Summary
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
A fundamental goal of genomics is to identify the complete set of expressed proteins. Automated annotation strategies rely on assumptions about protein-coding sequences (CDSs), e.g., they are conserved, do not overlap, and exceed a minimum length. However, an increasing number of newly discovered proteins violate these rules. Here we present an experimental and analytical framework, based on ribosome profiling and linear regression, for systematic identification and quantification of translation. Application of this approach to lipopolysaccharide-stimulated mouse dendritic cells and HCMV-infected human fibroblasts identifies thousands of novel CDSs, including micropeptides and variants of known proteins, that bear the hallmarks of canonical translation and exhibit comparable translation levels and dynamics to annotated CDSs. Remarkably, many translation events are identified in both mouse and human cells even when the peptide sequence is not conserved. Our work thus reveals an unexpected complexity to mammalian translation suited to provide both conserved regulatory or protein-based functions.
Introduction
Next-generation sequencing makes it possible to determine an organism's genomic sequence and complement of transcribed RNAs with relative ease. However, identifying the full set of protein coding regions has proven challenging. Of the vast number of open reading frames (ORFs) in any transcriptome, only a small fraction is translated by the ribosome. These protein-coding DNA sequences (CDSs) were traditionally identified with automated methods that applied a set of rational simplifying assumptions—for example, that CDSs initiate at AUG codons, span at least 100 codons, do not overlap each other, and/or exhibit phylogenetic conservation (Maeda et al., 2006). Although proteomic databases do include exceptions to these rules, such proteins are systematically underrepresented due to the difficulty of their identification (Andrews and Rothnagel, 2014; Frith et al., 2006; Pauli et al., 2014).
A range of observations argue that automated annotation approaches fail to capture the true complexity of physiologically important translation events. Ribosome-translated “micropeptides”, a class of proteins shorter than 100 amino acids, are well documented in prokaryotes and have recently been implicated in diverse eukaryotic functions such as development (Pauli et al., 2014; Savard et al., 2006), muscle contraction (Anderson et al., 2015; Magny et al., 2013), and DNA repair (Slavoff et al., 2014). In other cases, alternative initiation produces an N-terminally truncated or extended version of a protein that behaves differently from the canonical form (Acland et al., 1990; Brubaker et al., 2014). Translation may be functionally important independent of the sequence of the encoded polypeptide, for example by impacting the translation of downstream CDSs or modulating the stability of host RNAs (Morris and Geballe, 2000). Additionally, even peptides without clear functional roles can be immunogenic: peptides derived from the human cytomegalovirus (HCMV) “long noncoding” RNA P2.7, for example, were found to robustly stimulate T cell memory responses only in humans with a history of HCMV infection (Ingolia et al., 2014). Thus translation of previously neglected ORFs may contribute in important ways to cellular and organismal biology, emphasizing the need for an unbiased and comprehensive strategy to evaluate translation empirically.
Design
Recently, ribosome profiling, in which ribosome-protected mRNA fragments (RPFs) are isolated and sequenced (Ingolia et al., 2009), has enabled the empirical annotation of CDSs, leading to the identification of translated ORFs that do not always adhere to the length, start codon identity, and ORF organization rules characteristic of traditional ORFs. However, annotating CDSs from ribosome profiling data is nontrivial (Bazzini et al., 2014; Chew et al., 2013; Guttman et al., 2013; Ingolia et al., 2011; Pauli et al., 2015). Previous efforts have used peak detection and support vector machine (SVM) algorithms to search for translation initiation sites, but not all of the identified initiation sites lead to translated CDSs (Guttman et al., 2013; Pauli et al., 2015). Other methods have identified CDSs based on the heightened density (Aspden et al., 2014; Ingolia et al., 2009) or 3-nucleotide periodicity of RPFs along the body of a putative CDS (Bazzini et al., 2014; Smith et al., 2014), but these approaches have difficulty evaluating the translation of overlapping CDSs and therefore have typically been applied only to the longest AUG-initiated ORF in a region of interest, despite the fact that dually decoded regions occur in many genes (Michel et al., 2012). An important recent advance involved the use of an ensemble classifier that leverages a combination of features to identify CDSs (Chew et al., 2013). However, like other approaches this method was not engineered to identify short or overlapping CDSs.
Here we describe the ORF Regression Algorithm for Translational Evaluation of RPFs (ORF-RATER), which uses ribosome profiling data to identify and quantify translation from CDSs regardless of start codon, length, or overlap with other CDSs. ORF-RATER makes the assumption that translated ORFs display a pattern of ribosome occupancy that mimics that of annotated genes (e.g., initiation and termination peaks and phased elongation) and then queries all possible ORFs for evidence of translation by fitting them to expected profiles. ORF-RATER is based on linear regression, which naturally integrates multiple lines of evidence simultaneously and enables each ORF to be evaluated in the context of nearby and overlapping ORFs. The models are empirically constructed with reads mapping to annotated CDSs from the same dataset, so ORF-RATER is broadly applicable to ribosome profiling data from any species or cell type.
We applied ORF-RATER to two ribosome profiling datasets from mammalian primary cells undergoing dynamic physiological responses: mouse bone marrow-derived dendritic cells (BMDCs) stimulated with lipopolysaccharide (LPS) and human foreskin fibroblasts (HFFs) undergoing infection with HCMV (Stern-Ginossar et al., 2012). In both datasets, ORF-RATER identifies translation of annotated CDSs, but it also discovers thousands of novel CDSs. By providing a principled, comprehensive, and general solution to the challenge of CDS annotation, this work advances our understanding of the complexity of mammalian translation and facilitates comparisons of this complexity across growth conditions, cell types, and species. Cross-referencing of mouse BMDC and HFF translation reveals that many novel CDSs are robustly translated in both despite there being sequence conservation in only a subset of cases; thus, the newly identified translation events likely play conserved regulatory roles in addition to generating novel functional proteins.
Results
An integrated experimental and computational approach for the identification and quantification of translation
In order to develop an approach for the identification, validation, and quantification of translation events, we collected a multifaceted dataset that comprised ribosome profiling and mass spectrometry (MS) data from mouse BMDCs throughout 12 hours of LPS stimulation (Figure 1A), a treatment known to induce dynamic changes to the proteome (Jovanovic et al., 2015). At nine time points prior to and following LPS stimulation, cells were treated with one of three mechanistically distinct translation inhibitors (harringtonine [Harr], lactimidomycin [LTM], or cycloheximide [CHX]) or left untreated (no-drug [ND]) before isolation and sequencing of RPFs. These treatments were chosen to emphasize distinct phases of canonical ribosome-mediated protein synthesis. Specifically, translation initiation was highlighted by Harr (Ingolia et al., 2011) and LTM (Lee et al., 2012); translation elongation was highlighted by CHX and ND; and translation termination was highlighted by ND. For all treatments, we mapped RPFs to a BMDC-specific transcriptome assembly prepared from mouse BMDC RNA-seq data (Shalek et al., 2013). Data from the four treatment conditions showed the characteristic patterns of density at translated CDSs, readily apparent in “metagene” profiles from annotated CDSs (Figure 1B).
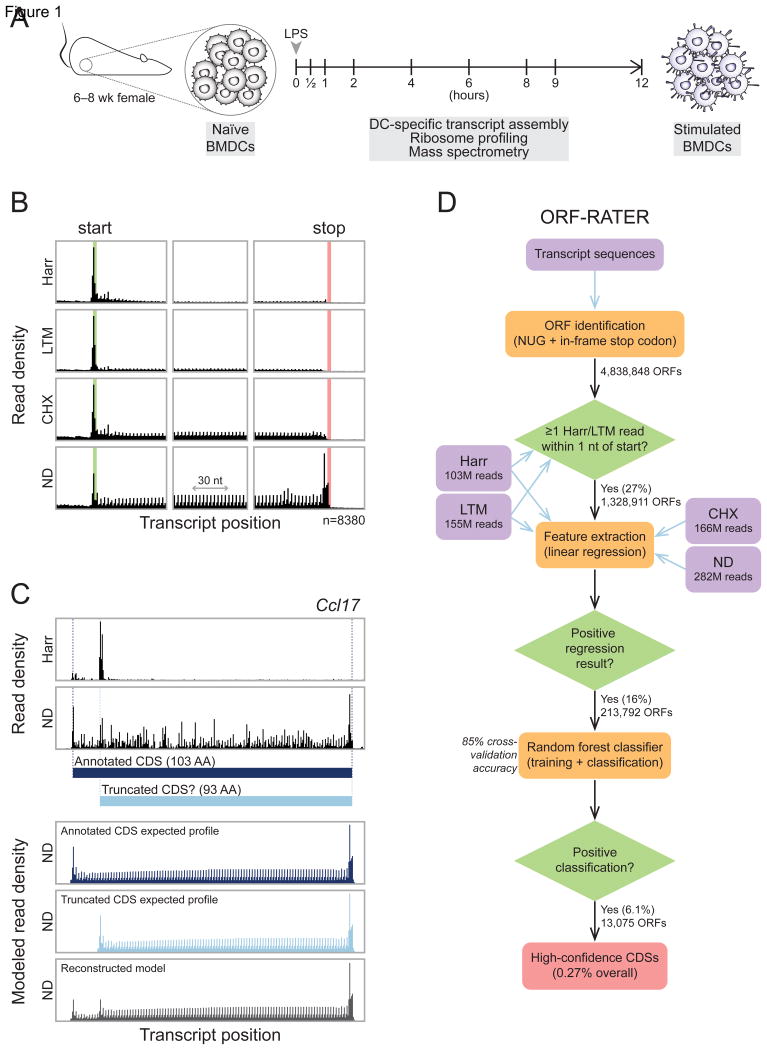
Figure 1. ORF-RATER identifies translated ORFs comprehensively in mouse BMDCs.
(A) Naïve BMDCs were isolated and stimulated with LPS for up to 12 hours. Ribosome profiling data sets were collected at the nine times indicated prior to or during stimulation, and mass spectrometry data sets were collected at 0, 2, 6, and 12 hours. (B) The average read density (“metagene”) profiles of the four BMDC ribosome profiling datasets near annotated start codons (left), at the center of annotated CDSs (center), and near annotated stop codons (right) reveal features of translation highlighted by each treatment (harringtonine [Harr], lactimidomycin [LTM], cycloheximide [CHX], or no-drug [ND]). The highlighted green and red regions indicate annotated start and stop codons, respectively. (C) Top, observed RPFs within the annotated CDS of chemokine ligand 17 (Ccl17). Ribosome density at an AUG codon 10 codons downstream from the canonical AUG following Harr treatment suggests that a truncated form lacking the N-terminal 10 amino acids may be translated in addition to the canonical form. Bottom, linear regression of the observed RPFs in the ND condition against the expected profiles of the two candidate ORFs suggests that both may be translated. (D) The ORF-RATER pipeline globally evaluates translation. NUG-initiated ORFs are identified from transcript sequences assembled from BMDC RNA-seq data and the Ensembl and UCSC Known Genes databases. After removing ORFs whose translation initiation sites lack ribosome density following Harr or LTM treatment, the remaining ORFs are analyzed by linear regression (C), the results of which are assayed for significance using a random forest classifier. See Figure S1 for the full distribution of scores.
We reasoned that bona fide translation at unannotated CDSs should show density patterns of RPFs similar to those of annotated CDSs. Additionally, if two or more overlapping ORFs are translated, the final read density would be expected to be the sum of the read densities of each independent ORF. Linear regression is, therefore, a natural tool to evaluate the contribution of each possible ORF to the observed read densities and is the basis of ORF-RATER (Figure 1C). To implement our algorithm, we first calculated metagene profiles of annotated CDSs (Figure 1B) to construct typical profiles of productive translation. Next, we used linear regression to find the level of translation of each ORF such that the sum of their expected read densities is most consistent with the observed read density. The regression fits read density in a read length-sensitive manner, enabling it to distinguish 80S footprints from those arising from non-ribosomal sources, akin to the recently described fragment length organization similarity score (Ingolia et al., 2014). A caveat of our approach is that it is likely to miss sites of non-canonical translation in which the RPF pattern does not match that at canonical CDSs. The extent to which such translation occurs remains to be addressed; here, our intention has been to identify those ORFs that are most convincingly translated, i.e., those whose RPF patterns are most similar to annotated CDSs. In particular, we do not assume that every RPF identified necessarily indicates translation.
The ORF-RATER pipeline (Figure 1D) first identifies all NUG-initiated ORFs within a transcriptome, to account for both traditional AUG, as well as the most common near cognate, initiation sites (Ingolia et al., 2011; Lee et al., 2012). ORFs lacking RPFs at the putative translation initiation site in either Harr or LTM datasets are immediately discarded. Following regression analysis, the vast majority of ORFs are assigned a translation level of zero. To determine which of the remainder represent true translation events rather than noise (e.g. a slightly elevated read density in part of a CDS, or a few reads corresponding to ribosomes scanning through a 5′ UTR), we applied a random forest classifier to the regression output (see Experimental Procedures). Briefly, we chose the random forest approach over other machine learning algorithms (e.g., SVM) because it uses minimal parameters and does not require imposition of an arbitrary distance metric. The classifier training set included all ORFs beginning with AUG codons that are at least 100 amino acids, a set that comprises 13,478 previously annotated CDSs which served as a positive set and the remaining 11,009 which served as the negative set. The random forest classifier achieved 85% cross-validation accuracy on the training set. The final scores ranged from 0 to 1, with higher values indicating greater confidence that the ORF was translated. We defined the “high-confidence” set of CDSs (i.e., translated ORFs) as those that received a score of at least 0.8, a threshold that is conservative yet captures the majority of expressed annotated CDSs (Figure S1).
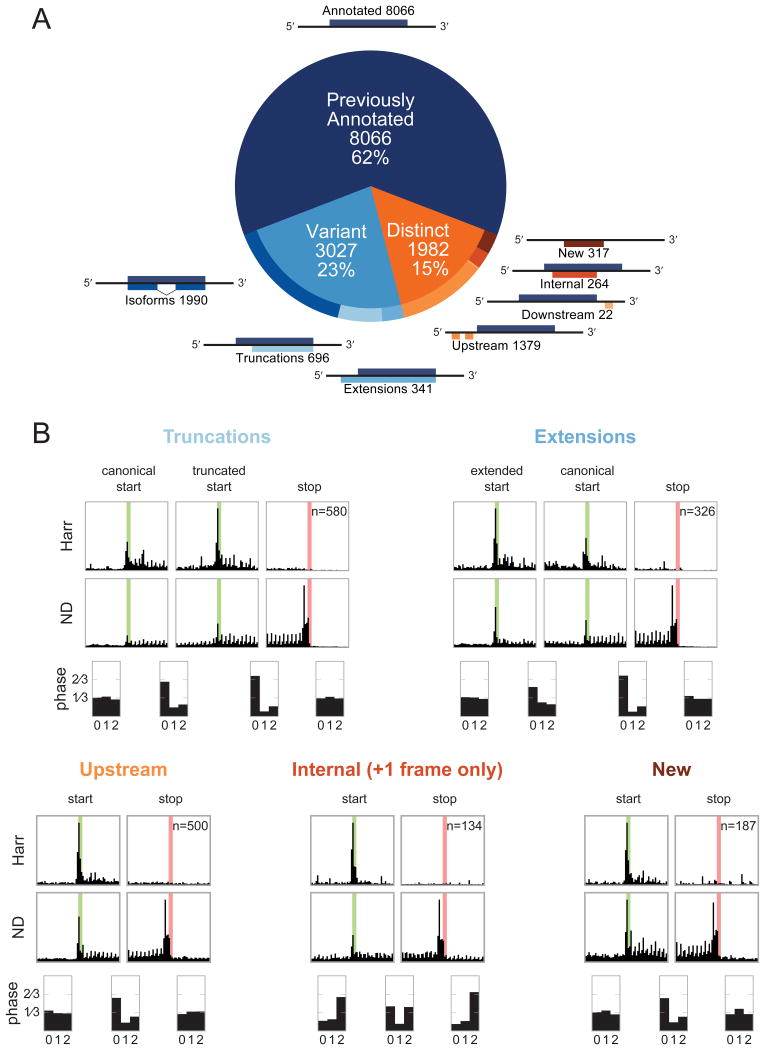
ORF-RATER identifies 13,075 high-confidence CDSs (Figure 2A and Table S1). The majority (62%) of these were previously annotated CDSs. Of the novel CDSs, 3,027 are “variants” of annotated CDSs such as splice isoforms or N-terminal extensions or truncations. The remaining 1,982 novel translated ORFs are “distinct” from annotated CDSs and consist primarily of “upstream” ORFs (uORFs) initiating in 5′ leaders. Distinct CDSs also include a set of “internal” ORFs that initiate in an alternative reading frame within canonical CDSs, and “new” ORFs on transcripts that previously lacked protein-coding annotations. Very few (22) “downstream” CDSs were identified in 3′ UTRs of annotated CDSs.
Figure 2. Previously unannotated translated CDSs in BMDCs fall into several classes, each of which displays patterns consistent with active translation.
(A) ORF-RATER identifies 13,075 high-confidence translated ORFs. The majority of these are previously annotated CDSs, and the majority of the remainder are variants of canonical CDSs that share portions of the coding sequence. ORFs distinct from annotated CDSs occur primarily in 5′ UTRs, though a sizable subset are found on transcripts without previously appreciated coding potential or in alternate frames of canonical CDSs. See Figure S1A for the distribution of ORF-RATER scores for each type, and Table S1 for a complete list of all high-confidence CDSs. (B) Metagene profiles of each class of new CDS display the hallmarks of translation, including peaks of density at newly identified start codons following Harr treatment, peaks of density at stop codons under ND treatment, and greater read density in between. Translated truncations (top left) and extensions (top right) display peaks of density at both the canonical and novel translation initiation sites, suggesting that both are used on average. The average read density in all translated regions show 3-nucleotide periodicity in the expected reading frame, with the exception of internal CDSs, for which the reading frame is on average a superposition of the canonical and alternative frames. Metagene profiles for the LTM and CHX datasets are plotted in Figure S2.
The metagene profiles of each type of newly identified CDS display patterns consistent with active translation (Figures 2B and S2; compare Figure 1B), including peaks of density at the start and stop codons and higher average density in between, which drops off in the regions immediately adjacent to the ORF. Importantly, reads within (but not outside of) novel CDSs display 3-nucleotide periodicity in the expected reading frame, with the notable exception that the average density within internal CDSs includes phased reads in both the canonical and alternative reading frames. This underscores the ability of ORF-RATER to identify overlapping CDSs. The average read density of translated N-terminal truncations and extensions includes peaks at both the canonical and alternative initiation codons, suggesting that both are typically used.
High-confidence translated ORFs identified by ORF-RATER are strongly (92%) enriched for AUG start codons (Table 1 and Figure S1B), consistent with the standard model of translation initiation. Of the various types of novel CDSs, all aside from N-terminal extensions are enriched for AUG-initiation. N-terminal extensions are specifically dis-enriched for AUG codons because annotation pipelines typically select the most upstream in-frame AUG as the canonical initiation site, so 5′ leaders almost never contain in-frame AUG codons. The ORF-RATER pipeline's enrichment for AUG codons does not result from an engineered bias; indeed, including only AUG-initiated ORFs in the training set for the random forest (in both positive and negative sets) avoids explicitly penalizing non-AUG-initiated ORFs. Nevertheless, prior experimental evidence has shown that translation can initiate from near-cognate codons (Mehdi et al., 1990; Peabody, 1989; Starck et al., 2012); ORF-RATER identifies 575 ORFs that initiate at CUG codons, 321 at GUG codons, and 107 at UUG codons. Except for extensions, each category of novel CDSs shows a preference for AUG > CUG > GUG ≥ UUG initiation codons, consistent with other empirical annotation efforts and in vitro results (Ingolia et al., 2011; Lee et al., 2012; Mehdi et al., 1990; Peabody, 1989).
Table 1. Translation initiation occurs predominantly at AUG codons.
| AUG | CUG | GUG | UUG | |
|---|---|---|---|---|
| Overall | 12072 [92.3%] | 575 [4.4%] | 321 [2.5%] | 107 [0.8%] |
| Annotated | 8061 [99.9%] | 5 [0.1%] | 0 [0.0%] | 0 [0.0%] |
| Variant | 2235 [73.8%] | 441 [14.6%] | 264 [8.7%] | 87 [2.9%] |
| Isoforms | 1875 [94.2%] | 67 [3.4%] | 42 [2.1%] | 6 [0.3%] |
| Truncations | 323 [46.4%] | 187 [26.9%] | 145 [20.8%] | 41 [5.9%] |
| Extensions | 37 [10.9%] | 187 [54.8%] | 77 [22.6%] | 40 [11.7%] |
| Novel | 1776 [89.6%] | 129 [6.5%] | 57 [2.9%] | 20 [1.0%] |
| Upstream | 1233 [89.4%] | 94 [6.8%] | 40 [2.9%] | 12 [0.9%] |
| Downstream | 18 [81.8%] | 2 [9.1%] | 2 [9.1%] | 0 [0.0%] |
| Internal | 239 [90.5%] | 15 [5.7%] | 5 [1.9%] | 5 [1.9%] |
| New | 286 [90.2%] | 18 [5.7%] | 10 [3.2%] | 3 [0.9%] |
See also Figure S1B.
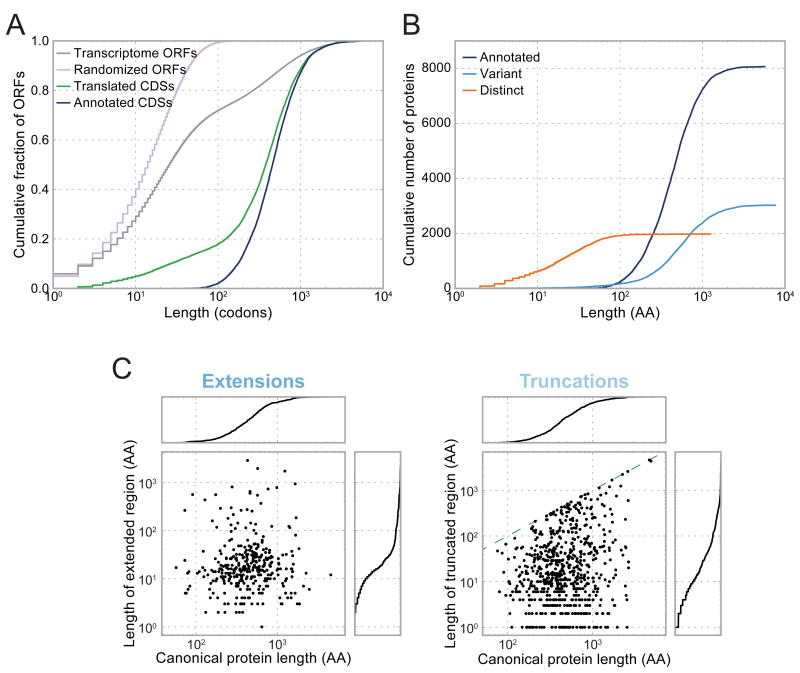
The set of translated CDSs are predominantly longer than 100 codons, but include a substantial fraction (18%) of shorter ORFs, well in excess of previous annotations (Figure 3A). Nonetheless, relative to the background length distribution of all ORFs on real or scrambled transcripts, CDSs identified by ORF-RATER are heavily skewed towards longer lengths. Long CDSs principally encode canonical proteins or their variants, whereas translated short CDSs almost all code for proteins whose amino acid sequences are distinct from annotated proteins (Figure 3B). Among variants of canonical proteins, N-terminal extensions and truncations add or remove a median of approximately 15 amino acids, which has the potential to impact the structure, targeting (Schatz and Dobberstein, 1996), or stability (Varshavsky, 1996) of the resulting protein (Figure 3C).
Figure 3. Novel CDSs include many short ORFs and variants of canonical proteins missed by prior annotations.
(A) Compared to the distribution of ORF sizes on real or scrambled transcripts, translated CDSs are highly enriched for long ORFs, but to a lesser extent than prior annotations. (B) Nearly all short translated CDSs are distinct from canonical proteins, and nearly all long translated CDSs are canonical proteins or their variants. (C) Length of extended (left) or truncated (right) regions is plotted as a function of the length of the canonical protein. Cumulative distributions are plotted to the right or above each scatter plot. For truncated CDSs, the dashed green line indicates the position beyond which the entire CDS would be removed.
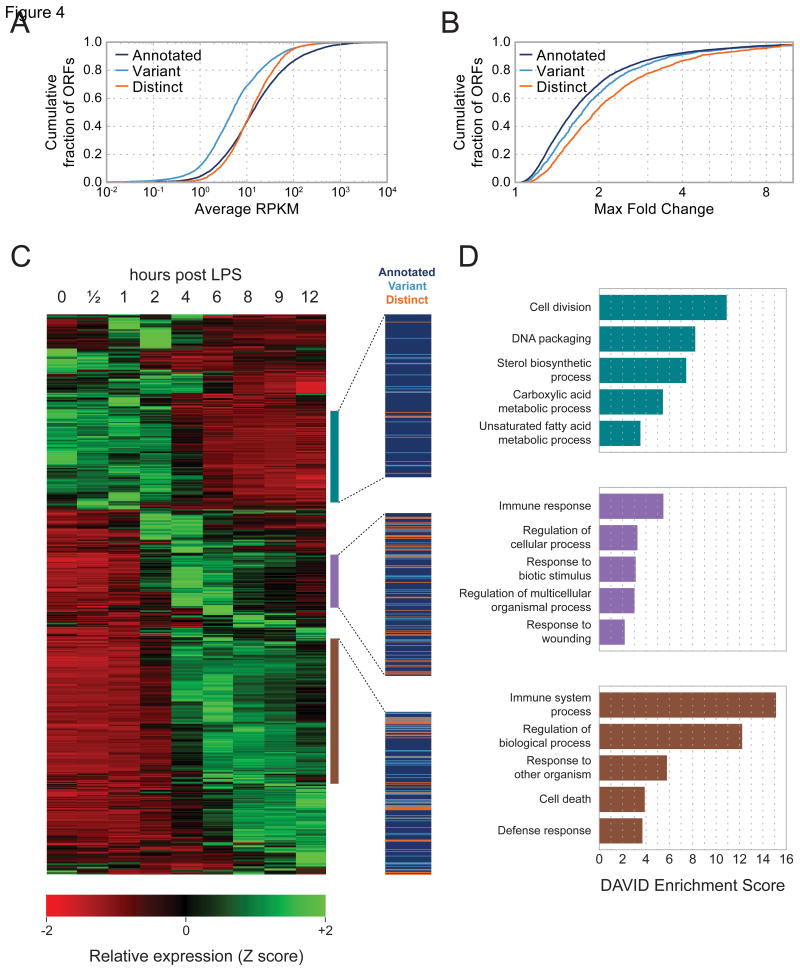
Newly identified CDSs are translated at comparable levels and with similar dynamics to annotated CDSs. To calculate translation rates, we applied a simplified regression strategy to all high-confidence ORFs that fractionally assigns reads in regions where multiple ORFs overlap. Distinct translated ORFs were translated with a narrower but nearly identically centered distribution of rates compared to previously annotated CDSs, whereas variants of canonical CDSs were translated at a somewhat lower (∼3-fold) rate (Figure 4A).
Figure 4. Novel CDSs are translated at similar levels and with similar dynamics to annotated CDSs in response to LPS stimulation.
(A) Cumulative distributions of translation rates for each class of translated CDS. (B) Cumulative distributions of maximal fold-change across the time course of LPS stimulation. (C) Hierarchically clustered heat map of dynamically regulated CDSs showing translation rates at indicated intervals of LPS stimulation. Each row represents one CDS. Three highlighted clusters show CDSs whose translation is maximal at early (top), intermediate (center), or late (bottom) time points. Each cluster contains a mixture of novel and annotated CDSs, indicated by the colored lines at right. RPKM values for all CDSs are included in Table S1. (D) GO term enrichments for the annotated genes contained in the three clusters highlighted in (C).
We calculated the maximal translational fold change across the LPS stimulation time series (see Experimental Procedures), and found that expression of all three ORF types is regulated to similar extents (Figure 4B). To identify groups of CDSs with correlated expression dynamics, we performed hierarchical clustering for the 2,896 CDSs with greater than 2-fold expression changes. We identify 3 major classes of expression profiles that are characterized by peak expression early (0–2 hours post LPS stimulation), mid (4–6 hours), and late (6 hours and later) in the time series. Each cluster is well populated by both annotated and novel CDSs, indicating that the translation of both groups is similarly regulated (Figure 4C). Gene ontology (GO) analysis of the annotated genes in these clusters reveals that BMDCs undergoing LPS stimulation downregulate housekeeping genes, and strongly activate expression of immune-related CDSs (Figure 4D). Interestingly, expression of many novel distinct CDSs peaks in the mid and late phases of the time series, mimicking the dynamics of known immune effectors, such as Irf8, Irf9, Stat1, Stat2, and Stat5a.
In this study, ORF-RATER benefits from the inclusion of four ribosome profiling datasets to search for patterns indicative of translation. In many cases, however, collection of all of these data may be infeasible. To investigate how much the additional datasets improve ORF-RATER's performance, we modified the algorithm to run on the ND dataset only. We estimate that loss of the drug-treated datasets increased the false positive rate by a factor of ∼3.5 (Supplemental Methods), leading to a lower rate of identification for annotated ORFs (82% vs 70%). The lack of translation initiation inhibitor-treated datasets (e.g. Harr or LTM) also greatly increases the number of ORFs to consider as possibly translated, increasing the algorithm's computational burden. If translation inhibitor-treated datasets are unavailable, a reasonable compromise might be to restrict analysis to only AUG- or AUG/CUG-initiated ORFs, which will decrease both the false positive rate and computational burden.
Conservation of mammalian translation events occurs in the absence of codon-level sequence conservation
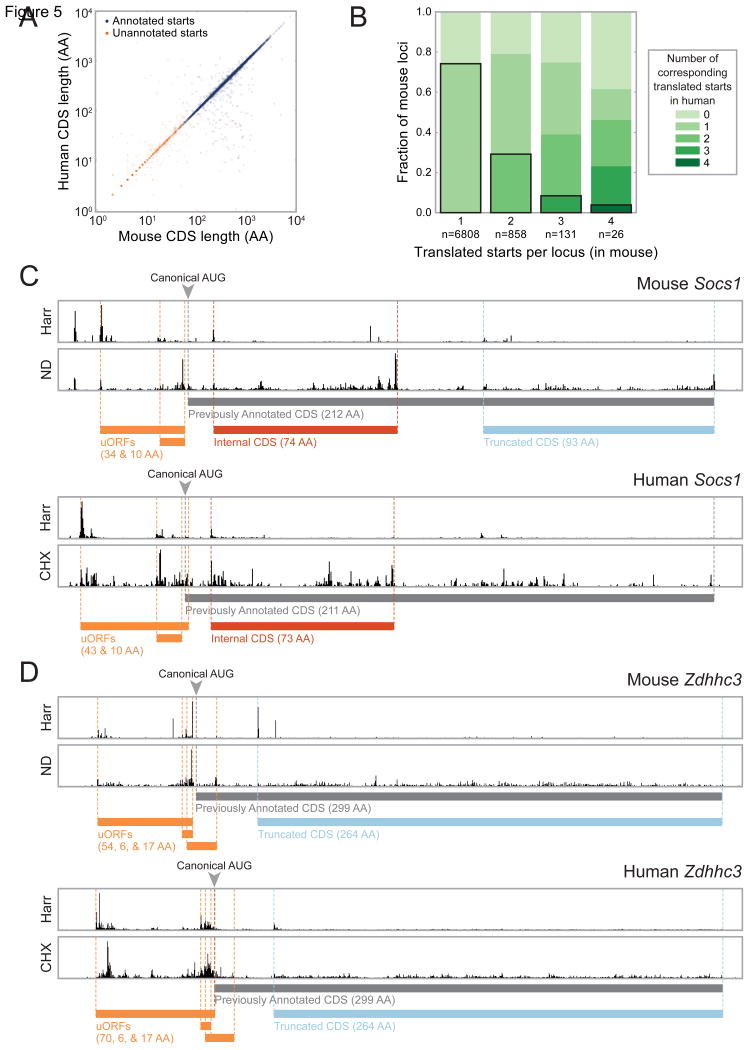
To enable cross-species comparison of translation, we applied ORF-RATER to a previously collected ribosome profiling dataset from HCMV-infected HFFs (Stern-Ginossar et al., 2012); the same four treatment conditions were applied to these cells. Of the observed sites of translation initiation in the mouse BMDC data for which a homologous human codon could be identified (see Experimental Procedures), 68% are also identified by ORF-RATER in the HFF data. Excluding those previously annotated as translation start sites in either species, 33% are identified by ORF-RATER in both. Remarkably, although translation of these unannotated CDSs appears to be conserved from mouse to human, the majority (∼60%) of them give rise to polypeptides lacking evidence of codon-level conservation (Figure S3A) (Lin et al., 2011). Instead, what appears to be maintained is the length of the open reading frame (Figure 5A), suggesting that in these cases translation may be conserved for regulatory purposes.
Figure 5. Many novel translated CDSs are seen in both human and mouse cells.
(A) Sites of productive translation initiation in both mouse BMDCs and HFFs encode proteins of similar length regardless of whether the protein had been previously annotated. Many of these previously unannotated proteins do not appear to be conserved at the level of protein sequence (Figure S3A). (B) Many loci encode multiple corresponding CDSs in both mice and humans. (C) RPF density at the Socs1 locus in mouse BMDCs (top) and HFFs (bottom) show similar organization of translated ORFs. (D) Zdhhc3 encodes four CDSs translated in both mouse BMDCs and HFFs. The longest translated uORF does not appear to be conserved for protein function despite being translated in both species; its multiple sequence alignment is included in Figure S3B. Reporter constructs indicate that the AUGs upstream of the one initiating the truncated form of Zdhhc3—including the canonical start codon—are repressive (Figure S4A).
We see many homologous loci that maintain the number (Figure 5B) and organization of translation events from mouse to human. Conserved polycistronic organization is readily apparent at the suppressor of cytokine signaling 1 (Socs1) gene (Figure 5C), where, in both mice and humans, ORF-RATER identifies two previously known uORFs that inhibit translation of the annotated CDS (Gregorieff et al., 2000; Schlüter et al., 2000). Instead of full-length Socs1, the ribosome profiling data suggest translation primarily of an internal out-of-frame CDS (in mice and humans) and a heavily truncated form of Socs1 (in mouse). Although the truncated form of Socs1 is not assigned a high confidence of translation by ORF-RATER in human cells, the locus does display some of the key features consistent with translation (such as a small peak of density with Harr treatment), suggesting that this may represent a false negative event, underscoring the conservative nature of the ORF-RATER algorithm.
A second example of conserved polycistronic organization is seen at the palmitoyltransferase Zdhhc3 gene (Figure 5D), where four CDSs are translated in both mice and humans: three uORFs and an N-terminal truncation missing the first 35 amino acids. Strikingly, the longest uORF in the Zdhhc3 gene shows little evidence of codon-level sequence conservation (Figure S3B), suggesting that the encoded polypeptide does not perform a conserved function despite being translated in both mammals. Using a series of fluorescent reporter constructs in which the N-terminal segment of Zdhhc3 is fused to eGFP, we confirmed that the truncated AUG is the predominant site of translation initiation (Figure S4A). Mutation of the AUGs initiating the three uORFs increased the fluorescence, suggesting that they play an inhibitory role that is likely conserved between mouse and human. Surprisingly, mutation of the canonical AUG also increased the fluorescence. We originally hypothesized that translation of Zdhhc3's uORFs might cause ribosomes to “bypass” the canonical AUG and instead reinitiate at the next one downstream; however, removal of the truncation-encoding AUG in conjunction with loss of the uORFs lead to a low fluorescence level, suggesting that even in the absence of the uORFs, the canonical AUG is bypassed or encodes an unstable protein.
A set of novel translation events display characteristics of functional proteins
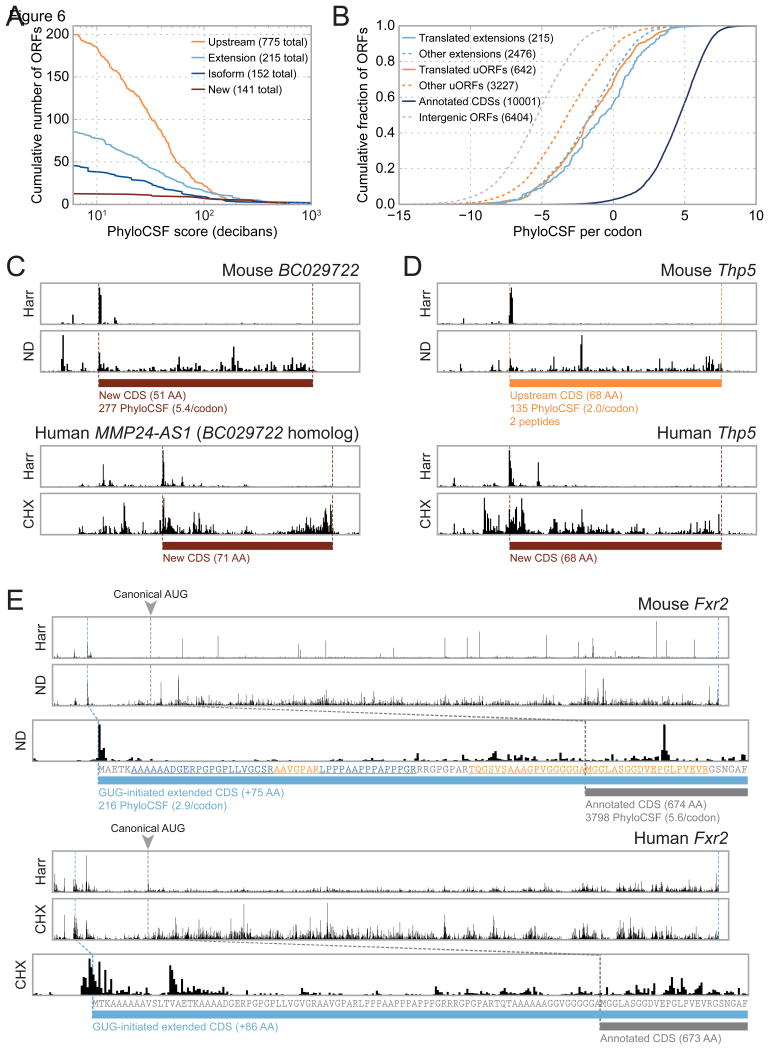
While some newly-identified CDSs are likely to play regulatory roles rather than producing stable protein products, multiple lines of evidence argue that a subset of novel CDSs encode functional proteins. First, many newly identified mouse CDSs show signatures of codon-level phylogenetic conservation. For this analysis, we excluded codons overlapping canonical CDSs and applied the PhyloCSF algorithm (Lin et al., 2011) to evaluate the likelihood that the remaining codons were protein-coding in the most recent common ancestor of the Euarchontoglires (the minimal phylogenetic clade including both mouse and human). Although the majority of novel CDSs received negative scores (likely in part due to depletion of conserved ORFs by prior annotation pipelines), hundreds received positive scores (Figure 6A). These phylogenetic conservation scores cannot be explained by the background level of conservation: translated uORFs and N-terminal extensions are significantly enriched for signatures of codon-level conservation relative to both intergenic ORFs and analogous non-translated ORFs in BMDCs (Figure 6B, see Experimental Procedures). Some highly conserved CDSs are also expressed in HFFs; for example, the BC029722 gene encodes a 51-amino acid protein in mouse (expanded to 71 amino acids and named MMP24-AS1 in human) whose sequence is strongly conserved across mammals (Figures 6C and S6A). A C-terminally eGFP-tagged MMP24-AS1 protein expressed in HeLa cells localized to the endoplasmic reticulum and the Golgi apparatus (Figure S6B), although the protein does not contain a known targeting sequence.
Figure 6. A significant subset of novel CDSs display signatures of codon-level conservation.
(A) For each threshold value, the number of novel CDSs of each type whose PhyloCSF score exceeds that threshold is plotted. PhyloCSF scores are calculated for only those codons non-overlapping with canonical CDSs. Scores indicate the log-likelihood that the ancestral locus was protein-coding; values of 10 or 20 correspond to 10:1 or 100:1 likelihood, respectively. The legend indicates the total number of ORFs for which a sequence alignment could be obtained, including those assigned negative PhyloCSF scores. (B) Cumulative distributions of per-codon PhyloCSF scores for translated uORFs and extensions of canonical CDSs. In both cases, PhyloCSF scores are significantly greater at translated CDSs relative to non-translated CDSs of the same type. Intergenic ORFs receive significantly lower scores and serve as negative controls. Because PhyloCSF scores vary linearly with the length of the sequence alignment, when comparing ORFs of different sizes, each score is normalized by the number of codons considered. See also Figure S3A. (C) RPF density at the mouse BC029722 (top) and human MMP24-AS1 (bottom) genes show translation of a previously unannotated CDS that is highly conserved phylogenetically. The multiple sequence alignment is shown in Figure S6A. A C-terminal eGFP fusion of human MMP24-AS1 was found to localize to the ER and Golgi apparatus (Figure S6B). (D) The Thp5 gene encodes a previously unannotated, conserved 68-amino acid protein in both mouse (top) and human (bottom). Two peptides from the mouse protein are identified by MS; full peptide and protein MS results are listed in Tables S2 and S3, and quality metrics are plotted in Figure S5. (E) Translation initiation of the Fxr2 gene occurs at an upstream GUG codon in both mouse BMDCs (top) and HFFs (bottom). In both cases, the canonical AUG initiation site appears to be unused. The translated region upstream of the canonical AUG appears to be highly conserved, and encodes multiple peptides detected by MS (peptide sequences highlighted in orange and blue). Translation initiation of Fxr2 via a GUG codon was confirmed via transient transfection with fluorescent reporter constructs (Figure S4B).
Shotgun proteomics identifies tryptic peptides corresponding to the polypeptide products of more than one hundred novel translation events (Table S2), confirming that they accumulate to appreciable levels in mouse BMDCs. For this analysis, only those peptides that do not map to canonical proteins are considered; standard peptide scoring metrics suggest that these peptides are only slightly less reliably identified than those matching canonical proteins (Figure S5). The large majority of the novel MS-confirmed proteins are variants of annotated proteins. Only a handful of CDSs encoding proteins distinct from canonical proteins are observed by MS; among these are translated uORFs also seen in other MS- and ribosome profiling-based surveys, such as those of Slc35a4, Smcr7l/Mief1 (Andreev et al., 2015; Vanderperre et al., 2013), and Polr2m/Grinl1a (Oyama et al., 2007). The relatively small number of detected peptides corresponding to the translation products of novel CDSs may reflect their rapid turnover or the difficulty of identifying them by MS. CDSs that are distinct from canonical proteins are challenging to detect because they are nearly all short (Figure 3B) and therefore encode fewer tryptic peptides than canonical proteins. Novel variants of canonical proteins are generally long enough to encode many tryptic peptides, but are challenging to distinguish from their canonical counterparts due to sequence identity. In total, 149,107 peptides were detected, corresponding by maximum parsimony to the translation products of 9,724 CDSs on 7,617 genes (Table S3).
The novel CDSs that are most amenable to immediate functional analyses are those supported by both phylogenetic and MS evidence. The mouse T helper cell-induced peptide 5 (Thp5) gene encodes one such CDS, which is translated in both mouse BMDCs and HFFs and produces a conserved 68-amino acid protein detectable by MS (Figure 6D). In mouse, this novel CDS occurs upstream of a non-conserved annotated protein that was previously found to play a role in T cell activation but that does not appear to be translated in BMDCs. Equally compelling is an N-terminal extension encoded in the fragile X mental retardation syndrome-related protein 2 (Fxr2) gene. In both the HFF and mouse BMDC datasets, Fxr2 is not translated from the annotated AUG initiation codon, but rather from a significantly upstream GUG codon (Figure 6E). The 75-amino acid extended region in mouse, which is enriched for alanine, proline, and glycine, receives a high PhyloCSF score and encompasses multiple peptides observed by MS. In HFFs, ORF-RATER suggests that translation initiates at a GUG codon even further upstream than the one corresponding to the one identified in mouse DCs. Manually inspecting the profile of read density, however, both GUGs appear to be plausible translation initiation sites; in particular, both have peaks of density following Harr treatment. To resolve this ambiguity, we prepared reporter constructs in which the first exon of Fxr2 was fused to eGFP, and in which each GUG codon or the annotated AUG codon was mutagenized to GGG or AGG, respectively (Figure S4B). The fluorescence intensity produced by transient transfection of these constructs in human embryonic kidney (HEK) 293T cells strongly suggests that the second GUG (i.e., the one homologous to that identified in mouse DCs) is the predominant site of translation initiation.
Conserved use of a GUG translation initiation site is exceedingly rare in mammals; one of the few well-documented examples is for the translation initiation factor Eif4g2 (Takahashi et al., 2005). Eif4g2 promotes cap-independent translation, including its own (Henis-Korenblit et al., 2000); Fxr2 is also known to serve as a translational regulator (Darnell et al., 2009; Guo et al., 2011), making its conserved use of a GUG translation initiation site particularly intriguing.
Discussion
Ribosome profiling—the deep sequencing of ribosome-protected mRNA footprints—offers the potential to identify all translated CDSs, but the comprehensive interpretation of these data remains challenging. Here we introduce a general framework for the empirical identification and quantification of translation, based on the application of the ORF-RATER algorithm to ribosome profiling data. ORF-RATER makes the minimal assumption that translated ORFs exhibit a pattern of ribosome density that displays the key features of protein synthesis, as reflected in the average profiles of previously annotated CDSs. The regression-based nature of ORF-RATER enables it to assess the protein-coding potential of all ORFs in a transcriptome, revealing translated CDSs that were disregarded by previous annotation pipelines, including those that are short, overlapping, or that do not initiate at AUG codons.
Application of ORF-RATER to two datasets from primary mammalian cells undergoing physiological processes illustrates the ability of this approach to expand our knowledge of the proteome. The majority of the novel translation events we identify produce variants of annotated proteins, such as isoforms or N-terminal truncations or extensions. ORF-RATER additionally identifies numerous translated ORFs upstream of or in a different reading frame relative to canonical CDSs. In many cases, a single transcript encodes multiple translated CDSs, indicating that, in addition to being commonplace in viruses and prokaryotes, polycistronic transcripts are well represented in mammalian transcriptomes. Finally, ORF-RATER identifies a set of translated ORFs on transcripts that do not encode canonical proteins.
Multiple lines of evidence support the ability of ORF-RATER to robustly identify and quantify bona fide and physiologically relevant translation. First, it captures the large majority of annotated CDS that are significantly expressed; conversely, a clear majority of all identified CDSs match previous annotations. Second, the translation start sites for novel CDS are highly enriched for AUG codons, despite having assigned equal a priori possibility to all NUG codons. Third, novel translated CDSs display features characteristic of ribosome-mediated polypeptide synthesis: peaks of ribosome density at translation initiation and termination codons, due to the slow kinetics of translation initiation and termination; marked drop off in density immediately upstream of the translation start site and downstream of the first in frame stop codon; 3-nucleotide periodicity of density within the CDS, reflecting the triplet nature of the genetic code; and peaks of density at translation initiation sites following treatment with inhibitors of translation initiation. Fourth, novel CDSs are translated at levels similar to canonical CDSs, and undergo similar dynamics in response to LPS stimulation. Fifth, mass spectrometry confirms that the protein products of some of these novel translation events accumulate to appreciable levels. Finally, an enriched fraction of the novel CDSs identified in mouse BMDCs show evidence of phylogenetic sequence conservation, and many are also translated in HFFs regardless of sequence conservation.
It should be straightforward to expand the ORF-RATER approach to an array of different species, conditions and cell types, thus enabling comprehensive comparison of translation. Given the antigenic potential of nonconventional translation events (Ingolia et al., 2014), such comprehensive maps could inform the design of immunomodulatory therapies, for example by identifying cancer specific antigens. On a technical level, the flexibility of the linear regression approach of the ORF-RATER algorithm facilitates its expansion to include additional features, such as the density of shorter footprints recently demonstrated to represent active translation (Lareau et al., 2014).
The outstanding challenge now is to define the functions of the newly identified translation events. Some undoubtedly produce functional micropeptides; however, many of the CDSs translated in both the mouse and human datasets do not appear to show conservation at the amino acid level, suggesting that it is the act of translation itself (rather than the resultant polypeptide) that is functionally important. Surgically mutating individual ORFs will enable their function(s) to be disambiguated from the functions of neighboring ORFs or of their host transcripts. Performing such experiments genome-wide is facilitated by CRISPR technology, which enables one to shut down transcription of whole transcripts or to introduce targeted mutations at particular ORFs. When paired with ORF-RATER, these tools will allow the function of each translation event to be mapped comprehensively—a key goal in deciphering the information content of the genome.
Limitations
The ORF-RATER pipeline identifies translated CDSs whose patterns of ribosome occupancy resemble those of annotated CDSs. The algorithm is tuned to indicate the highest-confidence sites of translation, at the expense of an increased false negative rate, so that in some cases, a translated ORF may be assigned a low score. Nonetheless, in cases in which prior evidence of translation exists, the ORF-RATER score may be used to supplement that knowledge, for example to prioritize among a set of possibly translated ORFs, even if all of those ORFs received low or moderate scores. ORF-RATER is additionally unsuited to the identification of translation events with non-canonical ribosome density. For example, the algorithm currently does not consider pause sequences, programmed frameshifts, or stop codon read-through, though it could be retrofitted to recognize such events. A final limitation is of a more technical nature. ORF-RATER performs its linear regression on each gene independently, but viral genomes may not be divisible into distinct genes, meaning that ORF-RATER as currently implemented would be applied to the entire genome simultaneously. This is computationally intractable, so the algorithm would need to be revised for application to such genomes.
Experimental Procedures
RPF and MS samples were prepared from mouse BMDCs with or without LPS stimulation and analyzed as previously described (Jovanovic et al., 2015; Mertins et al., 2013; Stern-Ginossar et al., 2012). RPF sequences were aligned to a BMDC-specific transcriptome and mapped to the ribosome P-site position. Metagene profiles for each treatment condition were assembled by averaging normalized read densities of canonical CDSs. For each possible NUG-initiated ORF within each gene, these profiles were scaled and fit to the observed read density using non-negative least-squares regression. A random forest classifier was used to combine the output of these regressions (across the multiple treatment conditions) into a final score for each ORF. Quantification of translation at different time points of LPS stimulation was achieved using a simplified regression procedure applied to ORFs that received a strongly positive score from the ORF-RATER pipeline (Supplemental Methods).
Conservation analysis was performed on portions of newly identified translated ORFs that were non-overlapping with annotated CDSs, as well as matched sets of non-translated ORFs. Alignments from 10 species of mammals were extracted and analyzed using PhyloCSF (Lin et al., 2011).
The ORF-RATER pipeline was also applied to previously collected RPF sequences from HFFs undergoing CMV infection. To enable comparison of mouse and human translation, translation initiation sites in mouse BMDCs were mapped to corresponding positions in the human genome using liftover software (Hinrichs et al., 2006).
Supplementary Material
Acknowledgments
We thank J. Dunn, M. Kampmann, G.-W. Li, M. Thomson, G. Brar, C. Jan, and L. Gilbert for helpful discussion and technical advice. We also thank Y. Chen, E. Chow, J. Lund, and the UCSF Center for Advanced Technology for help with sequencing, and C. Reiger and M. DeVera for administrative support. This research was supported by the Helen Hay Whitney Foundation postdoctoral fellowship (A.P.F.), the Howard Hughes Medical Institute (HHMI) Gilliam Fellowship for Advanced Studies (E.H.R.), the advanced postdoctoral fellowship of the Swiss National Science Foundation and the Marie Sklodowska-Curie International Outgoing Fellowship (M.J.), and HHMI (A.R. and J.S.W.).
Footnotes
Author Contributions: A.P.F., E.H.R., M.J., A.R., and J.S.W. were primarily responsible for the conception, design, and interpretation of the experiments. A.P.F., E.H.R., M.J., and R.R. conducted experiments. A.P.F., E.H.R., and B.J.H. performed computational analyses. A.P.F. and E.H.R. developed the ORF-RATER algorithm. E.H.R. and M.J. collected ribosome profiling data. M.J., P.M., and S.A.C. collected and analyzed mass spectrometry data. N.S.-G. contributed to the analysis and interpretation of HFF data. B.J.H. assembled the BMDC- and HFF-specific transcriptomes. R.R. isolated and differentiated BMDCs. N.H. contributed experimental methods and reagents. N.T.I. contributed to the design of the algorithm and the analysis and interpretation of the results.
Accession Numbers: Sequencing data have been submitted to the NCBI Gene Expression Omnibus under the accession code GSE74139. Original mass spectra have been submitted to MassIVE (http://massive.ucsd.edu) under the identifier MSV000079361.
References
- Acland P, Dixon M, Peters G, Dickson C. Subcellular fate of the lnt-2 oncoprotein is determined by choice of initiation codon. Nature. 1990;343:662–665. doi: 10.1038/343662a0. [DOI] [PubMed] [Google Scholar]
- Anderson DM, Anderson KM, Chang C, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, O'Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife. 2015;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MAS, Brocard M, Couso J. Extensive translation of small open reading frames revealed by Poly-Ribo-Seq. Elife. 2014;3:e03528. doi: 10.7554/eLife.03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, Giraldez AJ. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker SW, Gauthier AE, Mills EW, Ingolia NT, Kagan JC. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell. 2014;156:800–811. doi: 10.1016/j.cell.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development. 2013;140:2828–2834. doi: 10.1242/dev.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Darnell RB. Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs. Hum Mol Genet. 2009;18:3164–3177. doi: 10.1093/hmg/ddp255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Forrest AR, Nourbakhsh E, Pang KC, Kai C, Kawai J, Carninci P, Hayashizaki Y, Bailey TL, Grimmond SM. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2006;2:e52. doi: 10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Pyronnet S, Sonenberg N, Veillette A. Regulation of SOCS-1 expression by translational repression. J Biol Chem. 2000;275:21596–21604. doi: 10.1074/jbc.M910087199. [DOI] [PubMed] [Google Scholar]
- Guo W, Zhang L, Christopher DM, Teng Z, Fausett SR, Liu C, George OL, Klingensmith J, Jin P, Zhao X. RNA-binding protein FXR2 regulates adult hippocampal neurogenesis by reducing Noggin expression. Neuron. 2011;70:924–938. doi: 10.1016/j.neuron.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henis-Korenblit S, Strumpf NL, Goldstaub D, Kimchi A. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol Cell Biol. 2000;20:496–506. doi: 10.1128/mcb.20.2.496-506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–8. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M, Rooney MS, Mertins P, Przybylski D, Chevrier N, Satija R, Rodriguez EH, Fields AP, Schwartz S, Raychowdhury R, et al. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015;347:1259038. doi: 10.1126/science.1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, Brown PO. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife. 2014;3:e01257. doi: 10.7554/eLife.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci USA. 2012;109:E2424–32. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–82. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Kasukawa T, Oyama R, Gough J, Frith M, Engström PG, Lenhard B, Aturaliya RN, Batalov S, Beisel KW. Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS Genet. 2006;2:e62. doi: 10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, Couso JP. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341:1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- Mehdi H, Ono E, Gupta KC. Initiation of translation at CUG, GUG, and ACG codons in mammalian cells. Gene. 1990;91:173–178. doi: 10.1016/0378-1119(90)90085-6. [DOI] [PubMed] [Google Scholar]
- Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani D, Burgess MW, Gillette MA, Jaffe JD, Carr SA. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Choudhury KR, Firth AE, Ingolia NT, Atkins JF, Baranov PV. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res. 2012;22:2219–2229. doi: 10.1101/gr.133249.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama M, Kozuka-Hata H, Suzuki Y, Semba K, Yamamoto T, Sugano S. Diversity of translation start sites may define increased complexity of the human short ORFeome. Mol Cell Proteomics. 2007;6:1000–1006. doi: 10.1074/mcp.M600297-MCP200. [DOI] [PubMed] [Google Scholar]
- Pauli A, Valen E, Schier AF. Identifying (non-) coding RNAs and small peptides: Challenges and opportunities. Bioessays. 2015;37:103–112. doi: 10.1002/bies.201400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S, Mitchell A, Ma J, Dubrulle J, Reyon D, et al. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science. 2014;343:1248636. doi: 10.1126/science.1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- Savard J, Marques-Souza H, Aranda M, Tautz D. A segmentation gene in tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell. 2006;126:559–569. doi: 10.1016/j.cell.2006.05.053. [DOI] [PubMed] [Google Scholar]
- Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Schlüter G, Boinska D, Nieman-Seyde S. Evidence for translational repression of the SOCS-1 major open reading frame by an upstream open reading frame. Biochem Biophys Res Commun. 2000;268:255–261. doi: 10.1006/bbrc.2000.2109. [DOI] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff SA, Heo J, Budnik BA, Hanakahi LA, Saghatelian A. A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J Biol Chem. 2014;289:10950–10957. doi: 10.1074/jbc.C113.533968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JE, Alvarez-Dominguez JR, Kline N, Huynh NJ, Geisler S, Hu W, Coller J, Baker KE. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep. 2014;7:1858–1866. doi: 10.1016/j.celrep.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science. 2012;336:1719–1723. doi: 10.1126/science.1220270. [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Maruyama M, Tokuzawa Y, Murakami M, Oda Y, Yoshikane N, Makabe KW, Ichisaka T, Yamanaka S. Evolutionarily conserved non-AUG translation initiation in NAT1/p97/DAP5 (EIF4G2) Genomics. 2005;85:360–371. doi: 10.1016/j.ygeno.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Vanderperre B, Lucier J, Bissonnette C, Motard J, Tremblay G, Vanderperre S, Wisztorski M, Salzet M, Boisvert F, Roucou X. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PloS One. 2013;8:e70698. doi: 10.1371/journal.pone.0070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.