Significance
A major challenge in treating chronic infections is the lack of insight into microbial survival mechanisms in vivo. Many drugs require cells to be doubling rapidly to have their greatest effect, yet the in vivo pathogen growth rate is largely unknown. By labeling freshly expectorated mucus from cystic fibrosis patients with heavy water, we found that the effective growth rates of Staphylococcus aureus are at least two orders of magnitude slower, on average, than typically studied in the laboratory, and are extremely heterogeneous at the single-cell level. These findings underscore the need to study slow growth physiology to gain insight into pathogen survival mechanisms, motivated by the hope that such insight will ultimately help improve drug design and clinical outcomes.
Keywords: slow growth, infectious disease, metabolic heterogeneity, cystic fibrosis, hydrogen isotope labeling
Abstract
Effective treatment for chronic infections is undermined by a significant gap in understanding of the physiological state of pathogens at the site of infection. Chronic pulmonary infections are responsible for the morbidity and mortality of millions of immunocompromised individuals worldwide, yet drugs that are successful in laboratory culture are far less effective against pathogen populations persisting in vivo. Laboratory models, upon which preclinical development of new drugs is based, can only replicate host conditions when we understand the metabolic state of the pathogens and the degree of heterogeneity within the population. In this study, we measured the anabolic activity of the pathogen Staphylococcus aureus directly in the sputum of pediatric patients with cystic fibrosis (CF), by combining the high sensitivity of isotope ratio mass spectrometry with a heavy water labeling approach to capture the full range of in situ growth rates. Our results reveal S. aureus generation times with a median of 2.1 d, with extensive growth rate heterogeneity at the single-cell level. These growth rates are far below the detection limit of previous estimates of CF pathogen growth rates, and the rates are slowest in acutely sick patients undergoing pulmonary exacerbations; nevertheless, they are accessible to experimental replication within laboratory models. Treatment regimens that include specific antibiotics (vancomycin, piperacillin/tazobactam, tobramycin) further appear to correlate with slow growth of S. aureus on average, but follow-up longitudinal studies must be performed to determine whether this effect holds for individual patients.
Growth rate is arguably the simplest yet most profound phenotypic parameter that defines microbial existence. It integrates multiple aspects of a cell’s physiological state, and is often correlated with how cells respond to challenges presented by diverse stressors, including the immune system and antimicrobial drugs (1–4). Despite recognition that growth rate impacts microbial persistence, very few direct measurements of in vivo generation times exist, in large part because quantifying this parameter within a complex environment presents considerable technical challenges. Chronic infections are often assumed to comprise dormant pathogens, but whether they are truly dormant or merely growing slowly is unknown.
In this study, we focused our attention on the microbial populations within the lungs of patients with cystic fibrosis (CF). CF patients expectorate infected mucus daily, making the in vivo environment directly accessible to experimental investigation. Few prior estimates of microbial growth rates exist for CF sputum (5–7). These estimates are based on detecting the ribosomal RNA (rRNA) content of bacterial cells using fluorescence in situ hybridization (FISH). Kragh et al. (6), for example, showed that the majority of Pseudomonas aeruginosa cells in explanted CF lung samples display similar amounts of rRNA to cells in stationary phase, with an rRNA content below that detected for cells in the slowest exponentially growing batch cultures (∼0.35 divisions per hour). Although fluorescence intensity can assess growth when the cellular generation time is faster than ∼3 h, use of rRNA abundance as a quantitative proxy for slow growth in dynamically changing environments is limited because rRNA content can become completely decoupled from growth (8). This fundamental biological constraint necessitates a new approach to measuring in vivo growth rates.
Utility of Highly Sensitive Stable Isotope Tracers for Application in the Human Host
Stable isotope tracers (e.g., 13C, 2H, and 15N) have been used by geobiologists to study the growth and metabolism of slowly growing microbial populations in poorly accessible and energy-limited habitats at the seafloor (9, 10), and in the deep biosphere (11).
Unlike methods that count cells or particular biomolecules, isotope tracer methods can reveal growth rates that reflect population biosynthetic activity regardless of whether the population is expanding, at steady state, or declining. Isotopic enrichments thus provide a measure of biosynthetic turnover that is independent of total biomass (Fig. 1A), making their use well-suited to studies of infected human specimens like CF sputum, where the size of the population can vary due to interactions with other pathogens and the host immune system. Here, we used heavy water (2H2O) because of its advantages as a chemically and nutritionally passive isotope tracer when used at subtoxic concentrations, and measured 2H incorporation into microbial-specific fatty acids by gas chromatography/pyrolysis/isotope ratio mass spectrometry (GC/P/IRMS). Other 2H-based measures of microbial activity have been used successfully in laboratory cultures and animal models (12–14) but require high 2H incorporation from heavy water (detection limits 2–3 atom% 2H). GC/P/IRMS, however, can quantify isotopic enrichments well below 0.01 atom% 2H [and under ideal conditions as low as ∼0.0001 atom% 2H (15)] enabling much shorter incubation times as illustrated in Fig. 1B, making this approach particularly well-suited for application in samples from the human host environment.
Fig. 1.
Using stable isotope tracers to measure microbial activity. (A) Model of a microbial population illustrates the change in biomass and the corresponding isotope label incorporation over time after addition of an isotopic spike. In all three scenarios, the isotopic enrichment proceeds identically, allowing determination of the underlying growth rate from isotopic measurements, regardless of total biomass accumulation or decline. (B) The minimum incubation times required to detect microbial activity at analytical detection limits of 0.01, 0.1, or 1 atom% 2H above natural abundance for different average generation times of microbial populations exposed to a 10% 2H2O labeling solution. See Supporting Information for equations.
Our aim in this study was to develop a method to quantify average and single-cell growth rates for pathogens in chronic infections. For this purpose, we targeted measuring the growth rate of Staphylococcus aureus in CF sputum as a proof of principle. Although CF infections are polymicrobial and compositionally diverse, some bacterial species are common (16). S. aureus is one of the earliest and most prevalent bacteria detected in infants and children with CF, and has garnered attention in the last decade due to the rise of beta-lactam-resistant strains (methicillin-resistant S. aureus, or MRSA) (17). We therefore sought to measure the growth rate of S. aureus within freshly expectorated CF sputum both at the population and single-cell level. This approach has the potential to be extended to quantify slow growth rates of diverse organisms in many contexts, ranging from infectious diseases to industrial fermentations to the deep subsurface.
Approach: Methods, Calculations, and Controls
To confidently estimate the in vivo growth rate using an isotope labeling method, a number of factors must be taken into consideration. These include identification of an appropriate biological target molecule, controls to confirm biological compatibility of the isotope label, access of the label to the sample, determination of the extent of label incorporation into the target molecule during biosynthesis, and estimation of how much anabolic activity can be attributed to growth vs. maintenance. Fig. 2 presents an overview of all aspects of our approach and highlights the relevant figures for each step.
Fig. 2.
Schematic illustrating the experimental approach with clinical sample acquisition, biological considerations, and data processing steps. Four stages of the process are highlighted in color: sample labeling at the hospital (gray), incorporation of the isotope label by the cells (green), measurement of specific fatty acids (guava), and quantitative data processing (gold). Specific biological considerations and data reduction steps are listed and point to the relevant figures in the text and Supporting Information. In parentheses in blue, we provide what the relative overestimates (+)/underestimates (-) of the growth rates would be for the average clinical sample without the respective corrections. For the uncertainty assessment of all parameters involved in the growth rate calculations, please see Fig. S7. Also noted are the analytical difference and data processing steps required for single-cell measurements.
Identifying an appropriate biological target for analysis is of primary importance, and several biological considerations must be kept in mind (Fig. 2, marked in green). S. aureus synthesizes specific fatty acids (the anteiso methyl branched C14 and C16 saturated fatty acids, hereafter referred to by their total carbon numbers as a-C15:0 and a-C17:0, respectively) that can be distinguished from the human host and most other dominant CF pathogens (Fig. S1), and which are synthesized de novo without recycling (Fig. S2). The lack of fatty acid recycling by S. aureus is a critical aspect for our quantitative approach: If organisms build the targeted lipids from exogenous fatty acids, the amount of 2H incorporated from heavy water would be greatly reduced and therefore underestimate population growth rates. By targeting specifically a-C15:0 and a-C17:0 for isotopic analysis, we could focus on a suitable marker for S. aureus. Because heavy water can be toxic at high concentrations (18), we further tested the susceptibility of S. aureus to 2H2O to constrain its tolerance and found no adverse effects at doses as high as 35% 2H2O (Fig. S3).
Fig. S1.
S. aureus produces characteristic fatty acids distinct from host and competitors. Different pathogens have different fatty acid fingerprints that provide species-specific targets for growth rate measurements. However, only a subset is exclusively microbial (green shaded) or usually microbial (blue shaded), with several major fatty acids unsuitable for targeted analysis because they are also produced by host cells (orange shaded). We focused on S. aureus and targeted its most characteristic fatty acids (highlighted in red): a-C15:0 and a-C17:0, the anteiso methyl branched C14 and C16 saturated fatty acids.
Fig. S2.
S. aureus does not recycle exogenous fatty acids for the synthesis of methyl branched (a-C15:0, a-C17:0) fatty acids. We investigated whether S. aureus is capable of recycling exogenous fatty acids it might encounter in the lung environment to derive the biomarkers (a-C15:0 and a-C17:0) targeted in this study. The lack of fatty acid recycling is a critical aspect for this quantitative approach; if organisms build the targeted lipids from exogenous fatty acids, the amount of 2H incorporated from heavy water would be greatly reduced and therefore underestimate population growth rates. To test this, two perdeuterated precursor fatty acids (the naturally abundant octadecanoic acid and the microbially produced pentadecanoic acid, both entirely 2H-substituted in the hydrocarbon tail) were provided as an exogenous source of free fatty acids to test for recycling by S. aureus (Bottom and Middle, respectively). Although S. aureus is capable of elongating the exogenous fatty acids to produce longer-chain derivatives, it does not appear to partly break down the exogenous fatty acids and build them back up, unlike P. aeruginosa (no partly deuterated fatty acids shorter than C17/C20 could be detected in any analysis). This indicates that S. aureus’ methyl branched fatty acids (a-C15:0 and a-C17:0) measured in the lung environment are products of de novo synthesis and can be targeted for growth rate measurements.
Fig. S3.
S. aureus has a high tolerance for 2H2O in synthetic cystic fibrosis medium. We investigated the toxicity effects of increasing concentrations of 2H2O on S. aureus. This figure shows the semilog growth curves of S. aureus in the presence of varying amounts of 2H2O. Lines represent averages of at least four biological replicates; shaded area represents the maximal range of ODs in each condition.
For the isotope labeling of clinical samples (Fig. 2, marked in gray), CF sputum collected at Children’s Hospital Los Angeles (CHLA) was suspended in a prewarmed PBS isotope labeling solution (with 1% to maximally 30% vol/vol 2H2O to stay below the susceptibility of S. aureus to 2H2O toxicity) within 5–10 min of expectoration, and incubated at 37 °C for up to 60 min for bulk growth rate measurements, and up to 2 h for single-cell analyses. These relatively short incubation times were selected to preserve the in situ sputum environment (19) as closely as possible during the tracer incubation, while still ensuring long enough exposure to the isotope label to allow for measurable incorporation into slow-growing populations.
The resulting isotopic enrichment from growth of S. aureus in the presence of the 2H-spiked water was then used to model the apparent growth rate of the pathogen, taking into consideration several potentially confounding factors (Fig. 2, marked in gold). The first concerns the noninstantaneous penetration of the isotope label into a clinical sample. On the time scale of long environmental incubations lasting days or months, the time it takes for heavy water to mix into the sample would be negligible, but, due to the short incubation time (∼1 h), this effect becomes relevant, and we parametrized the mixing rate empirically by using isotope label equilibration times in clinical samples (Fig. S4). The second concerns the hydrogen metabolism of S. aureus during fatty acid biosynthesis. Although newly synthesized fatty acids follow the isotopic composition of water, this is generally not a 1:1 relationship, because of enzymatic kinetic isotope effects during biosynthesis and because not all H atoms are derived from water during heterotrophic growth. The appropriate value for the resulting water hydrogen assimilation by S. aureus growing in clinical samples was determined experimentally in synthetic CF sputum medium (SCFM) (20) following previously established methods (21) (Fig. S5). Finally, because our approach captures the overall anabolic activity, including both the specific growth rate (which signifies true cellular replication) and the maintenance turnover rate (which describes biosynthesis in excess of growth to compensate for degradation and material turnover), quantifying growth requires constraints on the maintenance contribution. We thus grew S. aureus in chemostat culture at a controlled, slow growth rate of 0.14 divisions per day (doubling time of 4.9 d) in SCFM, and measured 2H incorporation after a 2H2O spike to approximate the contribution of maintenance turnover to fatty acid 2H uptake (Fig. S6).
Fig. S4.
Parameterization of noninstantaneous water exchange. The isotopic composition of newly synthesized fatty acids after administration of the isotopic water spike depends on the isotopic composition of sputum water in the clinical samples. For this study, we conducted experiments with differently sized sputum samples (0.5−2.5 g) to derive an empirical relationship for the water equilibration time that would allow a functional parameterization of the average isotopic composition microorganisms experience in the sputum over time. (A) The measurements of saline solution around sputum over time: decreases as the label is exchanged into the sputum. Experiments are grouped by color, symbol sizes reflect sample weights, and dashed/dotted lines illustrate the best-fit equilibration curves for the heavy water spike/sputum water. (B) The expected sample weight dependence of the equilibration rate constant; i.e., as sputum samples become larger, it takes water from the isotopic labeling solution longer to exchange with water in the sputum sample. (C and D) The extent of the noninstantaneous water exchange effect for an average clinical sample. The solid lines show the modeled isotopic composition of the sputum water (C) and resulting fatty acid enrichment (D) if water exchange between the labeling solution and the sample were instantaneous. The dotted lines show the same metrics for a typical clinical sample (average weight of 0.86 g), based on the empirically derived water exchange model used in this study. The fatty acid enrichment for both conditions is modeled using the average growth rate measured in clinical sample.
Fig. S5.
Physiological parameters of water hydrogen assimilation. The isotopic composition of newly synthesized fatty acids after administration of the isotopic water spike depends partly on the physiology of hydrogen assimilation from water. The value of the water hydrogen assimilation constant () for S. aureus was determined from the slopes of the hydrogen isotope compositions of individual fatty acids (2Ffa) vs. medium water hydrogen isotope composition (2Fwater) in cultures grown in synthetic cystic fibrosis medium with variable water isotope compositions. (A) The regression lines for individual fatty acids with 95% confidence bands. (B) A summary of the water hydrogen assimilation constants () derived for individual fatty acids from regression analysis of A, and (C) their numerical values. The size of the symbols in B indicates the relative membrane abundance of the individual fatty acids. Error bars indicate 95% confidence intervals of the coefficients from the linear regression fit. The dashed horizontal line illustrates the average value for aw (0.41), determined from all fatty acids’ values weighted by the relative abundances of the individual fatty acids.
Fig. S6.
Chemostat constraints on maintenance vs. growth. Symbols show the time-dependent isotopic enrichment of S. aureus membrane fatty acids (weighted average isotopic composition of the different membrane components). The x axis records the time after spiking a steady-state culture of S. aureus growing at a controlled growth rate of 0.14 divisions per day (doubling time of ∼4.9 d) with heavy water. The relative fatty acid maintenance turnover rate was calculated from 2H uptake in excess of growth and was determined to amount to 56% of the growth rate.
Using these constraints, we could infer the apparent growth rate of S. aureus in clinical samples by numerically fitting the clinical data (2Ffa and 2Fw, the fractional isotope abundances of 2H measured in fatty acids and sputum water, respectively) to the following equation (full derivation in Supporting Information):
| [1] |
This takes into consideration the aforementioned incorporation of hydrogen from water by S. aureus (aω), rate of label exchange with the mucus (k), and turnover rate (ω). Uncertainties associated with these parameters are discussed in Supporting Information (Fig. S7). It is important to note that if the chemical environment within the sputum requires even higher relative rates of fatty acid maintenance/repair than the chemically stable environment of the chemostat, the clinical growth rates would be even lower than currently estimated.
Fig. S7.
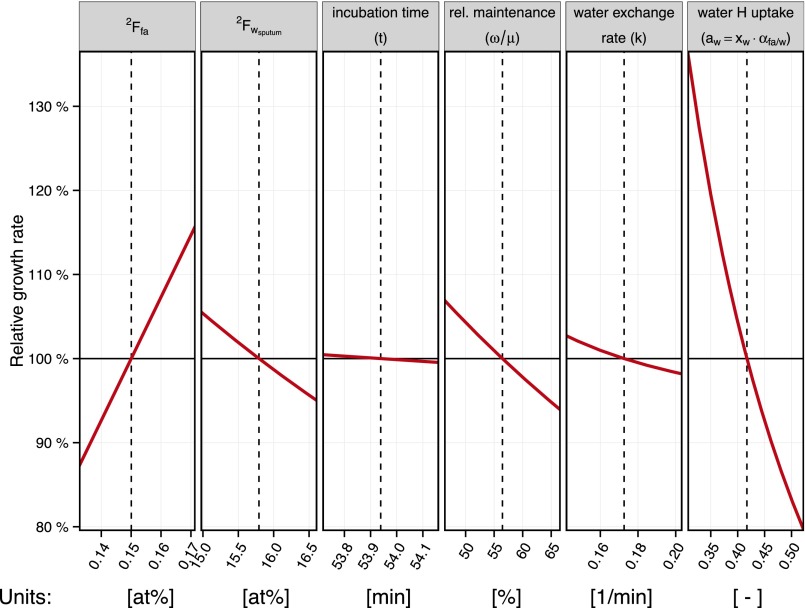
Sensitivity of growth rate calculations to parameter uncertainty. Various sources of uncertainty introduced by analytical constraints and mathematical approximations contribute to the uncertainty estimates for the growth rates presented in this study. This figure provides a visual summary of the sensitivities to the different error sources. Dashed vertical lines indicate the average value of each parameter for the clinical samples (fatty acid isotopic composition, spiked sputum water isotopic composition, incubation time, fatty acid maintenance turnover, water exchange rate, and water hydrogen assimilation constant). The sensitivities of the growth rate estimates to the uncertainty ranges considered for each parameter are illustrated in red. Because the growth equation in this system (Eq. S17) does not have an analytical solution for the growth rate, the upper and lower error bounds presented in Fig. 3 were estimated from the maximal combined uncertainties of all key parameters that offset the growth rate positively and negatively.
Results and Discussion
Growth Rates of S. aureus Populations in Clinical Samples.
Having performed these control experiments, we set out to collect a robust cross-sectional dataset from CF patients. Fig. 3 displays the average growth rate of S. aureus populations in 37 samples from 16 patients. The growth rate range for each sample is constrained by the a-C15:0 and a-C17:0 S. aureus specific fatty acid isotope enrichments, and the average rates are calculated from their mass balance. Major potential contributions to these specific fatty acids by other organisms (e.g., Stenotrophomonas maltophilia, Fig. S1) were ruled out on the basis of 16S rRNA gene sequencing, which indicated that Staphylococcaceae were far more abundant (54% on average, Table S1) than the entire family of Xanthomonadaceae to which Stenotrophomonas belongs (0.4%). Although the calculated in vivo growth rates of S. aureus vary between patients and even between samples from a single patient, all values are 2–100 times lower than the rates at which this organism is typically grown in the laboratory (22). The slow average growth rates suggest that S. aureus populations experience environmental constraints within the lung environment. Unlike in many aquatic environments in nature, carbon and nitrogen limitations are less likely to occur within the organic-rich CF sputum environment where millimolar concentrations of free amino acids and other potential nutrient sources have been measured (20). Research into the in situ concentrations and limits of oxygen penetration in sputum points to oxygen availability as a likely dominant growth constraint (19, 23), in addition to inhibition by microbial competitors in the polymicrobial environment (24), as well as effects from continuous antibiotic treatment in these patients and the activity of the immune system (6). Future work in a more expansive longitudinal study of sputum samples artificially amended with different nutrients and antibiotics will help shed light on the factors restricting pathogen growth.
Fig. 3.
Average growth rates of S. aureus in CF sputum are slow. S. aureus population growth rates calculated from the isotopic enrichment of S. aureus specific membrane components (a-C15:0△, a-C17:0,) and the abundance-weighted average isotopic composition for each data point (○/□). Growth rates are plotted on a logarithmic scale. Symbol size illustrates the relative abundance of the component within each sample. Error bars indicate the propagated error estimate on the population growth rate (Fig. S7). Dashed vertical lines connect the a-C15:0 and a-C17:0 data points of each sample. Samples are grouped by patient health status (well vs. sick) and color-coded by patient. Samples from sick patients are sorted in ascending order by their hospital stay day. Samples from patients whose treatment regimen at the time included any antibiotics active against S. aureus are marked with □ instead of ○. (Antibiotics are vancomycin, oxacillin, piperacillin/tazobactam, amoxicillin/clavulanic acid, trimethoprim/sulfamethoxazole, cefazolin, clindamycin, ciprofloxacin, tetracycline, linezolid, unless S. aureus resistance to the antibiotic was reported for the sample; see Table S1 for details.) Separate column (chemostat) shows a representative growth rate from a chemostat culture of S. aureus (Fig. S6), demonstrating that it is possible to model slow clinically relevant growth rates in the laboratory. Area highlighted in blue represents the typical range of growth rates studied in laboratory experiments with S. aureus.
Table S1.
Patient and treatment information, health indicators, microbial community, and growth rates
| Patient | Age | Sex | Race | BMI, kg/m2 | FEV1% | Pulmonary exacerbation | Hospital day no. | Active antibiotics* | P. aeruginosa present | Staphylo-coccaceae, % | Type | S. aureus clinical culture antibiotic resistance (R)/susceptability (S)† | Xanthomo-nadaceae | Incubation time, min | Sample weight, g | , atom% | , atom% | S. aureus growth rate, 1/d |
| 1 | 16 | male | Non-Hispanic White | 17.5 | 60 | no | n/a | COL, ETH, MOX | n/a | 41.2 | MSSA | n/a | 4.3% | 61 | 0.7 | 12.3 | 0.105 | 0.3 |
| 1 | 16 | male | Non-Hispanic White | 17.5 | 60 | no | n/a | COL, ETH, MOX | n/a | 42.4 | MSSA | n/a | 5.1% | 61 | 0.9 | 14.1 | 0.116 | 0.3 |
| 1 | 16 | male | Non-Hispanic White | 17.5 | 60 | no | n/a | COL, ETH, MOX | n/a | 48.9 | MSSA | n/a | 4.2% | 61 | 1.0 | 15.9 | 0.083 | 0.18 |
| 1 | 16 | male | Non-Hispanic White | 17.7 | 52 | yes | 4.0 | MOX, PIP, TOB | yes | 70.6 | MSSA | R: | S: OXA, CFZ, CIP, CLI, LIN, VAN, TET, TMP | none detected | 61 | 2.2 | 10.8 | 0.039 | 0.11 |
| 1 | 16 | male | Non-Hispanic White | 19.1 | 63 | no | n/a | MOX, PIP, TOB | n/a | 52.9 | MSSA | R: | S: OXA, CFZ, CIP, CLI, LIN, VAN, TET, TMP | none detected | 40 | 0.9 | 10.4 | 0.099 | 0.58 |
| 2 | 14 | male | Non-Hispanic White | n/a | 68 | no | n/a | DOX, LIN, TOB | no | 40 | MSSA | n/a | none detected | 60 | 1.4 | 11.2 | 0.069 | 0.22 |
| 2 | 14 | male | Non-Hispanic White | 15.9 | 67 | no | n/a | DOX, LIN, TOB | no | 41.6 | n/a | n/a | none detected | 61 | 2.6 | 24.3 | 0.235 | 0.49 |
| 3 | 20 | male | Non-Hispanic White | 18.2 | 38 | yes | 30.0 | CIP,‡ COL, TOB | yes | 65.3 | MSSA | R: CIP | S: OXA, CFZ, CLI, LIN, VAN, TET, TMP | none detected | 40 | 0.9 | 14.9 | 0.230 | 0.97 |
| 4 | 14 | male | Hispanic | 19.1 | 50 | no | n/a | TOB | no | 68.1 | MSSA | R: CLI | S: OXA, CFZ, LIN, VAN, TET, TMP | none detected | 63 | 0.8 | 20.4 | 0.461 | 0.88 |
| 4 | 14 | male | Hispanic | 20.1 | 36 | no | n/a | TOB | no | 66.6 | MSSA | R: CLI, TMP | S: OXA, CFZ, LIN, VAN, TET | none detected | 60 | 1.1 | 20.2 | 0.395 | 0.8 |
| 5 | 16 | female | Hispanic | 16.5 | 22 | yes | 10.0 | CFZ, TOB | no | NA | MSSA | R: | S: OXA, CFZ, CIP, CLI, LIN, VAN, TET, TMP | n/a | 62 | 0.7 | 28.0 | 0.618 | 0.87 |
| 5 | 16 | female | Hispanic | 16.5 | 25 | yes | 35.0 | CFZ, TOB | no | NA | MSSA | R: | S: OXA, CFZ, CIP, CLI, LIN, VAN, TET, TMP | n/a | 60 | 1.1 | 19.1 | 0.244 | 0.51 |
| 5 | 16 | female | Hispanic | 17.29 | 20 | no | n/a | AZT, CIP, TOB | yes | 41.5 | MSSA | n/a | none detected | 60 | 1.5 | 19.0 | 0.211 | 0.45 |
| 6 | 9 | female | Non-Hispanic White | 14.1 | 69 | yes | 1.0 | AZT, COL, PRI, TOB | no | NA | n/a | n/a | n/a | 65 | 0.6 | 19.6 | 0.217 | 0.39 |
| 7 | 14 | female | African-American | 17.7 | 42 | yes | 12.0 | AMI, AZT, CFP, TOB | yes | 61.7 | n/a | R: CIP | S: OXA, CFZ, CLI, LIN, VAN, TET, TMP | none detected | 64 | 0.8 | 21.1 | 0.054 | 0.075 |
| 8 | 11 | male | Hispanic | 19.6 | 118 | no | n/a | no | 60.7 | MSSA | R: | S: OXA, CFZ, CLI, LIN, VAN, TET, TMP | none detected | 63 | 1.0 | 22.4 | 0.157 | 0.25 | |
| 8 | 12 | male | Hispanic | 20.43 | 85 | no | n/a | no | 54 | MRSA | R: | S: OXA, CLI, LIN, VAN, TMP | none detected | 50 | 1.7 | 0.7 | 0.033 | 1.7 | |
| 8 | 12 | male | Hispanic | 21.32 | 94 | no | n/a | no | NA | MSSA | R: | S: OXA, CLI, LIN, VAN, TMP | n/a | 45 | 1.3 | 0.7 | 0.025 | 1.1 | |
| 9 | 14 | male | Hispanic | 19.6 | 84 | no | n/a | yes | 54.8 | MSSA | n/a | none detected | 63 | 1.0 | 21.3 | 0.399 | 0.72 | |
| 10 | 13 | female | Hispanic | 21.2 | 106 | yes | 4.0 | ITR, OXA | no | 47.9 | MSSA | R: | S: OXA, CFZ, CIP, CLI, LIN, VAN, TET, TMP | none detected | 62 | 0.6 | 21.2 | 0.100 | 0.16 |
| 11 | 17 | male | Hispanic | 17.3 | 67 | yes | n/a | COL, TOB | no | 70.8 | MSSA | R: CLI | S: OXA, CFZ, CIP, LIN, VAN, TET, TMP | none detected | 61 | 0.8 | 19.6 | 0.173 | 0.33 |
| 11 | 17 | male | Hispanic | 17.5 | 80 | yes | 8.0 | COL, PIP, TOB | no | 64.2 | MSSA | R: CLI | S: OXA, CFZ, CIP, LIN, VAN, TET, TMP | none detected | 60 | 0.8 | 19.9 | 0.101 | 0.18 |
| 11 | 17 | male | Hispanic | 17.3 | 64 | yes | 2.0 | AMI, COL, PIP, TOB | no | 62.6 | MSSA | R: CLI | S: OXA, CFZ, CIP, LIN, VAN, TET, TMP | none detected | 42 | 0.5 | 19.5 | 0.042 | 0.088 |
| 11 | 17 | male | Hispanic | 17.3 | 74 | yes | 6.0 | AMI, COL, PIP, TOB | n/a | 64.3 | n/a | R: CLI | S: OXA, CFZ, CIP, LIN, VAN, TET, TMP | none detected | 46 | 0.7 | 17.0 | 0.014 | 0.0088 |
| 12 | 12 | male | Hispanic | n/a | 72 | yes | n/a | AZT, CLI*, TOB | yes | 43.2 | MRSA | R: OXA, CFZ, CLI | S: LIN, VAN, TET, TMP | none detected | 63 | 0.8 | 19.9 | 0.541 | 1.1 |
| 12 | 12 | male | Hispanic | n/a | 70 | yes | 1.0 | AZT, PIP, TOB, VAN | yes | 69 | MRSA | R: OXA, CFZ, CLI | S: LIN, VAN, TET, TMP | none detected | 60 | 0.3 | 18.0 | 0.044 | 0.071 |
| 12 | 12 | male | Hispanic | n/a | 70 | yes | 1.0 | AZT, PIP, TOB, VAN | yes | 60.6 | MRSA | R: OXA, CFZ, CLI | S: LIN, VAN, TET, TMP | none detected | 61 | 0.5 | 20.2 | 0.067 | 0.11 |
| 12 | 12 | male | Hispanic | 16.8 | 71 | no | n/a | AZT, TET, TOB | yes | 49.8 | MRSA | R: OXA, CFZ, CLI | S: LIN, VAN, TET, TMP | none detected | 60 | 1.0 | 19.0 | 0.159 | 0.32 |
| 12 | 12 | male | Hispanic | 16.7 | 61 | no | n/a | AZT, TOB | yes | 46.8 | MRSA | R: OXA, CFZ, CLI | S: LIN, VAN, TET, TMP | none detected | 61 | 0.7 | 10.3 | 0.041 | 0.11 |
| 13 | 11 | male | Non-Hispanic White | 18.4 | 81 | no | n/a | no | 60.4 | MRSA | R: OXA, CFZ, CIP, CLI | S: LIN, VAN, TET, TMP | none detected | 61 | 0.5 | 19.2 | 0.556 | 1.2 | |
| 13 | 11 | male | Non-Hispanic White | 17.7 | 78 | no | n/a | no | 50.4 | MRSA | R: OXA, CFZ, CIP, CLI | S: LIN, VAN, TET, TMP | none detected | 63 | 0.6 | 19.5 | 0.210 | 0.39 | |
| 14 | 16 | male | African-American | 20.2 | 69 | yes | 2.0 | MER, TOB, VAN | no | 58.7 | MRSA | R: OXA, CFZ | S: CIP, CLI, LIN, VAN, TET, TMP | none detected | 47 | 1.0 | 22.8 | 0.013 | 0.0038 |
| 14 | 16 | male | African-American | 19.9 | 71 | yes | 6.0 | MER, TOB, VAN | no | NA | MRSA | R: OXA, CFZ | S: CIP, CLI, LIN, VAN, TET, TMP | n/a | 60 | 0.6 | 22.1 | 0.092 | 0.15 |
| 14 | 16 | male | African-American | 20.2 | 75 | yes | 14.0 | MER, TOB, VAN | no | 43.2 | MRSA | R: OXA, CFZ | S: CLI, LIN | none detected | 60 | 0.9 | 22.2 | 0.135 | 0.23 |
| 14 | 17 | male | African-American | 19.7 | 80 | no | n/a | TMP | no | 63.8 | MRSA | R: OXA, CFZ | S: CIP, CLI, LIN, VAN, TET, TMP | none detected | 60 | 0.9 | 11.3 | 0.122 | 0.4 |
| 15 | 13 | female | Hispanic | 18.6 | 47 | no | n/a | AZT, TOB | yes | 55.6 | n/a | n/a | none detected | 41 | 0.9 | 9.9 | 0.057 | 0.32 |
| 16 | 12 | female | Hispanic | 21.1 | 96 | no | n/a | AMO | no | 55.5 | MSSA | R: | S: OXA, CFZ, CIP, CLI, LIN, VAN, TET, TMP | none detected | 60 | 0.7 | 5.4 | 0.158 | 1.1 |
n/a indicates instances where no data was available.
Antibiotics the patient was getting at the time: AMI, amikacin; AMO, amoxicillin/clavulanic acid; AZT, inhaled aztreonam; CFP, cefepime; CFZ, cefazolin; CIP, ciprofloxacin; CLI, clindamycin; COL, colistin; DOX, doxycycline; ETH, ethambutol; ITR, itraconazole; LIN, linezolid; MER, meropenem; MOX, moxifloxacin; OXA, oxacillin; PIP, piperacillin/tazobactam; PRI, primaxin/imipenem/cilastatin; TET, tetracycline; TMP, trimethoprim/sulfamethoxazole; TOB, tobramycin; VAN, vancomycin.
Only antibiotics actually used for patient treatment during this study are listed (see abbreviations above). Also typically tested but not used in treatment and therefore excluded from the table for clarity were ampicillin/sulbactam, erythromycin, levofloxacin, cephalexin, rifampin, and benzylpenicillin, all of which had very high incidence of S. aureus resistance.
These antibiotics were reported as ineffective (R category) in the susceptibility data for the respective sample and were therefore not considered as being effective against S. aureus (e.g., in Fig. 3).
The data in Fig. 3 also reveal a systematically higher 2H enrichment of a-C15:0 over a-C17:0 fatty acids that is more pronounced than the equivalent pattern observed for S. aureus grown in chemostat culture in SCFM. Interestingly, S. aureus appears to shift its membrane fatty acid composition in response to slower growth (Fig. S8), a phenomenon also observed in other microorganisms (e.g., refs. 25 and 26). This could explain the systematically higher 2H enrichment of a-C15:0 if substantial heterogeneity exists in the underlying staphylococcal population, with slower-growing cells preferentially producing a-C17:0 and faster-growing cells producing more a-C15:0.
Fig. S8.
The membrane composition of S. aureus is growth rate-dependent. Lipid profile data from laboratory experiments at different growth rates suggest that slower growing populations of S. aureus shift their membrane composition to a higher a-C17:0 ratio (average a-C17:0 to a-C15:0 fatty acid ratio of S. aureus membranes are plotted as a function of growth rate; growth rate is plotted on a log scale for clarity). This could explain the observation in clinical samples that a-C15:0 systematically incorporates more isotope tracer than a-C17:0, potentially reflecting underlying heterogeneity in the microbial population with slower-growing cells producing a-C17:0 preferentially and faster-growing cells producing a-C15:0. Data are from Kopf et al. (27).
Heterogeneity of Microbial Activity.
To test the hypothesis that substantial population heterogeneity exists, we used FISH coupled to secondary ion mass spectrometry (FISH-NanoSIMS) to measure isotope incorporation at the single-cell level (27). Growth rates from single-cell 2H enrichments were calculated by converting thin-section NanoSIMS measurements to corresponding bulk membrane enrichments (details in Supporting Information). Fig. 4A shows representative FISH-NanoSIMS images for two cell clusters, illustrating the potential for significantly different in vivo single-cell growth activities on a small scale. The cluster on the left did not incorporate 2H above background, indicating that the cells were dormant or dead, whereas the cluster on the right incorporated a large amount of 2H within the 2-h incubation. In this visual field, clusters are microbial (identified by the bacterial FISH probe EUB338), but neither contains S. aureus cells.
Fig. 4.
NanoSIMS analysis reveals that single-cell growth rates are diverse. (A) (Top) Example of target identification and 2H enrichment in plastic sections. Frames shown are 10 × 10 µm, (Bottom) First column shows microscopy pictures with overlaid DAPI (blue), bacterial EUB338 FISH (red), and sample autofluorescence in the GFP channel (green); second column shows autofluorescence alone (contrast enhanced for mapping to ion image); third column shows the 14N12C− ion image; fourth column shows the fractional abundance image of 2H. (Scale bar in ion maps, 1 µm.) (B) Distribution of single-cell growth rates of S. aureus cells (in red) in four clinical samples using the approach described in Supporting Information. An unidentified group of bacteria (in blue) was captured in the fourth sample. Bars indicate the measured growth rates of individual cells. Curves illustrate smoothed density distribution function of the data. Growth rates plotted on logarithmic scale.
For the quantitative NanoSIMS analysis shown in Fig. 4B, however, we targeted cells that hybridized specifically with an S. aureus FISH probe (28). The abundance of Staphylococcaceae in the patients whose sputum was analyzed by NanoSIMS ranged from 42 to 56% (Table S1). The smoothed density distribution of the aggregated single-cell growth rates is illustrated for all measured S. aureus cells per patient sample. We observed substantial growth rate heterogeneity in all measured samples, with a particularly wide, apparently bimodal distribution observed in the third sample. Sample 4 also contained a large number of unidentified bacteria that were neither S. aureus nor P. aeruginosa, and appear to be consistently more active than the cooccurring S. aureus population (potential candidates from 16S data include members of the Moraxellaceae and Erythrobacteraceae, with sequence abundances of 13% and 9%, respectively). That S. aureus and other bacteria exhibit heterogeneous metabolic activity in vivo is consistent with a growing body of literature documenting the importance of heterogeneity in a wide range of systems (29–31). Moreover, this heterogeneity may help explain why antibiotic treatments are not fully effective: If a reservoir of slowly growing cells exists for any bacterial species being targeted by antibiotics whose efficacy requires fast growth rates, it is not surprising that 25% of pulmonary exacerbations fail to recover to baseline health after treatment with i.v. antibiotics (32).
Correlation with Clinical Parameters.
Despite this heterogeneity, we asked whether the average growth rate of S. aureus in CF sputum correlates with particular host characteristics and clinical metrics. There were no significant relationships between S. aureus growth rates and host age, sex, body mass index (BMI), or lung function (as measured by forced expiratory volume FEV1%) nor with the community abundance of Staphylococcaceae or presence of the pathogen P. aeruginosa (Table 1). Unexpectedly, S. aureus activity showed a statistically significant positive correlation with the day of the patient’s hospital stay (i.e., higher apparent growth rates the longer a patient had been in the hospital) with a transient decrease in growth rates early on. Additionally, S. aureus populations show significantly higher growth rates in samples from patients who were well (pulmonary function at baseline) compared with samples from patients who were acutely sick (suffering a pulmonary exacerbation; Fig. 3 and Fig. S9).
Table 1.
S. aureus growth rates vs. health and treatment indicators
| Clinical parameter | Samples | Data range | Statistic† | P value | Significance |
| Age | 37 | 9–20 y of age | Spearman (−0.23) | 0.2 | — |
| Sex | 37 | female (8), male (29) | MWW (female > male) | 0.3 | — |
| Race | 37 | Hispanic (21), Non-Hispanic White (11), African-American (5) | Spearman (−0.23) | 0.2 | — |
| BMI | 33 | 14.1–21.3 kg/m2 | Spearman (0.18) | 0.3 | — |
| FEV1% | 37 | 20–118% predicted | Spearman (0.06) | 0.7 | — |
| Pulmonary exacerbation | 37 | no (20), yes (17) | MWW (no > yes) | 0.004 | ** |
| Hospital day | 15 | day 1–30 | Spearman (0.53) | 0.04 | * |
| P. aeruginosa present‡ | 32 | no (21), yes (11) | MWW (no > yes) | 0.1 | — |
| Staphylococcacea‡ | 32 | 40–71% | Spearman (−0.12) | 0.5 | — |
| Vancomycin§ | 37 | treatment at the time included this antibiotic: no (32), yes (5) | MWW (no > yes) | 0.002 | ** |
| Piperacillin/tazobactam§ | 37 | treatment at the time included this antibiotic: no (30), yes (7) | MWW (no > yes) | 0.003 | ** |
| Tobramycin§ | 36 | treatment at the time included this antibiotic: no (11), yes (25) | MWW (no > yes) | 0.05 | * |
| Inhaled aztreonam§ | 36 | treatment at the time included this antibiotic: no (27), yes (9) | MWW (no > yes) | 0.1 | — |
| Colistin§ | 36 | treatment at the time included this antibiotic: no (27), yes (9) | MWW (no > yes) | 0.1 | — |
| Moxifloxacin§ | 36 | treatment at the time included this antibiotic: no (31), yes (5) | MWW (no > yes) | 0.3 | — |
Summary of statistical relationships between S. aureus average population growth rate and different health and treatment indicators by Mann−Whitney−Wilcoxon (MWW) test (for binary parameters) or Spearman rank analysis. Spearman’s P < 0 indicates positive correlation (i.e., faster growth rate with increase of the clinical parameter), and P < 0 indicates a negative correlation. See Table S1 for all data, and see Figs. S7 and S8 for correlation plots. Only antibiotics with at least five samples in each category (yes/no) were analyzed. —P > 0.05, *P ≤ 0.05, **P ≤ 0.01.
Evaluated by MWW test for binary parameters (sex, pulmonary exacerbation, and P. aeruginosa presence; each was evaluated for both alternative hypotheses; the most statistically significant alternative is given) and by Spearman rank analysis for all other parameters.
The presence of P. aeruginosa was tested in routine clinical selective plating screens. The abundance of Staphylococcaceae was assessed by 16S rRNA gene sequencing.
Vancomycin is used against Gram-positive pathogens including MRSA; piperacillin/tazobactam is used against P. aeruginosa and some S. aureus (not effective against MRSA); tobramycin, aztreonam, and colistin are mostly used against P. aeruginosa; moxifloxacin is a broad-spectrum antibiotic mostly used against mycobacteria.
Fig. S9.
S. aureus growth rates are correlated with few clinical parameters. This figure expands on correlations with clinical parameters discussed in Correlation with Clinical Parameters (Table 1) and illustrates the correlation between the clinical parameters and the measured bulk growth rates for S. aureus. (Top) Each panel denotes a different binary parameter with the respective P values of the MWW test for the alternative hypotheses indicated in the header. (Middle and Bottom) Each panel shows the respective Spearman correlation coefficient and P value for the correlation. The different colors indicate samples from different patients in the study. All growth rates (y axis) are plotted on a log scale for clarity. Not all clinical information was available for all data points (see Table S1 for data).
Intriguingly, on average, S. aureus growth rates were significantly lower when patients were taking the antibiotics vancomycin, piperacillin/tazobactam, or tobramycin at the time of sampling compared with when patients were not taking the respective antibiotic (Table 1 and Fig. S10), but no statistically significant trends were observed for the antibiotics aztreonam, colistin, and moxifloxacin. Although tobramycin is primarily used against P. aeruginosa, both piperacillin/tazobactam and vancomycin have antistaphylococcal activity. We also note, however, that we were unable to determine whether these correlations held statistical significance for individuals, due to the limited number of patient-specific longitudinal samples (Fig. S10). Generally, at the individual level, being on any antistaphylococcal antibiotics at the time of sampling exhibited no clear trend with respect to growth rate (Fig. 3). Intriguingly, the growth rate of S. aureus in some patients at the beginning of their hospital stay appeared to slow, with a return to baseline well before discontinuation of the treatment (e.g., red and teal, Fig. 3). Such dynamics make it tempting to speculate that early during a hospital stay, treatment with anti-Staph antibiotics depresses S. aureus growth rates, yet, because patients are simultaneously given many drugs that target various members of microbial community, as other bacteria are eradicated over time, S. aureus experiences resource expansion and/or reduced antagonism and begins to grow more quickly. We emphasize, however, that this speculative interpretation requires testing in dedicated longitudinal studies where multivariable regression models can help parse out the interplay between health indicators, treatment regimen, and pathogen growth rate.
Fig. S10.
S. aureus growth rates are correlated with specific antibiotic treatments, but more longitudinal data are required at the patient level. This figure expands on correlations with clinical parameters discussed in Correlation with Clinical Parameters (Table 1) and illustrates the correlation between antibiotic treatments and the measured bulk growth rates for S. aureus. (Top and Upper Middle) Each panel in the denotes a different antibiotic with the respective P values of the MWW test for the alternative hypotheses indicated in the header. Only antibiotics with at least five samples in each category (yes/no) are shown. (Lower Middle and Bottom) Each panel in the shows the same data only for patients with data in both categories (yes/no) and respective statistical tests broken down on a per-patient basis. Patients are distinguished by color. All growth rates (y axis) are plotted on a log scale for clarity. Not all clinical information was available for all data points (see Table S1 for data).
Conclusions.
Our studies of S. aureus in CF sputum reveal that pathogen growth rates can be far slower and more heterogeneous in human infections than previously assumed. Nevertheless, the range of growth rates that we observe here is largely accessible to experimental replication within laboratory models. Understanding metabolic diversity during slow growth, and how this impacts survival strategies for microorganisms, is a poorly developed but fundamental aspect of microbial physiology and ecology in any environment. What strategies do cells take to control core processes such as transcription, translation, and membrane integrity when persisting in a slow growth mode? How different are these strategies from those that promote cellular homeostasis when nutrients are plentiful and growth is fast? The answers to these and other questions in the context of pathogen behavior in chronic infections are important to understand for practical reasons. For example, Mycobacterium tuberculosis is known to persist under hypoxic, nonreplicating conditions (33); encouragingly, inhibition of membrane components that contribute to electron transport and/or proton translocation under these conditions shows promise for shortening the time of tuberculosis therapy (34, 35). Ultimately, a better understanding of how populations grow in vivo is crucial to developing representative in vitro tests of potential therapies for diverse pathogens that infect the human host (36).
Materials and Methods
Study Design.
All patients were recruited from CHLA and gave informed consent or assent in accordance with IRB CCI-13000211. Inclusion criteria were a positive diagnosis of CF, ability to expectorate sputum, and recent detection of an infection by S. aureus from clinical data.
Sample Collection.
Immediately upon expectoration (within 5–10 min), sputum samples were suspended at the hospital in a prewarmed PBS isotope labeling solution (with 1–30% vol/vol 2H2O), and incubated at 37 °C for up to 60 min. Microbial growth activity in samples for lipid analysis was arrested by flash-freezing in liquid nitrogen at the end of sample incubation, and samples were preserved at −80 °C. Typically, most pediatric patients could not expectorate more than 0.5–1 g of sputum, but when sufficiently large sputum samples (>0.6 g) were expectorated, the sample was divided before isotope labeling with a scalpel or by transfer with a large syringe. Samples for single-cell analysis were treated the same as bulk samples, except for extension of incubation times up to 2 h (with 10–15% 2H2O), and arrest of microbial activity at the end of incubation by transfer to a freshly thawed 1% formaldehyde solution in PBS. Just before sample preservation, the residual labeling solution was collected and filter-sterilized for water isotope analysis.
Isotope Notation.
Isotopic measurements were recorded in the conventional δ-notation () relative to the international reference VSMOW [(37)]. To allow consistent reporting and exact mass balance calculations, all measurements were converted to fractional abundances F () using the relation and are reported in atom percent (atom%). Data quantification and evaluation of analytical uncertainties are discussed in more detail in Supporting Information.
Water Isotope Analysis.
The water hydrogen isotopic composition () of all samples was measured using a Los Gatos Research DLT-100 liquid water isotope analyzer. Samples were analyzed in at least three replicate analyses with 10 injections each. Samples close to natural abundance isotopic composition were calibrated against four working standards ( values: −117‰, −11‰, +290‰, +1,270‰) that in turn were calibrated against the VSMOW, GISP, and SLAP international standards (38). Heavily enriched samples from isotope tracer solutions were beyond the linear response range of the instrument and were analyzed by dilution with water of known isotopic composition.
Fatty Acid Analysis.
Frozen sputum samples were lyophilized for at least 48 h. Homogenized dry powder was weighed into 40-mg aliquots (or the maximum available amount), and spiked with 10 µg 21:0 phosphatidyl choline as a phospholipid extraction standard. Samples were transesterified in the presence of a base catalyst (0.5M NaOH in anhydrous methanol) at room temperature for 10 min (39). Free fatty acids and aldehydes are not esterified under basic conditions, which prevents the derivatization of fatty acids from degraded materials in the sputum sample, as well as the derivatization of abundant aldehydes, which interfere chromatographically with target analytes. Derivatized fatty acid methyl esters (FAMEs) were extracted into hexane, and concentrated under a stream of N2 at room temperature. Because primary target analytes (a-C15:0 FA and a-C17:0 FA) typically constituted less than 1% of host-derived fatty acids, the high abundance of host material [primarily C18:1 FA, C18:2 FA, and longer chain (poly)-unsaturated fatty acids] interfered with compound-specific isotope ratio analysis of these compounds due to column overload. The low total amount of target analytes available from most sample sizes precluded the use of elaborate purification steps with low yields or risk of contamination. To remove several major host compounds and increase relative abundance of the analytes sufficiently for isotope ratio analysis, saturated FAMEs were separated from unsaturated FAMEs using Discovery Ag-Ion solid-phase extraction columns in combusted glass cartridges (Supelco; custom preparation to avoid contaminants from plastic columns). Extracted samples were dried to 1 mL and applied to preconditioned columns, followed by elution of saturated FAMEs in 0.125% acetone in hexane, and monosaturated and disaturated FAMEs in acetone. Separation of unsaturated and saturated fatty acids was quantitative using this procedure. Fractions were evaporated to dryness and resuspended in hexane before analysis.
FAMEs were analyzed by gas chromatography/mass spectrometry (GC/MS) on a Thermo-Scientific Trace DSQ equipped with a ZB-5ms column (30 m × 0.25 mm i.d., film thickness 0.25 µm) and PTV injector operated in splitless mode, using He as a carrier gas at 0.8 mL/min. The GC oven was held at 80 °C for 1 min, ramped at 20 °C/min to 130 °C, and ramped at 5 °C/min to a final temperature of 320 °C (held for 20 min). Peaks were identified by comparison of mass spectra and retention times to authentic standards and library data. Fatty acids are referred to using the nomenclature z-Cx:y, where x is the total number of carbons in the fatty acid skeleton (regardless of structure), y is the number of double bonds, and z- is a prefix describing additional structural features of the compound. The position and stereochemistry of the double bonds or cyclopropyl rings was not determined. The isotopic composition of the primary target analytes was measured in the saturated FAME fraction by GC/pyrolysis/isotope-ratio mass spectrometry on a Thermo-Scientific Delta+XP with methane of known isotopic composition as the calibration standard. A multicompound FAME standard was run every four to six samples to verify instrument accuracy and precision. Chromatographic conditions were identical to those from GC/MS analysis except for a thick-film column (ZB-5ms, 30 m × 0.25 mm i.d., film thickness 1.00 µm) and slight modifications to the temperature program to optimize chromatographic separation of i-C15:0 and a-C15:0/i-C17:0 and a-C17:0 despite peak broadening from isotope labeling and heavy column loading. Samples were injected in highly concentrated aliquots to obtain maximum signal for target analytes. Peaks were identified based on retention order and relative height based on the GC/MS analysis. High-abundance components in the saturate fraction (C16:0, C18:0, C20:0) that were too concentrated relative to target analytes were prevented from entering the source by appropriately timed backflush of the column effluent. Relative proportions of fatty acids for isotope mass balance calculations were determined from peak areas corrected for derivatization and isotopic composition.
Single-Cell Analysis.
Immediately upon receipt of samples from the hospital, formaldehyde-fixed sputum samples were transferred to excess PBS solution (10 mL) in a column above an 8-µm sterile filer. The sample was washed by slowly rinsing it with a total of 50 mL PBS solution that was added to the column in 10-mL aliquots and drained by gravimetric or slight vacuum flow through the filter. Samples were suspended in drops of molten noble agar [2% (wt/vol) Difco Agar Noble in 50 mM Hepes buffered filter-sterilized water], solidified by cooling at room temperature, and cut into 2-mm3 cubes. Fixed and washed agar cubes were incubated for at least 1 h at 30 °C in freshly prepared 1mg/mL lysozyme and 50 µg/mL lysostaphin (L2898; Sigma-Aldrich) in 10 mM Tris buffered water to digest S. aureus cell wall, washed once in PBS, resuspended in 50% (vol/vol) ethanol in PBS, and dehydrated in 100% ethanol over the course of three exchanges, with final resuspension in 100% for at least 1 h. Ethanol was then replaced twice with 100% Technovit 8100 infiltration solution (64709012; Heraeus Kulzer GmbH) to infiltrate the agar cubes overnight. Agar cubes were finally suspended in airtight 0.6-mL microcentrifuge tubes in Technovit 8100 infiltration solution amended with hardener II reagent and stored at 4 °C for overnight polymerization. Thin sections (1 µm thick) were cut using a rotary microtome. Each section was stretched on the surface of a water drop on polylysine-coated microscope slides and air-dried at room temperature.
FISH was conducted on the plastic thin sections using the universal bacterial probe EUB338 [5′ to 3′: GCT GCC TCC CGT AGG AGT (40)], which hybridizes to bacterial 16S rRNA, probe Sau [5′ to 3′: GAA GCA AGC TTC TCG TCC G (28)], which hybridizes to S. aureus 16S rRNA, and probe Psae [5′ to 3′: TCT CGG CCT TGA AAC CCC (41)], which hybridizes to P. aeruginosa 23S rRNA. Both probes were specifically designed and tested for use in microbial identification in CF sputum (41, 42). Additionally, probe Non338 [5′ to 3′: ACT CCT ACG GGA GGC AGC (43)], an oligonucleotide complementary to the probe EUB338, served as a negative control for nonspecific binding. Probes EUB338 and Non338 were labeled with the cyanine dye Cy3, probe Sau were labeled with the cyanine dye Cy5, and probe Psae were labeled with fluorescein. All probes were labeled on both the 5′ and 3′ end to increase fluorescence intensity (44). For hybridization, each section was covered with 20 µl of hybridization buffer [0.9 M NaCl, 20mM Tris⋅HCl at pH 8, 0.01% SDS, 20% (vol/vol) formamide (28, 42)] containing 50 ng of unlabeled oligonucleotide probe BET42a [5′ to 3′: GCC TTC CCA CTT CGT TT (45)] and preincubated for 10 min at 46 °C to reduce nonspecific binding of labeled oligonucleotide probes (41). Probes EUB338 (or Non338), Sau, and Psae were added (5 ng/µL), and samples were incubated in a moisture chamber at 46 °C for 3 h. Stringent washing was performed by incubating the samples in washing buffer at decreased NaCl concentration [225 mM NaCl with 5 mM EDTA, 20 mM Tris⋅HCl at pH 8, 0.01% SDS (46)] at 48 °C for 12 min. Finally, samples were dipped into ice-cold deionized water to rinse off the salt, air-dried, and mounted in the glycerol-based soft mount Vectashield (Vector Laboratories) (47) with 1.5 µg/mL 4′,6-diamidino-2-phenylindole (DAPI) as a DNA counterstain. Samples were routinely imaged using a Zeiss Axio Imager microscope, and mapped extensively for sample identification and target localization for FISH−NanoSIMS using a Keyence BZ-9000 microscope equipped with a mercury lamp and filter cubes for DAPI, GFP, Cy3, and Cy5. For secondary ion mass spectrometry, samples were made conductive by sputter-coating with a 50-nm layer of gold.
All samples were analyzed with a CAMECA NanoSIMS 50L (CAMECA) housed in the Division of Geological and Planetary Sciences at California Institute of Technology. Cells in plastic thin sections were analyzed using a 1.9- to 3.6-pA primary Cs+ beam, and were presputtered with a 200-pA primary Cs+ beam current () for 3–15 min (t), depending on the size of the presputtering area (A), to a cumulative charge density of 30 pC/µm2 (). Seven masses were collected in parallel (1H−, 2H−, 12C−, 13C−, 14N12C−, 15N12C−, and 28Si−) using the configuration reported in ref. 27. Target locations in individual samples were located using the NanoSIMS CCD camera, secondary electron image, and 14N12C− ion maps. For all analyses, the beam was rastered over a square region ranging from 5 × 5 µm2 to 50 × 50 µm2 for 5–30 min per analytical plane/frame, and images were collected in 256 × 256 pixel resolution up to 12 × 12 µm2 and 512 × 512 pixel resolution for larger areas. Presputtering was carried out on a region larger than the analytical frame by at least 2 µm on each side. Analytical parameters including primary beam focus, secondary beam centering, and mass resolution for all ions were verified every 30 min. To correct for instrument mass fractionation and matrix effects of the plastic polymer, we calibrated plastic-embedded single-cell measurements against bacterial isotope standards that were analyzed at the same time and were previously presented in detail (27). Data evaluation is discussed in more detail in Supporting Information.
The 16S Ribosomal RNA Gene Sequencing.
DNA was prepared from 1 mg lyophilized sputum samples treated with 2% vol/vol PBS/β-mercapto-ethanol for 2 h at room temperature and Sputolysin (Calbiochem) for 30 min at 37 °C, followed by lysis in 180 µL of buffer ATL (Qiagen Blood and Tissue DNeasy kit) amended with lysostaphin (final concentration 0.14 mg/mL) and lysozyme (final concentration of 2.9 mg/mL) (48) for 2 h at 37 °C, bead-beating with 0.1-mm glass beads for 10 min, and extraction using the Qiagen Animal Tissues Spin Column Protocol with 20 µL proteinase K during overnight incubation. High throughput sequencing was performed as previously described (49). Briefly, purified DNA was amplified in triplicate using 16S rRNA Illumina barcoded primers specific to the V4 region of the 16S rRNA gene. Samples were pooled, and paired-end sequencing was performed on an Illumina MiSeq platform at CHLA. Sequences were demultiplexed and trimmed, and paired-end reads were joined, trimmed according to quality score, and checked for chimeras. Sequences were assigned to de novo OTUs (operational taxonomic units) using uclust (50) to search sequences against the Greengenes database (2014 version), and were assigned if the sequence matched the database at 97% similarity or greater. If a read did not match a database sequence, it was considered unassigned. Taxonomy was assigned at the family level based on the Greengenes matching sequence. The genera Geobacillus, Rhodothermus, and Rhodocyclaceae were considered to be contaminants and were filtered out from the results. Table S1 includes the sequence abundances of the S. aureus family Staphylococcaceae, and the S. maltophilia family Xanthomonadaceae (the main organism with partly overlapping fatty acid profiles).
SI Introduction
Using Stable Isotope Tracers to Measure Microbial Activity.
This section provides an introduction to the equations describing isotopic enrichment in microbial populations exposed to an isotope tracer. Here we focus all nomenclature on the hydrogen isotope system only, although most equations are more broadly applicable.
The production and removal of biomass from a microbial population is governed by the specific growth rate μ (which signifies cellular replication), turnover rate ω (which describes biosynthesis in excess of growth to compensate for degradation and turnover of macromolecules), and death/degradation rate d. The set of differential equations describing the rate of change in total biomass () and the rate of change in new biomass () is the difference between the synthesis and removal fluxes,
| [S1] |
A differential equation for the fraction of new vs. total biomass is derived using the quotient rule
| [S2] |
Integration provides a solution for and its derivative ,
| [S3] |
Now, in the case of a population exposed to an isotope tracer (for example, water enriched in deuterium – ) starting at some time , all biomass that is newly produced after addition of the tracer will have a distinct hydrogen isotope composition () reflecting the tracer. Mass balance between the newly produced and original biomass determines the overall isotopic composition,
| [S4] |
In the most basic scenario where new biomass simply reflects the isotopic composition of the tracer (here the spiked water, ), and there is no maintenance turnover (), this yields the following link between isotopic enrichment and growth (27, 51):
| [S5] |
This relationship between the isotopic enrichment and growth is illustrated in Fig. 1A, which shows the evolution of total biomass, new biomass, and old biomass over the course of two generation times (the biomass is not necessarily doubling, so these are only the apparent generation times) for varying death/removal rates. As should be apparent from Eq. S5 and Fig. 1A, one important advantage of tracking growth by isotope tracer is that the effective pool size (B) of the microbial biomass does not affect the enrichment. This holds true even for the more complex scenarios that are relevant to this study and are derived in detail in Data processing for the analysis of clinical samples.
Minimum Incubation Times for Microbial Activity Detection.
The estimated minimum incubation time requirements illustrated in Fig. 1B are calculated from Eq. S5 as described previously (27), and are based on isotope labeling with 20% (vol/vol) 2H2O. Briefly, substituting the generation time for the growth rate and solving Eq. S5 for the time required to reach a final hydrogen isotope enrichment of after addition of isotopically heavy water yields
| [S6] |
with , and the isotopic composition of the original biomass before spiking approximated by natural abundance hydrogen isotope values . It is important to note that although the hydrogen isotopic composition of new biomass reflects the hydrogen isotope composition of source water in most experimental systems, it typically has a proportionality factor lower than unity (i.e., with ). Such lower values of α would lead to longer incubation time requirements than those reported in Fig. 1B, which reflect the idealized minimal incubation time requirements.
SI Materials and Methods
Laboratory Growth Media.
For all growth experiments in a simulated complex medium in the laboratory, S. aureus (MN8) (52) was grown in a defined SCFM. This medium is designed to mimic the average nutritional environment of the CF lung, and was chosen as a representative model medium that simulates a clinical context more closely. The medium was prepared as previously described (20); briefly, SCFM is a Mops buffered medium (adjusted to pH 6.8) that contains basic salts, a mixture of amino acids (about 19 mM equivalents in total), glucose (3.2 mM), and lactate (9.3 mM). Due to the high concentration of amino acids, microbial growth in this medium tends to raise the pH significantly (pH > 8), and the medium was buffered with 50 mM instead of 10 mM Mops in this study. SCFM was further amended with vitamins required by S. aureus: 100 g/L thiamine (B1), 100 g/L nicotinic acid (B3), and 10 g/L biotin (B7). All culturing experiments were conducted aerobically at 37 °C with agitation, and were inoculated from fresh cultures grown on the same medium. Growth was monitored by measuring optical density at 600 nm. Cells intended for lipid analysis were generally harvested by centrifugation at 5,000 × g for 10 min (at 4 °C), washed by resuspension in 1× PBS solution, repelleted, and frozen immediately at −80 °C until lipid extraction and analysis.
Biological Considerations for the Experimental Approach.
As outlined in Fig. 2, several biological considerations are important for evaluating the applicability of our experimental approach: the importance of biomarker specificity (consideration a), de novo biomarker synthesis (consideration b), and isotope label tolerance (consideration c). Here, we present data collected to address these aspects.
Evaluating biomarker specificity.
Quantifying the anabolic activity of a specific target organism in a complex environment is only possible if a suitable biomarker for the organism of interest can be identified. In the context of a polymicrobial infectious disease within the human host, this requires a biomarker that can be distinguished from molecules produced both by the host itself and by other dominant pathogens. The fatty acids targeted in this study (the anteiso methyl branched C14 and C16 saturated fatty acids, a-C15:0 and a-C17:0) are synthesized specifically by S. aureus and can be distinguished from the host and most other dominant CF pathogens (Fig. S1).
Testing for de novo biomarker synthesis.
Recycling exogenous sources is an important limitation to isotope labeling experiments in substrate-rich environments. If organisms can build their membranes from preexisting fatty acids, or fatty acid fragments, that are available in their environment, they no longer need to synthesize all fatty acids de novo. Consequentially, an isotopic tracer in the form of 2H-rich water would be incorporated into the total membrane at a much slower rate. Without knowledge of the ability to recycle and rate of recycling, isotopic enrichment could underestimate true growth of the population, because growth with recycled fatty acids does not incorporate the tracer.
Two examples for this process are provided by recent work on this topic in P. aeruginosa (53, 54). First, the organism appears capable of recycling exogenous free fatty acids, such as C16:0, by direct incorporation en bloc during phospholipid biosynthesis. Any fatty acid recycled by this mechanism would carry its original isotopic signature from when it was first synthesized rather than incorporate any water isotope label present at the time of en bloc incorporation. Second, P. aeruginosa appears capable of reelongating partially degraded exogenous fatty acids. Typically, the initiating step of fatty acid synthesis from acetyl-CoA is catalyzed by ß-acetoacetyl-ACP synthase (fabH in Escherichia coli). In P. aeruginosa, however, a closely related but distinct new class of synthases (now called fabY) catalyze this step (54). Additionally, a wide range of genetically very similar enzymes exist in P. aeruginosa, and, even with a deletion mutant of fabY, the organism is capable of growing. Ref. 53 discovered that in the absence of fabY, the preferred fatty acid synthesis pathway initiated from acetyl-CoA (which is no longer possible without the gene) can be replaced in the presence of exogenous fatty acids (C8, C12, C16, and undefined mixtures like LB). P. aeruginosa can simply shunt C8-CoA from -oxidation degradation of fatty acid metabolism back into fatty acid biosynthesis via the enzyme encoded by ORF PA3286, thereby skipping the de novo synthesis of the C8 precursor carbon skeleton. From the scavenged intermediate, the organism can produce all longer-chain cellular fatty acids, including both saturated and unsaturated fatty acids. Any fatty acid recycled by this mechanism would carry a mixed isotopic signature with the C8 tail maintaining its original isotopic composition, whereas the de novo elongated remainder of the molecule would reflect the water isotope label present at the time of reelongation.
We investigated whether S. aureus, the target organism of this study, is capable of recycling exogenous fatty acids it might encounter in the lung environment. Two commercially available perdeuterated precursor fatty acids (the naturally abundant octadecanoic acid and the only microbially produced pentadecanoic acid, both entirely 2H-substituted in the hydrocarbon tail) were provided as an exogenous source of free fatty acids to test for recycling by S. aureus. Experiments were carried out in batch culture in 250-mL flasks with 100 mL of a defined phosphate buffered minimal medium [pH 7.2, 2.5 g/L NaCl, 13.5 g/L K2HPO4, 4.7 g/L KH2PO4, 1 g/L K2SO4, 0.1 g/L MgSO47H2O, 10 mM NH4Cl, 10 mM glycerol, 100 g/L thiamine (B1), 100 g/L nicotinic acid (B3), 10 g/L biotin, 11.5 mg/L proline, and 10 mL/L 50× MEM Amino Acid solution (M5550; Sigma-Aldrich), with final amino acid concentrations 63.2 mg/L arginine, 15.6 mg/L cysteine, 21 mg/L histidine, 26.35 mg/L isoleucine, 26.2 mg/L leucine, 36.3 mg/L lysine, 7.6 mg/L methionine, 16.5 mg/L phenylalanine, 23.8 mg/L threonine, 5.1 mg/L tryptophan, 18.0 mg/L tyrosine, 23.4 mg/L valine]. The medium was amended with no exogenous fatty acids (control), 100g perdeuterated pentadecanoic acid (C15:0 FA), or 100g perdeuterated octadecanoic acid (C18:0) from 10 mg/mL stock solutions in DMSO (addition of 100L the solvent alone did not affect growth). The perdeuterated fatty acids were purchased from CDN Isotopes and are completely deuterated [CD3(CD2)13COOH and CD3(CD2)16COOH] with the sole exception of the carboxylic acid hydrogen (which exchanges too quickly in solution to retain a label). Cells were harvested in early stationary phase (), washed twice, and frozen immediately. Samples were transesterified in the presence of a base catalyst (0.5 M NaOH in anhydrous methanol) at room temperature for 10 min (39). Free fatty acids are not transesterified under these basic conditions, which prevented the derivatization of any remaining exogenous perdeuterated free fatty acids not consumed by the microorganisms or removed during washing steps. The resulting FAMEs from all samples were extracted into hexane after addition of a quantification standard (10 g C25:0 FAME), and concentrated under a stream of N2 at room temperature before analysis. All preparation, processing, and analysis of samples containing perdeuterated materials was performed in a separate laboratory to avoid any risk for cross-contamination of natural abundance or clinical isotope tracer experiments. Samples were analyzed on a Waters Micromass GCT Premier 6890N equipped with a Zebron-ZB-Wax column (29.5 m × 0.25 mm i.d., film thickness 0.25m) and PTV (programmable temperature vaporization) injector operated at a 10:1 split ratio, using He as a carrier gas at 15 mL/min. The GC oven was ramped at 2 C/min to 200 C, held for 1min, and ramped at 18 C/min to a final temperature of 250 °C (held for 7 min). Heavily deuterated fatty acids were detected as distinct peaks that eluted earlier than their respective natural abundance counterparts, and the degree of deuteration was determined from the mass shift of their molecular ions.
The data indicate that S. aureus is indeed capable of incorporating exogenous fatty acids en bloc, as can be seen from the presence of significant amounts of completely perdeuterated C15:0 and C18:0 in the intact polar lipids of the organism (Fig. S2). Additionally, it appears to be capable of elongating the exogenously provided free fatty acids by extension with 2 carbon units (acetyl-CoA), as witnessed by the presence of partly deuterated C17:0 and C19:0 when grown with perdeuterated C15:0 (the mass spectrum reveals a completely deuterated C15 skeleton extended with undeuterated C2/C4) and partly deuterated C20:0 when grown with perdeuterated C18:0 (again, a completely deuterated C18 skeleton extended with undeuterated C2). Curiously, none of these fatty acids are membrane components that S. aureus naturally produces in significant quantities (except some minor amounts of C18:0), yet they constitute a significant portion of the intact polar lipid membrane fraction when provided exogenously. It is striking that the major, naturally occurring component that is altered the most in its abundance is the longer chain a-C17:0 fatty acid, which is produced naturally by elongation of a-C15:0. This suggests either that chain elongation of the exogenous fatty acids directly competes for substrate (acetyl-CoA) with elongation of a-C15:0 to make a-C17:0 FA or that a regulatory response of the organism compensates for the presence and effect of the longer-chain fatty acids on the physical properties of the membrane by reducing a-C17:0 production.
The second observation, however, is that, unlike P. aeruginosa, S. aureus does not appear to partly break down the exogenous fatty acids and build them back up during fatty acid degradation (no partly deuterated fatty acids shorter than C17/C20 could be detected in any analysis). For the normally dominant components of its membrane (methyl branched a-C15:0 and a-C17:0 fatty acids), this is consistent with the known biosynthetic pathway of anteiso methyl branched fatty acids, which are built from an isoleucine primer in de novo fatty acid synthesis (55), and cannot be initiated from any partially degraded straight chain fatty acid by known biosynthetic mechanisms.
This indicates that S. aureus’ methyl branched fatty acids (a-C15:0 and a-C17:0) measured in the lung environment are pure products of de novo synthesis. For this reason, we focus on these biomarkers for this study. It is important to note that, for fatty acids potentially elongated directly from exogenous precursors (e.g., C18:0), the potential rate of fatty acid recycling must be considered carefully in any environment that experiences a heavy continuous influx of exogenous fatty acids from breakdown of organic matter. If the contribution of a precursor or component that can be incorporated en bloc is solely from other members of the same population, any isotopic spike still captures the activity of the population as a whole.
Testing for isotope label tolerance.
Because heavy water is known to be toxic at high concentrations (18), we tested the susceptibility of S. aureus to 2H2O by growing the organism in SCFM with varying concentrations of 2H2O up to 35%. The growth experiment was performed at 37 °C in 96 well plates with four replicates per culture condition, and was inoculated from an overnight culture that had not previously experienced elevated levels of 2H2O. Plates were shaken continuously, and optical density () was recorded every 5 min. The results (Fig. S3) show that there are no clear adverse effects of 2H2O doses as high as 35% on the growth of S. aureus in this medium. It is important to note, however, that the SCFM medium only approximates the nutritional environment of sputum and could fail to mimic other relevant chemical and structural features of sputum. To reduce the risk of the potentially toxic effects of high levels of 2H2O in sputum, we only used heavy water spikes well below the experimentally constrained 35%.
Data Processing for the Analysis of Clinical Samples.
In this study, the isotopic enrichment of 2H resulting from growth of S. aureus in the presence of 2H spiked water is traced by measuring the isotopic composition of the nonexchangeable hydrogen bound in membrane fatty acids. The most basic equation relating isotopic enrichment to growth was introduced in Eq. S5. However, Eq. S5 is based on the simplifying assumption that newly synthesized biomass directly reflects the isotopic composition of the heavy water tracer without taking into consideration additional effects such as the noninstantaneous penetration of the heavy water label into a clinical sample (consideration a), water hydrogen metabolism of the target organism (consideration b), and maintenance contributions to anabolic activity (consideration c). These effects and their magnitudes are outlined in the data processing roadmap in Fig. 2. The corresponding data corrections are derived in detail hereafter and are presented together with the data that we collected to constrain the individual parameters. In the context of fatty acid enrichment , the starting equation (without corrections, labeled ) becomes
| [S7] |
Accounting for label penetration into the sample.
Water provides an excellent isotope tracer due to its rapid self-diffusivity, which is well-understood in aqueous solutions ( for H2O at 35 °C, almost identical for H218O, lower for fully tritiated water 3H2O at ) (56), but more difficult to assess in the context of the irregularly shaped, highly viscous, biofilm-like sputum samples.
On long time scales (days to months), water diffusion would be negligible, but, due to the short incubation time scales required for quasi in situ experiments with expectorated sputum samples (∼1 h), this effect can become relevant (i.e., sputum water cannot be assumed to adopt the spiked isotopic composition instantaneously). The lack of data and complexity of sputum poses difficulty in calculating the diffusion of the isotopic label into the sputum water ab initio. Here, we instead conducted several experiments with differently sized sputum samples (from 0.5 g to 2.5g) to derive a simple empirical relationship of the form and estimate the time-dependent isotopic composition of sputum water [] that allows a functional parametrization of the average isotopic composition that the microorganisms in the sputum experience over time. In these experiments, the equilibration of the hydrogen isotopic composition of water in the isotope labeling solution [] from exchange with water contained within the suspended sputum sample [] (mixed about 2:1 wt/wt) was tracked over time, as illustrated in Fig. S4A. The hydrogen isotope composition of both end-member pools was described as exchanging toward the equilibrium isotope composition of the fully exchanged combined water pool with rate constant k,
| [S8] |
| [S9] |
where is the hydrogen isotope composition of the spiked isotope labeling solution (i.e., at ), and is the natural isotope composition of water contained within the sputum sample (i.e., at ). The data in Fig. S4A are fitted to Eq. S8 (fit of Eqs. S8 and S9 illustrated in dashed and dotted lines, respectively), to derive a measure of k for each sample.
Fig. S4B illustrates the derived values of k, which show a relationship with the sample weight as expected, and provides a model to estimate k for samples of differing weights. As sputum samples become larger, it takes water from the isotopic labeling solution longer to exchange with water in the sputum sample. To minimize the effect of this noninstantaneous water exchange, we aimed for an ideal clinical sample size of 0.8 g. Fig. S4 C and D illustrates the extent of this effect for a typical clinical sample.
For each isotopically labeled clinical sample, the hydrogen isotope composition of water in the labeling solution [] was measured in the residual solution at the conclusion of the experiment (at time ), and the corresponding equilibrium isotopic composition between labeling solution and sample water was calculated using the estimated value of k for the given sample weight,
| [S10] |
Based on these parameters, we can use Eq. S9 to model the time course of the sputum water isotopic composition that the microbial population experiences over the entire incubation time interval.
Assuming that newly synthesized fatty acids directly reflect the isotopic composition of the source water for now (see correction for this in Accounting for water hydrogen metabolism of the target organism), we can use to solve the original isotope mass balance (Eq. S4) from integral form, essentially adding a corrective term for the noninstantaneous water exchange to the starting equation [Eq. S7/],
| [S11] |
Accounting for water hydrogen metabolism of the target organism.
During the biosynthesis of fatty acids, the resulting macromolecules generally follow the isotopic composition of the source water the organism is growing in. However, because of isotopic fractionation during biosynthesis and because not all H atoms are derived from water, this is not a 1:1 relationship. The isotopic composition of newly synthesized fatty acids can be considered in terms of the combination of the mole fraction of water derived hydrogen () and associated net hydrogen isotope fraction (), and substrate-derived hydrogen () including metabolic water (57) with average substrate isotopic composition () and associated net isotope fractionation () (21),
| [S12] |
Isotopic fractionation factors are defined with respect to isotope ratios ; see Error from using fractionation factors in exact mass balance calculations for details on the use of fractionation factors with fractional abundances.
For growth under similar conditions (medium, temperature, etc.), the physiological parameters of hydrogen assimilation (, ) are assumed to be comparable. The appropriate value of the combined water hydrogen assimilation constant for S. aureus growing in clinical samples was determined in SCFM (20) following the approach of ref. 21. Briefly, S. aureus was grown in batch culture experiments with four different water isotopic compositions, and was determined for all major membrane fatty acids from the slopes of vs. (Fig. S5). For calculations involving bulk fatty acid hydrogen isotope compositions, an average value for (average ) was determined from all fatty acids’ values weighted by the relative abundances of the individual fatty acids. The substrate-dependent contribution to fatty acid hydrogen was determined from the fatty acids’ and sputum water’s natural isotopic compositions in clinical samples,
Combing these parameters with Eq. S9 (the time-dependent spiked water exchange), the expression for the isotopic composition of newly synthesized fatty acids (Eq. S12) becomes
| [S13] |
Finally, this equation can be used to once again solve the original isotope mass balance (Eq. S4) from integral form, adding a second term to Eq. S17 ,
| [S14] |
Accounting for maintenance contribution to anabolic activity.
Because our approach captures the overall anabolic activity including both the specific growth rate (which signifies true cellular replication), and the maintenance turnover rate (which describes biosynthesis in excess of growth to compensate for degradation and material turnover), quantifying growth () requires constraints on the maintenance contribution.
S. aureus MN8 was thus grown continuously in a chemostat system (Sartorius Biostat QPlus) in SCFM amended with 500 L/L Antifoam 204 (A6426; Sigma Aldrich) at 37 °C with agitation. The chemostat vessel with 550 mL working volume was inoculated from a single colony pregrown in the same medium, and continuous supply of medium was started upon reaching early stationary phase and maintained at a flow rate equivalent to a generation time of 4.9 d (0.14 divisions per day). Overflow from the vessel was continuously removed to maintain a fixed volume, and the exact dilution rate was confirmed gravimetrically from the total vessel content and medium flow rate. Redox potential, pH, and dissolved oxygen were monitored continuously, and optical density was measured in aliquots withdrawn aseptically from vessel overflow. After the monitored physiological parameters reached steady state (after approximately six generations), the chemostat vessel was spiked with 2 mL 70% 2H2O, and 2H incorporation into fatty acids was monitored over time.
At steady state, the growth rate of the culture (μ) must equal the known rate of dilution (d, corresponding to 4.9 d doubling time), which leaves the fatty acid maintenance rate ω as the only unconstrained parameter in this system. Unlike in clinical samples, the isotopic composition of the medium water decreases over time after the initial spike due to dilution of the tracer from the continuous supply of fresh medium (to maintain steady-state growth conditions). The water isotopic composition of the enriched medium is thus a function of the initial composition of the spiked medium (), and the dilution with fresh medium of original water isotopic composition () at dilution rate d,
| [S15] |
Using this equation with Eq. S13 [for ] to solve the original isotope mass balance (Eq. S4) from integral form but with yields the following solution for the chemostat system ():
| [S16] |
By fitting the time-dependent fatty acid enrichment from this isotope labeling experiment (Fig. S6) to Eq. S16, ω was calculated to amount to 56% of the growth rate (). Although maintenance turnover rates are sometimes considered to reflect a constant absolute rate of repair or basic energy requirement, this rate is likely variable, and depends on the actual population growth rate as discussed by ref. 58. In this study, we assumed that the maintenance rate scales with the growth rate and approximated the minimum fatty acid maintenance turnover rate for each clinical sample from the relative maintenance rate of 56% measured in our continuous culture experiment. If the chemical environment within the sputum requires higher relative rates of fatty acid maintenance/repair than the chemically stable environment of the chemostat, the clinical growth rates would be even lower than currently estimated (Fig. 3).
Final population growth rate calculations.
Finally, incorporating the equations and parameters for all three corrections—noninstantaneous water exchange (consideration a, Eq. S9), the hydrogen metabolism of S. aureus (consideration b, Eq. S13), and the maintenance turnover (consideration c, ω)—into the integral form of Eq. S4 yields the following equation for linking fatty acid hydrogen isotope composition to growth rates in the clinical samples:
| [S17] |
All microbial growth rates were estimated by numerically fitting the clinical data to this equation. The turnover rate ω was constrained with data from continuous culture experiments with S. aureus as described above. In addition to growth rate estimates from the isotopic enrichment of individual key fatty acids of S. aureus (a-C15:0 and a-C17:0), the overall growth rate of S. aureus populations was calculated from the average bulk membrane isotopic composition with relative total fatty acid abundance ratio .
Additional considerations for single-cell analysis.
The use of plastic thin sections for NanoSIMS analysis allowed accurate targeting and identification of individual microorganisms despite the small size of the pathogens, and the structurally complex nature of the sputum samples. The resin used in this study, Technovit 8100, polymerizes at cold temperatures ( 4 °C), precluding the need for extended exposure to relatively high heat (and the associated risk for structural changes). The structural support lent by the plastic matrix provided thin sections with a smooth surface that enabled high spatial resolution during NanoSIMS analysis due to the lack of topological features (27). However, as an acryl plastic (a combination of methyl methacrylate and glycol methacrylate), the Technovit resin contributed significant amounts of isotopically circumnatural carbon and hydrogen that dilute the isotopic signal from enriched cells. To correct for this effect, we calibrated both free and plastic-embedded single cell measurements against bacterial isotope standards in ref. 27.
Raw data from all acquired ion images were processed using the open-source MATLAB plugin Look@NanoSIMS (59). Ion images from multiple frames were corrected for dead time and QSA (quasi-simultaneous arrivals) effect and aligned, and the corresponding microscopy images were warped onto the 14N12C– ion image using functionality provided by Look@NanoSIMS. Discrete regions of interest were hand-drawn around individual microbes based on the 14N12C– ion image and the FISH+DAPI images. Fractional abundances of single cell analyses were calculated directly from raw ion counts and converted to bulk membrane equivalents using the reported calibration for hydrogen isotope measurements in embedded S. aureus cells (27). Single-cell growth rates were calculated using Eq. S17 after converting the thin section NanoSIMS measurements to their corresponding bulk membrane enrichments. Based on the findings of Musat et al. (60) that stationary phase cells do not experience significant isotope label dilution from mono FISH treatment, we considered this direct application of the reported calibration (which does not include FISH treatment) to be most appropriate given the relatively low single-cell growth rates measured throughout the samples in this study.
Uncertainty Assessment.
Here, we discuss the various sources of uncertainty introduced by analytical constraints and mathematical approximations that contribute to the uncertainty estimates for the growth rates presented in this study. Fig. S7 summarizes the sensitivity of the growth rate measurement to the uncertainty in key parameters.
Error from using fractionation factors in exact mass balance calculations.
Exact mass balance calculations that use fractional abundances F, instead of δ-values or isotope ratios R, are important in isotope labeling experiments due to the large error introduced by approximating mass balance at higher isotopic enrichments in δ-value space. However, for biosynthetic processes that can cause significant isotopic fractionation, exact mass balance calculations require an approximation regarding the use of fractionation factors. An isotope fractionation factor between two pools a and b is usually reported/known in terms of the ratio of isotope ratios between the two pools, . In the relevant case for this study, i.e., the biosynthetic incorporation of water hydrogen into fatty acids discussed in Materials and Methods, the formulation of is thus only an approximation of the exact calculation
| [S18] |
The relative error that this approximation introduces into the water assimilation constant can be calculated as follows:
| [S19] |
Although the combined assimilation constant can be measured experimentally (Materials and Methods), the two individual parameters that describe the assimilation of hydrogen from water, i.e., the fraction of fatty acid hydrogen derived from water, and the isotope fractionation associated with this biosynthetic pathway, cannot be experimentally separated for heterotrophic organisms. With current experimental approaches, the system is underconstrained. This limits the potential use of the exact formula above, which requires separate knowledge of and . However, ref. 21 estimated the range of possible values for for heterotrophic growth in complex media to fall between 0.8 (fractionation by −200‰) and 1.15 (+150‰). Based on these bounding estimates, we were able to constrain the error introduced by the approximation for each clinical sample using Eq. S19. This source of uncertainty (typically ) was taken into consideration in all growth rate calculations in addition to the experimentally determined confidence interval for .
Error propagation for analytical uncertainty.
The uncertainty in individual FAME measurements is constrained by the combined effects of the analytical uncertainty (sample SD ) and the maximal potential offset caused by hydrogen isotope memory effects (61) of 4% the difference in the isotope value to the preceding methane reference peaks. This provides a conservative upper bound on the error estimate. In reality, analysis of methane reference standards that follow analytical peaks suggests memory effects to be much lower ( 0.2% instead of 4%).
| [S20] |
The FAME uncertainty is propagated to the uncertainty in the fatty acids themselves, , by SE propagation of the derivatization correction equation (assuming that the error in the FAME and MeOH measurements are uncorrelated).
| [S21] |
| [S22] |
During the calculation of S. aureus average isotopic composition from the a-C15:0 and a-C17:0 fatty acids and their relative abundance , the uncertainties (, , and ) are propagated as well by SE propagation, taking into account the covariance between the quantities (, , and ), which are estimated from the data.
| [S23] |
The uncertainty in the isotopic composition of spiked sputum water () discussed in Materials and Methods was calculated similarly by SE propagation from uncertainties in the water exchange rate constant k, sample incubation time, and water isotopic compositions ( and ). Because Eq. S17 does not have an analytical solution for the growth rate μ, the upper and lower error bounds ( and ) were estimated from the maximal combined uncertainties of all key parameters that offset the growth rate positively and negatively. The sensitivities to the different errors are illustrated graphically in Fig. S7. Error bars in Fig. 3 indicate this conservative error estimate for the calculated bulk population growth rates.
Acknowledgments
We are grateful to Shawn McGlynn, Ryan Hunter, Abigail Green-Saxena, Yunbin Guan, Alejandro LaRiviere, Ian Booth, Nathan Dalleska, Fadi Asfour, Douglas Li, Kyle McCallin, Sally Ward, Thomas Keens, and patients of the Pulmonary CF Clinic at CHLA for supporting this study. We thank the editor and reviewers for constructive criticism that improved the manuscript. This research was supported by grants from the NIH (Grant 5R01HL117328-03) and the Howard Hughes Medical Institute (HHMI) (to D.K.N.). S.H.K. was an HHMI International Research Scholar, and D.K.N. is an HHMI Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512057112/-/DCSupplemental.
References
- 1.Gilbert P, Collier PJ, Brown MR. Influence of growth rate on susceptibility to antimicrobial agents: Biofilms, cell cycle, dormancy, and stringent response. Antimicrob Agents Chemother. 1990;34(10):1865–1868. doi: 10.1128/aac.34.10.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown MR, Collier PJ, Gilbert P. Influence of growth rate on susceptibility to antimicrobial agents: Modification of the cell envelope and batch and continuous culture studies. Antimicrob Agents Chemother. 1990;34(9):1623–1628. doi: 10.1128/aac.34.9.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baek SH, Li AH, Sassetti CM. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 2011;9(5):e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen D, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334(6058):982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L, et al. In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J Bacteriol. 2008;190(8):2767–2776. doi: 10.1128/JB.01581-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kragh KN, et al. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect Immun. 2014;82(11):4477–4486. doi: 10.1128/IAI.01969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolpen M, et al. Denitrification by cystic fibrosis pathogens —Stenotrophomonas maltophilia is dormant in sputum. Int J Med Microbiol. 2015;305(1):1–10. doi: 10.1016/j.ijmm.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. Evaluating rRNA as an indicator of microbial activity in environmental communities: Limitations and uses. ISME J. 2013;7(11):2061–2068. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wegener G, et al. Assessing sub-seafloor microbial activity by combined stable isotope probing with deuterated water and 13C-bicarbonate. Environ Microbiol. 2012;14(6):1517–1527. doi: 10.1111/j.1462-2920.2012.02739.x. [DOI] [PubMed] [Google Scholar]
- 10.Kellermann MY, et al. Autotrophy as a predominant mode of carbon fixation in anaerobic methane-oxidizing microbial communities. Proc Natl Acad Sci USA. 2012;109(47):19321–19326. doi: 10.1073/pnas.1208795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morono Y, et al. Carbon and nitrogen assimilation in deep subseafloor microbial cells. Proc Natl Acad Sci USA. 2011;108(45):18295–18300. doi: 10.1073/pnas.1107763108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berry D, et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc Natl Acad Sci USA. 2015;112(2):E194–E203. doi: 10.1073/pnas.1420406112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloehn J, Saunders EC, O’Callaghan S, Dagley MJ, McConville MJ. Characterization of metabolically quiescent Leishmania parasites in murine lesions using heavy water labeling. PLoS Pathog. 2015;11(2):e1004683. doi: 10.1371/journal.ppat.1004683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer CR, Bowen BP, Pan C, Northen TR, Banfield JF. Stable-isotope probing reveals that hydrogen isotope fractionation in proteins and lipids in a microbial community are different and species-specific. ACS Chem Biol. 2013;8(8):1755–1763. doi: 10.1021/cb400210q. [DOI] [PubMed] [Google Scholar]
- 15.Sessions AL. Isotope-ratio detection for gas chromatography. J Sep Sci. 2006;29(12):1946–1961. doi: 10.1002/jssc.200600002. [DOI] [PubMed] [Google Scholar]
- 16.Kahl BC. Impact of Staphylococcus aureus on the pathogenesis of chronic cystic fibrosis lung disease. Int J Med Microbiol. 2010;300(8):514–519. doi: 10.1016/j.ijmm.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Goss CH, Muhlebach MS. Review: Staphylococcus aureus and MRSA in cystic fibrosis. J Cyst Fibros. 2011;10(5):298–306. doi: 10.1016/j.jcf.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Kushner DJ, Baker A, Dunstall TG. Pharmacological uses and perspectives of heavy water and deuterated compounds. Can J Physiol Pharmacol. 1999;77(2):79–88. [PubMed] [Google Scholar]
- 19.Cowley ES, Kopf SH, LaRiviere A, Ziebis W, Newman DK. Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. MBio. 2015;6(4):e00767. doi: 10.1128/mBio.00767-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189(22):8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Gillespie AL, Sessions AL. Large D/H variations in bacterial lipids reflect central metabolic pathways. Proc Natl Acad Sci USA. 2009;106(31):12580–12586. doi: 10.1073/pnas.0903030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dengremont E, Membré JM. Statistical approach for comparison of the growth rates of five strains of Staphylococcus aureus. Appl Environ Microbiol. 1995;61(12):4389–4395. doi: 10.1128/aem.61.12.4389-4395.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Worlitzsch D, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002;109(3):317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filkins LM, et al. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol. 2015;197(14):2252–2264. doi: 10.1128/JB.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill CO, Suisted JR. The effects of temperature and growth rate on the proportion of unsaturated fatty acids in bacterial lipids. J Gen Microbiol. 1978;104(1):31–36. doi: 10.1099/00221287-104-1-31. [DOI] [PubMed] [Google Scholar]
- 26.Cohen Z, Vonshak A, Richmond A. Effect of environmental conditions on fatty acid composition of the red alga Porphyridium cruentum: Correlation to growth rate. J Phycol. 1988;24(3):328–332. [Google Scholar]
- 27.Kopf SH, et al. Heavy water and 15N labelling with NanoSIMS analysis reveals growth rate-dependent metabolic heterogeneity in chemostats. Environ Microbiol. 2015;17(7):2542–2556. doi: 10.1111/1462-2920.12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kempf VA, Trebesius K, Autenrieth IB. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J Clin Microbiol. 2000;38(2):830–838. doi: 10.1128/jcm.38.2.830-838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiviet DJ, et al. Stochasticity of metabolism and growth at the single-cell level. Nature. 2014;514(7522):376–379. doi: 10.1038/nature13582. [DOI] [PubMed] [Google Scholar]
- 30.Stewart MK, Cummings LA, Johnson ML, Berezow AB, Cookson BT. Regulation of phenotypic heterogeneity permits Salmonella evasion of the host caspase-1 inflammatory response. Proc Natl Acad Sci USA. 2011;108(51):20742–20747. doi: 10.1073/pnas.1108963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenwood D. Phenotypic resistance to antimicrobial agents. J Antimicrob Chemother. 1985;15(6):653–655. doi: 10.1093/jac/15.6.653. [DOI] [PubMed] [Google Scholar]
- 32.Sanders DB, et al. Failure to recover to baseline pulmonary function after cystic fibrosis pulmonary exacerbation. Am J Respir Crit Care Med. 2010;182(5):627–632. doi: 10.1164/rccm.200909-1421OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y. Persisters, persistent infections and the Yin-Yang model. Emerging Microbes Infect. 2014;3(1):e3. doi: 10.1038/emi.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao SPS, Alonso S, Rand L, Dick T, Pethe K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2008;105(33):11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yano T, et al. Reduction of clofazimine by mycobacterial type 2 NADH:quinone oxidoreductase: A pathway for the generation of bactericidal levels of reactive oxygen species. J Biol Chem. 2011;286(12):10276–10287. doi: 10.1074/jbc.M110.200501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrison F. Microbial ecology of the cystic fibrosis lung. Microbiology. 2007;153(Pt 4):917–923. doi: 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- 37.de Laeter JR, et al. Atomic weights of the elements. Review 2000 (IUPAC Technical Report) Pure Appl Chem. 2003;75(6):683–800. [Google Scholar]
- 38.Coplen TB. Normalization of oxygen and hydrogen isotope data. Chem Geol Isot Geosci Sect. 1988;72(4):293–297. [Google Scholar]
- 39.Griffiths MJ, van Hille RP, Harrison STL. Selection of direct transesterification as the preferred method for assay of fatty acid content of microalgae. Lipids. 2010;45(11):1053–1060. doi: 10.1007/s11745-010-3468-2. [DOI] [PubMed] [Google Scholar]
- 40.Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hogardt M, et al. Specific and rapid detection by fluorescent in situ hybridization of bacteria in clinical samples obtained from cystic fibrosis patients. J Clin Microbiol. 2000;38(2):818–825. doi: 10.1128/jcm.38.2.818-825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tajbakhsh S, et al. Detection of Pseudomonas aeruginosa in sputum samples by modified fluorescent in situ hybridization. Afr J Biotechnol. 2008;7(5):553–556. [Google Scholar]
- 43.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14(2):136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 44.Stoecker K, Dorninger C, Daims H, Wagner M. Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl Environ Microbiol. 2010;76(3):922–926. doi: 10.1128/AEM.02456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manz W, Amann R, Ludwig W, Wagner M, Schleifer KH. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: Problems and solutions. Syst Appl Microbiol. 1992;15(4):593–600. [Google Scholar]
- 46.Lathe R. Synthetic oligonucleotide probes deduced from amino acid sequence data. Theoretical and practical considerations. J Mol Biol. 1985;183(1):1–12. doi: 10.1016/0022-2836(85)90276-1. [DOI] [PubMed] [Google Scholar]
- 47.Florijn RJ, Slats J, Tanke HJ, Raap AK. Analysis of antifading reagents for fluorescence microscopy. Cytometry. 1995;19(2):177–182. doi: 10.1002/cyto.990190213. [DOI] [PubMed] [Google Scholar]
- 48.Zhao J, et al. Impact of enhanced Staphylococcus DNA extraction on microbial community measures in cystic fibrosis sputum. PLoS One. 2012;7(3):e33127. doi: 10.1371/journal.pone.0033127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 51.Chen SS, Sperling E, Silverman JM, Davis JH, Williamson JR. Measuring the dynamics of E. coli ribosome biogenesis using pulse-labeling and quantitative mass spectrometry. Mol Biosyst. 2012;8(12):3325–3334. doi: 10.1039/c2mb25310k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kreiswirth BN, et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305(5936):709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 53.Yuan Y, Leeds JA, Meredith TC. Pseudomonas aeruginosa directly shunts β-oxidation degradation intermediates into de novo fatty acid biosynthesis. J Bacteriol. 2012;194(19):5185–5196. doi: 10.1128/JB.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan Y, Sachdeva M, Leeds JA, Meredith TC. Fatty acid biosynthesis in Pseudomonas aeruginosa is initiated by the FabY class of β-ketoacyl acyl carrier protein synthases. J Bacteriol. 2012;194(19):5171–5184. doi: 10.1128/JB.00792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaneda T. Iso- and anteiso-fatty acids in bacteria: Biosynthesis, function, and taxonomic significance. Microbiol Rev. 1991;55(2):288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Easteal AJ, Edge A, Woolf LA. Isotope effects in water. Tracer diffusion coefficients for O in ordinary water. J Phys Chem. 1984;88(24):6060–6063. [Google Scholar]
- 57.Kreuzer-Martin HW, Lott MJ, Ehleringer JR, Hegg EL. Metabolic processes account for the majority of the intracellular water in log-phase Escherichia coli cells as revealed by hydrogen isotopes. Biochemistry. 2006;45(45):13622–13630. doi: 10.1021/bi0609164. [DOI] [PubMed] [Google Scholar]
- 58.van Bodegom P. Microbial maintenance: A critical review on its quantification. Microb Ecol. 2007;53(4):513–523. doi: 10.1007/s00248-006-9049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polerecky L, et al. Look@NanoSIMS—;A tool for the analysis of NanoSIMS data in environmental microbiology. Environ Microbiol. 2012;14(4):1009–1023. doi: 10.1111/j.1462-2920.2011.02681.x. [DOI] [PubMed] [Google Scholar]
- 60.Musat N, et al. The effect of FISH and CARD-FISH on the isotopic composition of 13C- and 15N-labeled Pseudomonas putida cells measured by nanoSIMS. Syst Appl Microbiol. 2014;37(4):267–276. doi: 10.1016/j.syapm.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y, Sessions AL. Memory effects in compound-specific D/H analysis by gas chromatography/pyrolysis/isotope-ratio mass spectrometry. Anal Chem. 2008;80(23):9162–9170. doi: 10.1021/ac801170v. [DOI] [PubMed] [Google Scholar]