Significance
The integrated stress response (ISR) is an adaptation pathway that integrates multiple stress signals to decrease translation rates and promote specific transcriptional programs. Modulation of this pathway is emerging as a possible therapeutic strategy in pathologies associated with defects in protein homeostasis. In this paper, the antiviral drug Nelfinavir is found to be a strong and unconventional inducer of the ISR both in vitro and in vivo. This study uncovers an atypical mechanism that can initiate this pathway and provides insights on possible mechanisms underlying the metabolic deregulations observed in Nelfinavir-treated patients. Nelfinavir has been used for years to treat HIV; its repositioning in diseases that would benefit from decreased translation speed could therefore be of interest.
Keywords: ER stress, translation initiation, Nelfinavir, HIV protease inhibitors, PPP1R15B
Abstract
Inhibitors of the HIV aspartyl protease [HIV protease inhibitors (HIV-PIs)] are the cornerstone of treatment for HIV. Beyond their well-defined antiretroviral activity, these drugs have additional effects that modulate cell viability and homeostasis. However, little is known about the virus-independent pathways engaged by these molecules. Here we show that the HIV-PI Nelfinavir decreases translation rates and promotes a transcriptional program characteristic of the integrated stress response (ISR). Mice treated with Nelfinavir display hallmarks of this stress response in the liver, including α subunit of translation initiation factor 2 (eIF2α) phosphorylation, activating transcription factor-4 (ATF4) induction, and increased expression of known downstream targets. Mechanistically, Nelfinavir-mediated ISR bypassed direct activation of the eIF2α stress kinases and instead relied on the inhibition of the constitutive eIF2α dephosphorylation and down-regulation of the phophatase cofactor CReP (Constitutive Repressor of eIF2α Phosphorylation; also known as PPP1R15B). These findings demonstrate that the modulation of eIF2α-specific phosphatase cofactor activity can be a rheostat of cellular homeostasis that initiates a functional ISR and suggest that the HIV-PIs could be repositioned as therapeutics in human diseases to modulate translation rates and stress responses.
In mammalian cells, metabolic and environmental stresses, such as viral infection, nutrient deprivation, and perturbation of endoplasmic reticulum (ER) homeostasis, converge on the phosphorylation of the α subunit of translation initiation factor 2 (eIF2α) to trigger an adaptation program known as the integrated stress response (ISR) (1). Phosphorylation of eIF2α occurs on serine 51 and negatively regulates the guanine nucleotide exchange factor eIF2B, impairing the recycling of eIF2 to its active GTP-bound form (2). This signaling event inhibits the initiation steps in mRNA translation, leading to a decrease of global translation rates. Concomitantly, the phosphorylation of eIF2α selectively increases the translation of a subset of genes, including the activating transcription factor-4 (ATF4) (3). Together, translational and transcriptional reprogramming orchestrates the stress response and homeostasis restoration.
The initiation of the ISR relies on four evolutionarily related eIF2α kinases that each senses specific insults and signals by phosphorylating eIF2α (4). On the other hand, dephosphorylation of eIF2α and subsequent restoration of the translational capacity is emerging as a key event that controls the complete recovery from stress and ISR termination. Two cellular cofactors of the protein phosphatase-1 (PP1c) can specifically reverse the phosphorylation of eIF2α. The first one is GADD34, which is induced by the ISR to specifically direct PP1c to dephosphorylate eIF2α (5–7), allowing termination of the response and restoration of the homeostatic pace of translation (7, 8). The second one is the protein CReP (Constitutive Repressor of eIF2α Phosphorylation), also known as PPP1R15B, that is expressed ubiquitously in unstressed cells and was identified as a key factor maintaining low levels of eIF2α phosphorylation (9, 10). Molecules that promote the activation of the eIF2α kinases or modulate the dephosphorylation of eIF2α have been identified and are being considered as potential therapeutics in human diseases characterized by loss of cellular homeostasis including cancer, metabolic deregulation, and neurodegenerative diseases (11–13).
It is becoming more and more evident that even the most thoughtfully designed drugs elicit promiscuous interaction profiles within cells (14), and accordingly, many biological effects of these drugs lack a compelling molecular explanation (14, 15). This is particularly true for molecules such as Nelfinavir that may interact with multiple off-targets (16). Beyond understanding the off-target effects of the drugs, the study of these molecular mechanisms provides an opportunity to identify cellular pathways of biological relevance. Nelfinavir is an HIV protease inhibitor (HIV-PI) that was developed in the 1990s, and its incorporation into highly active antiretroviral therapy (HAART) led to remarkable suppression of HIV replication in patients. The HIV-PIs were among the first drugs to reach the clinic that were developed with computer technology applied to compound design based on the X-ray structures. Although these drugs were designed to specifically inhibit the HIV aspartyl protease, they display a wide range of activities independently of their ability to target HIV (17, 18). Clinical trials are underway to evaluate the effectiveness of HIV-PIs on suppressing tumor progression (17, 19). It was proposed that ER stress contributes to their antineoplastic activity (20–22). However, a detailed characterization of the stress responses triggered by the HIV-PIs has not been reported.
Here we investigated the mechanisms leading to activation of cellular stress responses in the presence of the HIV-PIs. We found that these inhibitors, such as Nelfinavir, only triggered a partial ER-stress response characterized by the absence of detectable PKR-like endoplasmic reticulum kinase (PERK) or ATF6 activation or accumulation of misfolded proteins within the ER. However, Nelfinavir affected proteostasis by eliciting a robust ISR. It did so by modulating constitutive dephosphorylation of eIF2α, therefore bypassing direct activation of the eIF2α kinases. These findings highlight the relevance of the modulation of eIF2α-specific phosphatase cofactor activity as a mechanism that can sense and respond to perturbations of cellular homeostasis.
Results
The HIV-PIs Trigger a Limited Unfolded Protein Response.
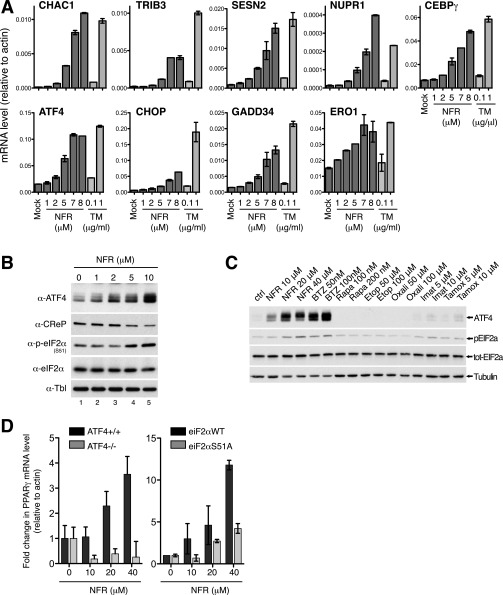
To identify stress pathways engaged by the HIV-PIs, we carried out RNA sequencing (RNA-seq) on HeLa cells treated for 6 h with Nelfinavir or its carrier DMSO (Dataset S1). We used a drug concentration in accordance with the levels found in Nelfinavir-treated patients, which can reach 10 μM in the plasma and can be 4–5-fold higher in peripheral blood mononuclear cells (23, 24). A heat map of mRNA expression revealed a group of genes that were significantly induced upon treatment with Nelfinavir (Fig. 1A). We then investigated the functional impact of Nelfinavir, using AmiGO software to search for Gene Ontology term enrichment (25). Response to ER stress was the most significant functional category enriched among Nelfinavir up-regulated genes (Fig. 1B and Dataset S2). This observation is consistent with previous studies reporting the activation of ER-stress–related genes such as GADD34 and CHOP in cells treated with Nelfinavir (20, 21, 26, 27). To get more insight on the type of response engaged by Nelfinavir, we compared this signature with a dataset dissecting the ER-stress response in mouse embryonic fibroblasts (MEFs) (28). We found that among the 32 genes up-regulated in both studies, 30 were previously identified in the ER-stress study as targets of the transcription factor ATF4 (28) (Fig. 1C and Dataset S3). Moreover, of the two exceptions, STC2 has also been reported to be an ATF4-induced gene in another study (29). To confirm these results, we interrogated the expression of a panel of ER-stress–induced genes upon treatment with Nelfinavir. The expression of ATF4-dependent genes such as GADD34, TRIB3, and SESN2 was increased to levels comparable to the expression observed in the presence of Tunicamycin, a glycosylation inhibitor that triggers a classical ER-stress response (Fig. 1D). In contrast, when we extended this analysis to additional ER-stress genes previously shown to be ATF4-independent, such as EDEM, ERdj4, or HERP, we found that Nelfinavir, compared with Tunicamycin, induced no or only weak expression of these transcripts (Fig. 2A). Moreover, in the presence of Nelfinavir, we could not detect activation of the ER-stress sensor ATF6, as monitored by liberation of its cleaved fragment (Fig. 2B). We did not observe an effect of Nelfinavir on Tunicamycin-mediated ATF6 cleavage; thus, Nelfinavir does not inhibit the ATF6-specific proteases S1P and S2P (Fig. 2B, lanes 5 and 6). Similarly, Nelfinavir did not promote the accumulation of high-molecular weight complexes containing the ER chaperone BiP (Fig. 2C), which is usually associated with the presence of misfolded proteins (13, 30). These findings suggest that Nelfinavir initiates an ATF4-dependent transcriptional program rather than a typical ER-stress response.
Fig. 1.
Nelfinavir triggers an ATF4 transcriptional response. (A–C) Triplicates of HeLa cells treated 6 h with DMSO or 20 μM NFR were analyzed for gene expression by RNA-seq. (A) Heat map showing the 153 Nelfinavir up-regulated genes with a P value < 0.01. The genes are listed in order of statistical significance. The z score is based on the mean of the six samples; green color indicates high expression and red low expression. The Right panel shows the 20 most significantly induced genes. Each column represents the data of one experiment; purple and pink dots refer to ATF4 only and ATF4 and CHOP targets, respectively, as reported in C. (B) Gene ontology analysis for the genes identified in A. (C) Venn diagram showing the overlap between the NFR-induced genes (with a P value < 0.05) in HeLa cells (yellow) and the published tunicamycin (TM)-triggered ER-stress signature (orange) identified in MEFs (28). Note that 30 of the 32 overlapping genes were reported to be ATF4 only (purple) or ATF4 and CHOP (pink) target genes (28). (D) ATF4 WT and ATF4 −/− MEFs treated for 6 h with the indicated concentration of NFR or TM were analyzed for expression of indicated genes by real-time PCR relative to β-actin (data are presented as fold change compared with untreated cells, and mean and SEM of technical triplicates of one representative experiment are shown).
Fig. 2.
Nelfinavir does not trigger a classical UPR. (A) WT MEFs were treated for 6 h with indicated doses of NFR (dark gray bars) or Tunicamycin (TM) (light gray bars). Induction of XBP-1/ATF6–dependent genes Calreticulin, BiP, EDEM, p58IPK, HRD1, Grp94, ERdJ4, and HERP was measured by real-time PCR relative to β-actin (mean and SEM of technical triplicates of one representative experiment are shown). (B) Immunoblot of ATF6 in WT MEFs treated for 6 h with the indicated concentration of NFR, TM (10 μg/mL), or a combination of NFR and TM. Arrows represent full-length (ATF6) and cleaved ATF6 (cATF6) as indicated. Asterisk shows unglycosilated ATF6. Tubulin (Tbl) is used as loading control. (C) Immunoblot of BiP in aggregated insoluble fraction and in total extracts of HeLa cells treated with TM (2 μg/mL) or indicated doses of NFR for 16 h. Tubulin (Tbl) is used as the loading control.
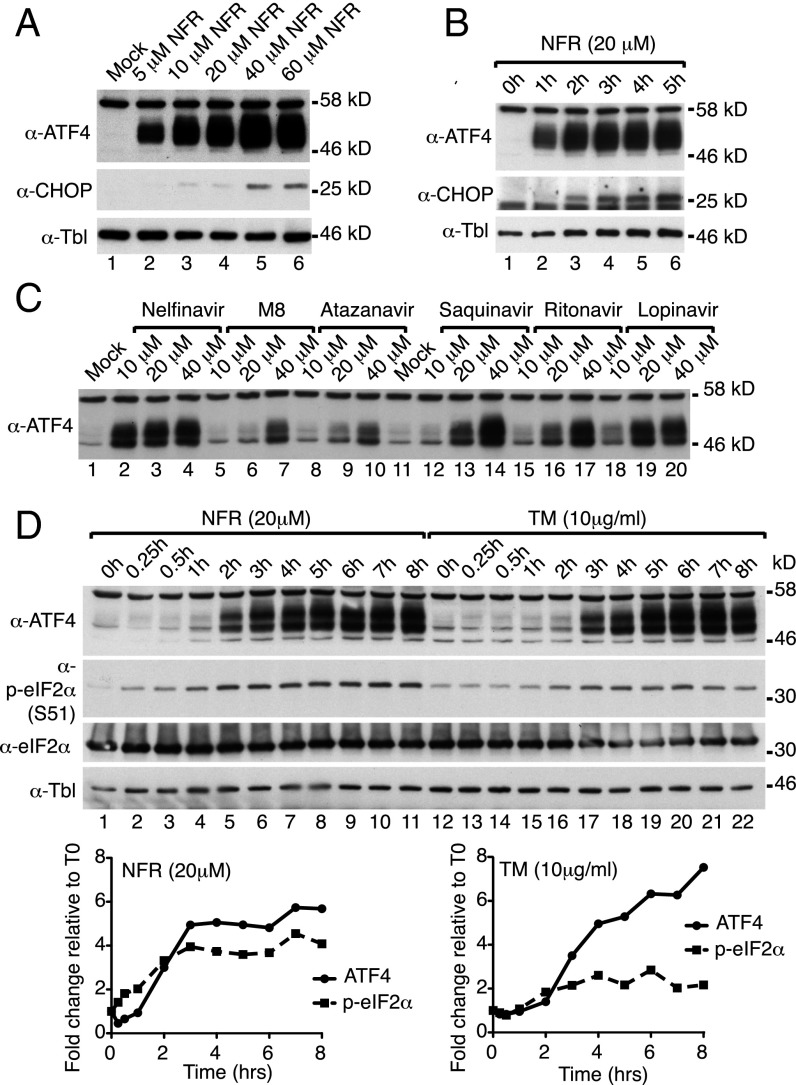
We assessed the protein expression of ATF4 and its downstream target CHOP in MEFs treated for 6 h with increasing doses of Nelfinavir. ATF4 expression was efficiently induced at Nelfinavir concentrations above 5 μM (Fig. 3A). In a time course experiment using 20 μM Nelfinavir, induction of ATF4 occurred as early as 1 h after treatment, indicating that the activation of this stress pathway is a rapid event (Fig. 3B). This response is sustained over time, as shown in HeLa cells treated for 14 h with increasing concentration of Nelfinavir (Fig. S1 A and B). Gene induction and ATF4 production was robust and observed above concentration around 2–5 μM. Investigation of a panel of HIV-PIs used in the clinic as well as hydroxy-t-butylamidenelfinavir (M8), the active Nelfinavir metabolite, showed that most HIV-PIs trigger robust ATF4 expression (Fig. 3C). Nevertheless, we consistently found that among the HIV-PIs, Nelfinavir initiates the most robust ATF4 activation in all tested mouse and human cell types. Altogether these data indicate that Nelfinavir triggers a transcriptional response mostly characterized by the selective activation of ATF4.
Fig. 3.
Nelfinavir promotes the production of the ATF4 transcription factor. (A and B) WB analysis of ATF4 and CHOP expression in WT MEFs treated for 6 h with increasing doses of NFR (A) or with 20 μM NFR for the indicated times (B). (C) HeLa cell response to increasing doses of different HIV-PIs and NFR metabolite M8. Cells were treated for 6 h and analyzed by WB for ATF4 expression. (D) WB analysis (Upper panel) and quantification (Lower panels) of a time course of NFR (20 μM) or TM (10 μg/mL) treatment in HeLa cells. Immunoblot for p-eIF2α shows a robust and sustained eIF2α phosphorylation over time with NFR that correlates with ATF4 expression. Tubulin (Tbl) is used as the loading control.
Fig. S1.
ISR activation by NFR (related to Figs. 4 and 5). (A and B) HeLa cells treated for 14 h with the indicated concentration of NFR or TM were analyzed for expression of indicated genes by real-time PCR relative to β-actin (A) (mean and SEM of technical triplicates of one representative experiment are shown). Samples were also analyzed by WB to monitor ATF4 induction, eIF2α phosphorylation, as well as CReP level (B). Tubulin (Tbl) is used as the loading control. (C) HeLa cells treated for 6 h with the relevant concentration of Nelfinavir (NFR), Bortezomib (BTZ), Rapamycin (Rapa), Etoposide (Eto), Oxaliplatin (Oxali), Imatinib (Imat), and (Z)-4-Hydroxytamoxifen (Tamox) were analyzed by WB to monitor ATF4 induction and eIF2α phosphorylation. Tubulin (Tbl) is used as the loading control. (D) PPARγ is induced by the ISR. ATF4−/− and ATF4+/+ MEFs (Left panel) and eIF2αWT and eIF2α S51A MEFs (Right panel) were treated for 6 h with indicated doses of NFR. PPARγ mRNA level was measured by real-time PCR relative to β-actin (fold change of untreated cells; mean and SEM of technical triplicates of one representative experiment).
Nelfinavir Induces a Robust ISR.
Expression of ATF4 is usually dependent on the phosphorylation of eIF2α at serine 51, the effector branch of the ISR (31). Compared with the response observed in the presence of ER-stress inducer Tunicamycin, we found that treatment with Nelfinavir triggered a strong and sustained eIF2α phosphorylation and concomitant ATF4 expression (Fig. 3D). Accordingly, Nelfinavir-mediated ATF4 and CHOP induction was impaired in MEFs homozygous for the phosphorylation-deficient mutant of eIF2α, eIF2α(S51A) (Fig. 4A). To evaluate the contribution of eIF2α phosphorylation to the Nelfinavir-mediated transcriptional program, we carried out RNA-seq on eIF2α(WT) or eIF2α(S51A) MEFs treated with DMSO or Nelfinavir. We found that 646 genes were up-regulated at least twofold by Nelfinavir in MEFs (Fig. 4B and Dataset S4). Most of these genes (84%) were found to require eIF2α phosphorylation for optimal induction by Nelfinavir (Fig. 4C and Dataset S5), demonstrating that the ISR is the major transcriptional program initiated upon treatment with Nelfinavir. Similar results were obtained by real-time PCR analysis of eIF2α(WT) or eIF2α(S51A) MEFs treated with increasing amounts of Nelfinavir or Tunicamycin (Fig. 4D). In tumors, eIF2α phosphorylation is often detected in the context of chemotherapy- and radiotherapy-mediated cell death (32). We compared Nelfinavir-mediated eIF2α phosphorylation and ATF4 activation with other treatments used or considered as anticancer therapeutics. Compared with the drugs tested, short-term treatment with Nelfinavir elicits a stronger response similar to Bortezomib, an inhibitor of the proteasome that can cause ER stress (Fig. S1C). Collectively, these findings demonstrate that Nelfinavir is a potent activator of the ISR.
Fig. 4.
NFR-mediated ATF4 transcriptional signature is controlled by eIF2α phosphorylation. (A) EIF2αWT and eIF2α S51A MEFs treated for 6 h with the indicated concentration of NFR or TM (10 μg/mL) and analyzed by WB with the indicated antibodies. In the absence of eIF2α phosphorylation, ATF4 and CHOP induction are completely abolished. Tubulin (Tbl) is used as the loading control. (B) Heat map comparing gene up-regulation in eIF2αWT and eIF2α S51A MEFs treated for 6 h with 20 μM NFR. Genes that were up-regulated >twofold in eIF2αWT were included and listed in order of fold induction. Each row corresponds to a single gene. Right panel shows the 30 highly induced genes in eIF2αWT compared with eIF2α S51A MEFs. (C) Pie chart of the 646 genes induced by NFR >2-fold in eIF2αWT MEFs (identified in B), of which 544 (84%) showed a reduced or no induction in eIF2α S51A MEFs. (D) eIF2αWT and eIF2α S51A MEFs treated for 6 h with the indicated concentration of NFR or TM were analyzed for expression of indicated genes by real-time PCR relative to β-actin (data are presented as fold change compared with untreated cells, and mean and SEM of technical triplicates of one representative experiment are shown).
Nelfinavir Promotes the Activation of ATF4 and the ISR in the Liver of Treated Mice.
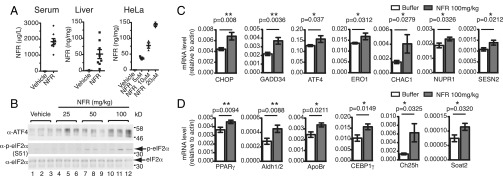
To test whether Nelfinavir triggers the ISR response in vivo, we injected Nelfinavir intraperitoneally as described previously (33). Mice treated with 100 mg/kg were killed and Nelfinavir concentrations were measured in the serum and liver (Fig. 5A). We found that serum concentration was in the range of 1,000 μg/L (1.67 μM) to 3,000 μg/L (5 μM), with a median concentration of 1,757 μg/L. A study in treated HIV patients reported median concentrations of Nelfinavir of 1,632 μg/L for patients with Nelfinavir administrated 1,250 mg twice daily and 1,862 μg/L for patients under a 750 mg thrice daily regimen (34). This indicates that our mouse model recapitulates the concentrations observed in patients. Moreover, Nelfinavir concentrations in the liver reflected the amount found in HeLa cells treated for 6 h with 5–10 μM Nelfinavir (Fig. 5A), suggesting that in the liver, Nelfinavir can reach concentrations that can trigger an ISR in vitro. We therefore tested ISR activation in treated mice. Compared with vehicle-treated mice, NFR treatment augmented the ATF4 protein level and eIF2α phosphorylation in the liver (Fig. 5B). Expression of known ISR-dependent genes such as CHOP, CHAC1, as well as ATF4 was significantly increased in the liver of mice treated with 100 mg/kg of Nelfinavir (Fig. 5C). Through expression studies in MEFs, we found that PPARγ, a nuclear receptor involved in lipid metabolism, previously shown to be under the control of ATF4 in the liver (35), was induced by Nelfinavir treatment in an ATF4-dependent manner (Fig. S1D and Dataset S5). Increased PPARγ was also observed in the liver of Nelfinavir-treated mice (Fig. 5D). We analyzed a few additional metabolic regulators that were identified in the RNA-seq experiments (Datasets S1 and S5) and found increased expression of genes involved in lipid metabolism such as the Apolipoprotein B Receptor (ApoBr), Cholesterol 25-Hydroxylase (Ch25h), and Sterol O-Acyltransferase 2 (Soat2). Together these results indicate that Nelfinavir-mediated ISR can affect gene expression and metabolic responses in vivo.
Fig. 5.
NFR induces expression of ATF4 target genes in mouse liver. (A) NFR concentration measured in serum and liver of mice injected i.p. for 3 consecutive days with 100 mg/kg or vehicle (n = 8) and intracellular concentration observed in HeLa cells treated for 6 h with the indicated amount of NFR or vehicle (n = 3). (B) Immunoblot for ATF4, p-eIF2α and total eIF2α level in liver protein extracts of mice injected i.p. for 3 consecutive days with the indicated doses of NFR (three mice per condition). (C and D) Real-time PCR analysis of the indicated mRNA isolated from the liver of mice injected i.p. for 3 consecutive days with NFR (100 mg/kg; n = 10) or with the vehicle only (n = 10). P values were determined by a one-tailed unpaired Student’s t test (**P < 0.01, *P < 0.05).
ISR Activation Bypasses Direct Activation of the eIF2α Kinases.
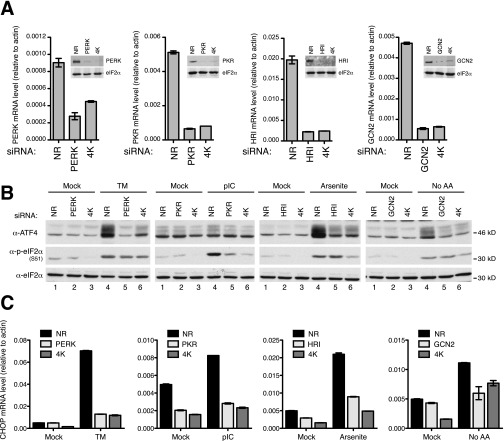
In the context of ER stress, the ISR component of the ER-stress response is mediated by the eIF2α-kinase PERK. To interrogate PERK involvement in mediating Nelfinavir-induced ISR, we treated PERK-sufficient and -deficient MEFs with Nelfinavir or Tunicamycin. As expected, Tunicamycin induced expression of ATF4 and CHOP in a PERK-dependent manner (Fig. 6A, compare lane 5–6 and 11–12, and Fig. 6B). In contrast, Nelfinavir-mediated ISR was unaffected by PERK deficiency (Fig. 6 A and B and Fig. S2A). These data are consistent with the observation that Nelfinavir triggers only a partial ER-stress–like response and raised the question of the possible involvement of other eIF2α kinases. We analyzed MEFs deficient for PKR, GCN2, and HRI and found that deletion of any of the three eIF2α kinases does not alter Nelfinavir-mediated ISR responses, as measured by Western blot detection of ATF4 and CHOP expression and eIF2α phosphorylation (Fig. 6C) or by quantitative measurement of CHOP mRNA (Fig. 6D). As expected, the response induced by the well-known ISR stimuli poly(I:C) lipofection, lack of amino acids, or arsenite treatment required PKR, GCN2, and HRI, respectively (Fig. 6C, compare lanes 11 and 12 for each panel). Note that in this experimental setup, poly(I:C) does not induce a detectable increase in ATF4 expression but elicits a strong eIF2α phosphorylation that requires PKR (36). Next, we silenced the expression of the individual eIF2α kinase in HeLa cells using specific siRNAs. Each siRNA was tested and selected for its ability to diminish eIF2α kinase protein and mRNA expression (Fig. S3A) and to yield a significant decrease in eIF2α phosphorylation and subsequent ATF4 expression as well as in CHOP mRNA induction upon treatment with the corresponding activating stress signal (Fig. S3 B and C). Consistent with the observations made in knockout cells, silencing of each eIF2α kinase individually did not affect Nelfinavir responses, including ATF4 expression, eIF2α phosphorylation (Fig. 6E), and CHOP transcription (Fig. 6F).
Fig. 6.
NFR triggers eIF2α phosphorylation and downstream effectors independently of the eIF2α kinases PERK, PKR, HRI, and GCN2. (A) PERK+/+ and PERK−/− MEFs treated for 6 h with the indicated concentration of NFR or TM and analyzed by WB with indicated antibodies. (B) CHOP mRNA level in PERK+/+ and PERK−/− MEFs treated for 6 h with the indicated concentration of NFR or TM was measured by real-time PCR. (C) PKR−/−, GCN2−/−, and HRI−/− MEFs and their respective WT control were treated for 6 h with the indicated concentration of NFR or TM (10 μg/mL) and analyzed by WB with the indicated antibodies. Each cell type was also treated with kinase-specific stimuli; PKR cells were lipofected with p(I:C) (10 μg/mL) for 6 h, GCN2 cells were cultured 6 h in a medium depleted of amino acid, and HRI cells were treated for 1 h with 50 μM arsenite. (D) CHOP mRNA level in PKR−/−, GCN2−/−, and HRI−/− MEFs and their respective WT control treated for 6 h with the indicated concentration of NFR or TM (10 μg/mL) was measured by real-time PCR. (E) HeLa cells were transfected with PKR, HRI, GCN2, PERK, or nonrelevant (NR) siRNA for 48 h and treated for 6 h with NFR (20 μM) or TM (10 μg/mL). Immunoblots were performed with ATF4, p-eIF2α, and total eIF2α antibodies. (F) CHOP mRNA level was measured by real-time PCR in HeLa cells transfected for 48 h with indicated siRNA and left untreated (Mock; black bars) or treated for 6 h with 20 μM NFR (gray bars). (G) PERK, PKR, GCN2, and HRI activation was measured using phos-tag SDS/PAGE and specific antibodies in HeLa cells subjected to the indicated treatments: 6 h NFR or TM to activate PERK; No AA, 6 h medium without amino acids to activate GCN2; 50 μM arsenite for 1 h to activate HRI; p(I:C) (10 μg/mL) was lipofected for 6 h to activate PKR. Tubulin (Tbl) is used as loading control. (H) HeLa cells were transfected with a mixture of PKR, HRI, GCN2, and PERK siRNA (4K) or nonrelevant (NR) siRNA. At 48 h after transfection, cells were treated for 6 h with NFR or left untreated (Mock). Immunoblots were performed for the indicated antibodies. (I) CHOP mRNA level was measured by real-time PCR in HeLa cells transfected for 48 h with the indicated siRNA and left untreated (Mock; black bars) or treated for 6 h with 20 μM NFR (gray bars). Real-time PCRs are represented as mean and SEM of technical triplicates of one representative experiment.
Fig. S2.
NFR-mediated ATF4 response is PERK and IRE1-independent (related to Fig. 6 A and B). (A) PERK+/+ and PERK−/− MEFs were treated for 6 h with indicated doses of NFR or TM and analyzed for expression of indicated genes by real-time PCR relative to β-actin (data are presented as fold change compared with untreated cells, and mean and SEM of technical triplicates of one representative experiment are shown). (B and C) IRE1+/+ and IRE1−/− MEFs were treated for 6 h with indicated doses of NFR or TM and analyzed by WB for ATF4 and CHOP expression (B) or for induction of CHOP mRNA by real-time PCR relative to β-actin (data are presented as fold change compared with untreated cells, and mean and SEM of technical triplicates of one representative experiment are shown) (C). (D) CReP protein level was analyzed by WB in IRE1−/− MEFs treated for 6 h with indicated doses of NFR or TM (10 μg/mL). Untreated CReP+/+ and CReP−/− MEFs were used as the positive control for antibody specificity. Asterisk shows an unspecific band, and tubulin is used as the loading control.
Fig. S3.
PERK, PKR, HRI, and GCN2 siRNA are efficient after 48 h transfection (related to Fig. 4 E, F, H, and I). (A) WB analysis and mRNA-level quantification of PERK, PKR, GCN2, and HRI 48 h after HeLa cell transfection with specific siRNA or the combination of four siRNAs (4K) (for the mRNA level, data are presented as mean and SEM of technical triplicates of one representative experiment). (B) HeLa cells were transfected with the indicated siRNA for 48 h and left untreated (Mock) or treated with kinase-specific stimuli: TM was used at 10 μg/mL for 6 h to activate PERK; No AA, complete medium was replaced with medium without amino acids for 6 h to activate GCN2; arsenite was used at 50 μM for 1 h to activate HRI; p(I:C) (10 μg/mL) was lipofected for 6 h to activate PKR. After treatment, cells were analyzed by WB for eIF2α phosphorylation and ATF4 induction. (C) Cells treated as in B and analyzed by quantitative real-time PCR for CHOP mRNA level relative to β-actin (mean and SEM of technical triplicates of one representative experiment).
Activation of the eIF2α kinase can be detected by monitoring its phosphorylation status. To detect active eIF2α kinases, we used a Phos-tag reagent, which selectively binds to phosphorylated amino acid residues (37, 38). Compared with stress signals that specifically triggered activation of each of the eIF2α kinases, we did not detect increased phosphorylation of any of the eIF2α kinases in the presence of Nelfinavir (Fig. 6G). Intriguingly, these data indicate that Nelfinavir does not promote eIF2α phosphorylation by directly augmenting the activity of the upstream kinases. To test whether Nelfinavir-induced ISR required the eIF2α kinases at all, we silenced all four eIF2α kinases using a mixture of specific siRNAs that was, as for the individual siRNA, tested for its ability to decrease ISR induction by specific stimuli (Fig. S3, 4K conditions). Reducing the expression of the four eIF2α kinases together reduced the Nelfinavir-mediated ISR (Fig. 6 H and I). These results indicate that Nelfinavir-induced ISR relies on the redundant and basal activity of eIF2α kinases, without significantly increasing their activity.
eIF2α Dephosphorylation Is Modulated by Nelfinavir.
Previous studies identified CReP as a constitutive repressor of eIF2α phosphorylation that mediates basal dephosphorylation of eIF2α by recruiting the phosphatase PP1 (9). We investigated PP1 and CReP expression levels in Nelfinavir- or Tunicamycin-treated samples. Although we observed no differences in CReP expression upon Tunicamycin treatment over time, Nelfinavir triggered a decrease of CReP protein expression as early as 1 h after treatment (Fig. 7A). Nelfinavir-induced CReP down-regulation was dose-dependent (Fig. 7B) and correlated with ATF4 activation.
Fig. 7.
Nelfinavir modulates constitutive eIF2α dephosphorylation. (A) Time course of NFR (20 μM) or TM (10 μg/mL) treatment in HeLa cells. Immunoblot for indicated antibodies shows CReP down-regulation in NFR-treated cells, which correlates with ATF4 induction. PP1 level is not affected. Lower panel showed a WB quantification for CReP and ATF4 protein level over time in NFR- and TM-treated HeLa cells. (B) HeLa cells were treated for 6 h with increasing doses of NFR and analyzed by WB using the indicated antibodies. NFR induced CReP down-regulation in a dose-dependent manner. (C) CReP mRNA level in HeLa cells treated for 6 h with the indicated concentration of NFR (dark gray bars) or with TM (10 μg/mL; light gray bars) was measured by real-time PCR relative to β-actin (mean and SEM of technical triplicates of one representative experiment). (D) EIF2αWT and eIF2αS51A MEFs treated for 6 h with the indicated concentration of NFR or with TM (10 μg/mL) and analyzed by WB with the indicated antibodies. Untreated CReP+/+ and CReP−/− MEFs were used as positive control for antibody specificity. Asterisk shows an unspecific band, and PP1 is used as loading control. (E) HEK293T cells stably expressing a doxycycline (DOX)-inducible version of VSV-eIF2α were treated as indicated (DOX 1 μg/mL for 24 h followed by NFR 20 or 40 μM for 6 h). VSV-eIF2α, PP1, and CReP protein levels were analyzed by WB using specific antibodies after anti-VSV immunoprecipitation (IP; Upper panel) and in total cell lysates (Input; Lower panel). Association of PP1 and CReP with VSV-eIF2α is impaired in NFR-treated samples. (F) HeLa cells were treated for 24 h with the indicated concentration of NFR, Salubrinal (Sal), or Guanabenz (Gua) and analyzed by WB for CReP, ATF4, p-eIF2α, and total eIF2α expression level. Tubulin (Tbl) is used as the loading control.
CReP mRNA has recently been shown to be a target of the regulated IRE1α-dependent decay (RIDD) process (39). We therefore tested Nelfinavir-mediated ISR induction in IRE1α-deficient MEFs. ATF4 and CHOP induction as well as CReP down-regulation was not affected in IRE1α-deficient cells compared with control, suggesting that RIDD pathway is not involved in NFR-mediated CReP decrease (Fig. S2 B–D). Accordingly, the level of CReP mRNA was not affected by Nelfinavir treatment (Fig. 7C), indicating that CReP regulation is likely posttranscriptional.
Importantly, Nelfinavir-mediated CReP reduction was maintained in MEF eIF2αS51A, showing that it occurs upstream of eIF2α phosphorylation and did not depend on subsequent inhibition of protein translation (Fig. 7D). In line with this result, we found that Nelfinavir decreased CReP levels in the presence of cycloheximide, a general inhibitor of protein translation (Fig. S4A). Moreover, proteasome inhibition with MG132 did not significantly affect Nelfinavir-mediated CReP protein decrease (Fig. S4A). Similarly we found that down-regulation of βTRCP1 and βTRCP2, two E3 ligases that bind and regulate CReP protein levels (40), increased basal CReP levels but did not affect Nelfinavir-mediated ATF4 induction or CReP decrease (Fig. S4B). These findings indicate that the Nelfinavir-dependent regulation of CReP is posttranslational and does not involve the classical proteasome-dependent degradation pathway.
Fig. S4.
NFR disrupts PP1/CReP association without affecting PP1 phosphatase activity (related to Fig. 7). (A) HeLa cells were treated for 4 h with 20 μM NFR with or without cycloheximide (CHX, 25 μM) and MG132 (5 μM) as indicated. Immunoblot shows CReP, p-eIF2α, and total eIF2α levels (Upper panel). Lower panel is a WB quantification of CReP protein level in NFR-treated samples (gray bars) expressed in percentage of the level in samples without NFR (black bars) (bars represent mean and SEM of quantification of three independent experiments). (B) Down-regulation of βTRCP1 and βTRCP2 does not affect Nelfinavir-mediated ATF4 induction or CReP declining. HeLa cells were treated for 6 h with the indicated concentration of NFR or TM (10 μg/mL) 48 h after transfection with specific siRNAs targeting βTRCP1 and βTRCP2. Immunoblot shows CReP, ATF4, p-eIF2α, and total eIF2α levels. mRNA quantification confirmed βTRCP1 and βTRCP2 knock-down efficiency (Lower panel). (C) PP1 phosphatase activity is not affected by NFR. HeLa cells were treated with indicated doses of NFR or TM (10 μg/mL) for 6 h or with CaliculinA (CalA, 10 nM; PP1 specific inhibitor) or Okadaic Acid (OkA, 200 mM; PP2 specific inhibitor) for 1 h. Following treatments, PP1 was immunoprecipitated and its activity measured as described in Materials and Methods. Phosphatase activities are given as percent change relative to control (Mock-treated cells). Results shown are the mean and SEM of three independent experiments. P values were determined by a one-tailed unpaired Student’s t test (***P < 0.001, **P < 0.01, *P < 0.05). Lower panel shows one representative immunoblot of PP1 after immunoprecipitation. Asterisk shows unspecific band. (D) H2AX phosphorylation and PP1 total protein levels were assessed by immunoblot. Phosphorylation of H2AX was induced in HeLa cells after treatment with the PP1-specific inhibitor Caliculin A (CalA) for 2 h but not after 6 h NFR. Tubulin (Tbl) is used as loading control. (E) HeLa cells were treated for 2 h with Caliculin A (CalA) at indicated doses. Immunoblot shows that PP1 inhibition triggers eIF2a and H2AX phosphorylation and ATF4 induction. Tubulin (Tbl) is used as the loading control.
PP1 is a key cellular phosphatase that catalyzes dephosphorylation of many proteins in a highly regulated and specific manner (41). We tested whether Nelfinavir altered PP1 generic activity. First, we did not detect PP1 protein changes upon treatment with Nelfinavir (Fig. 7 A and D). Further, PP1 phosphatase activity (42) was not affected in cell extracts of Nelfinavir-treated samples (Fig. S4C). Finally, Nelfinavir did not enhance the phosphorylation of the PP1 target histone H2AX (Fig. S4D). In contrast, treatment with the PP1 inhibitor Caliculin A affected PP1 activity and triggered phosphorylation of both eIF2α and H2AX (Fig. S4 D and E). These data indicate that Nelfinavir does not alter the generic activity of PP1. To interrogate the possibility that Nelfinavir regulates the recruitment of the PP1 complex to eIF2α, we generated a cell line expressing an inducible VSV-tagged eIF2α protein. Immunoprecipitation of VSV-eIF2α revealed decreased PP1 binding to eIF2α in the presence of Nelfinavir (Fig. 7E). Interestingly, neither Salubrinal nor Guanabenz (two drugs that have been shown to affect eIF2α phosphorylation by targeting PP1 recruitment to eIF2α) (12, 13) affected CReP protein levels (Fig. 7F). Thus, the previously described experiments all point to inhibition of eIF2α dephosphorylation that correlates with decreased CReP protein levels, as a specific hallmark of the Nelfinavir-mediated stress response.
CReP Is Required for Nelfinavir-Mediated ISR.
In accordance with previous results that showed ISR activation in cells expressing an shRNA construct targeting CReP (9) or isolated from CReP-deficient mice (10), we observed increased ATF4 and CHOP expression in unstressed cells transfected with CReP-specific siRNAs, as well as in CReP-deficient MEFs (Fig. 8 A–D). The transcriptional program triggered by CReP deficiency recapitulates the ISR elicited by Nelfinavir; factors such as PPARγ, CHAC1, and SESN2 were induced, whereas BiP and ERdj4 were not (Fig. 8C). Moreover, the ISR mediated by CreP deficiency was reduced when expression of the four eIF2α kinases was silenced (Fig. S5). These data further demonstrate that basal phosphorylation of eIF2α is a default program in unstressed cells that is constitutively inhibited by CReP–PP1 complexes to maintain homeostasis. Interestingly, when we monitored Nelfinavir responses in CReP-silenced or -deficient cells, we found that Nelfinavir still partially increased ATF4 expression (Fig. 8 B and D), suggesting that beyond CReP, Nelfinavir may affect an alternative ISR regulatory pathway. Because GADD34 was increased by CReP deficiency (Fig. 8C), we analyzed ISR activation in GADD34-deficient MEFs transfected with CReP siRNA (Fig. 8E) as well as in HeLa cells transfected with a combination of GADD34 and CReP siRNAs (Fig. S6). Nelfinavir no longer increased ATF4 expression in cells deficient for both CReP and GADD34, whereas Tunicamycin treatment, which drives ISR through PERK activation, still induced ATF4 under the same conditions (Fig. 8E). This result suggests that Nelfinavir affects the GADD34-mediated negative feedback loop and is consistent with the sustained and robust ISR activation observed with Nelfinavir.
Fig. 8.
CReP regulation is sufficient to trigger ISR. (A) ATF4, CHOP, and CReP mRNA level in HeLa cells transfected for 24 h with nonrelevant (NR) siRNA or two different CReP-specific siRNAs were measured by real-time PCR relative to β-actin (mean and SEM of technical triplicates of one representative experiment). (B) HeLa cells were transfected for 24 h with the indicated siRNA and treated or not with 20 μM NFR. Immunoblot shows that CReP silencing is sufficient to induce a robust eIF2α phosphorylation, leading to ATF4 and CHOP induction. (C) CReP−/− MEFs (gray bars) and WT control (CReP+/+; black bars) were compared for the mRNA level of indicated genes by real-time PCR relative to β-actin (mean and SEM of technical triplicates of one representative experiment). (D) CReP−/− MEFs and WT control (CReP+/+) were treated for 6 h with NFR at the indicated concentration and TM (10 μg/mL) and analyzed by WB for ATF4 expression and eIF2α phosphorylation. (E) GADD34 ΔC/ΔC MEF and WT control cells were transfected for 24 h with nonrelevant (NR) or CReP-specific siRNA and treated for 6 h with NFR or TM (10 μg/mL). WB analysis using indicated antibodies shows that both CReP and GADD34 silencing are required to abolish NFR-mediated eIF2α phosphorylation and ATF4 induction, whereas in the same condition TM is still able to increase the ATF4 level. (F) Quantification of newly synthesized proteins at 0, 2, 4, and 6 h after 10 μg/mL TM (Left panels) or 20 μM NFR (Right panels) treatment in indicated MEFs (eIF2αWT, eIF2α S51A, CReP+/+, and CReP−/−). Treated cells were labeled for 15 min with [35S]methionine and visualized by SDS/PAGE and subsequent autoradiography. Autoradiography was quantified, and results show percentage of translation compared with untreated cells. The mean and SEM of three independent metabolic labeling experiments are shown. P values were determined by a one-tailed unpaired Student’s t test (***P < 0.001, **P < 0.01, *P < 0.05).
Fig. S5.
CReP controls basal level of eIF2α phosphorylation maintained by constitutive activity of eIF2α-specific kinases (related to Fig. 8). HeLa cells were transfected with a mixture of siRNAs targeting PERK, PKR, HRI, and GCN2. Twenty-four hours later, a second transfection with nonrelevant (NR) or two different CReP-specific siRNAs was performed. After 24 h, cells were analyzed by WB with the indicated antibodies. Results show that in the absence of the four stress kinases PERK, PKR, HRI, and GCN2, CReP silencing-mediated ATF4 induction is abolished. Tubulin (Tbl) is used as the loading control.
Fig. S6.
Both CReP and GADD34 silencing are required to completely abolish NFR-mediated ATF4 induction (related to Fig. 8). HeLa cells were transfected with indicated siRNA for 24 h and treated for 6 h with NFR at the indicated concentration. Immunoblot shows that both CReP and GADD34 silencing are required to completely abolish NFR-mediated ATF4 induction (lanes 4–6).
Because inhibition of translation initiation is a key feature of the ISR and is controlled by the eIF2α phosphorylation status, we quantified the role of CReP in NFR- or Tunicamycin-mediated ISR activation by monitoring protein synthesis. As expected, the mutation in the eIF2α phosphorylation site rescued translation inhibition in the presence of Tunicamycin or Nelfinavir (Fig. 8F). However, CReP deficiency only affected Nelfinavir-mediated translation inhibition without affecting Tunicamycin-mediated decrease in translation rates (Fig. 8F). These findings further demonstrate the specific role of CReP in initiating Nelfinavir-induced ISR and identify CReP modulation as a specific mechanism of ISR activation.
Discussion
EIF2α phosphorylation and the ISR are mostly initiated by stress insults that prime specific eIF2α kinases. EIF2α dephosphorylation events, on the other hand, are considered as negative feedback loops that regulate the intensity and duration of the response. Here we identify modulation of eIF2α dephosphorylation and CReP down-regulation as the initiating event driving Nelfinavir-dependent transcriptional reprogramming. Both Nelfinavir and CReP deficiency trigger a comparable response that is only impaired in the absence of the four eIF2α kinases. These results exclude the hypothesis that Nelfinavir activates a unique eIF2α kinase. Instead, they suggest that the four known eIF2α kinases (i.e., PERK, GCN2, HRI, and PKR) have basal and redundant activity by default and that CReP is a key guardian of cellular integrity that keeps the ISR switched off. The finding that the CReP level can be pharmacologically modulated raises the possibility that other cellular mechanisms may regulate CReP activity to trigger this specific cellular stress response. Here we show that Nelfinavir is associated with decreased CReP protein levels, however whether this is the direct cause of ISR activation is unclear. Indeed, our result showing that PP1 association with eIF2α decreases upon NFR treatment (Fig. 7E) suggests that NFR-mediated CReP decline could be an indirect consequence of a functional regulation of the eIF2α phosphatase complex. In line with this idea, we showed that upon NFR treatment, GADD34, the inducible eIF2α-specific cofactor of PP1, is expressed but cannot compensate for the loss of CReP, suggesting that NFR interferes with GADD34 function. Accordingly, GADD34 silencing impaired the remaining NFR-mediated ISR activity observed in CReP-deficient cells. It is likely that identification of Nelfinavir’s cellular targets will help define the mechanisms controlling CReP levels and phosphatase complex activity. If this mechanism is probably complex and may rely on binding to multiple targets (16), the common biological properties shared by the HIV-PIs may also reflect their common specific chemical properties. Most HIV-PIs are peptidomimetics that were designed based on a synthetic analog of the peptide bond between phenylalanine and proline at positions 167 and 168 of the gag–pol polyprotein, a target of the HIV protease (43). Therefore, we cannot exclude that these common structures shared by HIV-PIs could engage a specific proteostasis sensor to initiate the ISR.
Increased eIF2α phosphorylation is associated with several pathophysiological conditions including neurodegeneration, cancer, diabetes, and obesity (44–46). Long-term treatment with the HIV-PIs is associated with adverse effects such as hyperlipidemia or hypolipidemia, body fat redistribution, osteopenia and osteoporosis (47), as well as insulin resistance and susceptibility to type II diabetes (48–51). Interestingly, loss-of-function mutation in CReP protein leading to decreased PP1 binding and sustained eIF2α phosphorylation has recently been shown to drive β-cell dysfunction and metabolic defects that can lead to diabetes in human (52). Elevated levels of ISR markers (such as ATF4 or eIF2α phosophorylation) in the liver of NFR-treated mice suggest that activation of this pathway may contribute to the side effects observed in HIV-PI–treated patients. Indeed, ATF4 is known to play a key role in osteoblast differentiation (53) and its transgenic expression in osteoclasts promotes severe osteopenia (54). Therefore, bone diseases found in HIV-PI–treated patients could be linked to the deregulation of this transcription factor. In addition, these patients present defects in metabolic pathways including lipid metabolism where ATF4 was shown to play an important role (55, 56). Studies in ATF4-deficient mice have shown that the ATF4 transcriptional program may affect lipolysis and expression of lipogenic genes (57). Moreover, it was previously reported that mice with enforced expression of an active C-terminal fragment of GADD34 to attenuate eIF2α phosphorylation have decreased expression of key hepatic transcriptional regulators of intermediary metabolism including PPARγ (58). In line with these findings, we observed that PPARγ is up-regulated in liver of mice upon NFR treatment. PPARγ, is a master regulator of key proteins involved in lipid metabolism, vascular inflammation, and proliferation that can affect multiple cell types, including hepatocytes, macrophages, endothelial cells, and vascular smooth muscle cells. Its possible contribution to side effects in HIV patients treated with HIV-PIs is therefore likely multifaceted and may involve different pathways possibly beyond lipid metabolism. We identified other genes related to lipid metabolism whose expression is up-regulated in the liver of NFR-treated mice, including Soat2 (also abbreviated as Acat2), which was also found among the top genes up-regulated in MEFs in an eIF2α phosphorylation-dependent manner. This enzyme contributes to cholesterol ester synthesis in the small intestine and liver and therefore can promote hypercholesterolaemia and atherosclerosis (59), two metabolic disturbances often observed in HIV-PI–treated patients. All together these data further show that the ISR is a key metabolic regulator that can affect multiple pathways related to lipid metabolism. It is therefore likely that sustained ISR activation may account for some of the metabolic deregulations associated with the use of HIV-PIs in patients. In this context, ISRIB, a small molecule that potently inhibits the effects of eIF2α phosphorylation (60–62), could become an interesting therapeutic option to alleviate ISR-associated metabolic alterations in HIV-PI–treated patients.
Another important question is whether the HIV-PI–induced ISR could contribute to their antitumoral activity. Numerous clinical trials are underway to address the efficacy of these drugs in a variety of human tumors, but so far the underlying mechanisms are unknown (19). Whereas induction of ISR by blocking dephosphorylation of eIF2α is an adaptation program that increases the survival of stressed cells (9), strategies aimed at increasing eIF2α phosphorylation were found to limit cancer cell proliferation and tumor growth (12, 63). Similarly, in a genome-wide functional screen, CReP down-regulation was identified to increase sensitivity to tamoxifen (64). The outcome of fine-tuning the eIF2α phosphorylation level under intratumoral stresses is likely to affect the balance between death and survival (44, 65, 66). It is therefore tempting to speculate that sustained pharmacological activation of ISR could modify the cell fate decision process and tilt the balance in favor of death. Thus, a possible contribution for the ISR in mediating Nelfinavir anticancer properties is a plausible hypothesis that needs to be tested.
In addition to promoting ATF4 expression, the ISR decreases translation rates—a feature that could be relevant in human diseases characterized by perturbation of cellular proteostasis, such as protein misfolding and aggregation diseases. Slowing down translation rates can significantly improve protein folding, therefore contributing to reestablishment of homeostasis in these diseases (67–69). Drugs such as Salubrinal (11) and Guanabenz (13) or Sephin 1 (70), which were described as selective inhibitors of cellular complexes that dephosphorylate eIF2α, have been considered toward that purpose and showed promising results in animals. Nelfinavir has the advantage of being a rather safe compound with well-known pharmacology; it has been tested and used in thousands of cancer and HIV patients over many years. Our data show that repositioning this compound to promote the ISR and down-regulate translation rates in patients with misfolding and aggregation diseases should be considered and may represent a new approach to restore folding and homeostasis in these pathologies.
Materials and Methods
High-throughput sequencing, statistical analysis, lentivirus production, cell line infection, BiP aggregation assay, metabolic labeling, siRNA transfection, immunoprecipitation, and phosphatase activity assay are described in SI Materials and Methods.
Cell Culture and Drug Treatment.
All cell lines were cultured in DMEM supplemented with 10% (vol/vol) FBS, antibiotics (1% PSN; penicillin 5 mg/mL, streptomycin 5mg/mL, and neomycin 10mg/mL from Gibco-Life Technologies), 1% nonessential amino acids (Gibco-Life Technologies) and 1% L‐glutamine (AMIMED, Bioconcept). Cells were not tested for mycoplasma contamination during the study. Each knockout or transgenic MEF cell line was compared with littermate control. Table S1 shows the origin and provider of every MEF cell lines used in these study.
Table S1.
List of knockout cells used in the study
| MEFs | Origin | Provider | Reference |
| Eif2αWT/Eif2αS51A | R.J. Kaufman, Center for Neuroscience, Aging, and Stem Cell Research, Sanford Burnham Medical Research Institute, La Jolla, CA | R.J. Kaufman, Center for Neuroscience, Aging, and Stem Cell Research, Sanford Burnham Medical Research Institute, La Jolla, CA | (76) |
| XBP1+/+/XBP1−/− | L. Glimcher, Weill Cornell Medical College, New York, NY | L. Glimcher, Weill Cornell Medical College, New York, NY | (77) |
| IRE1+/+/IRE1−/− | D. Ron, Cambridge Institute for Medical Research (CIMR), University of Cambridge, Cambridge, UK | L. Glimcher, Weill Cornell Medical College, New York, NY | (78) |
| ATF4+/+/ATF4−/− | D. Ron, CIMR, University of Cambridge, Cambridge, UK | A. Bruhat, Institut National de la Recherche Agronomique (INRA), Saint-Genes-Champanelle, France | (1) |
| PERK+/+/PERK−/− | D. Ron, CIMR, University of Cambridge, Cambridge, UK | L. Glimcher, Weill Cornell Medical College, New York, NY | (79) |
| GCN2+/+/GCN2−/− | D. Ron, CIMR, University of Cambridge, Cambridge, UK | A. Bruhat, INRA, Saint-Genes-Champanelle, France | (1) |
| PKR+/+/PKR−/− | C. Weissmann, Department of Infectology, Scripps Florida, Jupiter, FL | J. Pavlovic, Institute of Medical Virology, Zürich, Switzerland | (80) |
| HRI+/+/HRI−/− | J.J. Chen, Institute for Medical Engineering and Science, Cambridge, MA | M. Sierant, Department of Cytokine Biology, The Forsyth Institute, Cambridge, MA | (81) |
| CReP+/+/CReP−/− | D. Ron, CIMR, University of Cambridge, Cambridge, UK | D. Ron, CIMR, University of Cambridge, Cambridge, UK | (10) |
| GADD34WT/WT/GADD34DC/DC | D. Ron, CIMR, University of Cambridge, Cambridge, UK | P. Pierre, Centre d′Immunologie de Marseille-Luminy (CIML), Marseille, France | (82) |
Nelfinavir Mesylate (CAS 159989–65-8) was from Axon Medchem; Ritonavir, Atazanavir, Lopinavir, and Saquinavir were obtained from The NIH AIDS Reagent Program; and Nelfinavir hydroxy-tert-butylamide (M8) and Oxaliplatin were from Santa-Cruz. Tunicamycin, sodium arsenite solution, Rapamycin, and Cycloheximide were from Enzo-Life Sciences. Guanabenz, Salubrinal, Doxycycline, (Z)-4-Hydroxytamoxifen, Etoposide, and MG132 were from Sigma-Aldrich. Calyculin A, Bortezomib, and Imatinib were from LC-Laboratories, and Okadaic acid was from Santa-Cruz. Poly(I:C) HMW (Invivogen) was lipofected using Lipofectamin2000 (Life Technology).
Mice.
Animal experiments were approved by the Veterinary Office of the Canton de Vaud and the Animal Ethics Committee (authorization 2390). WT C57BL/6J mice were housed at the University of Lausanne in accordance with local and national guidelines. Female mice 6–8 wk old were randomly distributed in two groups and injected intraperitoneally for 3 consecutive days either with vehicle [4% (vol/vol) DMSO, 5% (vol/vol) PEG, 5% (vol/vol) Tween 80 in saline] or with 100 mg/kg NFR as described previously (33, 71). Mice were killed 6 h after the last injection, and livers were harvested for protein and mRNA analysis. Experiments were repeated three times with 3–10 mice per group. One representative experiment with three mice per group is shown for Western blot (WB) analysis. One representative experiment with 10 mice per group is shown for mRNA level quantification by real-time PCR.
Measurement of NFR Concentration.
The quantification of nelfinavir in plasma, liver tissue, and cells has been performed with the stable isotope labeled internal standard method using an adaptation of the assay by liquid chromatography tandem mass spectrometry developed in our laboratory (72).
RNA Extraction and RT-PCR.
Total RNA from cells and tissues was extracted with PeqGOLD TriFast (PeqLAB) according to the manufacturer’s instructions, and cDNA was synthesized with a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). SYBR Green fluorescent reagent and LightCycler480 Real Time PCR System from Roche were used for quantitative RT-PCR. The relative amount of mRNA was calculated by the comparative threshold cycle method with β-actin as control. Primer sequences are described in Table S2.
Table S2.
Real-time primers used for mRNA quantification
| Target | Sequences |
| Human PERK | F, 5′-ACGATGAGACAGAGTTGCGAC-3′ |
| R, 5′-AATCCCACTGCTTTTTACCATGA-3′ | |
| Human PKR | F, 5′-ATGATGGAAAGCGAACAAGGA-3′ |
| R, 5′-TCCATAGTCTTGCGAAGAAAT-3′ | |
| Human HRI | F, 5′-ACCCCGAATATGACGAATCTGA-3′ |
| R, 5′-CAAGTGCTCCAGCAAAGAAAC-3′ | |
| Human GCN2 | F, 5′-TGGTAAACATCGGGCAAACTC-3′ |
| R, 5′-GGACCCACTCATACAACAAGA-3′ | |
| Human CHOP | F, 5′-GGAAACAGAGTGGTCATTCCC-3′ |
| R, 5′-CTGCTTGAGCCGTTCATTCTC-3′ | |
| Human CreP | F, 5′-CAGACTCGGTACAGCGTGAC-3′ |
| R, 5′-TTCGTTTCTGGAACCTGCAT-3′ | |
| Human ATF4 | F, 5′-TCCGAATGGCTGGCTGTGG-3′ |
| R, 5′-AGTGTAGTCTGGCTTCCTATCTCC-3′ | |
| Human βTRCP1 | F, 5′-TGCTCTATGCCCAGGTCTCT-3′ |
| R, 5′-AGGGGGTTCGCCATTATTAC-3′ | |
| Human βTRCP2 | F, 5′-AGAGGGTGGGATCAGTACCT-3′ |
| R, 5′-TCCACACCGCCAGTTAGATT-3′ | |
| Mouse GADD34 | F, 5′-CGGAGAGAAGCCAGAATCAC-3′ |
| R, 5′-CTTCGATCTCGTGCAAACTG-3′ | |
| Mouse ERO1 | F, 5′-TCAGTGGACCAAGCATGATGA-3′ |
| R, 5′-TCCACATACTCAGCATCGGG-3′ | |
| Mouse CHOP | F, 5′-GTCCCTAGCTTGGCTGACAGA-3′ |
| R, 5′-TGGAGAGCGAGGGCTTTG-3′ | |
| Mouse ATF4 | F, 5′-AGACACCGGCAAGGAGGATG-3′ |
| R, 5′-ACGTGGTCAAGAGCTCATCTG-3′ | |
| Mouse Calreticulin | F, 5′-GCAGACCCTGCCATCTATTTC-3′ |
| R, 5′-TCGGACTTATGTTTGGATTCGAC-3′ | |
| Mouse BiP | F, 5′-TCATCGGACGCACTTGGAA-3′ |
| R, 5′-CAACCACCTTGAATGGCAAGA-3′ | |
| Mouse EDEM | F, 5′-AAGCCCTCTGGAACTTGCG-3′ |
| R, 5′-AACCCAATGGCCTGTCTGG-3′ | |
| Mouse p58IPK | F, 5′-GGCGCTGAGTGTGGAGTAAAT-3′ |
| R, 5′-GGCTAAGAAGACGGTAGCTCTC-3′ | |
| Mouse HRD1 | F, 5′-CTCATGCCTACTACCTCAAACAC-3′ |
| R, 5′-TGCCCGAAGAACACCTTGC-3′ | |
| Mouse Grp94 | F, 5′-GTTCGTCAGAGCTGATGATGAA-3′ |
| R, 5′-GCGTTTAACCCATCCAACTGAAT-3′ | |
| Mouse ErDJ4 | F, 5′-GGCGCACAGGTTATTAGAAATG-3′ |
| R, 5′-TCGCTCTGAGGCAGACTTTG-3′ | |
| Mouse HERP | F, 5′-GCAGTTGGAGTGTGAGTCG-3′ |
| R, 5′-TCTGTGGATTCAGCACCCTTT-3′ | |
| Mouse PPARg | F, 5′-CCCTGGCAAAGCATTTGTAT-3′ |
| R, 5′-GAAACTGGCACCCTTGAAAA-3′ | |
| Mouse XBP1 | F, 5′-ACACGCTTGGGAATGGACAC-3′ |
| R, 5′-CCATGGGAAGATGTTCTGGG-3′ | |
| Human and mouse actin | F, 5′-TACCACCATGTACCCAGGCA-3′ |
| R, 5′-CTCAGGAGGAGCAATGATCTT-3′ |
siRNA Transfection.
Specific and nonrelevant siRNA were purchased from Qiagen and listed in Table S3. Cells were transfected with Lipofectamine RNAiMAX Transfection Reagent (Life Technology) according to the manufacturer's instructions.
Table S3.
SiRNA sequence used for silencing
| Target mRNA | Target sequences | QIAGEN reference |
| Human CreP (#1) | 5′-ATGCTATTGGATATTGCTTGA-3′ | SI03050257 |
| Human CreP (#2) | 5′-CCGAATAAGTGTAGTTGATTA-3′ | SI03079923 |
| Human PERK | 5′-CGGCAGGTCATTAGTAATTAT-3′ | SI02223725 |
| Human PKR | 5′-ACGGAAAGACTTACGTTATTA-3′ | SI02223011 |
| Human HRI | 5′-CTGATTAAGGGTGCAACTAAA-3′ | SI00105784 |
| Human GCN2 | 5′-CACCGTCAAGATTACGGACTA-3′ | SI03058629 |
| Human GADD34 (#1) | 5′-GTGGATAGTGAGGATAAGGAA-3′ | SI03106936 |
| Human GADD34 (#2) | 5′-GACCAACTGGTTTGCCTATAA-3′ | SI04439197 |
| Human βTRCP1 | 5′-CGGAAGAGUUUUUCGACUATT-3′ | (40) |
| Human βTRCP2 | 5′-GGUUGUUAGUGGAUCAUCATT-3′ | (40) |
| Human βTRCP1/2 | 5′-GUGGAAUUUGUGGAACAUCTT-3′ | (40) |
| Mouse CReP | 5′-CTCCAGGCTAACAAAGGGCAA-3′ | SI04925214 |
| Nonrelevant | 5′-AGGTAGTGTAATCGCCTTG-3′ | SI1027280 |
Immunoblot Analysis.
Every WB shown in the study is representative of at least three independent experiments performed in the same conditions.
Cells and liver protein extracts were prepared with RIPA (radioimmunoprecipitation assay) buffer (50 mM NaCl, 50 mM Tris pH 7,4, 1 mM EDTA, 0.1% SDS, 1% Nonidet P-40, 1% Sodium Deoxycholate) supplemented with protease inhibitors mixture (Roche) and 5 μM MG132 (Sigma-Aldrich). Extracts were separated by SDS/PAGE and transferred to nitrocellulose blotting membranes (Amersham). PERK, PKR, GCN2, and HRI phosphorylation was monitored by Phos-tag SDS/PAGE (38). The following antibodies were used for immunoblot analysis: anti-ATF4 (Santa Cruz; sc-200), anti–phospho-eIF2α (Cell Signaling; #3597S), anti–total-eIF2α (Cell Signaling; #9722S), anti-IRE1α (Cell Signaling; #3294), anti-PERK (Cell Signaling; #3192), anti-HRI (Santa Cruz; sc-30143), anti-GCN2 (Cell Signaling; #3302), anti-PKR (Santa Cruz; sc-6282), anti-CHOP (Cell Signaling; #2895S), anti-Tubulin (Adipogen; F2C), anti-HsCReP (Proteintech Group; 14634–1-AP), anti-GADD34 (Proteintech Group; 10449–1-AP), anti-mouse CReP (kindly provided by David Ron, University of Cambridge, Cambridge, UK) (10), anti-VSV (Sigma-Aldrich; Clone P5D4), anti-PP1 (Santa Cruz; sc-6108), anti–phospho-H2AX (Upstate; 07–164), polyclonal anti-ATF6 (obtained from Laurie H. Glimcher and Ann-Hwee Lee, Weill Cornell Medical College, New York, NY) (73), and anti-BiP (Cell Signaling; #3177S).
SI Materials and Methods
High-Throughput Sequencing.
High-throughput sequencing was performed at the Lausanne Genomics Technologies Facility (University of Lausanne). RNA-seq libraries were prepared using 500 ng of total RNA and the Illumina TruSeq Stranded mRNA reagents (Illumina) on a Caliper Sciclone liquid handling robot (PerkinElmer) using a Caliper-developed automated script. Cluster generation was performed with the resulting libraries using the Illumina TruSeq PE Cluster Kit v3 reagents and sequenced on the Illumina HiSEq. 2500 using TruSeq SBS Kit v3 reagents. Sequencing data were processed using the Illumina Pipeline Software version 1.82. Heat map picture was visualized using gitools (www.gitools.org). Gene ontology was performed using the Term Enrichment tool in AmiGO version 2.1.4 (geneontology.org).
Lentivirus Production and Cell Line Infection.
Lentiviral pINDUCER21 plasmid was obtained from Steve Elledge, Harvard Medical School, Boston, MA (74). Human eIF2αWT was amplified by PCR from pCDNA-eIF2S1 plasmid from Addgene (#21807) using specific oligos allowing addition of an N-terminal VSV-tag and cloning into pENTR vector using SalI/XhoI restriction sites (forward primer, ctaGTCGACATGtatacagacatagagatgaaccgacttggaaagCTCAGAATGCCGGGTCTAAG; reverse primer, gatCTCGAGGCTCTGTGTTCCTTAAATTGGAC). Recombination was performed as described to obtained the p21-VSV-eIF2αWT lentiviral plasmid.
Lentiviruses were produced as previously described (75). The 293T cells were infected, and GFP-positive cells were FACS-sorted 96 h after infection. Cells were cultured in complete medium, and VSV-eIF2αWT expression was induced using doxycycline (1 μg/mL for 24 h).
BiP Aggregation.
BiP aggregation upon NFR or TM treatment was determined as previously described (13, 30). Briefly, 4 × 106 cells were plated in 15 cm2 plates. The day after, cells were treated with the indicated concentration of NFR or TM for 16 h. Cells were washed in PBS, harvested with PBS–5 mM EDTA, incubated for 5 min with ice-cold PBS containing 20 mM N-Ethylmaleimide (Sigma-Aldrich), and washed one time more with PBS. After centrifugation, cell pellet was lysed in 150 μL lysis buffer [0.5% Triton X-100, 20 mM Hepes pH 7.5, 250 mM sucrose, 100 mM NaCl, 2.5 mM CaCl2, and EDTA-free protease inhibitor mixture (Roche)] and incubated 1 h on ice. After 10 min of centrifugation at 15,000 × g, supernatants were harvested and total protein content was measured using a bicinchoninic acid (BCA) protein assay kit (Thermo-Scientific). Samples were adjusted to a final volume of 170 μL containing 700 μg of total proteins and 0.8% SDS. We loaded 150 μL on the top of 850 μL of buffer cushion [20% glycerol, 20 mM Hepes (pH 7.5), 0.5% TritonX-100, 0.8% SDS] in 1 mL thick-wall polycarbonate tubes (Beckman-Coulter) and centrifuged it at 35,000 rpm for 1 h using TLS-55 rotor. The pellet was resuspended in 40 μL of urea loading buffer (9.6 M urea, 12% glycerol, 1.36% SDS, 40 mM Tris at pH 6.8, 0.002% bromophenol blue) and incubated at room temperature for 30 min and at 95 °C for 10 min. Samples were loaded on 8% acrylamide gel together with total protein extracts.
Immunoprecipitation and Phosphatase Activity Assay.
For VSV-eIF2αWT immunoprecipitation, 4 × 106 293T transduced with p21-VSV-eIF2αWT lentiviruses were treated for 24 h with doxyclycline (1 μg/mL) to induce VSV-eIF2αWT expression. Cells were treated for 6 h with NFR (20 and 40 μM), washed with PBS, and lysed at 4 °C in 1% Nonidet P-40, 10 mM Tris·HCl, pH 7.6, 100 mM NaCl, 5 mM MgCl2, which was freshly supplemented with complete protease inhibitor mixture (Roche), and 5 μM MG132 (Sigma-Aldrich). Nuclei were removed by centrifugation. Soluble cell lysates were mixed with 10 μL of Anti–VSV-G Agarose Conjugate (A1970; Sigma-Aldrich) and rocked overnight. Beads were washed four times with lysis buffer. Immunoprecipitated material was eluted in SDS sample buffer for 5 min at 100 °C and separated with SDS/PAGE.
Phosphatase activity was determined on immunoprecipitates of the phosphatase PP1 (42). Briefly, 4 × 106 HeLa cells were treated for 6 h with NFR (10, 20, and 40 µM) or TM (10 μg/mL) or for 2 h with Calyculin A (10 nM) or Okadaic acid (200 nM). After washing with PBS, cells were lysed for 15 min on ice in PP1 lysis buffer (20 mM Tris·HCl, pH 7.5, 1% Triton X-100, 10% glycerol, 132 mM NaCl, freshly supplemented with Roche complete protease inhibitor). Total protein content was measured using BCA protein assay kit (Thermo-Scientific), and 500 μg proteins were immunoprecipitated overnight at 4 °C with 2 μg of anti-PP1γ antibody (Santa Cruz sc-6108) and then incubated with Protein A/G plus-Sepharose (Santa-Cruz). Immunoprecipitates were washed three times in lysis buffer, followed by resuspension in phosphatase assay buffer (50 mM Tris·HCl, pH 7.0, 0.2 mM MnCl2, 0.1 mM CaCl2, 125 μg/mL BSA, 0.05% Tween 20), supplemented with 100 μM 6,8-difluoro-4-methyl-umbelliferyl phosphate (Invitrogen/Molecular Probes). Precipitates were allowed to react with substrate for 1 h at 37 °C, and 6,8-difluoro-4-methyl-umbelliferyl phosphate fluorescence was measured on a microplate reader (360 nmEx/460 nmEm). Phosphatase activities are given as percent change relative to the control (Mock-treated cells).
Metabolic Labeling.
We plated 75,000 cells in 24-well plates. The day after, cells were treated for 2, 4, and 6 h with 20 μM NFR or 10 μg/mL TM and incubated for the last 20 min with a mixture of l-(35)S-Methionine/Cysteine (5 μCi/mL). Cells were washed with PBS and lysed with RIPA buffer for SDS/PAGE separation. Proteins were transferred onto a PVDF membrane and analyzed by autoradiography and anti-Tubulin immunoblot to check for equal loading. ImageJ software was used for autoradiography quantification, and values were normalized as percentage of Mock-treated cells.
Statistical Analysis.
Where indicated, statistical significance was ascertained by performing unpaired Student’s t tests. Significant differences were indicated by asterisks: *P ≤ 0.05, **P ≤ 0.01, or ***P ≤ 0.001. For the RNA-seq analysis, we used a moderated t test from the R bioconductor package “limma” (R version 3.1.1, limma version 3.20.8). The “adjusted P values” reported in Dataset S1 correspond to the P values corrected for multiple testing using the Benjamini–Hochberg method, which controls for false discovery rate. For animal studies, no statistical method was used to predetermine sample size. Animals were randomly distributed in different groups, and the investigators were not blinded to allocation during experiments and outcome assessment. Data were considered as outliers and excluded from analysis when they deviated from the mean by more than three times the SD.
Supplementary Material
Acknowledgments
We thank D. Ron, C. Weissmann, J. Pavlovic, L. H. Glimcher, A. Bruhat, R. J. Kaufman, J. J. Chen, M. Sierant, P. Pierre, S. Elledge, and The NIH AIDS Reagent Program for sharing key reagents. We thank Laurie Glimcher, David Ron, Kendle Maslowski, and Margot Thome for critical reading of the manuscript. We thank Keith Harshman, Leonore Wigger, and the staff of the Lausanne Genomics Technologies Facility for high-throughput sequencing. We are indebted to Thomas Mercier, a chemist engineer, for excellent analytical work. The nelfinavir mass spectrometry assay has been developed and maintained thanks to the support of the Swiss National Fund. L.A.D. is supported by the Swiss National Science Foundation (SNF) Grant 324730-141234 and has received a REQUIP Grant SNF 326000-121314/1 for the acquisition of the LC-MS/MS instrumentation. F.M. is supported by European Research Council Starting Grant 281996, Human Frontier Science Program Career Development Award CDA00059/2011, and Swiss National Science Foundation Grant 31003A-130476. B.B. is supported by a fellowship of the Institut Suisse de Recherches Experimentales sur le Cancer foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514076113/-/DCSupplemental.
References
- 1.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 2.Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108(4):545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 3.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6(5):1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 4.Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2α kinases: Their structures and functions. Cell Mol Life Sci. 2013;70(19):3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol Cell Biol. 2001;21(20):6841–6850. doi: 10.1128/MCB.21.20.6841-6850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He B, Chou J, Liebermann DA, Hoffman B, Roizman B. The carboxyl terminus of the murine MyD116 gene substitutes for the corresponding domain of the gamma(1)34.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J Virol. 1996;70(1):84–90. doi: 10.1128/jvi.70.1.84-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153(5):1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Y, Hendershot LM. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J Biol Chem. 2003;278(37):34864–34873. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- 9.Jousse C, et al. Inhibition of a constitutive translation initiation factor 2alpha phosphatase, CReP, promotes survival of stressed cells. J Cell Biol. 2003;163(4):767–775. doi: 10.1083/jcb.200308075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harding HP, et al. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2alpha) dephosphorylation in mammalian development. Proc Natl Acad Sci USA. 2009;106(6):1832–1837. doi: 10.1073/pnas.0809632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyce M, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307(5711):935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 12.Chen T, et al. Chemical genetics identify eIF2α kinase heme-regulated inhibitor as an anticancer target. Nat Chem Biol. 2011;7(9):610–616. doi: 10.1038/nchembio.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsaytler P, Harding HP, Ron D, Bertolotti A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 2011;332(6025):91–94. doi: 10.1126/science.1201396. [DOI] [PubMed] [Google Scholar]
- 14.Yildirim MA, Goh KI, Cusick ME, Barabási AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25(10):1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 15.Bauer-Mehren A, et al. Automatic filtering and substantiation of drug safety signals. PLOS Comput Biol. 2012;8(4):e1002457. doi: 10.1371/journal.pcbi.1002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie L, Evangelidis T, Xie L, Bourne PE. Drug discovery using chemical systems biology: Weak inhibition of multiple kinases may contribute to the anti-cancer effect of nelfinavir. PLOS Comput Biol. 2011;7(4):e1002037. doi: 10.1371/journal.pcbi.1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chow WA, Jiang C, Guan M. Anti-HIV drugs for cancer therapeutics: Back to the future? Lancet Oncol. 2009;10(1):61–71. doi: 10.1016/S1470-2045(08)70334-6. [DOI] [PubMed] [Google Scholar]
- 18.Boesecke C, Cooper DA. Toxicity of HIV protease inhibitors: Clinical considerations. Curr Opin HIV AIDS. 2008;3(6):653–659. doi: 10.1097/COH.0b013e328312c392. [DOI] [PubMed] [Google Scholar]
- 19.Gantt S, Casper C, Ambinder RF. Insights into the broad cellular effects of nelfinavir and the HIV protease inhibitors supporting their role in cancer treatment and prevention. Curr Opin Oncol. 2013;25(5):495–502. doi: 10.1097/CCO.0b013e328363dfee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta AK, et al. The HIV protease inhibitor nelfinavir downregulates Akt phosphorylation by inhibiting proteasomal activity and inducing the unfolded protein response. Neoplasia. 2007;9(4):271–278. doi: 10.1593/neo.07124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gills JJ, et al. Nelfinavir, a lead HIV protease inhibitor, is a broad-spectrum, anticancer agent that induces endoplasmic reticulum stress, autophagy, and apoptosis in vitro and in vivo. Clin Cancer Res. 2007;13(17):5183–5194. doi: 10.1158/1078-0432.CCR-07-0161. [DOI] [PubMed] [Google Scholar]
- 22.Martinon F. Targeting endoplasmic reticulum signaling pathways in cancer. Acta Oncol. 2012;51(7):822–830. doi: 10.3109/0284186X.2012.689113. [DOI] [PubMed] [Google Scholar]
- 23.Ford J, et al. Intracellular and plasma pharmacokinetics of nelfinavir and M8 in HIV-infected patients: Relationship with P-glycoprotein expression. Antivir Ther. 2004;9(1):77–84. [PubMed] [Google Scholar]
- 24.Hennessy M, et al. Intracellular accumulation of nelfinavir and its relationship to P-glycoprotein expression and function in HIV-infected patients. Antivir Ther. 2004;9(1):115–122. [PubMed] [Google Scholar]
- 25.Carbon S, et al. AmiGO Hub Web Presence Working Group AmiGO: Online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brüning A, et al. Nelfinavir induces the unfolded protein response in ovarian cancer cells, resulting in ER vacuolization, cell cycle retardation and apoptosis. Cancer Biol Ther. 2009;8(3):226–232. doi: 10.4161/cbt.8.3.7339. [DOI] [PubMed] [Google Scholar]
- 27.Zhou H, et al. HIV protease inhibitors activate the unfolded protein response and disrupt lipid metabolism in primary hepatocytes. Am J Physiol Gastrointest Liver Physiol. 2006;291(6):G1071–G1080. doi: 10.1152/ajpgi.00182.2006. [DOI] [PubMed] [Google Scholar]
- 28.Han J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15(5):481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito D, et al. Characterization of stanniocalcin 2, a novel target of the mammalian unfolded protein response with cytoprotective properties. Mol Cell Biol. 2004;24(21):9456–9469. doi: 10.1128/MCB.24.21.9456-9469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marciniak SJ, et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18(24):3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutkowski DT, Kaufman RJ. All roads lead to ATF4. Dev Cell. 2003;4(4):442–444. doi: 10.1016/s1534-5807(03)00100-x. [DOI] [PubMed] [Google Scholar]
- 32.Kepp O, et al. eIF2α phosphorylation as a biomarker of immunogenic cell death. Semin Cancer Biol. 2015;33:86–92. doi: 10.1016/j.semcancer.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Utkina-Sosunova IV, et al. Nelfinavir inhibits intra-mitochondrial calcium influx and protects brain against hypoxic-ischemic injury in neonatal mice. PLoS One. 2013;8(4):e62448. doi: 10.1371/journal.pone.0062448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goujard C, et al. COPHAR1-ANRS 102 Study Group High variability of indinavir and nelfinavir pharmacokinetics in HIV-infected patients with a sustained virological response on highly active antiretroviral therapy. Clin Pharmacokinet. 2005;44(12):1267–1278. doi: 10.2165/00003088-200544120-00005. [DOI] [PubMed] [Google Scholar]
- 35.Xiao G, et al. ATF4 protein deficiency protects against high fructose-induced hypertriglyceridemia in mice. J Biol Chem. 2013;288(35):25350–25361. doi: 10.1074/jbc.M113.470526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woo CW, Kutzler L, Kimball SR, Tabas I. Toll-like receptor activation suppresses ER stress factor CHOP and translation inhibition through activation of eIF2B. Nat Cell Biol. 2012;14(2):192–200. doi: 10.1038/ncb2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11(5):411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sha H, et al. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 2009;9(6):556–564. doi: 10.1016/j.cmet.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.So JS, Cho S, Min SH, Kimball SR, Lee AH. IRE1α-dependent decay of CReP/Ppp1r15b mRNA increases eukaryotic initiation factor 2α phosphorylation and suppresses protein synthesis. Mol Cell Biol. 2015;35(16):2761–2770. doi: 10.1128/MCB.00215-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coyaud E, et al. BioID-based identification of Skp Cullin F-box (SCF)β-TrCP1/2 E3 ligase substrates. Mol Cell Proteomics. 2015;14(7):1781–1795. doi: 10.1074/mcp.M114.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: Designed to create specificity. Trends Biochem Sci. 2010;35(8):450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drexler HC. Synergistic apoptosis induction in leukemic cells by the phosphatase inhibitor salubrinal and proteasome inhibitors. PLoS One. 2009;4(1):e4161. doi: 10.1371/journal.pone.0004161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Randolph JT, DeGoey DA. Peptidomimetic inhibitors of HIV protease. Curr Top Med Chem. 2004;4(10):1079–1095. doi: 10.2174/1568026043388330. [DOI] [PubMed] [Google Scholar]
- 44.Koromilas AE. Roles of the translation initiation factor eIF2α serine 51 phosphorylation in cancer formation and treatment. Biochim Biophys Acta. 2015;1849(7):871–880. doi: 10.1016/j.bbagrm.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 45.Fullwood MJ, Zhou W, Shenolikar S. Targeting phosphorylation of eukaryotic initiation factor-2α to treat human disease. Prog Mol Biol Transl Sci. 2012;106:75–106. doi: 10.1016/B978-0-12-396456-4.00005-5. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura T, et al. A critical role for PKR complexes with TRBP in Immunometabolic regulation and eIF2α phosphorylation in obesity. Cell Reports. 2015;11(2):295–307. doi: 10.1016/j.celrep.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown TT, Qaqish RB. Antiretroviral therapy and the prevalence of osteopenia and osteoporosis: A meta-analytic review. AIDS. 2006;20(17):2165–2174. doi: 10.1097/QAD.0b013e32801022eb. [DOI] [PubMed] [Google Scholar]
- 48.Carr A, et al. A syndrome of peripheral lipodystrophy, hyperlipidaemia and insulin resistance in patients receiving HIV protease inhibitors. AIDS. 1998;12(7):F51–F58. doi: 10.1097/00002030-199807000-00003. [DOI] [PubMed] [Google Scholar]
- 49.Ena J, Benito C, Llácer P, Pasquau F, Amador C. [Abnormal body fat distribution and type of antiretroviral therapy as predictors of cardiovascular disease risk in HIV-infected patients] Med Clin (Barc) 2004;122(19):721–726. doi: 10.1016/s0025-7753(04)74368-7. [DOI] [PubMed] [Google Scholar]
- 50.Yarasheski KE, et al. Insulin resistance in HIV protease inhibitor-associated diabetes. J Acquir Immune Defic Syndr. 1999;21(3):209–216. doi: 10.1097/00126334-199907010-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bradbury RA, Samaras K. Antiretroviral therapy and the human immunodeficiency virus--Improved survival but at what cost? Diabetes Obes Metab. 2008;10(6):441–450. doi: 10.1111/j.1463-1326.2007.00760.x. [DOI] [PubMed] [Google Scholar]
- 52.Abdulkarim B, et al. A missense mutation in PPP1R15B causes a syndrome including diabetes, short stature, and microcephaly. Diabetes. 2015;64(11):3951–3962. doi: 10.2337/db15-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franceschi RT, Ge C, Xiao G, Roca H, Jiang D. Transcriptional regulation of osteoblasts. Cells Tissues Organs. 2009;189(1-4):144–152. doi: 10.1159/000151747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao H, et al. Activating transcription factor 4 regulates osteoclast differentiation in mice. J Clin Invest. 2010;120(8):2755–2766. doi: 10.1172/JCI42106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang C, Guo F. Effects of activating transcription factor 4 deficiency on carbohydrate and lipid metabolism in mammals. IUBMB Life. 2012;64(3):226–230. doi: 10.1002/iub.605. [DOI] [PubMed] [Google Scholar]
- 56.Volmer R, Ron D. Lipid-dependent regulation of the unfolded protein response. Curr Opin Cell Biol. 2015;33:67–73. doi: 10.1016/j.ceb.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C, et al. ATF4 regulates lipid metabolism and thermogenesis. Cell Res. 2010;20(2):174–184. doi: 10.1038/cr.2010.4. [DOI] [PubMed] [Google Scholar]
- 58.Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7(6):520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willner EL, et al. Deficiency of acyl CoA:cholesterol acyltransferase 2 prevents atherosclerosis in apolipoprotein E-deficient mice. Proc Natl Acad Sci USA. 2003;100(3):1262–1267. doi: 10.1073/pnas.0336398100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sidrauski C, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife. 2013;2:e00498. doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sidrauski C, et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife. 2015;4:e07314. doi: 10.7554/eLife.07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sekine Y, et al. Stress responses. Mutations in a translation initiation factor identify the target of a memory-enhancing compound. Science. 2015;348(6238):1027–1030. doi: 10.1126/science.aaa6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aktas BH, et al. Small-molecule targeting of translation initiation for cancer therapy. Oncotarget. 2013;4(10):1606–1617. doi: 10.18632/oncotarget.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mendes-Pereira AM, et al. Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc Natl Acad Sci USA. 2012;109(8):2730–2735. doi: 10.1073/pnas.1018872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leprivier G, Rotblat B, Khan D, Jan E, Sorensen PH. Stress-mediated translational control in cancer cells. Biochim Biophys Acta. 2015;1849(7):845–860. doi: 10.1016/j.bbagrm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 66.Clarke HJ, Chambers JE, Liniker E, Marciniak SJ. Endoplasmic reticulum stress in malignancy. Cancer Cell. 2014;25(5):563–573. doi: 10.1016/j.ccr.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 67.Lindquist SL, Kelly JW. Chemical and biological approaches for adapting proteostasis to ameliorate protein misfolding and aggregation diseases: Progress and prognosis. Cold Spring Harb Perspect Biol. 2011;3(12):a004507. doi: 10.1101/cshperspect.a004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sherman MY, Qian SB. Less is more: Improving proteostasis by translation slow down. Trends Biochem Sci. 2013;38(12):585–591. doi: 10.1016/j.tibs.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 69.Schneider K, Bertolotti A. Surviving protein quality control catastrophes—From cells to organisms. J Cell Sci. 2015;128(21):3861–3869. doi: 10.1242/jcs.173047. [DOI] [PubMed] [Google Scholar]
- 70.Das I, et al. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science. 2015;348(6231):239–242. doi: 10.1126/science.aaa4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kawabata S, et al. Synergistic effects of nelfinavir and bortezomib on proteotoxic death of NSCLC and multiple myeloma cells. Cell Death Dis. 2012;3:e353. doi: 10.1038/cddis.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Colombo S, et al. Intracellular measurements of anti-HIV drugs indinavir, amprenavir, saquinavir, ritonavir, nelfinavir, lopinavir, atazanavir, efavirenz and nevirapine in peripheral blood mononuclear cells by liquid chromatography coupled to tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;819(2):259–276. doi: 10.1016/j.jchromb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 73.Hur KY, et al. IRE1α activation protects mice against acetaminophen-induced hepatotoxicity. J Exp Med. 2012;209(2):307–318. doi: 10.1084/jem.20111298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meerbrey KL, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci USA. 2011;108(9):3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bagnis C, Bailly P, Chapel-Fernandes S. Using an EGFPmeter to evaluate the lentiviral vector production: Tricks and traps. Methods Mol Biol. 2009;515:151–163. doi: 10.1007/978-1-59745-559-6_10. [DOI] [PubMed] [Google Scholar]
- 76.Scheuner D, et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7(6):1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 77.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24(24):4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Urano F, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287(5453):664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 79.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397(6716):271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 80.Yang YL, et al. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14(24):6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han AP, et al. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 2001;20(23):6909–6918. doi: 10.1093/emboj/20.23.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Novoa I, et al. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 2003;22(5):1180–1187. doi: 10.1093/emboj/cdg112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.