Significance
Nonsteroidal antiinflammatory drugs (NSAIDs) work by inhibiting cyclooxygenase-2 (COX-2) induced at sites of inflammation. They are among the most widely used drugs worldwide, but their cardiovascular side effects are a major concern for patients, regulators, and industry. NSAID side effects are mediated by inhibition of constitutively expressed COX-2 present in discrete regions, including the kidney. However, the pathways driving constitutive COX-2 remain poorly understood. The work presented here defines these pathways and importantly shows constitutive COX-2 expression in the kidney occurs through pathways distinct to those driving COX-2 in inflammation. These data therefore highlight the potential that targeting COX-2 at the transcriptional level may provide a way to dissociate antiinflammatory benefits of NSAIDs from their treatment-limiting cardiovascular side effects.
Keywords: cyclooxygenase, nonsteroidal antiinflammatory drugs, prostacyclin, cardiovascular, Vioxx
Abstract
Cyclooxygenase-2 (COX-2) is an inducible enzyme that drives inflammation and is the therapeutic target for widely used nonsteroidal antiinflammatory drugs (NSAIDs). However, COX-2 is also constitutively expressed, in the absence of overt inflammation, with a specific tissue distribution that includes the kidney, gastrointestinal tract, brain, and thymus. Constitutive COX-2 expression is therapeutically important because NSAIDs cause cardiovascular and renal side effects in otherwise healthy individuals. These side effects are now of major concern globally. However, the pathways driving constitutive COX-2 expression remain poorly understood. Here we show that in the kidney and other sites, constitutive COX-2 expression is a sterile response, independent of commensal microorganisms and not associated with activity of the inflammatory transcription factor NF-κB. Instead, COX-2 expression in the kidney but not other regions colocalized with nuclear factor of activated T cells (NFAT) transcription factor activity and was sensitive to inhibition of calcineurin-dependent NFAT activation. However, calcineurin/NFAT regulation did not contribute to constitutive expression elsewhere or to inflammatory COX-2 induction at any site. These data address the mechanisms driving constitutive COX-2 and suggest that by targeting transcription it may be possible to develop antiinflammatory therapies that spare the constitutive expression necessary for normal homeostatic functions, including those important to the cardiovascular-renal system.
Cyclooxygenase (COX) converts arachidonic acid to prostanoids, which include prostaglandins (PGs), prostacyclin, and thromboxane. Prostanoids are important mediators that regulate diverse functions in the cardiovascular, gastrointestinal, urogenital, and nervous systems, as well as playing critical roles in immunity, inflammation and resolution of inflammation. There are two COX isoforms: COX-1 and COX-2. COX-1 is a constitutive, housekeeping enzyme that is ubiquitously expressed and is responsible for prostanoid production in most tissues (1). In contrast, COX-2 is an inducible enzyme, expressed at sites of inflammation, infection, and cancer (2) that generates prostanoids that drive disease pathogenesis. COX-2 is the therapeutic target for antiinflammatory drugs, including ibuprofen, naproxen, and diclofenac, as well as newer COX-2 selective inhibitors such as Celebrex (celecoxib; Pfizer); collectively these drugs are known as nonsteroidal antiinflammatory drugs (NSAIDs). NSAIDs are among the most commonly taken antiinflammatory pain medications globally; they are the first-line therapy for patients with arthritis and can prevent cancer. However, COX-2 is also expressed constitutively in areas not associated with inflammation, including the brain, thymus, gut, and kidney (3). Constitutive COX-2 is increasingly recognized to play a major role in homeostatic function, for example, having defined roles in the developing brain (4) and adult kidney (5) and gut (6). As such, inhibition of constitutive COX-2 by NSAIDs causes adverse effects that limit their use. Upper gastrointestinal side effects associated with NSAID use are due to the combined inhibition of COX-1 and -2 (6) but are relatively well managed by the use of NSAIDs selective for COX-2 and concurrent use of proton pump inhibitor drugs. However, NSAID-associated cardiovascular and renal side effects remain a major clinical concern. This concern has resulted in a series of regulatory events including (i) the withdrawal of the blockbuster drug Vioxx (rofecoxib; Merck) in 2004, (ii) the introduction of “black box” warnings on some NSAIDs from 2005 and on all drugs in this class since 2015, (iii) the withdrawal of Onsenal (celecoxib; Pfizer) for the prevention of cancer in 2011, and (iv) the reclassification of the over-the-counter medication diclofenac as prescription only in 2015 (United Kingdom). Now the fear of cardiovascular events caused by NSAIDs has resulted in the cautious prescribing of COX-2–selective drugs (7) in favor of older style medications that are more toxic to the gut and a failure to realize the full clinical potential of NSAIDs in the prevention of cancer (8). Because side effects associated with NSAIDs are a result of inhibition of constitutive COX-2, at sites important to homeostatic regulation, it is imperative that we establish what drives expression at these sites in the absence and presence of inflammation.
Our recent work demonstrated that, in the kidney, constitutive COX-2 expression in the renal medulla works as a powerful transcriptional brake on hundreds of genes, including those regulating cardiovascular pathways (5). Constitutive COX-2 in the kidney and other sites occurs in the absence of inflammation, suggesting that distinct mechanisms exist whereby COX-2 can be regulated in two very different settings: constitutively in health and in an inducible manner in disease. Detailed studies identifying and comparing the pathways responsible for constitutive vs. inducible COX-2 expression have not yet been reported, but are of critical importance to both our understanding of COX-2 biology and the search for better, safer ways to therapeutically target the COX-2 pathway.
COX-2 gene [Ptgs2 (prostaglandin-endoperoxide synthase 2)] transcription is driven by a number of pathways, with binding sites in the promoter region for several transcription factor complexes including, most notably, NF-κB (9), and, of direct relevance to renal COX-2, nuclear factor of activated T cells (NFAT) (10). For both NF-κB and NFAT transcription pathways, bacteria, including some forms of commensal bacteria (11), are key driving factors. These observations suggest that the microbiome could be a source of direct stimuli for constitutive COX-2 expression, particularly in the gut and other barrier tissues. Indeed, studies using germ-free mice have demonstrated that commensal bacteria influence a range of pathological processes impacting on the cardiovascular (12) and nervous systems (13).
Here, we directly address the pathways by which COX-2 is constitutively expressed, under noninflammatory conditions, across a range of tissues and, for comparison, in the same tissues where inflammation has been robustly induced using LPS. Based on the rationale above, we focused our studies of constitutive COX-2 expression on the role of (i) commensal bacterial, (ii) NF-κB, and (iii) NFAT.
Results and Discussion
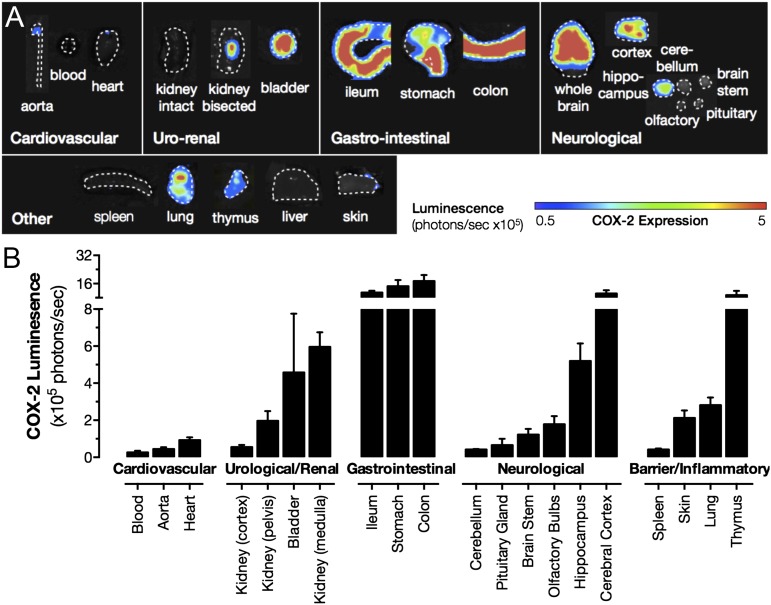
Although COX-1 is considered the archetypal constitutive form of the enzyme and is widely expressed throughout the body, COX-2 is also expressed constitutively, but restricted to discrete regions (3). Here we confirm and extend our previous observations (3), using luciferase COX-2 reporter mice (Fig. 1) and quantitative PCR (qPCR) (SI Appendix, Fig. S1) to compare constitutive COX-2 expression throughout the body. As we reported previously (3), COX-2 expression is essentially absent in cardiovascular tissues, including the heart and aorta, but is highly expressed in the renal medulla and renal pelvis, the gastrointestinal tract, lung, thymus, and brain, with the cerebral cortex showing the highest expression (Fig. 1). Here we also show that the COX-2 gene is highly expressed in the bladder (Fig. 1), which may be important in understanding the contribution of COX-2 to urinary prostanoid metabolites, which are commonly used indicators of their circulating counterparts (14).
Fig. 1.
Tissue distribution of luciferase activity driven from the endogenous Cox2 gene. Basal expression from the endogenous Cox2 gene promoter was visualized by bioluminescent imaging of tissue from Cox2fLuc/+ mice (A). Quantification of images, as maximal luminescence from each tissue, demonstrates high expression in the renal medulla, gastrointestinal (GI) tract, and selected regions of the brain and thymus (B). Data are means ± SEM; n = 8.
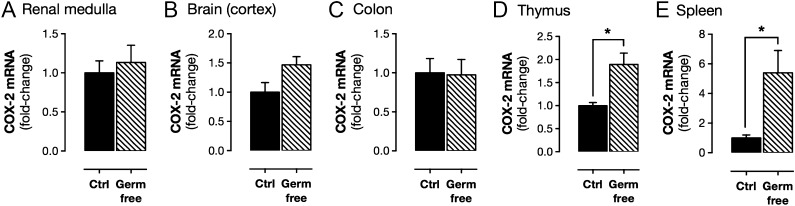
Because COX-2 is readily induced by inflammatory insults such as bacterial LPS, we studied the possibility that commensal bacteria might regulate constitutive COX-2 expression. Germ-free mice are devoid of a microbiome, including gut bacteria. We found that these mice display gastrointestinal abnormalities, reduced body weight, and reduced basal levels of the IFN response cytokine CXCL10 (SI Appendix, Fig. S2). Our observations are consistent with those previously reported for germ-free mice, which have developmental constraints in the gut and altered immune function (15). Despite these differences, constitutive COX-2 in the renal medulla, cerebral cortex, and gastrointestinal tract was not affected in germ-free mice (Fig. 2 and SI Appendix, Table S1). However, constitutive expression in the spleen and thymus was increased (Fig. 2 and SI Appendix, Table S1). Elevated COX-2 expression in the spleen and thymus may reflect altered T-cell function in these mice (15). Germ-free mice responded like control mice to pathogenic LPS derived from Escherichia coli with increased expression of COX-2 and the related inducible iNOS gene [Nos2 (nitric oxide synthase 2, inducible)] in the spleen and elevated plasma levels of CXCL10 (SI Appendix, Fig. S2). These observations allow us to effectively rule out products of bacterial or other microbial drivers for constitutive COX-2 expression.
Fig. 2.
Constitutive COX-2 mRNA expression in germ-free mice. Basal expression of COX-2 mRNA, measured by qPCR, was not different between control mice and germ-free mice in the renal medulla (A), cerebral cortex (B), or colon (C), and was increased in the thymus (D) and spleen (E) of germ-free animals. Data are means ± SEM; n = 12. *P < 0.05 by unpaired t test.
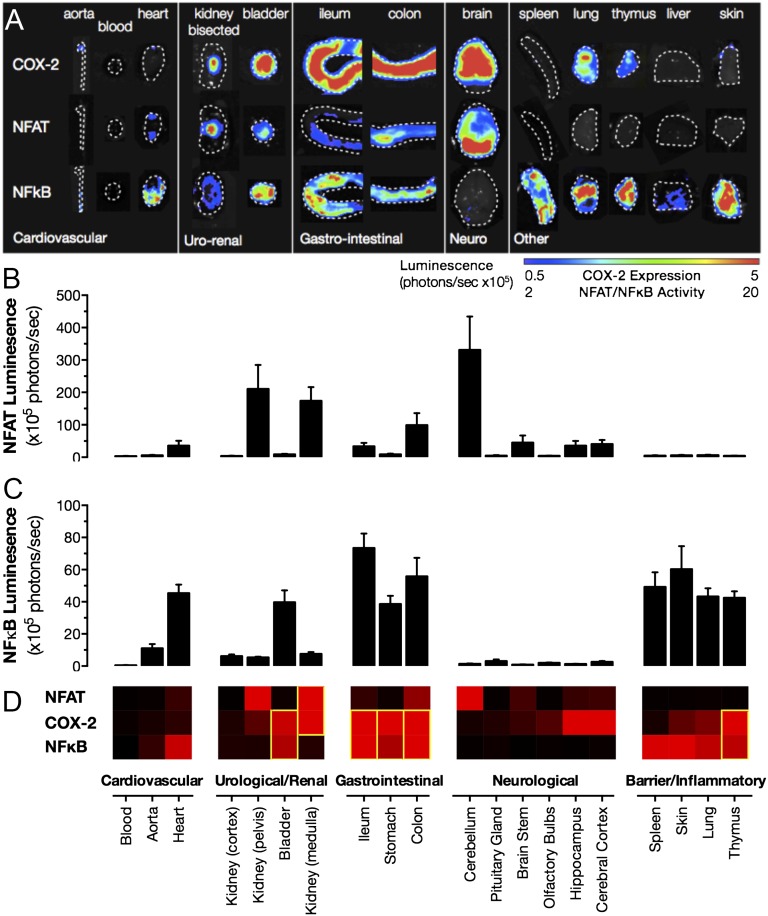
If not through pathogenic stimuli, then how is constitutive COX-2 expression regulated? Multiple transcriptional pathways have been implicated in driving inducible COX-2 expression. Growth factors predominantly act through the mitogen-activated protein kinase-signaling cascades to induce COX-2 in proliferating cells. During inflammation, the two best-studied transcriptional pathways driving inducible COX-2 expression are NFAT and NF-κB (16). We focused on these inflammation-associated transcriptional pathways and sought to establish whether they also play a role in sterile, constitutive COX-2 expression. Using specific NFAT (17) and NF-κB (18) luciferase reporter mice to map transcription factor activity, we found colocalization in the kidney for NFAT-driven transcriptional activity and endogenous constitutive COX-2 promoter-driven reporter gene expression (Fig. 3). Constitutive NFAT activity, like constitutive COX-2 expression, was essentially absent in the renal cortex but abundant in the renal medulla and renal pelvis regions (Fig. 3). In these regions, there was a striking overlap between NFAT activity and COX-2 expression. Additional, strong NFAT activity was present in the distal renal pelvis where COX-2 is present but less abundant than the inner medulla (Fig. 3), suggesting complex regulation of COX-2 by NFAT in the kidney. In contrast to the pattern of NFAT activity, NF-κB activity was detected only at a low level in all studied regions of the kidney and did not spatially map to COX-2 expression (Fig. 3). In other regions of high COX-2 expression, notably in the brain and the gut, NFAT activity was relatively high, but the distribution did not match that of COX-2 expression (Fig. 3). Constitutive NF-κB activity was surprisingly high in the heart (Fig. 3), with some activity present in the aorta. However, as described above, these cardiovascular structures do not constitutively express COX-2. NF-κB transcriptional activity was also high in the bladder, lung, thymus, and gut tissues, which do express constitutive COX-2 (Fig. 3). Although spleen, skin, and liver showed among the highest levels of constitutive NF-κB activity, these organs do not highly express COX-2, indicating that the presence of NF-κB (or NFAT) activity alone in these organs is not sufficient to drive COX-2 gene expression, and additional layers of regulation must exist.
Fig. 3.
Distribution of constitutive luciferase activity driven from NFAT and NF-κB transcriptional response elements. Constitutive activity of NFAT and NF-κB and expression from the endogenous Cox2 gene promoter (COX-2), visualized using bioluminescent imaging of tissue from NFAT, NF-κB, and COX-2 luciferase reporter mice (A). Quantification demonstrates that constitutive NFAT activity is greatest in the renal medulla/pelvis, colon, and cerebellum (B) and that constitutive NF-κB activity is greatest in the heart, bladder, gastrointestinal tract, liver, as well as barrier and inflammatory tissues (C). Comparison of these patterns with those for constitutive COX-2 expression shows strong overlap between NFAT and COX-2 in the renal medulla and between NF-κB and COX-2 in the gastrointestinal tract and thymus (D). Heat map gradients from black (baseline) to red (maximal tissue) are individually scaled for each target. Data are means ± SEM; n = 10–16.
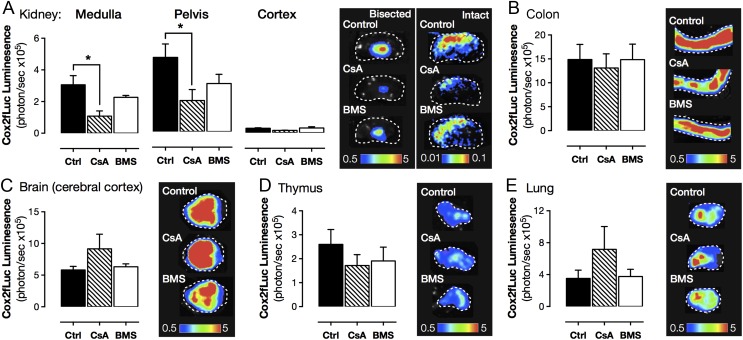
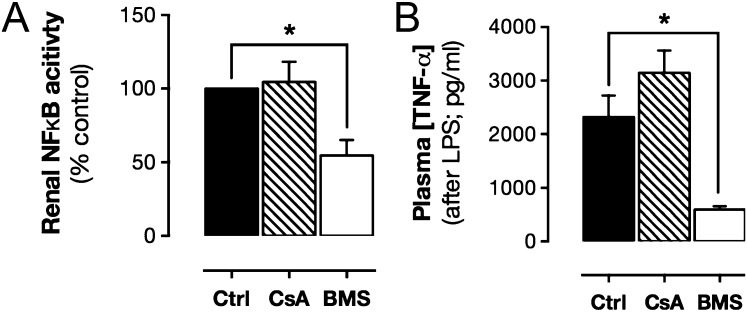
Although mapping transcription factor activity provided a compelling case for NFAT, but not NF-κB, as the transcriptional driver of COX-2 expression in the kidney, colocalization does not prove cause and effect. To test the roles of the NFAT and NF-κB requirement in constitutive COX-2 expression in the kidney (and elsewhere), we used a pharmacological approach; examining the modulatory effects of cyclosporin A, which inhibits calcineurin-dependent NFAT activation (19), and BMS-345541, which inhibits IκB kinase-2–dependent NF-κB activation (20). COX-2 expression (Fig. 4 and SI Appendix, Table S2) was significantly reduced in the renal medulla and renal pelvis of mice treated with cyclosporin A but not in mice treated with BMS-345541 (Fig. 4 and SI Appendix, Table S2), consistent with a lack of constitutive NF-κB activation in this area (Figs. 3 and 4). The selective effect of cyclosporin A on COX-2 observed in luciferase reporter mice was confirmed at the level of mRNA in renal medulla (control, 1.00 ± 0.59; BMS, 1.29 ± 0.66; P = 0.75, n = 6; cyclosporin A; Fig. 5 and SI Appendix, Table S3). To demonstrate the functional significance of these changes in COX-2 expression, we measured prostaglandin formation in the isolated renal medulla and found a profound reduction in COX-2–dependent PGE2 formation in mice treated with cyclosporin A (SI Appendix, Table S3). The inability of BMS-345541 to influence constitutive COX-2 expression was not due to inactivity of the drug/dosing regime as BMS-345541 reduced renal NF-κB activity and LPS-induced plasma TNFα accumulation (Fig. 5). Further, in agreement with an NFAT, rather than an NF-κB transcriptional pathway, constitutive COX-2 expression was not inhibited by dexamethasone (SI Appendix, Fig. S3). For all other regions studied, neither cyclosporin A nor BMS-345541 altered constitutive expression from the COX-2 promoter (Fig. 4 and SI Appendix, Table S2), suggesting constitutive COX-2 at these sites is not driven by classical inflammatory COX-2 pathways or commensal microorganisms, but rather through distinct mechanisms. In the gut, for example, COX-2 may be driven through proliferative pathways such as MAPK as a byproduct of continual epithelial proliferation. Alternatively, in the brain, COX-2 may be constitutively induced through neuronal activity and associated transcriptional pathways including cAMP response element-binding protein (CREB), whose pattern of constitutive activity bears striking similarity to that of COX-2 (21), and thyroid-transcription factor-1 (TTF-1), which has been linked to COX-2 in certain brain regions (22).
Fig. 4.
Effect of NFAT and NF-κB pathway inhibitors on luciferase activity driven from the endogenous Cox2 gene. Constitutive COX-2 expression, measured using bioluminescent imaging of tissue from Cox2fLuc/+ mice, was reduced by cyclosporin A (CsA; 4 d; 20 mg/kg per day; s.c.) an inhibitor of calcineurin/NFAT activation in the renal medulla and pelvis (A) but not the renal cortex (A), colon (B), brain (C), thymus (D), or lung (E). BMS-345541 (BMS; 4 d; 10 mg/kg per day; i.v.), an inhibitor of NF-κB activation had no effect in any studied tissue (A–E). Data are means ± SEM; n = 4. *P < 0.05 by one-way ANOVA with Dunnett's posttest.
Fig. 5.
Effect of CsA and BMS-345541 on renal NF-κB activity and LPS-induced cytokine levels. Treatment of NF-κB-luc mice with BMS-345541 (BMS; 4 d; 10 mg/kg per day; i.v.) reduced renal medulla NF-κB activity (A), whereas treatment with CsA (4 d; 20 mg/kg per day; s.c.) had no effect (A). BMS but not CsA treatment suppressed LPS (10 mg/kg; 4 h)-induced plasma levels of TNFα (B). Data are means ± SEM; n = 3–6. *P < 0.05 by one-way ANOVA with Dunnett’s posttest.
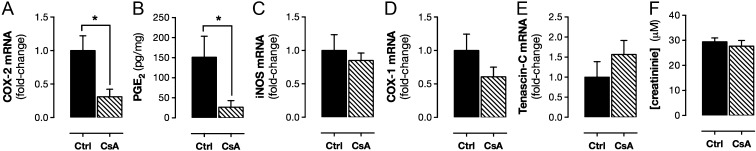
Cyclosporin A is used clinically as an immunosuppressant. In patients, as well as animal models, it has been associated with renal toxicity (23), which may reflect its ability of suppress renal COX-2. Nonetheless, cyclosporin A at the dose and duration used here did not alter serum creatinine levels (Fig. 6), suggesting its effects on COX-2 expression in our study were not a result of fundamentally compromised renal function. Cyclosporin A also did not cause nonspecific effects on gene transcription, because it did not alter expression of iNOS or constitutive COX-1 [Ptgs1 (prostaglandin-endoperoxide synthase 1)] (Fig. 6). In the renal medulla, COX-2 is highly localized to interstitial fibroblasts, where it colocalizes with the cell selective marker tenascin C (24). Cyclosporin A suppression of renal COX-2 expression was not due to loss of interstitial fibroblasts, because cyclosporin A did not alter tenascin C levels (Fig. 6).
Fig. 6.
Effect of CsA on renal gene expression and function. Constitutive COX-2 mRNA expression, measured using qPCR, was reduced in the renal medulla (A) by treatment with CsA (4 d; 20 mg/kg per day; s.c.). CsA treatment also reduced COX-2–dependent PGE2 formation by isolated renal medulla (B). In WT mice, CsA treatment did not alter constitutive expression of iNOS (C), COX-1 (D), or the renal interstitial cell marker tenascin C (E) and did not alter plasma levels of creatinine (F). Data are means ± SEM; n = 3–6. *P < 0.05 by unpaired t test.
NFAT has five isoforms, four (NFATc1–c4) of which are typically dependent on calcineurin for activation and thus sensitive to cyclosporin A (25). Cyclosporin A can also affect transcriptional function of the fifth isoform, NFAT5, by inhibiting its translocation to the nucleus (26), although NFAT5 can also be activated through cyclosporin A-insensitive pathways. In the renal medulla, mRNAs for all NFAT isoforms (NFATc1–c4, NFAT5) were detectable by qPCR (Ct values < 30; SI Appendix, Fig. S4). However, it is currently beyond the scope of this study to establish the isoform(s) of NFAT that regulate constitutive COX-2 expression, due to development defects (including to the kidney) (27, 28) arising from specific NFAT deletions, redundancy among NFAT isoforms, and a lack of isoform-specific pharmacological tools. We suggest that identification of the NFAT isoform responsible for constitutive COX-2 expression will only be possible with the development of mice lacking the multiple NFAT isoforms, specifically in the kidney and/or specific inhibitors. Nevertheless, our initial objective was to establish the transcriptional pathway by which COX-2 is expressed constitutively in the renal medulla and our data establish NFAT as the predominate mechanism.
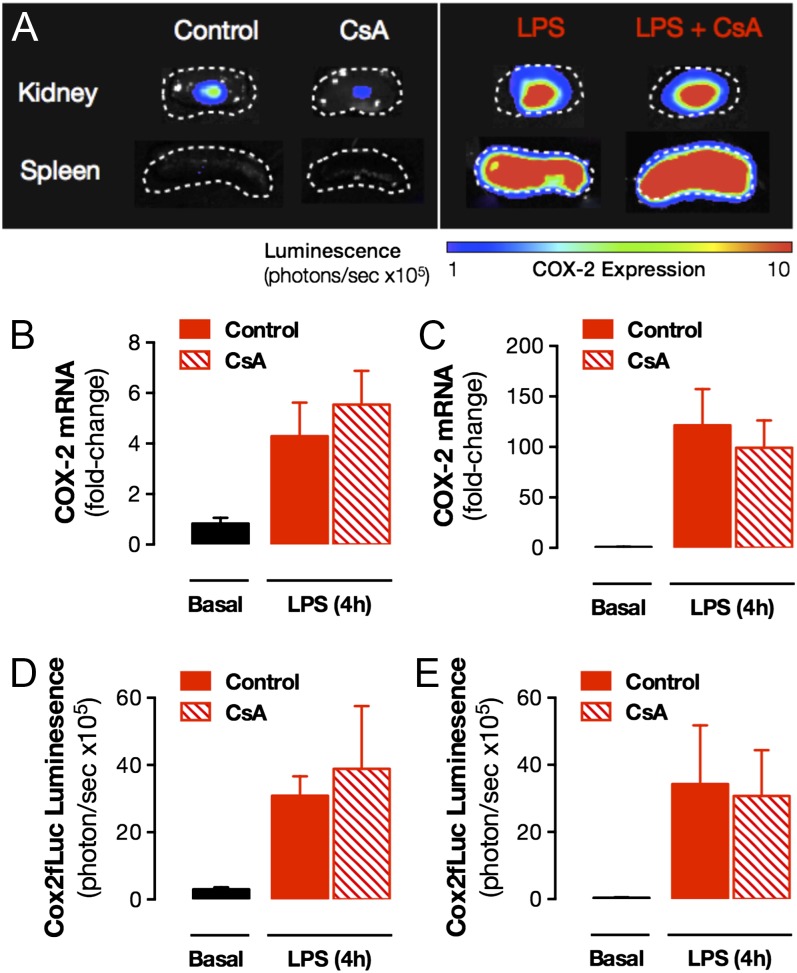
Last, we sought to determine whether NFAT also drives expression of COX-2 in the kidney, and other structures, at times of overt inflammation. We regard this as an important issue; if we are able to dissociate constitutive COX-2 expression in the kidney from inducible COX-2 at sites of inflammation, we will create future therapeutic opportunities to target inflammation-driven COX-2 expression (at the transcriptional level) while sparing the kidney, thereby avoiding NSAID cardio-renal side effects. As we have shown previously (29), in mice treated with LPS, COX-2 is readily induced in the renal medulla, the spleen (Fig. 7), and a full range of other tissues (SI Appendix, Table S4). In direct contrast to results seen for constitutive expression, cyclosporin A-sensitive NFAT pathways did not drive inflammatory COX-2 expression in the renal medulla or, indeed, in any tissue tested (SI Appendix, Table S4).
Fig. 7.
Effect of CsA on luciferase activity driven from the endogenous Cox2 gene in LPS-treated mice. LPS (10 mg/kg; 4 h; i.p.) increased COX-2 expression in renal medulla and spleen, visualized using bioluminescent imaging of tissue from Cox2fLuc/+ mice (A). Quantification of imaging data indicated no effect of CsA treatment (4 d; 20 mg/kg per day; s.c.) on LPS-induced COX-2 expression in either the renal medulla (B) or spleen (C). This result was confirmed by qPCR analysis of renal medulla (D) and spleen (E) from WT mice. Data are means ± SEM; n = 3–6. *For all panels, effect of CsA, P > 0.05 by unpaired t test.
Summary and Conclusions
COX-2 is expressed constitutively in discrete tissue regions, including the renal medulla. This constitutive COX-2 expression is not mediated by host pathogenic stimuli because no effect was seen on levels in tissues from germ-free mice. NFAT, but not NF-κB, transcription regulates constitutive COX-2 expression in the renal medulla; however, neither transcriptional pathway contributes to constitutive expression in other tissues. This indicates that constitutive COX-2 across different tissues is driven through distinct, locally relevant pathways (e.g., NFAT, MAPK, CREB, TTF-1) rather than a broad regulation through classical inflammatory pathways/commensal microorganisms. In addition, the transcriptional regulation of constitutive COX-2 in the renal medulla is distinct from pathways that drive inflammatory COX-2, which is the therapeutic target of NSAIDs.
These findings address a fundamental and long-standing question in the field of COX-2 biology and have direct relevance to the therapeutic targeting of COX-2 to spare activity and expression at sites of homeostatic control and thereby avoid the side effects associated with NSAIDs. First, we showed that cyclosporin A selectively suppresses COX-2 expression in the kidney without altering constitutive COX-2 at other sites. We previously suggested the kidney to be a major driver of NSAID cardiovascular side effects, where constitutive COX-2 in the renal medulla produces profound regulation of cardioprotective gene networks (5). As such, cyclosporin A may provide an important tool to probe the contribution of renal COX-2 to the cardiovascular side effects attributed to NSAIDs use. In this regard, it is interesting to note that in animal models (30) and in humans (31), cyclosporin A, as with COX-2 inhibitors, increases circulating levels of the naturally occurring eNOS inhibitor ADMA; a mechanism that we recently highlighted as relevant to the cardiovascular side effects associated with NSAIDs (5). Second and perhaps more importantly, our observations lay groundwork for the idea that targeting COX-2 at the transcriptional level could dissociate antiinflammatory benefit of NSAIDs from their cardio-renal side effects. Although this will require a more complete understanding of the drivers of COX-2 in inflammation and cancer, these data indicate that, in the future, any therapies directed at COX-2 transcriptional pathways that spare NFAT signaling would avoid the renal pathways responsible for driving the cardiovascular side effects that currently limit NSAID use.
Materials and Methods
Additional methodological information can be found in SI Appendix.
Animals.
Studies were performed on male and female 8- to 10-wk-old COX-2 luciferase knock-in reporter mice (Cox2fLuc/+) (32), transgenic luciferase reporter mice for NFAT (NFAT-luc) (17) or NF-κB (NFκB-RE-luc) activity (18), germ-line COX-1 KO mice (Cox1−/−) (33), or WT C57BL/6 mice (Charles River/Taconic) raised under normal or germ-free conditions (34) (Farncombe Gnotobiotic Unit). All procedures were carried out in accordance with local rules and ethical review by the University of California Los Angeles Animal Research Committee (Protocol no.: 1999-066-32; Cox2fLuc experiments), Animal Care Committee of the Faculty of Health Sciences at McMaster University (NFκB-RE-luc and germ-free experiments), Malmö/Lund Animal Care and Use Committee (Permit no.: M29-12; NFAT-luc experiments), or the Imperial College Ethical Review Panel (PPL 70/7013; all other experiments). Mice were treated with cyclosporin A (20 mg/kg; Novartis), BMS-345541 (10 mg/kg; Sigma), dexamethasone (3 mg/kg; Organon), or vehicle for 4 d. In some cases, inflammation was induced by LPS (10 mg/kg; from E. coli serotype 055:B5; Sigma) 4 h before euthanasia.
Bioluminescent Imaging.
Ex vivo bioluminescent imaging of tissue from luciferase reporter strains was performed as we previously described (3) using an IVIS imaging system (Xenogen).
qPCR.
RNA was extracted from tissue homogenates using a silica bead kit (Life Technologies) and reverse transcribed (Life Technologies), and gene expression was measured with TaqMan (Life Technologies) or SYBRGreen assays (Bio-Rad). Data were analyzed using the comparative Ct method, with reference to housekeeping genes [18S and Gapdh (glyceraldehyde 3-phosphate dehydrogenase)].
PGE2 Synthesis Bioassay.
COX-2–dependent prostaglandin synthesis was measured in tissue segments from Cox1−/− mice stimulated with A23187 calcium ionophore (50 μM; Sigma) for 30 min at 37 °C. PGE2 accumulation was measured by immunoassay (Cisbio).
Plasma Cytokine and Creatinine Measurement.
Plasma levels of CXCL10 (R&D Systems) and TNFα (Meso Scale Discovery) were measured by immunoassay. Creatinine was measured by a commercial biochemistry service (IDEXX Laboratories).
Statistics and Data Analysis.
Data were analyzed using Prism 6.0 software (GraphPad Software) and are presented as mean ± SE. Details of statistical tests are given in figure legends.
Supplementary Material
Acknowledgments
This research was supported by Wellcome Trust Program Grant 0852551Z108/Z (to J.A.M.), National Institutes of Health-National Cancer Institute P50 Award CA086306 (to H.R.H.), and grants from the Swedish Research Council and Swedish Heart and Lung Foundation (20130700 and 2014-3352; to M.F.G.). N.S.K. is the recipient of an Imperial College junior research fellowship. A.K.Z. is the recipient of an American Society of Hematology Scholar award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517642113/-/DCSupplemental.
References
- 1.Kirkby NS, et al. Cyclooxygenase-1, not cyclooxygenase-2, is responsible for physiological production of prostacyclin in the cardiovascular system. Proc Natl Acad Sci USA. 2012;109(43):17597–17602. doi: 10.1073/pnas.1209192109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nandakishore R, Yalavarthi PR, Kiran YR, Rajapranathi M. Selective cyclooxygenase inhibitors: Current status. Curr Drug Discov Technol. 2014;11(2):127–132. doi: 10.2174/1570163811666140127123717. [DOI] [PubMed] [Google Scholar]
- 3.Kirkby NS, et al. LC-MS/MS confirms that COX-1 drives vascular prostacyclin whilst gene expression pattern reveals non-vascular sites of COX-2 expression. PLoS One. 2013;8(7):e69524. doi: 10.1371/journal.pone.0069524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amateau SK, McCarthy MM. Induction of PGE2 by estradiol mediates developmental masculinization of sex behavior. Nat Neurosci. 2004;7(6):643–650. doi: 10.1038/nn1254. [DOI] [PubMed] [Google Scholar]
- 5.Ahmetaj-Shala B, et al. Evidence that links loss of cyclo-oxygenase-2 with increased asymmetric dimethylarginine: Novel explanation of cardiovascular side effects associated with anti-inflammatory drugs. Circulation. 2014;131(7):633–642. doi: 10.1161/CIRCULATIONAHA.114.011591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace JL, McKnight W, Reuter BK, Vergnolle N. NSAID-induced gastric damage in rats: Requirement for inhibition of both cyclooxygenase 1 and 2. Gastroenterology. 2000;119(3):706–714. doi: 10.1053/gast.2000.16510. [DOI] [PubMed] [Google Scholar]
- 7.Scarpignato C, et al. International NSAID Consensus Group Safe prescribing of non-steroidal anti-inflammatory drugs in patients with osteoarthritis--an expert consensus addressing benefits as well as gastrointestinal and cardiovascular risks. BMC Med. 2015;13:55. doi: 10.1186/s12916-015-0285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia Rodriguez LA, Cea-Soriano L, Tacconelli S, Patrignani P. Coxibs: Pharmacology, toxicity and efficacy in cancer clinical trials. Recent Results Cancer Res. 2013;191:67–93. doi: 10.1007/978-3-642-30331-9_4. [DOI] [PubMed] [Google Scholar]
- 9.Kang YJ, Mbonye UR, DeLong CJ, Wada M, Smith WL. Regulation of intracellular cyclooxygenase levels by gene transcription and protein degradation. Prog Lipid Res. 2007;46(2):108–125. doi: 10.1016/j.plipres.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugimoto T, et al. Endothelin-1 induces cyclooxygenase-2 expression via nuclear factor of activated T-cell transcription factor in glomerular mesangial cells. J Am Soc Nephrol. 2001;12(7):1359–1368. doi: 10.1681/ASN.V1271359. [DOI] [PubMed] [Google Scholar]
- 11.Fukata M, Abreu MT. Role of Toll-like receptors in gastrointestinal malignancies. Oncogene. 2008;27(2):234–243. doi: 10.1038/sj.onc.1210908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stepankova R, et al. Absence of microbiota (germ-free conditions) accelerates the atherosclerosis in ApoE-deficient mice fed standard low cholesterol diet. J Atheroscler Thromb. 2010;17(8):796–804. doi: 10.5551/jat.3285. [DOI] [PubMed] [Google Scholar]
- 13.Amaral FA, et al. Commensal microbiota is fundamental for the development of inflammatory pain. Proc Natl Acad Sci USA. 2008;105(6):2193–2197. doi: 10.1073/pnas.0711891105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flavahan NA. Balancing prostanoid activity in the human vascular system. Trends Pharmacol Sci. 2007;28(3):106–110. doi: 10.1016/j.tips.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsatsanis C, Androulidaki A, Venihaki M, Margioris AN. Signalling networks regulating cyclooxygenase-2. Int J Biochem Cell Biol. 2006;38(10):1654–1661. doi: 10.1016/j.biocel.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 17.Wilkins BJ, et al. Calcineurin/NFAT coupling participates in pathological, but not physiological, cardiac hypertrophy. Circ Res. 2004;94(1):110–118. doi: 10.1161/01.RES.0000109415.17511.18. [DOI] [PubMed] [Google Scholar]
- 18.Carlsen H, Moskaug JO, Fromm SH, Blomhoff R. In vivo imaging of NF-kappa B activity. J Immunol. 2002;168(3):1441–1446. doi: 10.4049/jimmunol.168.3.1441. [DOI] [PubMed] [Google Scholar]
- 19.Campbell PM, Pimm J, Ramassar V, Halloran PF. Identification of a calcium-inducible, cyclosporine sensitive element in the IFN-gamma promoter that is a potential NFAT binding site. Transplantation. 1996;61(6):933–939. doi: 10.1097/00007890-199603270-00016. [DOI] [PubMed] [Google Scholar]
- 20.Burke JR, et al. BMS-345541 is a highly selective inhibitor of I kappa B kinase that binds at an allosteric site of the enzyme and blocks NF-kappa B-dependent transcription in mice. J Biol Chem. 2003;278(3):1450–1456. doi: 10.1074/jbc.M209677200. [DOI] [PubMed] [Google Scholar]
- 21.Böer U, et al. CRE/CREB-driven up-regulation of gene expression by chronic social stress in CRE-luciferase transgenic mice: Reversal by antidepressant treatment. PLoS One. 2007;2(5):e431. doi: 10.1371/journal.pone.0000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yun CH, et al. TTF-1 action on the transcriptional regulation of cyclooxygenase-2 gene in the rat brain. PLoS One. 2011;6(12):e28959. doi: 10.1371/journal.pone.0028959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao Z, et al. Mechanisms of cyclosporine-induced renal cell apoptosis: A systematic review. Am J Nephrol. 2013;37(1):30–40. doi: 10.1159/000345988. [DOI] [PubMed] [Google Scholar]
- 24.He W, et al. Generation of a tenascin-C-CreER2 knockin mouse line for conditional DNA recombination in renal medullary interstitial cells. PLoS One. 2013;8(11):e79839. doi: 10.1371/journal.pone.0079839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller MR, Rao A. NFAT, immunity and cancer: A transcription factor comes of age. Nat Rev Immunol. 2010;10(9):645–656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- 26.Sheikh-Hamad D, et al. Cyclosporine A inhibits the adaptive responses to hypertonicity: A potential mechanism of nephrotoxicity. J Am Soc Nephrol. 2001;12(12):2732–2741. doi: 10.1681/ASN.V12122732. [DOI] [PubMed] [Google Scholar]
- 27.López-Rodríguez C, et al. Loss of NFAT5 results in renal atrophy and lack of tonicity-responsive gene expression. Proc Natl Acad Sci USA. 2004;101(8):2392–2397. doi: 10.1073/pnas.0308703100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ranger AM, et al. The transcription factor NF-ATc is essential for cardiac valve formation. Nature. 1998;392(6672):186–190. doi: 10.1038/32426. [DOI] [PubMed] [Google Scholar]
- 29.Kirkby NS, et al. Differential COX-2 induction by viral and bacterial PAMPs: Consequences for cytokine and interferon responses and implications for anti-viral COX-2 directed therapies. Biochem Biophys Res Commun. 2013;438(2):249–256. doi: 10.1016/j.bbrc.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renner B, et al. Cyclosporine induces endothelial cell release of complement-activating microparticles. J Am Soc Nephrol. 2013;24(11):1849–1862. doi: 10.1681/ASN.2012111064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Namjud N, Chariyavilaskul P, Townamchai N, Wittayalertpanya S. A reduction of asymmetric dimethylarginine in renal transplant recipients receiving sirolimus-based regimen. J Med Assoc Thai. 2015;98(Suppl 1):S9–S13. [PubMed] [Google Scholar]
- 32.Ishikawa TO, Jain NK, Taketo MM, Herschman HR. Imaging cyclooxygenase-2 (Cox-2) gene expression in living animals with a luciferase knock-in reporter gene. Mol Imaging Biol. 2006;8(3):171–187. doi: 10.1007/s11307-006-0034-7. [DOI] [PubMed] [Google Scholar]
- 33.Langenbach R, et al. Prostaglandin synthase 1 gene disruption in mice reduces arachidonic acid-induced inflammation and indomethacin-induced gastric ulceration. Cell. 1995;83(3):483–492. doi: 10.1016/0092-8674(95)90126-4. [DOI] [PubMed] [Google Scholar]
- 34.Slack E, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325(5940):617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.