trends Microbiol 2012; 20:485-93; PMID:22819099; http://dx.doi.org/ 10.1016/j.tim.2012.06.004 [DOI] [PubMed] [Google Scholar]
- 17.Adamou JE, Heinrichs JH, Erwin AL, Walsh W, Gayle T, Dormitzer M, Dagan R, Brewah YA, Barren P, Lathigra R, et al.. Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect Immun 2001; 69:949-58; PMID:11159990; http://dx.doi.org/ 10.1128/IAI.69.2.949-958.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waldemarsson J, Areschoug T, Lindahl G, Johnsson E. The streptococcal Blr and Slr proteins define a family of surface proteins with leucine-rich repeats: camouflaging by other surface structures. J Bacteriol 2006; 188:378-88; PMID:16385027; http://dx.doi.org/ 10.1128/JB.188.2.378-388.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunitomo E, Terao Y, Okamoto S, Rikimaru T, Hamada S, Kawabata S. Molecular and biological characterization of histidine triad protein in group A streptococci. Microbes Infect 2008; 10:414-23; PMID:18403236; http://dx.doi.org/ 10.1016/j.micinf.2008.01.003 [DOI] [PubMed] [Google Scholar]
- 20.Bersch B, Bougault C, Roux L, Favier A, Vernet T, Durmort C. New Insights into Histidine Triad Proteins: Solution Structure of a Streptococcus pneumoniae PhtD Domain and Zinc Transfer to AdcAII. PLoS One 2013; 8:e81168; PMID:24312273; http://dx.doi.org/ 10.1371/journal.pone.0081168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol 2001; 11:725-32; PMID:11751054; http://dx.doi.org/ 10.1016/S0959-440X(01)00266-4 [DOI] [PubMed] [Google Scholar]
- 22.Chen C, Tang J, Dong W, Wang C, Feng Y, Wang J, Zheng F, Pan X, Liu D, Li M, et al.. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS One 2007; 2:e315; PMID:17375201; http://dx.doi.org/ 10.1371/journal.pone.0000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotter G, Weedle R, Kavanagh K. Monoclonal antibodies directed against extracellular matrix proteins reduce the adherence of Candida albicans to HEp-2 cells. Mycopathologia 1998; 141:137-42; PMID:9755505; http://dx.doi.org/ 10.1023/A:1006940203962 [DOI] [PubMed] [Google Scholar]
- 24.Bober M, Morgelin M, Olin AI, von Pawel-Rammingen U, Collin M. The membrane bound LRR lipoprotein Slr, and the cell wall-anchored M1 protein from Streptococcus pyogenes both interact with type I collagen. PLoS One 2011; 6:e20345; PMID:21655249; http://dx.doi.org/ 10.1371/journal.pone.0020345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonazzi M, Lecuit M, Cossart P. Listeria monocytogenes internalin and E-cadherin: from structure to pathogenesis. Cell Microbiol 2009; 11:693-702; PMID:19191787; http://dx.doi.org/ 10.1111/j.1462-5822.2009.01293.x [DOI] [PubMed] [Google Scholar]
- 26.Esgleas M, Lacouture S, Gottschalk M. Streptococcus suis serotype 2 binding to extracellular matrix proteins. FEMS Microbiol Lett 2005; 244:33-40; PMID:15727818; http://dx.doi.org/ 10.1016/j.femsle.2005.01.017 [DOI] [PubMed] [Google Scholar]
- 27.Brassard J, Gottschalk M, Quessy S. Cloning and purification of the Streptococcus suis serotype 2 glyceraldehyde-3-phosphate dehydrogenase and its involvement as an adhesin. Vet Microbiol 2004; 102:87-94; PMID:15288930; http://dx.doi.org/ 10.1016/j.vetmic.2004.05.008 [DOI] [PubMed] [Google Scholar]
- 28.Dodds PN, Lawrence GJ, Ellis JG. Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 2001; 13:163-78; PMID:11158537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Y, Zhang H, Ma Y, Gao GF. Uncovering newly emerging variants of Streptococcus suis, an important zoonotic agent. Trends Microbiol 2010; 18:124-31; PMID:20071175; http://dx.doi.org/ 10.1016/j.tim.2009.12.003 [DOI] [PubMed] [Google Scholar]
- 30.Beckmann C, Waggoner JD, Harris TO, Tamura GS, Rubens CE. Identification of novel adhesins from Group B streptococci by use of phage display reveals that C5a peptidase mediates fibronectin binding. Infection and immunity 2002; 70:2869-76; PMID:12010974; http://dx.doi.org/ 10.1128/IAI.70.6.2869-2876.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kallio A, Sepponen K, Hermand P, Denoel P, Godfroid F, Melin M. Role of Pht proteins in attachment of Streptococcus pneumoniae to respiratory epithelial cells. Infect Immun 2014; 82:1683-91; PMID:24491577; http://dx.doi.org/ 10.1128/IAI.00699-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang A, Chen B, Li R, Mu X, Han L, Zhou H, Chen H, Meilin J. Identification of a surface protective antigen, HP0197 of Streptococcus suis serotype 2. Vaccine 2009; 27:5209-13; PMID:19596417; http://dx.doi.org/ 10.1016/j.vaccine.2009.06.074 [DOI] [PubMed] [Google Scholar]
- 33.Plumptre CD, Ogunniyi AD, Paton JC. Surface association of Pht proteins of Streptococcus pneumoniae. Infect Immun 2013; 81:3644-51; PMID:23876799; http://dx.doi.org/ 10.1128/IAI.00562-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens TL, Bossie A, Sanders VM, Fernandez-Botran R, Coffman RL, Mosmann TR, Vitetta ES. Regulation of antibody isotype secretion by subsets of antigen-specific helper T cells. Nature 1988; 334:255-8; PMID:2456466; http://dx.doi.org/ 10.1038/334255a0 [DOI] [PubMed] [Google Scholar]
- 35.Bowes T, Wagner ER, Boffey J, Nicholl D, Cochrane L, Benboubetra M, Conner J, Furukawa K, Furukawa K, Willison HJ. Tolerance to self gangliosides is the major factor restricting the antibody response to lipopolysaccharide core oligosaccharides in Campylobacter jejuni strains associated with Guillain-Barre syndrome. Infect Immun 2002; 70:5008-18; PMID:12183547; http://dx.doi.org/ 10.1128/IAI.70.9.5008-5018.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L, Cheng G, Wang C, Pan X, Cong Y, Pan Q, Wang J, Zheng F, Hu F, Tang J. Identification and experimental verification of protective antigens against Streptococcus suis serotype 2 based on genome sequence analysis. Curr Microbiol 2009; 58:11-7; PMID:18839251; http://dx.doi.org/ 10.1007/s00284-008-9258-x [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Martinez G, Gottschalk M, Lacouture S, Willson P, Dubreuil JD, Jacques M, Harel J. Identification of a surface protein of Streptococcus suis and evaluation of its immunogenic and protective capacity in pigs. Infect Immun 2006; 74:305-12; PMID:16368985; http://dx.doi.org/ 10.1128/IAI.74.1.305-312.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M, Wang C, Feng Y, Pan X, Cheng G, Wang J, Ge J, Zheng F, Cao M, Dong Y, et al.. SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS One 2008; 3:e2080; PMID:18461172; http://dx.doi.org/ 10.1371/journal.pone.0002080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takamatsu D, Osaki M, Sekizaki T. Construction and characterization of Streptococcus suis-Escherichia coli shuttle cloning vectors. Plasmid 2001; 45:101-13; PMID:11322824; http://dx.doi.org/ 10.1006/plas.2000.1510 [DOI] [PubMed] [Google Scholar]
- 40.Zheng F, Ji H, Cao M, Wang C, Feng Y, Li M, Pan X, Wang J, Qin Y, Hu F, et al.. Contribution of the Rgg transcription regulator to metabolism and virulence of Streptococcus suis serotype 2. Infect Immun 2011; 79:1319-28; PMID:21149588; http://dx.doi.org/ 10.1128/IAI.00193-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Y, Cao M, Shi J, Zhang H, Hu D, Zhu J, Zhang X, Geng M, Zheng F, Pan X, et al.. Attenuation of Streptococcus suis virulence by the alteration of bacterial surface architecture. Sci Rep 2012; 2:710; PMID:23050094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joseph jaffe, Shira Natanson-yaron G.Caparon M. Protein F2,a novel fibronectin-binding protein from Streptococcus pyogenes,possesses two binding domains. Mol Microbiol 1996; 21:373-84; PMID:8858591; http://dx.doi.org/ 10.1046/j.1365-2958.1996.6331356.x [DOI] [PubMed] [Google Scholar]