Significance
Amyloid fibril formation is a key process accompanying many neurodegenerative diseases. Oligomers formed in the early stages of aggregation have been thought to play a key role in disease effects, but their studies are challenging. We use site-specific measurements of surface water diffusion, protein segmental dynamics, and interstrand packing to track early tau protein aggregation processes in situ. Our study reveals that tau aggregation is accompanied by a dramatic structural transformation within minutes of initiating aggregation, followed by the formation of partially structured aggregation intermediates and their rearrangement into stable aggregate species with β-sheet signatures and fibrils. Our findings suggest that therapeutic intervention may focus on disrupting the earliest aggregation events occurring in solution.
Keywords: biological water, soluble oligomers, amyloid formation, site-directed spin labeling, Alzheimer's disease
Abstract
Protein aggregation plays a critical role in the pathogenesis of neurodegenerative diseases, and the mechanism of its progression is poorly understood. Here, we examine the structural and dynamic characteristics of transiently evolving protein aggregates under ambient conditions by directly probing protein surface water diffusivity, local protein segment dynamics, and interprotein packing as a function of aggregation time, along the third repeat domain and C terminus of Δtau187 spanning residues 255–441 of the longest isoform of human tau. These measurements were achieved with a set of highly sensitive magnetic resonance tools that rely on site-specific electron spin labeling of Δtau187. Within minutes of initiated aggregation, the majority of Δtau187 that is initially homogeneously hydrated undergoes structural transformations to form partially structured aggregation intermediates. This is reflected in the dispersion of surface water dynamics that is distinct around the third repeat domain, found to be embedded in an intertau interface, from that of the solvent-exposed C terminus. Over the course of hours and in a rate-limiting process, a majority of these aggregation intermediates proceed to convert into stable β-sheet structured species and maintain their stacking order without exchanging their subunits. The population of β-sheet structured species is >5% within 5 min of aggregation and gradually grows to 50–70% within the early stages of fibril formation, while they mostly anneal block-wisely to form elongated fibrils. Our findings suggest that the formation of dynamic aggregation intermediates constitutes a major event occurring in the earliest stages of tau aggregation that precedes, and likely facilitates, fibril formation and growth.
The question of whether there is a unified mechanism for amyloid formation that contributes to the progression of neurodegenerative diseases, including Alzheimer’s disease (AD), is a subject of intense debate (1). This view is fueled by the observation that a wide range of amyloidogenic proteins or peptides with different primary sequences can ultimately aggregate into highly structured amyloid fibrils with similar morphologies (2). It has been recognized that a common feature of neurodegenerative diseases is associated with the accumulation and deposits of misfolded proteins that affect various cell signaling processes (3). Among them, the pathological form of a microtubule-associated protein, tau, can dissociate from microtubules and aggregate, resulting in the deposition of insoluble neurofibrillary tangles in neuronal cells (3, 4). Several lines of evidence suggest that small aggregated protein intermediates, that may constitute soluble oligomers and precede the formation of highly structured fibrils, are primary toxic species contributing to the early onset of neurodegenerative diseases (4–6). For example, the presence of granular tau oligomers with prefilamentous structures in transgenic mice was found to correlate with the earliest sign of cognitive decline and memory loss (6). Although early aggregation intermediates formed before fibrils may represent effective targets for common therapeutic intervention, knowledge about their structure and properties is only now emerging. The studies of early intermediates—many of which currently focus on amyloid-β, and comparably few on tau oligomers—are challenging, given the transient nature and complex equilibria involving monomer and oligomer species (4, 7, 8). Recently, several computational studies have identified structural and dynamic features of intermediate aggregates at the molecular level (9, 10), as well as possible driving forces at different aggregation stages (10, 11). Characterization of freeze-trapped amylospheroids by solid-state NMR showed that intermediate structures of amyloid-β aggregates are composed of single conformers containing parallel β-sheets (12). Still, direct experimental observations of transient aggregation and/or folding intermediates remain sparse (5, 12–15).
We have previously established a broadly applicable spectroscopic method, Overhauser dynamic nuclear polarization-enhanced NMR relaxometry (ODNP) (16, 17), to probe changes in translational diffusivity of local water within 1 nm of nitroxide radical-based electron spin labels tethered to specific protein residues, and successfully reported on the study of protein folding, protein aggregation, and conformational changes of globular and membrane protein segments (18–21). ODNP has revealed increased heterogeneity, i.e., dispersion, in local water diffusivity on the surface of a folded protein, compared with its unfolded counterpart or folding intermediate (20). The basic connection between surface water diffusion and protein structural transformation is that conformational changes, locally or globally, are modulated by the shift in balance between protein surface−water versus protein−protein interactions. The varying stability of this local hydration shell will be collectively reflected in varying retardation of water diffusivity within about 1 nm of the protein surface of interest (22, 23). It has been reported that this surface water dynamics not only varies from surface to surface but is also dramatically heterogeneous from region to region on a given protein surface (20, 22). Terahertz measurements have shown that structural rearrangements of a protein at different folded states exhibit different Terahertz absorption energies that were attributed to changes in the coupling dynamics between the protein surface and hydration water (23). However, the heterogeneous water dynamics landscape of a structurally evolving or interacting protein in situ and in dilute solution has been elusive to probe to date. This study, aided by ODNP, sets out to exactly do that, namely, to probe the structural transition of tau in the early stages of aggregation in solution.
Tau monomers belong to a family of intrinsically disordered proteins (IDPs) that, upon misfolding and aggregation, form highly structured amyloid fibrils (24). In this work, we studied the tau variant truncated between residues 255 and 441 (also known as Δtau187, Fig. 1A) of the longest isoform of the human tau protein (25) that includes all four microtubule-binding repeat domains (MTBD) and the C-terminal region (25, 26) (Fig. 1A). This and related truncated variants of tau have been shown to have greatly accelerated aggregation kinetics to form mature fibrils after approximately >12 h of aggregation (24–26). The known hydrophobic hexapeptide stretches, 275VQIINK280 (PHF6*) and 306VQIVYK311 (PHF6), found at the beginning of the second and third repeat segment are highly amyloidogenic, and form core regions of the tau fibrils (27, 28) with propensities for parallel in-register β-sheet structures reported (28–30). To induce tau fibrillization in vitro, polyanions, such as heparin or arachidonic acid micelles, are typically added to the solution (31). Although the molecular basis for this process is not fully understood, the structure of tau fibrils induced by heparin in vitro displays similar propensities to those found in AD patient tissues (31). Furthermore, heparin ultimately does not remain in the mature fibril core (32). Thus, heparin-induced tau aggregation has served as an accepted model for pathological tau fibrillization (30–32).
Fig. 1.
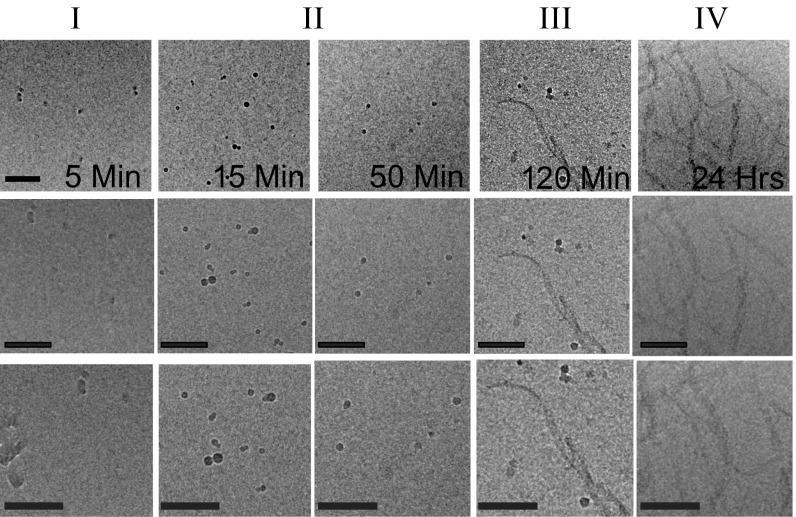
Visualization of Δtau187 aggregates at different aggregation time (). (A) Δtau187 consists of four microtubule-binding domains indicated as 1–4, where the amyloidogenic hexapeptide region of the third repeat is marked as a black box. The positions of the spin labels at mutated cysteine residues are indicated. (B) Cryo-TEM micrographs display the morphologies of Δtau187 (322C) aggregates after various aggregation times. (Scale bar, 100 nm.) (C) Cross-linked Δtau187 (322C) aggregates at different aggregation time analyzed by SDS/PAGE. (D) A schematic representation of the hypothesized Δtau187 species evolving at five aggregation stages. (E) Aggregation time dependence of τ of hydration water near different spin-labeled residues of Δtau187.
In this study, we focused on capturing the structural transformation of the heparin-induced aggregation of Δtau187 in situ under ambient solution conditions and as a function of aggregation time. To achieve this, we used ODNP to probe the translational diffusion dynamics of surface water on protein surfaces and concurrently continuous wave (cw) electron spin resonance (ESR) line shape analysis (33, 34) to monitor protein side-chain mobility and interprotein contacts using the same protein sample (16, 17). Both methods rely on the site-directed mutagenesis and spin-labeling of the protein at a single-cysteine site with an (1-oxyl-2,2,5,5-tetramethylpyrrolidin-3-yl) methyl methanethiosulfonate (MTSL)-based nitroxide label, whose side chain is commonly termed “R1” (35). Δtau187 was singly R1 spin-labeled across several key residues of its third repeat MTBD and C terminus. The ODNP measurement was used to derive (as discussed in SI Overview of the ODNP Technique in detail) the translational correlation time, τ, of water within ∼1 nm of the R1 label tethered to the protein surface, together with two relaxivity parameters termed “” and “” that report on contributions from freely diffusing water near the protein surface and bound water to the protein surface on the timescale of a few nanoseconds or longer, respectively. Thus, when a characteristic dispersion, i.e., an increased heterogeneity, of surface water diffusivity develops on the Δtau187 surface, as it transforms from monomeric species to oligomeric and larger fibrillar species, this indirectly shows that Δtau187 is undergoing structural transformation (18). However, ODNP analysis alone cannot differentiate between intraprotein or interprotein structural changes, nor does it offer population information. The cw ESR concurrently measures the structural and dynamic features of the same R1 spin-labeled protein species, as the backbone rearranges and interprotein contacts form during protein assembles. Specifically, we carried out quantitative ESR line shape simulation to determine the spin label mobility, and the population (percent) of the spin label in the respective motional states, and as embedded in parallel β-sheet arrangements. The spin label may experience distinct motional environments—slow vs. fast—that can be attributed to a local protein environment in which R1 is embedded at an intertau interface vs. one in which R1 is freely rotating in the solvent. Their evolving populations with increased aggregation time can be quantified. If R1 of Δtau187 at a specific position gets embedded in β-sheet structures, the evolving content of the β-sheets in which R1 labels of adjacent protein strands stack in parallel can be estimated by ESR line shape analysis. This is because, when packing in parallel β-sheet structures, the same R1-labeled sites from different protein strands come into intimate contact within 5–8 Å, yielding a characteristic single-line ESR spectral feature due to spin exchange between overlapping electron spin orbitals of the R1 labels (29, 36–41). We can verify the assignment of this spectral signature to parallel β-sheet stacking by systematic “spin dilution” to yield a mixture where only 20% of the tau strands are labeled with paramagnetic R1 labels and 80% are labeled with the diamagnetic analog of R1 labels, designated here as R1’, at a given site of Δtau187. This approach allows us to differentiate between the spin-exchanged and simply immobile spectral features, thereby confirming the origin of the characteristic spectral component as caused by β-sheet packing (36–41). Single-line, spin-exchanged, ESR spectra of protein fibrils have been previously assigned to parallel β-sheet structures (29, 36–41), including the mature fibrils of full-length tau proteins that were found to stack in parallel and in-register particularly around the PHF6 region (29).
By combining ODNP and cw ESR measurements performed in situ and in solution together with conventional techniques of turbidimetry, Thioflavin T (ThT) staining fluorescence, transmission electron microscopy (TEM), and chemical cross-linking to probe aggregation of Δtau187, we aim to answer the following key questions: (i) At what stage of aggregation do structural transformations of tau proteins and their assembly occur? (ii) What are the structural and dynamic properties of the aggregation intermediates, and are they “on pathway” toward fibril formation? (iii) At what stage do stable β-sheet structures form, and what are their populations? (iv) Does fibril growth proceed mainly by elongation of preassembled oligomers or fibrils, or predominantly by monomer recruitment?
Results and Discussions
Verifying Fibril Formation of Δtau187 by Global Measurement.
We attached one R1 per Δtau187 molecule at a time at select residues along the aggregation-prone third repeat region (i.e., residues 313C, 316C, and 322C) and the C terminus (i.e., residues 400C and 404C) (see Fig. 1A). The third repeat domain is thought to undergo large structural transformation (24, 29, 32, 42) to eventually form parallel β-sheets and bury inside the aggregated structure, whereas residues 400C and 404C at the C terminus are expected to remain disordered and solvent-exposed during the entire aggregation process (24, 29, 42). Therefore, the C terminus serves as an important control in this study. The spin-labeled Δtau187 samples were prepared either as undiluted samples, i.e., 100% R1-labeled, or as spin-diluted samples, i.e., by mixing 20% R1-labeled and 80% R1’-labeled Δtau187, all labeled at the same site (36–41).
First, we have verified that each spin-labeled variant forms stable fibrils with comparable morphology after 12–24 h of aggregation by negative staining TEM (Fig. S1). Next, we monitored the fibril formation from the monomer of each spin-labeled Δtau187 variant using turbidimetry at 350 nm, whose absorbance amplitude increases with increasing aggregate quantity and size. We confirmed that the global aggregation kinetics of heparin-induced R1-labeled Δtau187 variants display a characteristic lag phase, followed by an elongation phase, until the aggregation proceeds to its steady state after 200–600 min (Fig. S1). Notably, the turbidity (absorbance at 350 nm) through aggregating samples of 313C, 316C, and 322C variants increases and reaches its maximal level within 400 min of aggregation, whereas the 400C and 404C variants exhibit a longer lag phase and slower aggregation kinetics (Fig. S1). Therefore, these two different groups of Δtau187 variants will be compared with care by only discussing stark contrast between early and late aggregation stages. ThT fluorescence was used to monitor the formation of β-sheet-rich aggregates, verifying that all five tau variants form β-sheets within hours of heparin-induced tau aggregation. Importantly, a combination of turbidimetry, TEM, and ThT fluorescence analysis verified that a series of spin dilution for a given spin-labeled tau mutant does not affect the global aggregation kinetics (Fig. S2A), and confirmed the formation of β-sheet fibrillar species within a few hours of aggregation (Fig. S2 B and C).
Fig. S1.
R1 spin-labeled Δtau187 at positions 313, 316, 322, 400, and 404 show variable kinetics of fibrillization that all lead to qualitative formation of mature fibrils. Time course of the increase in turbidity of five representative heparin-induced Δtau187 mutants, monitored by turbidimetry at 350 nm of 100 μM Δtau187 immediately after mixing with 25 μM heparin at 25 °C. Electron micrographs (TEM) to the right of each graph show mature fibrils derived from the respective spin-labeled variants of Δtau187, following 24 h after incubation with heparin. (Scale bar, 100 nm.)
Fig. S2.
(A) Spin-labeling has a negligible effect on the rate of fibrillization and formation of mature fibrils of Δtau187 variant (R1 label at position 322) following 24 h after incubation with heparin, confirmed by TEM and turbidimetry. (Scale bar, 100 nm.) (B) Comparison of turbidimetry and ThT fluorescence of Δtau187 with 322S (cysteineless) as a function of aggregation time upon heparin addition (protein:heparin = 4:1). (C) ThT staining fluorescence was used to monitor the β-sheet formation as a function of time following incubation with heparin for 20%, 50%, and 100% R1-labeled Δtau187 at position 322.
Morphologies and Accumulation of Aggregation Intermediates and Aggregates.
To observe the morphological evolution of aggregates and persistent aggregation intermediates of Δtau187 as a function of aggregation time, cryo-TEM was carried out at various aggregation times of the R1 spin-labeled 322C variant (Fig. 1B). The cryo-TEM images with different magnifications are presented in Fig. S3. We initiated the aggregation of the 322C variant with heparin, took aliquots of the aggregation solution at chosen aggregation time () after heparin addition (i.e., = 5 min, 15 min, 50 min, 120 min, and 24 h), and flash-froze them for cryo-TEM analysis. Samples collected at = 5 min, 15 min, and 50 min reveal sparsely distributed, nonfibrous, spherical assemblies with 10- to 20-nm diameters that persist during this aggregation period (stages I and II in Fig. 1B and Fig. S3). In contrast, the samples collected at = 120 min (stage III) begin to exhibit short, but clearly fibrillar, aggregates that coexist with the spherical assemblies. The growth of tau fibrils continues thereafter, and the amyloid fibrils appear mature at = 24 h. Such mature fibrils can span several micrometers in length, with, importantly, the spherical oligomers no longer visible at this stage (stage IV in Fig. 1B and Fig. S3). The proteinaceous content of the observed spherical and fibrillar species was validated by intentional overexposure to higher-intensity electron beams, and verified that their appearance is not due to contaminants, damage to the samples, or artifacts in the images. Our observations suggest that the Δtau187 monomer may initially assemble into stable spherical assemblies that prevail in solution for an extended period after aggregation (15−50 min, stage II). Their spherical shape suggests these to be protein droplets. Importantly, their eventual disappearance at stage IV together with the appearance of elongated fibrils imply these assemblies are intermediate states toward fibril formation. This leads us to hypothesize that structural rearrangements of Δtau187 are occurring within these assemblies to transition from spherical to fibrillar aggregates (i.e., stages I−III), eventually achieving the structural order found in mature fibrils after several hours of aggregation.
Fig. S3.
Cryo-TEM micrographs display the aggregation species of Δtau187 variant, R1 spin label at position 322, after 5 min, 15 min, 50 min, 120 min, and 24 h, following heparin addition. The different magnifications of the micrographs serve as a visual demonstration of either the relative population density (Top) and/or the relative size (Middle and Bottom) of the protein aggregates at the different time periods of aggregation. The black scale bars in the top row represents 100 nm length scale. The images in the middle and bottom row are 1.3× and 1.7× magnified images in the top row. Spherical aggregates were formed at 5 min through 50 min of aggregation. Fibrils together with spherical oligomers are seen after 120 min of aggregation. Mature fibrils are predominant after 24 h of aggregation.
To verify the accumulation of quaternary structures of R1 spin-labeled Δtau187 variants at early oligomeric states, chemical cross-linking experiments were performed using a water-soluble cross-linker, bis(sulfosuccinimidyl) suberate (BS3). Each subunit of oligomers at various aggregation times can be covalently cross-linked using BS3, allowing them to be analyzed on denaturing gels (i.e., SDS/PAGE), as shown in Fig. 1C. Using this approach, we observed that dimer, tetramer, and other high-order oligomers of R1-labeled 322C variants start to form as early as = 5 min (Fig. 1C), whereas such bands are absent in the control sample of Δtau187 without heparin addition. These results clearly prove the formation and existence of oligomers within 5–10 min of initiating aggregation, but do not offer quantitative insights into the population of Δtau187 oligomeric species. This is because the cross-linker may link oligomers or monomers by collision, yielding artificial higher-order aggregates (43), and the SDS/PAGE may break apart any Δtau187 aggregates that are not efficiently cross-linked into smaller oligomers and monomers. This could explain why a larger monomer population is still seen, whereas the oligomer population does not apparently increase at longer aggregation times. Still, the finding that higher-order oligomeric species do form at = 5–10 min presents a key insight to support the subsequent discussion of the ODNP and ESR data.
In summary, based on the presented data, we categorize tau aggregation into five stages to aid our further discussion, as schematically summarized in Fig. 1D. Stage 0 presents Δtau187 before the addition of heparin that have been confirmed to be predominantly monomers. Stage I represents early aggregation events at = 5–10 min, where spherical aggregates first appear in the cryo-TEM, and higher-order oligomers are present according to the cross-linking experiment. Stage III includes the intermediate aggregation events at = 100–200 min, which is also the stage where short fibrillar aggregates first appear. Stage II represents the mixed and transitional events between stage I and stage III. Finally, stage IV represents the late aggregation events at ≈ 14–24 h, where the majority of Δtau187 has transformed into mature fibrils.
Homogeneous Hydration Shell Around Δtau187 at Stage 0.
To examine the structural state of Δtau187 at stage 0, the site-specific hydration dynamics on the protein surface was measured by ODNP to extract the translational correlation time, τ, of water at five select sites of Δtau187 (18) (see SI Overview of the ODNP Technique). Notably, at stage 0, the disordered Δtau187 monomers are surrounded by a homogeneous hydration shell as represented with similar τ values in the range of 95–105 ps around all five R1-labeled tau sites (see Figs. 1E and 2A). The τ value of 95–105 ps corresponds to a threefold to fourfold retardation of surface water diffusivity compared with that of bulk water, and is characteristic of hydration water on solvent-exposed protein surfaces. This finding suggests that the third repeat region and C terminus of Δtau187 at stage 0 are solvent-exposed, without being a part of defined structure.
Fig. 2.
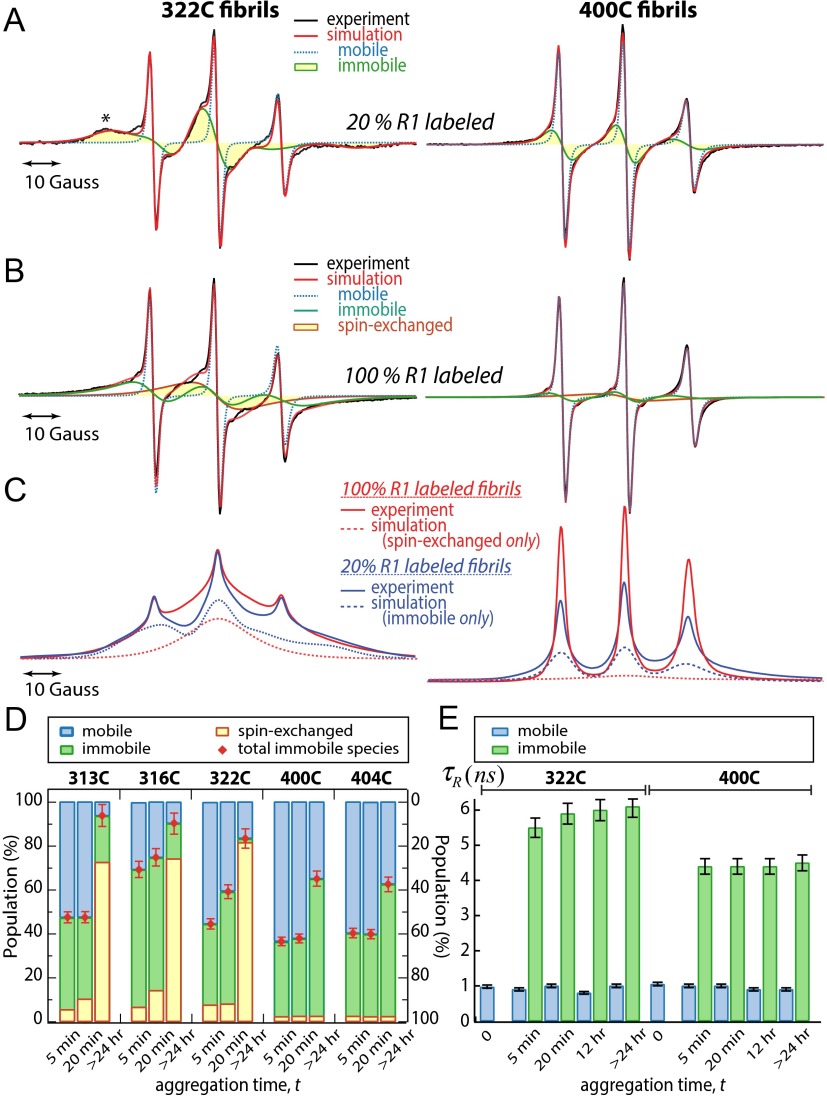
The site-specific changes of fast and slow dynamics of hydration water and side-chain mobilities accompany Δtau187 aggregation. The bar graphs of (A) τ, (B) cross-relaxivity parameter , and (C) self-relaxivity parameter of water for each R1 spin-labeled variant of Δtau187 at various aggregation times determined by ODNP. (D) The ESR peak amplitude of the resolved peak on the low-field resonance line at position * of 20% R1-labeled Δtau187 variants measured at various aggregation times. The error bars represent SD from two to three repeated measurements.
The contribution of water that is freely diffusing near the protein vs. bound to the protein surface was separately evaluated by a detailed analysis of the ODNP data to extract both the and relaxivities. These ODNP parameters have been introduced previously and applied to a membrane protein and DNA (17, 19, 21). The analysis of these ODNP-derived relaxivities is particularly informative to account for the contribution of tightly or loosely bound water in the heterogeneous environment of a structured protein (19), as described in SI Overview of the ODNP Technique and the literature (17, 21). Briefly, the cross-relaxivity depends on the fluctuations of interactions at the ESR frequency (9.8 GHz), and therefore reports on fast water dynamics on the picosecond timescale. On the other hand, the self-relaxivity contains the information from fluctuation at the NMR frequency (14.8 MHz), and is strongly weighted by the contribution of bound water moving on slower motional timescales on the order of a few nanoseconds. At stage 0, the value found to be on the order of ∼0.035 mM−1⋅s−1 (Fig. 2B), and the value is on the order of ∼0.2 mM−1⋅s−1 for all five R1-labeled Δtau187 sites (Fig. 2C). This observation of the highest value and the lowest value at stage 0 implies that the Δtau187 surface is surrounded by fast-diffusing water (according to ) and experiences minimal contribution from bound water (according to ) at stage 0. This is consistent with what is expected from an IDP with predominantly monomeric populations. These initial values serve as references for assessing changes in these parameters throughout the aggregation stages.
Structural Transformation of Δtau187 During Aggregation According to ODNP Analysis.
Next, we used ODNP to examine the structural transformation of Δtau187 by monitoring changes in site-specific hydration dynamics on the protein surface as a function of aggregation time. Fig. 1E exhibits the aggregation time dependence of the translational correlation time, τ, of surface hydration water around select residues of Δtau187, and the τ values at given aggregation stages are presented in Fig. 2A.
During stages I−III, the water diffusivity continuously slows around sites 313, 316, and 322 compared with that at stage 0, whereas there is negligible change around the sites 400 and 404 (Fig. 1E). This dispersion in τ values occurs immediately at stage I, which is the earliest detectable stage of aggregation. The time course of τ reveals rapidly and progressively diverging surface hydration dynamics around select residues in the third repeat domain, implying that structural transformations are occurring in the third repeat region but not in the C terminus. Specifically, the steepest increase in τ values (∼20−55%) are observed around residues 313, 316, and 322 during the earliest aggregation stages of I and II, corresponding to sites that are close to the β-sheet interface of tau fibrils (24, 29, 32, 42). This observation suggests that the majority of the early aggregation intermediates formed at stages I and II are already prestructuring toward fibril formation, as will be further discussed.
Toward the end of stage III, the τ values around sites 313, 316, and 322 approach steady state at stage IV. This finding suggest that the third repeat region of Δtau187 experiences a local environment at the end of stage III as found in mature fibrils in stage IV (Fig. 1E), whereas the elongated fibrils have just begun to form and evolve according to cryo-TEM (Fig. 1B). With this ODNP data alone, we cannot differentiate whether all populations of tau aggregates undergo a gradual structural transformation or whether multiple populations of tau aggregates with heterogeneous local environments around the third repeat region eventually converge to one dominant population. As will be discussed in Population and Structural Properties of the Evolving Aggregation Species According to cw ESR Analysis, quantitative ESR line shape analysis will reveal the latter to be the case. Crucially, the converging of the local structural transformation of the third repeat region toward equilibrium, as observed by ODNP, occurs at a stage when the transition from spherical oligomers to fibrillar aggregates or small fibrils is observed by cryo-TEM.
Population and Structural Properties of the Evolving Aggregation Species According to cw ESR Analysis.
To gain insight into the population and structural properties of the aggregated species that form as a function of aggregation time, including that of β-sheet structured species, we carried out quantitative ESR line shape analysis (see SI ESR Simulation). At stage 0, the ESR spectra of all tau variants displayed three sharp lines due to isotropic hyperfine coupling between the electron spin and the 14N nuclear spin of the nitroxide radical, yielding a rotational correlation time of R1, = 0.9 0.1 ns (Table S1). This value is consistent with what is expected to be IDPs, monomers, or small oligomers in solution (35). This will be referred to as the “mobile component” when we identify spectral components of a more complex ESR spectrum.
Table S1.
Calculated parameters for cw ESR spectral simulation of Δtau187 mutants contained 20% and 100% R1 spin labels at various aggregation time
| R1 spin label | Mutants | Aggregation time | , % | , ns | , % | , ns | , % | S |
| 20%* | 313C | 5 min | 52.9 | 0.9 | 47.1 | 5.4 | — | 0.54 |
| 20 min | 52.7 | 1.1 | 47.3 | 5.5 | — | 0.56 | ||
| 12 h | 37.1 | 1.0 | 62.9 | 5.6 | — | 0.67 | ||
| >24 h | 6.5 | 1.1 | 93.5 | 5.7 | — | 0.84 | ||
| 316C | 5 min | 30.7 | 0.9 | 69.3 | 5.4 | — | 0.61 | |
| 20 min | 25.5 | 1.0 | 74.5 | 5.7 | — | 0.66 | ||
| 12 h | 11.5 | 1.0 | 88.5 | 5.6 | — | 0.72 | ||
| >24 h | 9.9 | 1.0 | 90.1 | 5.7 | — | 0.77 | ||
| 322C | 0 min† | — | 0.9 | — | — | — | — | |
| 5 min | 55.7 | 1.0 | 44.3 | 5.5 | — | 0.59 | ||
| 20 min | 40.8 | 0.8 | 59.2 | 5.9 | — | 0.61 | ||
| 12 h | 21.3 | 1.0 | 78.7 | 6.0 | — | 0.63 | ||
| >24 h | 16.8 | 1.0 | 83.2 | 6.1 | — | 0.78 | ||
| 400C | 5 min | 63.7 | 1.0 | 36.3 | 4.4 | — | 0.58 | |
| 20 min | 62.4 | 1.0 | 37.6 | 4.4 | — | 0.56 | ||
| 12 h | 51.1 | 0.9 | 48.9 | 4.4 | — | 0.66 | ||
| >24 h | 35.2 | 0.9 | 64.8 | 4.5 | — | 0.70 | ||
| 404C | 5 min | 60.0 | 0.9 | 40.0 | 4.3 | — | 0.58 | |
| 20 min | 60.3 | 0.9 | 39.7 | 4.3 | — | 0.55 | ||
| 12 h | 39.9 | 0.8 | 60.1 | 4.4 | — | 0.68 | ||
| >24 h | 37.0 | 0.8 | 63.0 | 4.5 | — | 0.65 | ||
| 100%‡ | 313C | 5 min | 36.2 | 0.8 | 57.4 | 2.1 | 6.1 | 0.65 |
| 20 min | 38.0 | 0.8 | 51.6 | 2.2 | 10.4 | 0.62 | ||
| >24 h | 7.2 | 0.9 | 20.1 | 2.3 | 72.7 | 0.63 | ||
| 316C | 5 min | 35.6 | 1.1 | 57.7 | 2.1 | 6.7 | 0.61 | |
| 20 min | 35.4 | 1.0 | 50.3 | 2.1 | 14.3 | 0.66 | ||
| >24 h | 7.9 | 0.9 | 17.8 | 2.3 | 74.3 | 0.64 | ||
| 322C | 5 min | 37.4 | 0.8 | 55.0 | 2.2 | 7.6 | 0.63 | |
| 20 min | 37.3 | 0.9 | 54.5 | 2.2 | 8.2 | 0.66 | ||
| >24 h | 7.1 | 0.9 | 11.2 | 2.3 | 81.7 | 0.66 | ||
| 400C | 5 min | 52.1 | 1.0 | 46.1 | 2.0 | 1.8 | 0.64 | |
| 20 min | 50.2 | 1.1 | 47.4 | 2.0 | 2.4 | 0.62 | ||
| >24 h | 38.7 | 0.9 | 58.7 | 1.9 | 2.6 | 0.61 | ||
| 404C | 5 min | 50.4 | 1.0 | 47.4 | 2.0 | 2.2 | 0.61 | |
| 20 min | 50.7 | 0.9 | 47.4 | 2.0 | 1.9 | 0.62 | ||
| >24 h | 40.8 | 1.0 | 56.5 | 1.9 | 2.7 | 0.61 |
The order parameter S applies on and/or .
Two-component fit.
One-component fit.
Three-component fit.
Upon initiation of aggregation with heparin (stage I), the ESR spectra generally become broadened due to the superposition of several spectral components that can be deconvoluted by computer simulations. We found that the ESR spectra of the 20% R1-labeled tau samples can be reliably simulated with a two-component model (33, 34). Two representative spectra of 20% R1-labeled tau samples that are R1-labeled at position 322 and 400, respectively, are shown in Fig. 3A, with the experimental spectrum displayed in black and the simulated one displayed in red. The two spectral components derived from the spectral simulation are separately displayed and overlaid on the total spectrum in Fig. 3A: a “fast” component with = 1.0 0.1 ns that was assigned to mobile R1 (blue dotted line in Fig. 3A), i.e., freely rotating and facing the solvent, and a “slow” component with = 6.1 0.5 ns that was assigned to immobilized R1 (green line in Fig. 3A) embedded at a protein interface. Crucially, we found that only the population, not the value, of these two components changes with increasing aggregation time (Fig. 3E). It is unlikely that the origin of the slow spectral component is due to immobilization at an intratau fold as there is only a single slow value identified, whose population increases over the course of hours of aggregation. This characteristic is consistent with the slow value originating from immobilization of R1 at an intertau interface. Taken together, we can assign the two spectral components to two distinct physical states of Δtau187 as represented by the motional state of R1: mobile, solvent-exposed on monomers or small oligomers, and immobilized at an intertau interface. The results of the ESR spectral analysis, as summarized in Fig. 3D, revealed that as tau aggregation progresses with time, the population of the mobile component assigned to monomers or small oligomers decreases, and, concurrently, the population of the immobile component assigned to species embedded in oligomer and aggregate interfaces increases.
Fig. 3.
ESR line shape analysis at various Δtau187 sites at different aggregation stages revealed through spin dilution ESR experiments. (A) Representative ESR spectra of 20% R1-labeled Δtau187 fibrils at residues 322 and 400. The simulated ESR spectra display the presence of mobile and immobile components. The position of the characteristic resolved peak at low-field region is indicate as *. (B) Representative ESR spectra of 100% R1-labeled Δtau187 fibrils at residues 322 and 400. The simulated ESR spectra suggest the presence of mobile, immobile, and spin-exchanged, single-line components. (C) The comparison of absorption ESR spectra of 20% and 100% R1-labeled Δtau187 fibrils at residues 322 and 400. The simulated spectra contributed from spin exchange (dashed red) and immobilization (dashed blue) are separately presented. Spectral width is 100 G in all cases. (D) Population of each component extracted from ESR spectral analysis of Δtau187 variants at various aggregation time. (E) of mobile and immobile components in 20% R1-labeled Δtau187 at residues 322 and 400 at various aggregation times.
In contrast to the ESR line shape of 20% R1-labeled Δtau187 shown in Fig. 3A, the ESR spectral simulation of the 100% R1-labeled Δtau187 samples requires a three-component model: (i) a mobile component, (ii) an immobile component, and (iii) a spin-exchanged (single-line) component. Fig. 3B shows two representative experimental spectra (in black), overlaid with the simulated ESR spectra (in red) resulting from the three-component analysis of 100% R1-labeled tau samples at positions 322 and 400. As discussed in the Introduction, the single-line, spin-exchanged component can be reliably assigned to tau strands in a parallel β-sheet arrangement, where the respective R1-labeled sites on adjacent protein strains come within 5–8 Å of each other. The clear single-line feature of this component can be seen when displayed and highlighted separately in Fig. 3B (orange line, filled in yellow), and contrasted to the distinctly different immobile spectral component highlighted in Fig. 3A (green line, filled in yellow). In other words, the interpretation of the single-line component of the 100% R1-labeled Δtau187 sample is validated by its clear absence in the spectrum of the 20% R1-labeled, i.e., spin-diluted, Δtau187 sample. Consistently, the of the mobile component from the three-component analysis matches that of the mobile component from the two-component analysis. However, the value of the immobile component from the three-component analysis is not meaningful. This is because the contribution from the spin-exchanged components, that are by nature also immobilized, is not considered in the immobile component derived from the 100% R1-labeled sample, whereas it is included in that of the 20% R1-labeled sample. Therefore, the population of mobile and immobile components as summarized in Fig. 3D are derived from the two-component ESR spectral simulation and the population of the spin-exchanged component from the three-component ESR spectral simulation. All relevant parameters obtained from the two- and three-component spectral simulation can be found in Table S1.
It is important to note that the total observed population of R1-labeled Δtau187 remains constant as a function of aggregation time in all of the samples studied, as verified by a constant double integral for the first derivative ESR signal of the R1-labeled Δtau187 throughout the entire aggregation time. Thus, we can exclude the possibility that a decrease in total tau population, e.g., due to sedimentation of tau aggregates or the broadening of ESR signal, dominates the changes in the ESR or ODNP results.
Rapid Assembly of Δtau187 to Partially Structured Aggregation Intermediates at Stage I.
We concluded earlier, from ODNP analysis of surface hydration dynamics, that Δtau187 is undergoing structural transformation at early aggregation stages I−III toward fibrils formation, but direct information about the structural properties of the evolving aggregation species and their population was missing. With combined ODNP data and quantitative cw ESR interpretation at hand, we set out to more directly examine the nature of the aggregation intermediates that form in the earliest aggregation stage (stage I).
At stage I, there is minimal increase in the turbidity and fluorescence intensity upon ThT staining as part of a lag phase (Figs. S1 and S2B), although only small quantities of spherical oligomers start to appear according to cryo-TEM (Fig. 1B). In contrast, ODNP reports on significant retardation in local hydration dynamics around the third repeat domain of Δtau187 at stage I (Fig. 1E). Specifically, the values that account for contributions from bound water increase by about a factor of 1.5 around the third repeat residues (at 313, 316, and 322 sites) at stage I (Fig. 2C), whereas the values that report on diffusively mobile water decrease by 20–40% at stage I (Fig. 2B). No changes in and are observed around residues 400 and 404 at any aggregation stages.
Concurrently, a broadening of the ESR spectrum (Fig. S4A) of the 20% R1-labeled Δtau187 sample at position 322 is observed with increasing aggregation time, together with a concomitant increase of a characteristic low-field peak as marked with * in the first derivative ESR spectra shown in Fig. 3A and Fig. S4A. When evaluated against the simulated spectral components as overlaid on the total ESR spectrum in Fig. 3A, it becomes clear that this low-field peak qualitatively captures only the immobile spectral component (green line, filled in yellow), with minimal contributions from the mobile spectral component (blue dotted line). Notably, the peak amplitude at position * increases most dramatically at stages I and II, but only for the third repeat residues, not for the C-terminal sites, as summarized in Fig. 2D. Here, the two-component ESR spectral simulation revealed that the rotational correlation times of R1 in these two spectral components described by two distinct values do not change with aggregation time (Fig. 3E); only their populations change (Fig. 3D). It was found that the population of the immobile component that was attributed to R1 embedded at the oligomer and aggregate interface constitutes a majority (40–70%) of the total ensemble of R1 at stage I (t = 5 min, Fig. 3D), where it was nonexistent at stage 0. This analysis suggests that the ESR spectral broadening seen at stage I (Fig. S5) is due to population conversion from mobile to immobile component, not due to a gradual slowing of the mobile component along a continuum.
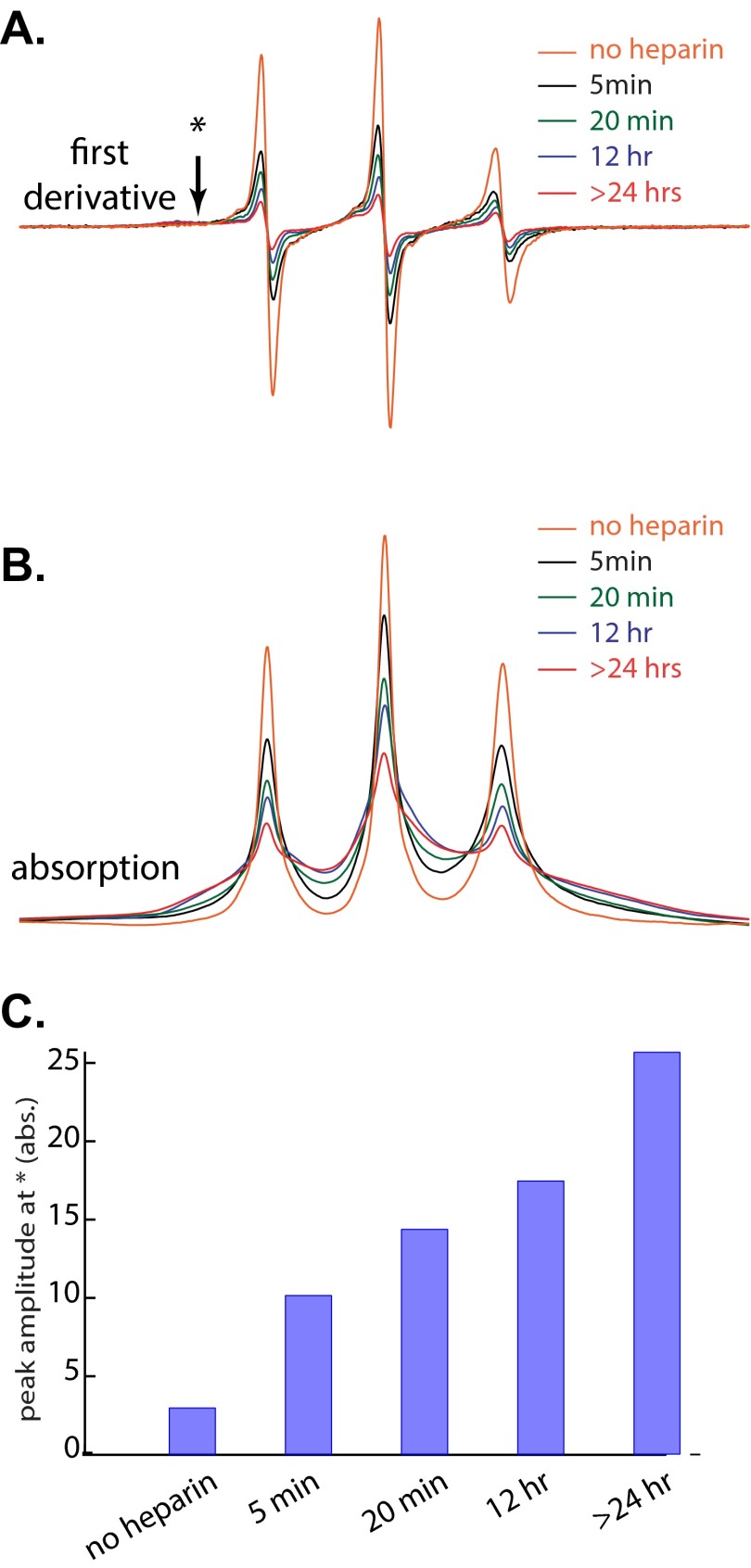
Fig. S4.
Cw ESR spectra in first derivative (A) and absorption (B) modes of 20% R1 spin-labeled Δtau187 (322C) at different aggregation states. (C) Peak amplitude of position * indicated in the first derivative spectra shown as a function of aggregation time. The spectra in A and B are normalized to their double integral values.
Fig. S5.
(A) The cw ESR spectra of 20% R1 labeled Δtau187 (322C) before and after heparin addition for 5 min. Experimental and simulated cw ESR spectrum of 20% R1-labeled Δtau187 (322C) before heparin addition (B) and after 5 min of heparin addition (C).
Taken together, we summarize that residues 313, 316, and 322 experience accumulation of bound water (increased value) and a reduced contribution of fast-diffusing water (decreased value) on the protein surfaces during tau aggregation. However, these changes do not occur at sites 400 and 404, suggesting that the C terminus stays fully solvent-exposed as fringes of the forming tau assemblies. In contrast, the population of these embedded tau species increases to 40–70% for all tau variants at stage I (Fig. 3D), with only small differences rendered for the τR values of the immobile component between the third repeat and C-terminal residues (Fig. 3E). This result can be reconciled with the formation of a dominant population of aggregation intermediates in the earliest aggregation stage (stage I) that is partially structured, judging by the distinct differences in surface water dynamics between the third repeat and C-terminal residues. In stages I and II, there are no fibrillar species visible, but a sustained population of spherical assemblies is observed by cryo-TEM (Fig. 1B).
In summary, these early aggregation intermediates constitute a major population of tau assemblies that are partially structured, with their site-specific hydration and protein dynamics, according to ODNP data (Fig. 2) and ESR spectral simulations, resembling those found in mature tau fibrils (Fig. 3 D and E). Our findings offer support for these intermediates to be on-pathway species toward fibril formation.
Structural Rearrangement as Reflected in the Accumulation of Bound Water at Stages I−IV.
With supporting data for the hypothesis at hand that the aggregation intermediates constitute a major population at stage I and are on pathway toward fibrils, we next examined the site-specific details of the structural rearrangement occurring throughout the stages I−IV by observing changes in the local water properties at selected sites of Δtau187 by ODNP analysis. Through stages I−IV, the values around the 313, 316, and 322 sites significantly decrease by about 75%, whereas the changes in around the 400 and 404 sites are negligible throughout all stages. When separately evaluating (Fig. 2B), we found that the trend in qualitatively correlates with the trend in 1/τ (Fig. 2A). In contrast, we observed that all three sites in the third repeat domain begin to accumulate bound water at stage I, as evaluated by increased value (Fig. 2C), whereas their values only slightly change further throughout stages II−IV. Again, the values around sites 400 and 404 remain unaltered throughout all stages. It is intriguing that the contribution from bound water () around site 313 initially increases from stage 0 to stage I, and subsequently gradually decreases from stage I through stage IV. Based on this observation, we speculate that bound water is first accumulated around the third repeat region upon forming aggregation intermediates at stage I, and then is expelled only from the interface near the 313 site that is closest to the hydrophobic PHF6 region in this study. This finding is in contrast to the continued increase of the value near sites 316 and 322, which are flanking a more hydrophilic/polar region of Δtau187.
We conclude that the formation of the dynamic aggregates at stage I is accompanied by the accumulation of bound water around the third repeat domain of Δtau187, followed by the expulsion of this bound water near (and likely of) the PHF6 region, and in an otherwise region-specific way as tau undergoes structural rearrangements throughout stages II−IV.
Structural Rearrangement to Form β-Sheets at Stages I−IV According to cw ESR Analysis.
Although we can infer structural rearrangements of Δtau187 within the aggregates formed during stages I−III from the results discussed so far, it is critical to concurrently obtain direct structural information. This is achieved with systematic ESR spectral analysis of 20% and 100% R1-labeled tau samples, with the details of the analysis method discussed in Population and Structural Properties of the Evolving Aggregation Species According to cw ESR Analysis. Specifically, we determined the β-sheet content from the population of the single-line, spin-exchanged component as derived from the three-component ESR analysis of 100% R1-labeled tau (see SI ESR Simulation). The assignment of spin-exchanged spectral features to parallel β-sheet arrangement, in which the R1-labeled sites of adjacent protein strands approach each other within 5–8 Å interspin label distance, has been established in the literature (29, 36–41), and, specifically, has been demonstrated on mature fibrils of full length tau (29). However, this method has not been used to track tau aggregation intermediates in situ and in solution as performed in this study.
The ESR spectral analysis, as summarized in Fig. 3D and Fig. S6, shows that the population of Δtau187 strands arranged in parallel β-sheets gradually increases throughout stages I−III. Notably, more than 5% of Δtau187 strands are arranged in β-sheets along sites 313, 316, and 322 at stage I, which content increases to more than 10% at stage II (t = 20 min), and eventually reaches 70% at the final stage IV (t > 24 h). In contrast, the spin-exchanged component at sites 400 and 404 is negligible throughout the aggregation stages. The finding of significant β-sheet arrangements of the third repeat domain in the earliest aggregation stage (stage I) is striking. It is also notable that the single-line feature representing parallel β-sheets is virtually absent at sites 400 and 404 (Fig. 3B), whereas the population of immobile R1 still increases from 0% to 40% and up to 60% around these C-terminal sites throughout the aggregation stages (Fig. 3D). This result not only confirms that the C terminus stays disordered and flexible throughout the aggregation stages but also supports a one-dimensional growth pattern of tau aggregates along the fibril axis at the earliest to latest aggregation stages. Clearly, the early aggregation intermediates formed at stage I are not globally collapsed aggregates but are partially structured species. In summary, 40–70% of the total tau population is embedded in oligomers and aggregation intermediates at stage I, and their population gradually reaches a plateau value of 80–90% at stage IV. Meanwhile, 5–10% of the total tau population arranges in β-sheets at stages I and II and eventually reaches 70–80% at stage IV, but only around sites 313, 316, and 322 (Fig. 3D). These observations suggest that the majority of aggregation intermediate species formed at stage I are transforming relatively efficiently into parallel β-sheet structures, and thus offer important support for the earlier posed model that the majority of aggregation intermediates formed at stage I are on pathway toward fibril formation.
Fig. S6.
Parallel stacking of Δtau187 regions at different aggregation stages revealed through spin dilution ESR experiments. Representative ESR spectra of Δtau187 variant 316 display the presence of exchange-narrowed component in 100% R1-labeled (black) mature fibrils, in contrast to the clearly resolved immobile component (arrows) in spin-diluted (red) 20% R1-labeled tau fibrils. The parallel stacking of tau proteins in mature fibrils is based on the presence of the exchange-broadened component in 100% R1-labeled and the resolved immobile spectral component in 20% R1 spin-diluted tau fibrils. Experimental (black) and simulated (red) cw ESR spectra of Δtau187 mutants contained 20% R1 (A) and 100% R1 (B) spin labels at various aggregation times. The cw ESR spectra were fit using the two-component fit for A and the three-component fit for B.
Mechanism of Fibril Formation and Elongation.
The end of the structural transformations observed by ODNP at stage III marks the time when elongated fibrils begin to appear according to cryo-TEM, but also where the majority of the tau proteins are already embedded in a parallel β-sheet arrangement involving their third repeat domain residues according to in situ ESR line shape analysis. Between stages III and IV, cryo-TEM analysis shows that the fibrils of Δtau187 are maturing and elongating (Fig. 1B). To better understand the seeding and growth mechanism of Δtau187 fibrils, it is important to further explore whether the main mode of fibril elongation is by block-wise addition of β-sheet structured intermediates and fibrils or by the addition of monomers that are breakdown products of oligomers and aggregates. We investigated this question by examining the propensity for monomer or subunit exchange, and hence stability, of the β-sheet structured species.
To identify how oligomers assemble into mature fibrils in solution, we used quantitative ESR spectral analysis of the mature fibrils formed from mixtures of R1 and R1’ spin-labeled Δtau187 322C variant. As a control, we performed turbidimetry on 0, 20%, 50%, and 100% R1-labeled Δtau187 (322C), and found that their aggregation kinetics are comparable (Fig. S2A), suggesting that mixing different ratio of R1- and R1’-labeled Δtau187 does not affect the aggregation kinetics. The experimental design is illustrated in Fig. 4A. First, we separately initiated the aggregation of a 100% R1-labeled and 100% R1’-labeled monomeric Δtau187 variant (322C) by the addition of heparin and incubation of these two samples with heparin for a set amount of time, Δtinc, ranging from 5 min to 15 h (stages I−IV). We then mixed aliquots of these R1- and R1’-labeled tau samples in a 1:4 ratio (by volume) after a series of Δtinc. These mixtures that contain 20% R1 spin-labeled and 80% R1’-labeled tau aggregates were further incubated for 24 h to ensure quantitative fibril formation. We then acquired ESR spectra of the samples that contained these final aggregated products. The goal of this experiment was to determine the monomer or subunit exchange propensity of the β-sheet structured species formed at the given incubation time Δtinc. If the oligomers and β-sheet structured species rapidly exchange monomers at the stage marked by Δtinc, the final fibrils grown from this mixture after 24 h will be effectively spin-diluted, thus resulting in a resolved hyperfine peak in the low-field region at position *, as illustrated with an exemplary ESR spectrum shown in Fig. 4B of 20% R1-labeled, i.e., spin-diluted, Δtau187 after 24 h of aggregation. In this case, there will be a minimal contribution from spin exchange interaction in the corresponding ESR spectrum. Thus, if the monomer exchange is prevalent and the final aggregates are effectively spin-diluted, the result would be characterized by a two-component model, as was the case with the ESR spectrum of 20% R1-labeled Δtau187 fibrils at position 322 (Fig. 3A). However, if the exchange of monomers or subunits between the β-sheet structured species is slowed or diminished at Δtinc, the final fibrils grown from these mixtures are not effectively spin-diluted (Fig. 4A). As a result, the spin-exchanged component will be apparent in the resulting spectrum, similar to that represented by the ESR spectrum of 100% R1-labeled fibrils at position 322 in Fig. 3B. Furthermore, the resolved hyperfine peak in the low-field region at position * would be broadened out, as illustrated with an exemplary ESR spectrum of 50% and 100% R1-labeled Δtau187 (322C) after 24 h of aggregation shown in Fig. 4B.
Fig. 4.
Structural transformation of the early tau aggregates studied by tracking the exchange of tau within aggregated subunits via ESR spin dilution. (A) Experimental design. We prepared two sets of undiluted (100%) R1- and R1’-labeled Δtau187 (322C) samples, and separately initiated their aggregations with heparin. Both sets of samples were allowed a given incubation time, Δtinc, to aggregate. After Δtinc, both samples were mixed in a 1:4 ratio of R1-:R1’-labeled Δtau187 and continuously aggregated for an additional 24 h. The resultant mixed samples were analyzed by ESR spectroscopy. (B) ESR spectra of mature tau fibrils, composed of 20%, 50%, or 100% R1-labeled Δtau187 (322C) variant, representing the possible outcomes depending on the percentage of dilution of the R1-labeled Δtau187 in individual aggregates. (C) ESR spectra of the mixed R1- and R1’-labeled Δtau187 (322C) fibrils at various incubation times, Δtinc, measured 24 h after mixing. Noted the decrease in the peak amplitude at position * of resolved hyperfine components of the ESR spectra as Δtinc increased. Spectral width is 100 G in all cases. (D) Population of each component extracted from ESR spectral analysis of R1-labeled Δtau187 (322C) fibrils at various incubation times, Δtinc. (E) The of mobile and immobile components in R1-labeled Δtau187 (322C) fibrils at various incubation times, Δtinc.
The resulting ESR spectra of this experiment are presented in Fig. 4C, and their spectral analyses are summarized in Fig. 4 D and E. The data from the 100% R1-labeled Δtau187 at stage IV and 20% R1-labeled Δtau187 at stage IV mixed at Δtinc = 0 serve as references for maximally vs. minimally spin-exchanged samples. Fig. 4C reveals that the resolved hyperfine peak in the low-field region at position * gradually decreases with increasing Δtinc, suggesting that the majority of aggregated species are not exchanging monomers during tau aggregation. A more detailed analysis of the growing population of nonexchanging β-sheet structured species was obtained by determining the population of the single-line, spin-exchanged, ESR spectral component that remains after mixing, as illustrated in Fig. 4A, to generate an aggregated sample at stage IV that nominally contains 20% R1-labeled Δtau187. This information was obtained from a three-component ESR spectral analysis of the spectra as presented in Fig. 4C and Fig. S7, with the results summarized in Fig. 4D and Table S2. Moreover, when comparing the population of the immobile component in the ESR spectra of the final tau fibrils formed at various Δtinc, all samples consistently revealed a comparable population of ∼90% (red diamond, Fig. 4D). Most notably, we observed that the population of the spin-exchanged component grows from 5% to 10% at stage I, to 20–40% at stage II, to 50–70% at stage III, and eventually approaches ∼80% at stage IV. It becomes clear that these populations, obtained from Δtau187 fibrils formed after mixing and spin dilution at Δtinc (shown in Fig. 4D), are comparable to or higher than that obtained from the ESR spectra of Δtau187 aggregated in situ after a similar aggregation time as Δtinc (Fig. 3D). These results suggest that parallel β-sheet structured species, once formed, do not readily exchange monomeric tau units. Crucially, the β-sheet population is not only comparable between the in situ aggregated Δtau187 samples (Fig. 3D) and the aggregated Δtau187 samples after spin dilution with species formed at stage III but is also comparable with species of earlier aggregation stages, including stage I (Fig. 4D). The β-sheet population at t = 5 min (stage I) involving the 322 site in the third repeat domain is a little over 5%, which stunningly remains at a similar value of ∼5% in the aggregates formed by 20% R1-labeled Δtau187, mixed at Δtinc = 5 min after heparin addition, i.e., at stage I (Fig. 3D). Similar trends can be extracted from the population of parallel β-sheet structures formed at t = 20 min to t > 24 h after aggregation. Looking at these results from a different angle, the majority of the partially structured aggregation intermediates at sites 313, 316, and 322 that are not yet embedded in stable parallel β-sheet arrangements still constitute 40–60% of all tau strands at stages I and II and are dynamic species, in the sense that these sites are immobilized at an intertau interface, with strands likely already prearranged but not yet locked in a stably structured arrangement, as schematically illustrated in Fig. 5.
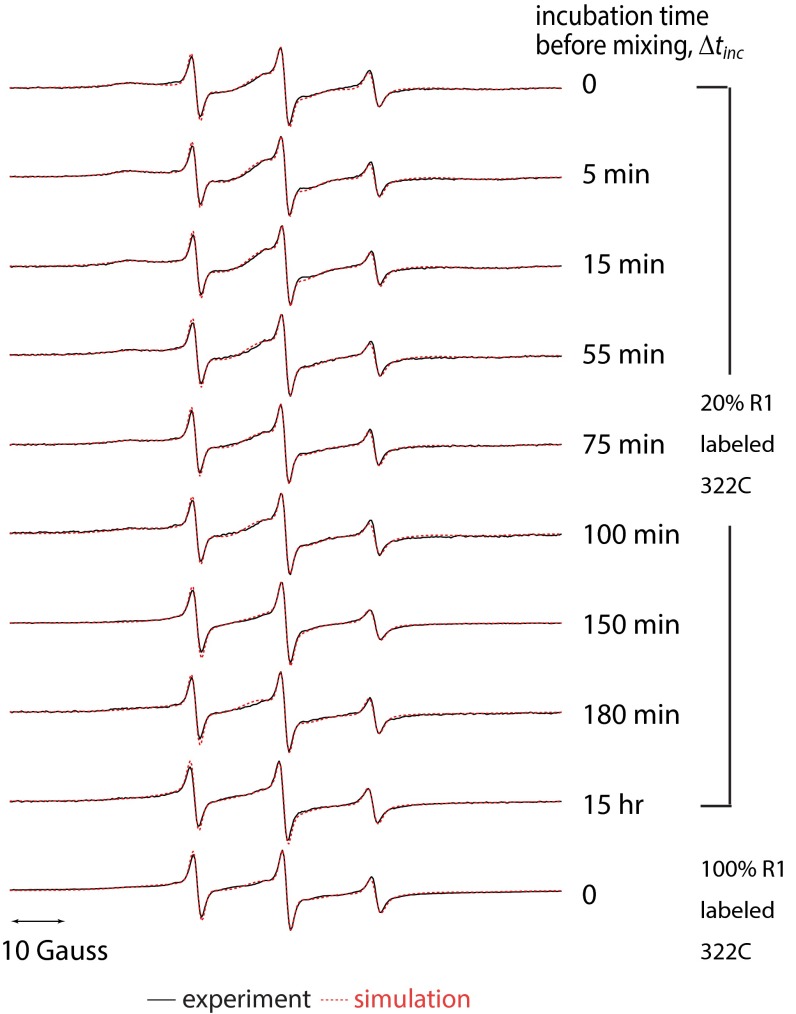
Fig. S7.
Experimental (black) and simulated (red) cw ESR spectra of Δtau187 322C mutants contained 20% R1 spin labels at various incubation times for spin dilution experiments in Fig. 4. The cw ESR spectra were fit using the three-component model.
Table S2.
Calculated parameters for cw ESR spectral simulation of Δtau187 (322C) contained 20% R1 spin labels for spin dilution experiments using the three-component fit presented in Fig. 4 C and D
| R1 spin-labeled content | Aggregation time | , % | , ns | , % | , ns | , % | S |
| 20% | 0 min | 10.1 | 1.1 | 87.5 | 3.6 | 2.4 | 0.64 |
| 5 min | 9.5 | 0.9 | 82.7 | 3.8 | 7.8 | 0.62 | |
| 15 min | 7.4 | 1.0 | 75.4 | 3.4 | 17.2 | 0.65 | |
| 55 min | 8.7 | 1.0 | 65.9 | 3.6 | 25.4 | 0.66 | |
| 75 min | 9.4 | 0.9 | 57.8 | 3.5 | 32.8 | 0.62 | |
| 100 min | 8.5 | 0.8 | 50.6 | 2.9 | 40.9 | 0.65 | |
| 150 min | 8.7 | 0.8 | 34.5 | 2.8 | 56.8 | 0.63 | |
| 180 min | 8.3 | 0.9 | 18.6 | 2.7 | 73.1 | 0.61 | |
| 14 h | 7.9 | 0.9 | 15.8 | 2.6 | 76.5 | 0.60 | |
| 100% | 0 min | 7.1 | 0.9 | 11.2 | 2.3 | 81.7 | 0.66 |
The order parameter S applies on and/or .
Fig. 5.
Mechanism of tau aggregation based on our experimental results.
The fibrils formed from aggregated mixtures at stage III (Δtinc = 180 min) resulted in the ESR spectra with the maximum signatures of spin-exchanged component (73%). This shows that 70–80% of all Δtau187 species are embedded in parallel β-sheets, stacked along the site of 322 that is R1- or R1’-labeled, whereas they undergo minimal exchange of monomers between their R1- and R1’-labeled Δtau187 aggregates. However, we know from cryo-TEM data and literature reports (27) that Δtau187 fibrils mature and elongate throughout stages III and IV. Thus, the β-sheet structured aggregates that include 70–80% of all Δtau187 species at stage III must be elongating predominantly by a block-wise addition to form mature fibrils at stage IV, as illustrated in Fig. 5. Although it is still possible that monomer addition to the growing aggregated species contribute to tau aggregation, we claim, based on our experimental observation, that the stacking of fibrillar and/or oligomeric blocks is the major contributing mechanism of tau fibril elongation.
Discussion and Conclusion
Overall, we postulated several key hypotheses and questions in the Introduction, each of which we have addressed experimentally, as summarized here and illustrated in Fig. 5. The core finding of our study is that early structural changes and the formation of dynamic aggregation intermediates are dominant events involving the majority of the tau population within what is conventionally regarded as the lag phase of tau aggregation. Following the rapid assemblies of dynamic and partially structured aggregation intermediates, they gradually, in a rate-limiting process, transform into fibrils with parallel β-sheet structured order.
Although our finding that quantitative conversion into parallel β-sheet structured fibrils is a rate-limiting process and plateaus at the later aggregation stages is consistent with studies of amyloid-β, tau, and other fibrillizing proteins, there are diverging results and opposite views in the literature regarding the structural order and the role of the aggregation intermediates. We found that the β-sheet content of Δtau187 is >5% at stage I, rapidly increases to 20–40% at stage II, and reaches 50–70% at stage III, suggesting that the aggregation intermediates constituting the majority species already at stage I are on pathway to converting to fibrils. This model of direct conversion and growth is further supported by the finding that the β-sheet structured species, once formed, are not significantly exchanging their subunits, and thus maintain their stacking order. Remarkably, this applies to all β-sheet structured species, even to the 5–10% population formed at stage I. It follows that the nucleation of aggregation may be implied in the earliest events, even preceding the formation of the dynamic aggregation intermediates, which therapeutic efforts to prevent fibril formation will have to focus on. This is supported by our observation that the parallel β-sheet structured species are stable entities that can elongate to form fibrils, but do not effectively exchange monomers throughout the entire aggregation process, suggesting that these intermediates may not be effective vehicles to recruit more tau monomers for aggregation.
What is the driver and nucleus of the aggregation then? In order for monomeric Δtau187 to stay soluble in the absence of heparin, the hydrophobic region, such as the PHF6 hexapeptide segment, must be initially shielded from the bulk solvent. The wetting of this hydrophobic segment is enabled by the stable hydration shell of neighboring polar or charged residues that bridge or encompass this hydrophobic segment as part of a specific or partially structured protein assembly (44, 45). Perturbing the protein conformation, altering the effective local charges by mutation, or the addition of strongly adsorbing counter ions can weaken this hydration shell that protects an initially stable state of Δtau187, subsequently altering the balance to favor protein−protein over protein−water interactions. Such changes will facilitate the approach of the initially shielded hydrophobic hexapeptide segments toward other hexapeptide segments of adjacent tau strands. We, indeed, experimentally observed the dewetting around site 313 that is closest to the hydrophobic hexapeptide PHF6 in this study, as signified by an initial increase of at stage I, followed by a marked decrease of at stage II (Fig. 2C). This is consistent with the expectation that this stretch including site 313 gets involved in β-sheet packing, whose formation must be necessarily preceded by the dewetting of this interface. The hypothesis that the dewetting of hydrophobic segments (44) of unfolded proteins facilitates protein aggregation is prevalent. However, the direct experimental observation of segment-specific dewetting, i.e., shedding of bound water, as observed here around site 313 by decreasing values is unprecedented. Crucially, our experimental approach to measure the site-specific local diffusion dynamics of hydration water and spin label dynamics at the protein interface sheds light on the mechanism of protein aggregation, at stages well before where the formation of stable and elongated β-sheet structured fibrils become visible to conventional characterization techniques. Detailed future studies are needed to investigate the local changes involving the hydrophobic regions of tau, the timescale and extent of dewetting, and, more broadly, the nature and role of the earliest conformational or structural changes of Δtau187 that precede the formation of the dynamic aggregation intermediates that mark stage I.
Materials and Methods
Preparation of Spin-Labeled Δtau187.
The cloning, expression, and purification of Δtau187 have been previously described (25). The single-cysteine variants 313C, 316C, 322C, 400C, and 404C were engineered into Δtau187 by first replacing two native cysteine residues by serines (291S/322S). Proteins were expressed in Escherichia coli BL21 (DE3) cells and purified using Ni-affinity chromatography. Purified protein samples were resuspended in assay buffer (20 mM sodium phosphate pH 7, 100 mM NaCl, 0.1 mM EDTA). Spin-labeled variants were prepared by incubation of a 10-fold molar excess of paramagnetic (1-oxyl-2,2,5,5-tetramethylpyrrolidin-3-yl) methyl methanethiosulfonate MTSL (R1) spin label (Toronto Research Chemicals) or diamagnetic analog (1-Acetyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl methanethiosulfonate (dMTSL, R1’) (Toronto Research Chemicals) with the single-cysteine Δtau187 variants. The detailed procedures of protein preparation and spin labeling are given in Supporting Information.
X-Band cw ESR and ODNP 1H NMR Spectroscopy.
ESR spectra were acquired at room temperature using a Bruker EMX X-band (9.8 GHz) spectrometer equipped with a dielectric cavity (model ER 4123D). The ESR and ODNP 1H NMR spectra were acquired on protein samples of 3.5 μL volume placed into a quartz capillary of 0.6 mm inner diameter (VitroCom). For ODNP, the sample was secured inside a homebuilt U-shaped NMR coil, which was then mounted in the ESR cavity. ESR spectra were acquired using signal averaging of ∼6–16 scans over a field sweep of 150–200 G with a microwave power of >18 mW and modulation amplitudes of 0.25–0.35 G. Δtau187 concentrations for all ESR and ODNP experiments were 500 ± 100 μM, with the concentration of heparin (18 kDa; Sigma Aldrich) adjusted accordingly to produce a 6:1 protein:heparin molar ratio to induce the aggregation reaction.
During ODNP, the sample was continuously irradiated at ESR frequency using a homebuilt 8- to 10-GHz microwave amplifier and 1H NMR signal recorded as a function of microwave power. The sample was continuously cooled down throughout the ODNP experiment by flowing a dry gas over the sample. T1 and T10 measurements were conducted using a typical inversion-recovery pulse sequence operated by a Bruker Avance spectrometer and a 0.35 T electromagnet. The data analysis of , , and values is summarized in Supporting Information. Additional experimental details, including ODNP, ESR, TEM, cross-linking experiment, turbidimetry, and fluorescence spectroscopy, are summarized in Supporting Information.
SI Overview of the ODNP Technique
Experimentally and directly probing the site-specific hydration dynamics around proteins in solution and under ambient conditions has proven to be challenging. The emerging technique of ODNP relaxometry enables the direct measurement of translational diffusivity of hydration water that rapidly moves with correlation times on the order of tens of picoseconds to subnanoseconds relative to the precession frequency of an electron spin, tethered to a specific site of the protein (16, 17). This method relies on the selective amplification of the 1H NMR signal of the local water within 5–10 Å proximity of the spin-labeled protein sites (16). This selectivity arises from the efficiency of polarization transfer from the unpaired electron of the spin label to the water protons, mediated through the electron−1H dipolar cross-relaxation that critically relies on the presence of water dynamics with timescales close to or faster than the reciprocal of the electron spin precession frequency, here 1/9.8 GHz ≈10 ps at the magnetic field of 0.35 tesla. The translational diffusion coefficient of surface water, , that moves within a closest distance, b, from the spin label near the protein surfaces with a correlation time, , between the water proton and the electron of the spin label satisfies , if random Brownian motion can be assumed within the relevant short length and timescales (17).
Briefly, the efficiency of coupling between the electron spin and the water proton can be expressed by the coupling factor, , which contains key information about hydration dynamics and is defined by two fundamental proton relaxivity parameters, . Here, represents the paramagnetic contributions to the relaxation rates of water driven by dipolar interaction with the spin label, , which can be determined by measuring the spin lattice relaxation time of water proton in the presence, , and absence, , of the paramagnetic spin label at a known spin-labeled concentration, (17). Given that includes the dipolar self-relaxation efficiency of water protons at 15 MHz, the contribution of tightly bound water near the spin label around the protein surface, moving with a few nanoseconds or longer correlation times, is strongly weighted. On the other hand, captures the cross-relaxation between the electron and 1H spins driven by electron spin flip excitation at ∼10 GHz, and thus is exclusively sensitive to characteristics of fast-moving, loosely bound, surface water with tens of picoseconds to subnanoseconds correlation times with respect to the spin label. The value for can be experimentally obtained by measuring the maximum NMR signal enhancement extrapolated to infinite microwave power (Emax) and value, following .
From determined by the ratio of and , the translational correlation time, , can be extracted, with methods described elsewhere that rely on assumptions proven to be valid for spin label–1H water interactions that are dominated by random diffusion processes of free or loosely bound water molecules. For nitroxide radicals free in water, as well as nitroxide radicals tethered on the macromolecular surfaces (e.g., protein or liposome), the force-free hard-sphere model has been proved to give a good fit to the field-cycling relaxometry data, and can be used to model the spectral density function. This method becomes convenient when the translational diffusion is the dominant modulator of the electron spin-mediated 1H nuclear spin relaxation that is driven via dipolar coupling between electron and 1H spins. Therefore, this model can be used in our systems, as free diffusing water interacts with radicals tethered on the protein surfaces. In this case, the coupling factor is given by the following spectral density function:
| [S1] |
with the following spectral density function
| [S2] |
More theoretical description and data analysis of the ODNP method can be found elsewhere (16, 17).
Specifically, a monomeric tau free in solution is highly soluble and disordered under native conditions (24), and its hydration shell, constituting two to four layers of water (5−10 Å), evenly surrounds the entire protein surface. The water within these hydration layers, often termed “biological water,” is physically different from that in the bulk water, and typically displays a twofold to fourfold retarded (local) diffusion coefficient compared with that of bulk water (i.e., 2.3 × 10−9 m2/s at 25 °C) (16). This translates into an additional activation energy barrier for water diffusion of 0.7- to 1.6-fold of the thermal energy, kT, compared with that in bulk water. Similar to what has been observed in protein folding, and protein−membrane interactions, when protein−protein interaction is favored over protein−water interaction, the water structure and dynamics at the protein interface become significantly modulated (44). Specifically, the interaction between hydration water and specific protein segments at the interprotein contacts may be initially weakened, which lowers the energetic barrier to rearrange or expel the repulsive hydration water at the interfaces (44). We propose that this is directly reflected in an initially increased diffusivity of the interfacial surface water, and a favoring of protein−protein over protein−solvent interactions. Once the interprotein contacts have begun to form, as part of a dimer or high-order oligomer arrangement, the role of water in mediating the close-up molecular interactions may become much more complex (10, 44), and likely quite specific to the particular protein under study. In this work, we provide experimental evidence of oligomeric species in the early events in the aggregation process of Δtau187 and its assembly mechanism under ambient solution conditions. The focus of this study is to elucidate, in situ, the early aggregation pathway by which this dramatic structural transformation occurs in solution.
SI ESR Simulation
The cw ESR spectra recorded in this work were analyzed by single- or multiple-component ESR spectral simulations. The simulations were performed using the MultiComponent program developed by Christian Altenbach (University of California, Los Angeles), where a microscopic order macroscopic disorder (MOMD) model was used as previously described for the anisotropic motion of the nitroxide radical (33, 34). A least-squares fit of the user-defined spectral parameters was performed using the Levenberg−Marquardt algorithm (33). For all ESR spectral fits in this work, the values of the magnetic tensors A and g were used as constraints, which were previously determined for R1 spin label in protein systems (35). These values are Axx = 6.2 G, Ayy = 5.9 G, Azz = 37.0 G, and gxx = 2.0078, gyy = 2.0058, and gzz = 2.0022.
A detailed description of the MOMD model can be found in the literature. Briefly, in the MOMD model, three coordinate frames are used to represent the internal motion of the nitroxide in the protein with respect to a fixed macroscopic director. In the molecular-fixed magnetic tensor frame (XM, YM, ZM), ZM lies along the 2p orbital of nitroxide, XM lies along the nitroxide NO bond axis, and YM is selected for a right-handed coordinate system, calculated as ZM XM. This molecular frame is the principle frame for the nitroxide A and g tensors. The second coordinate frame is the rotation diffusion tensor frame (XR, YR, ZR). The orientations of these axes with respect to the magnetic frame are specified by three Euler angles , , and . The rate of nitroxide motion around each of these axes and their orientation with respect to the molecular frame indicated with spin label side chain, R1, depend critically on the hydrodynamic properties, geometry, and protein backbones, and they contribute to motional averaging in the EPR spectrum. Moreover, anisotropic motion restrictions are simulated in the MOMD model by introducing a restoring (ordering) potential that constrains the diffusion about ZR within a core defined by the angle between ZR and the symmetry axis of the potential. The fitted EPR spectra of spin-labeled proteins use the simple restoring potential , where is a scaling coefficient. This symmetry axis of the potential defines the z axis of the third coordinate frame, the director axis (ZD). The resorting potential results in an anisotropic motional and can be characterized by the order parameter , where the brackets indicate the spatial averaging.
In the rotational diffusion tensor, Rx, Ry, and Rz are the principal components in Cartesian coordinate in a modified spherical form as . In this study, the mobile component of the spin-labeled protein site at room temperature was assumed to have isotropic motion with , whereas the immobile and spin-exchanged components were assumed to have axially symmetric anisotropic motion with tilt angles of the diffusion tensor 0°, 36°, and 0° and subject to an ordering potential described by order parameter S (20). The number of fit parameters was kept at a minimum, which includes population for each component (p), rotational diffusion constant (R), and order parameter (S). Rotational correlation time () was calculated using . The fit parameters used in the simulation are summarized in Table S1.
The best single-component fit was obtained from the ESR spectra in tau monomer (at stage 0 without heparin addition) presented here, assuming rotational diffusion without a restoring potential. Therefore, the ESR spectra of monomers were fit without a restoring potential to reduce the number of fitting parameters. The overlaps between multiple components of simulated ESR spectra may introduce significant uncertainty for each fit parameter in the fitting process. To address this issue, the single-component ESR spectrum of monomer tau in the absence of heparin was initially fit, and the parameter of rotational correlation rate R from this fit was used as the initial input parameter for the mobile component during the subsequent two- or three-component fits. For the three-component fit, we included Heisenberg exchange frequency () of 140 MHz in the third component (i.e., single-line, spin-exchanged component), and the parameters p, R, and S were allowed to vary. Notably, to see the single-line, spin-exchanged component in the simulated ESR spectrum, the exchange frequency must be greater than the hyperfine interaction, which means the exchange frequency needs to be >100 MHz. To minimize the number of fit parameters, we empirically optimized and used a fix value of = 140 MHz in our simulation. We found that a three-component model using five fit parameters (i.e., , , and S, including a constraint of ) to fully R1-labeled samples gives a better fit than using the two-component model. To fine-tune the fit parameters, if necessary, the p or R value in the immobile and/or spin-exchanged component can be varied again, and this process can be iteratively repeated until a satisfactory fit is obtained. Once these parameters were optimized and a good fit was established, the Gaussian inhomogeneous broadening, initially set to 0 G, was allowed to vary slightly to obtain the best fit. The quality of fit was visually evaluated using the difference between the experimental and theoretical spectra.
A two-component analysis yields a mobile and immobile component, whereas a three-component analysis yields a mobile, spin-exchange, and residual immobile component. Although the population of the spin-exchanged component can be accurately determined from the three-component fit (p3), the population (p2), correlation time (), and order parameters (S) for the immobile components have to be extracted from the two-component fit. This is because these parameters include the contribution of the population of the spin-exchanged component, which is a subpopulation of the immobilized component. Thus, we set as boundary conditions that the rotational correlation time () of the spin label and order parameter (S) for the spin-exchanged components (from the three-component analysis) be the same as that of the immobile component from the two-component analysis. This is a logical boundary condition because the spin-exchanged component would be, by definition, at an interface and immobilized; therefore, both components are set to have the same and S values. The order parameter applies to p2 and p3, as indicated in the captions of Tables S1 and S2. The fit parameters for the ESR simulation are summarized in Tables S1 and S2, and have been described above.
SI Materials and Methods
Cloning, Expression, and Purification.
The cloning, expression, and purification of Δtau187 have been previously described (18, 25). The single-cysteine variants 313C, 316C, 322C, 400C, and 404C of Δtau187 were generated using QuikChange mutagenesis kit (Stratagene). E. coli BL21 (DE3) cells previously transfected with each variant were cultured from a frozen glycerol stock overnight in 10 mL luria broth (LB). This culture was used to inoculate 1 L of fresh LB and grown at 37 °C with shaking. At OD600 optical density of 0.6–0.8, Δtau187 variant expression was induced by incubation with 1 mM isopropylß-d-thiogalactoside (MP Biomedicals) for 2–3 h. Cells were harvested by centrifugation for 20 min at 4500 × g (Beckman J-10; Beckman Instruments, Inc.), and the pellets were stored at −80 °C until further use.
Cells were resuspended in 20 mL lysis buffer (20 mM sodium phosphate, pH 7.4, 500 mM NaCl, 10 mM imidizole, 0.1 mM EDTA, and 1 mM DTT), supplemented with PMSF (1 mM), leupeptin (0.5 μg/mL), aprotinin (2 μg/mL), and pepstatin A (0.7 μg/mL) to inhibit proteases. After incubation for 30 min on ice with lysozyme (2 mg/mL), DNase (20 μg/mL), and MgCl2 (10 mM), the suspensions were sonicated using a probe sonicator (Branson Sonifier 250; Branson Ultrasonic Co.) for 2 min with 50% duty cycle and five-output control. The lysate was cleared by centrifugation at 13,000 × g for 30 min. After addition of PMSF to a final concentration of 1 mM, the cleared lysate was heated for 15 min in a 65 °C water bath to denature bacterial proteins. After adding another 1 mM PMSF, the lysate was cooled and cleared again by centrifugation, as described above. The supernatant was then loaded onto Ni-NTA agarose resin (Qiagen) preequilibrated with lysis buffer for 1 h at 4 °C. The loaded column was washed with 20–100 mL of lysis buffer followed by 40–100 mL lysis buffer containing 1 M NaCl to remove contaminating nucleotides and nonspecific binding proteins. Finally, the column was washed again with lysis buffer, and subsequently Δtau187 was eluted with lysis buffer containing 300 mM imidazole. Elution fractions were analyzed by SDS/PAGE, and the fractions containing pure tau proteins were pooled. The eluted protein was reduced with 5 mM DTT, and monomeric tau was precipitated by addition of an equal volume of ice-cold methanol and incubation at −20 °C overnight. The precipitated tau protein was then collected by centrifugation at 14,000 × g. The protein was resuspended in the assay buffer (20 mM sodium phosphate, pH 7, 100 mM NaCl, 0.1 mM EDTA) for experimental use.
Preparation of Spin-Labeled Δtau187 Samples.
Precipitated Δtau187 was solubilized in 6 M guanidinium hydrochloride, and the labeling reaction was initiated with the addition of a 10-fold molar excess of paramagnetic (1-oxyl-2,2,5,5-tetramethylpyrrolidin-3-yl) methyl methanethiosulfonate MTSL (R1) spin label (Toronto Research Chemicals), or diamagnetic analog 1-Acetyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl methanethiosulfonate (dMTSL, R1’) (Toronto Research Chemicals). After incubating for 1–2 h at room temperature with gentle shaking, excess spin label was removed by repeated buffer exchange using PD-10 columns (Amersham Pharmacia Biotech) preequilibrated in storage buffer (20 mM sodium phosphate, pH 7, 100 mM NaCl, 0.1 mM EDTA). The protein was concentrated using an Amicon centrifugal filter (MWCO 3 kDa; Millipore). The final spin-labeled protein was >95% pure as determined by SDS/PAGE. Finally, protein concentration was determined by absorbance at OD274 using an extinction coefficient ε274 = 2.8 cm−1⋅mM−1. Protein samples were stored at −20 °C until further use. To determine the extent of labeling, 20 μM labeled Δtau187 was reacted with an equivalent volume of 20 μM 2,2'-dithiodipyridine, and quantified by absorbance at OD343 (>95%).
Before ODNP and ESR or kinetic experiments, the thawed stock tau samples were centrifuged at 14,000 × g for 10 min to remove any aggregated species, and the protein concentration was confirmed spectroscopically as above. In all experiments in this study, the tau aggregation process was induced by addition of heparin (average molecular weight = 18 kDa) at a 6:1 (Δtau187:heparin) molar ratio, unless stated otherwise. These experimental conditions ensure relatively fast aggregation of Δtau187 (compared with full-length human tau), with bona fide tau fibrils formed in liquid solution after ∼16 h of incubation with heparin (18, 25).
X-Band cw ESR Spectroscopy.
First derivative ESR spectra were acquired on a Bruker EMX X-band (9.8 GHz) spectrometer at room temperature equipped with a high-sensitivity dielectric DER102 cavity. The spectra were acquired on protein samples of 3.5 μL volume, placed into quartz capillaries of 0.6 mm inner diameter (VitroCom), which was then held by a Teflon sample holder and mounted inside the microwave cavity. ESR spectra were acquired using signal averaging of ∼6–16 scans over a field sweep of 150–200 G, a microwave power of >18 mW, and modulation amplitudes of 0.25–0.35 G, which was optimized to the natural line width of the spectra.
ESR Time-Dependent Experiments and Analysis of Δtau187 Aggregation.
Δtau187 concentrations for all ESR experiments were 500 ± 100 μM, with the concentration of heparin adjusted accordingly to produce a 6:1 protein:heparin molar ratio to induce the aggregation reaction. This optimal heparin:protein ratio for the resulting aggregation kinetics was verified by turbidity measurements of Δtau187. Time-resolved ESR spectra were initiated immediately after heparin addition and continued over a time period of 800 min. Consecutive cw ESR spectra were recorded, by averaging of >6 scans per spectra, into a 2D array. To assure that the sample was not overheating over the course of the experiment, we purged a gas stream of dry air into the cavity, with a 10 mL/min flow rate. ESR spectra were collected for each Δtau187 variant in a mixture of R1 and R1’ labeled at position 322, with 100% and 20% R1 paramagnetic spin-labeled Δtau187, in the monomeric state and after heparin addition, i.e., at 5–10 min, 20–30 min, and 14–16 h and/or >24 h.
ODNP 1H NMR Spectroscopy.
ODNP relaxometry was carried out using the ESR instrument components as described in X-Band cw ESR Spectroscopy, with the following modifications. The spectra were acquired on protein samples of 3.5 μL volume and placed into quartz capillaries of 0.6 mm inner diameter that were then secured inside the home-built U-shaped NMR coil made of 0.013-inch-diameter Teflon-coated silver wire. These were mounted inside the dielectric DER102 ESR cavity. The NMR coil was, in turn, connected via a coaxial cable to a NMR tuning box (14.8 ± 0.5 MHz), and connected to a broadband channel of a Bruker Avance 300 NMR spectrometer to allow the detection of 1H NMR signal at 14.8 MHz. The sample temperature was kept constant throughout the ODNP experiment by flowing a dry gas over the sample, to prevent overheating of the samples upon microwave irradiation. The measurements of the spin lattice relaxation times, T1 and T10, were performed using the inversion-recovery pulse sequence with optimized delay times and 90-degree pulse. The measurements of the 1H NMR signal enhancement with increasing microwave power were performed by applying a saturation pulse train before NMR signal acquisition, allowing shortening of the duration of these time-sensitive aggregation kinetic measurements.
ODNP Time-Dependent Experiments and Analysis of Δtau187 Aggregation.
For 1H ODNP experiments, the final Δtau187 samples concentrations were 500 ± 100 μM, unless stated otherwise, with the concentration of heparin adjusted accordingly to produce a 6:1 protein:heparin molar ratio for the aggregation reaction. Aggregation of Δtau187 was induced by mixing R1-labeled Δtau187 and heparin (18 kDa; Sigma-Aldrich) at a molar ratio of 6:1 under ambient conditions. All of the ODNP and ESR time-dependent measurements were carried out in a consecutive manner, within the same ∼48 h, using the same stock protein sample solution, to assure comparable behavior of the samples.
Time-resolved ODNP relaxometry data were recorded, upon constant microwave irradiation immediately after heparin addition, and continuously measured over a time period of 800 min. A pulse sequence with a saturation pulse train following an optimized 90° pulse was used, where consecutive NMR spectra were averaged over four scans per spectra. To avoid overheating samples, while maintaining sufficient electron saturation, microwave powers of 150 ± 50 mW with a constant dry gas purged into the sample cavity were used for ODNP experiments. The ODNP time courses were repeated several times for each R1-labeled Δtau187 variant, varying the batch of the stock protein sample and/or varying the total concentration of aggregation samples, i.e., 100-μM or 500-μM tau samples preserving the 6:1 protein:heparin ratio, to assure that total protein concentration did not drastically change the observed profiles of ODNP detected aggregation kinetics.
Quantitative analysis of relaxivity and and values were determined from Emax, T1, and T10 measurements recorded for different Δtau187 samples at chosen times before and/or during aggregation. The Emax and T1 of water 1H measured in the presence of paramagnetic R1-labeled Δtau187 and T10 in the presence of diamagnetic analog of MTSL (R1’) attached to the surface of Δtau187 in its monomeric state and at time points 5–15 min, 60–120 min, and 14–16 h (and/or ≥24 h) following heparin addition. The T1 measurements for each sample at each time point were made before and after each Emax measurement at that time point, to assure that T1 values were not changing while the measurements were being taken for a single reported time point or due to the heating of the sample during the Emax measurement. T10 values of the monomeric and aggregating Δtau187 samples at each time point were measured at varying protein concentrations, and, eventually, the T10s used for calculations of values were extrapolated from these measurements to minimize the error due to slight variations in concentrations of nonparamagnetic R1’ relative to paramagnetic R1 protein samples. Thus, the average value of the two T10 measurements and/or two T1 measurements (that agree within error) and a value of one Emax measurement were used for calculations of and at a given time point. We note that the error in calculated values could potentially increase (beyond our predictions), due to inconsistencies arising from T10 measurements for different aggregation species of different Δtau187 variants. However, the measured values of and Emax are bound to be consistent for a given spin-labeled tau mutant with respect to its aggregation stages because these measurements do not have T10 dependencies. Alternatively, in some cases, the ODNP measurements immediately after heparin addition were repeated several times, alternating the order in which Emax and T1 measurement were carried out on the same samples (first Emax followed by T1 or, vice versa, first T1 followed by Emax measurement). This is to minimize the artifacts in the measurement of parameters produced by rapid aggregation kinetics and to develop knowledge and estimate the experimental error limits on .
Generating the τ vs. Time Graphs.
The τ vs. time graphs presented in this paper were generated by first recording the ODNP enhancement (Emax) of 1H NMR signal vs. time of aggregation, followed by “numeric translation”/”numeric scaling” of the measured enhancement vs. time into ξ vs. time graphs, and then determining the corresponding τ values from the extrapolated ξ values by assuming the appropriate spectral density function (Eq. S2). This was done by the separately measured ξ parameters at the beginning and end of the aggregation period and extrapolating those values onto the whole Emax vs. time graph, resulting in the corresponding ξ vs. time graph, which could then be converted into a τ vs. time graph from the density function.
Turbidimetry.
These measurements were conducted to confirm the aggregation potential of each variant. Immediately after adding heparin (6:1 tau:heparin molar ratio) to 100 μM Δtau187 diluted in an assay buffer, optical density at a 350-nm wavelength was monitored at room temperature continuously over >14 h, using a Shimadzu UV-1601 spectrophotometer (Shimadzu Inc.) equipped with an automated multicell changer. As controls, assay buffer alone and tau sample without heparin were measured concurrently to monitor the background signal throughout the experiment. In the turbidity measurement, similar results were obtained in three separate experiments (error within 5%). We note that the sample concentrations had to be reduced relative to those used in the ODNP and ESR measurements to ensure that the detector would not saturate during the course of the experiment. However, the purpose of turbidity measurements was to determine the appearance of aggregates and not to compare the kinetics to ODNP and ESR time course measurements. Therefore, the difference in concentration does not affect the outcome of our conclusions.
Fluorescence Spectroscopy.
These measurements were conducted to confirm β-sheet formation during aggregation of Δtau187. First, 25 μM of Δtau187 diluted in an assay buffer was incubated with 20 μM ThT dye (Sigma-Aldrich). This was followed by mixing Δtau187 samples with heparin (18 kDa), in a 6:1 molar ratio, and monitoring changes in fluorescence intensity, with excitation at 450 nm and emission at 484 nm, over a time period of >14 h at room temperature with a Varian Cary Eclipse Fluorimeter (Agilent Technologies) equipped with a four-cell cuvette sample holder (1 mL cuvette volume). Excitation and emission slit widths were set to 5 nm. Δtau187 samples in assay buffer without heparin, but incubated with ThT dye, were used to obtain the background signal throughout the measurements. The concentrations of fluorescence samples were reduced from those used in ODNP and ESR measurements to prevent the signal detector from saturation during the course of the experiment.
TEM.
TEM experiments were performed as described previously (18). Briefly, 2 μL of Δtau187 fibrillar solution (aggregation induced >14 h prior) was applied to a 300 mesh Formvar/carbon-coated copper grid, and then the excess liquid was removed by blotting on filter papers. Samples were stained with 10 μL of 1.5% (vol/vol) uranyl acetate followed by removal of the excess uranyl acetate by blotting. Finally, the samples were washed with 10 μL of deionized water and were air-dried. Samples were analyzed using a JOEL JEM-1230 TEM microscope (JEOL) at room temperature.
Cryo-TEM.
For cryo-TEM experiments, we induced the aggregation of P1-labeled Δtau187 at position 322 with heparin at a molar ratio of 6:1 and then took aliquots of the ongoing reaction and froze them onto the cryo-TEM grids with liquid hexane 5 min, 15 min, 50 min, 120 min, and 24 h after aggregation initiation. As aggregation proceeded, aliquots collected at different aggregation time points upon adding heparin into Δtau187 solution were frozen with liquid hexane near liquid nitrogen temperature (77 K). The required concentration of protein for cryo-TEM experiments was ∼20 μM. The aggregation reaction was carried out at total tau concentration of 20 μM, where aliquots were taken out and directly used for cryo-TEM analysis, or, alternatively, the aggregation reaction was carried out with total concentration of tau of 100 μM, followed by taking out the aliquots and diluting tau samples to 20 μM concentration. These cryo-TEM imaging experiments were carried out by the National Center for Macromolecular Imaging facility, Baylor College of Medicine, Houston.
Chemical Cross-Linking.
The oligomeric status of Δtau187 was determined using the water-soluble bis(sulfosuccinimidyl) suberate (BS3; Thermo Fisher Scientific), which has an ∼11.4-Å tether that reacts with primary amines of lysines (there are 22 lysines in Δtau187). Then, 100 μM of Δtau187 (R1-labeled 322C) was first mixed with 25 μM heparin, and aliquots were sampled and frozen into liquid nitrogen at various aggregation times. The samples were then incubated at room temperature in the presence of a 10-fold molar excess of BS3 for 5 min. Cross-linking was stopped by the addition of 10 μL of 2× Laemmli sample buffer. After cross-linking, proteins were separated on a 10–20% tricine SDS/PAGE (Life Technologies) under a reducing condition (1% mercaptoethanol), then stained using a SilverQuest silver staining kit (Life Technologies).
Acknowledgments
We gratefully acknowledge Dr. Ilia Kaminker for advice on ESR data analysis and simulation. We acknowledge the use, for cw ESR spectral simulation, of the MultiComponent software developed by Dr. Christian Altenbach (University of California, Los Angeles). We thank Lauryn Keeler for performing the staining TEM experiments. This work was supported by the 2011 NIH Director New Innovator Award, the Cluster of Excellence RESOLV (EXC 1069) funded by the Deutsche Forschungsgemeinschaft, and a generous gift by Paul and Cathie Slavik. This work made use of the Material Research Laboratory (MRL) Central Facilities supported by the National Science Foundation (NSF) through the Materials Research Science and Engineering Center under Grant DMR 1121053. MRL is a member of the NSF-funded Materials Research Facilities Network (www.mrfn.org). Cryo-TEM experiments were carried out by the National Center for Macromolecular Imaging facility, Baylor College of Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504415113/-/DCSupplemental.
References
- 1.Glabe CG. Structural classification of toxic amyloid oligomers. J Biol Chem. 2008;283(44):29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 3.Avila J, Lucas JJ, Perez M, Hernandez F. Role of tau protein in both physiological and pathological conditions. Physiol Rev. 2004;84(2):361–384. doi: 10.1152/physrev.00024.2003. [DOI] [PubMed] [Google Scholar]
- 4.Lasagna-Reeves CA, et al. Identification of oligomers at early stages of tau aggregation in Alzheimer’s disease. FASEB J. 2012;26(5):1946–1959. doi: 10.1096/fj.11-199851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laganowsky A, et al. Atomic view of a toxic amyloid small oligomer. Science. 2012;335(6073):1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda S, et al. Increased levels of granular tau oligomers: An early sign of brain aging and Alzheimer’s disease. Neurosci Res. 2006;54(3):197–201. doi: 10.1016/j.neures.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed M, et al. Structural conversion of neurotoxic amyloid-beta(1-42) oligomers to fibrils. Nat Struct Mol Biol. 2010;17(5):561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgado I, et al. Molecular basis of β-amyloid oligomer recognition with a conformational antibody fragment. Proc Natl Acad Sci USA. 2012;109(31):12503–12508. doi: 10.1073/pnas.1206433109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Straub JE, Thirumalai D. Toward a molecular theory of early and late events in monomer to amyloid fibril formation. Annu Rev Phys Chem. 2011;62:437–463. doi: 10.1146/annurev-physchem-032210-103526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong SH, Ham S. Impact of chemical heterogeneity on protein self-assembly in water. Proc Natl Acad Sci USA. 2012;109(20):7636–7641. doi: 10.1073/pnas.1120646109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmit JD, Ghosh K, Dill K. What drives amyloid molecules to assemble into oligomers and fibrils? Biophys J. 2011;100(2):450–458. doi: 10.1016/j.bpj.2010.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parthasarathy S, et al. Structural insight into an Alzheimer’s brain-derived spherical assembly of amyloid β by solid-state NMR. J Am Chem Soc. 2015;137(20):6480–6483. doi: 10.1021/jacs.5b03373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchanan LE, et al. Mechanism of IAPP amyloid fibril formation involves an intermediate with a transient β-sheet. Proc Natl Acad Sci USA. 2013;110(48):19285–19290. doi: 10.1073/pnas.1314481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stroud JC, Liu C, Teng PK, Eisenberg D. Toxic fibrillar oligomers of amyloid-β have cross-β structure. Proc Natl Acad Sci USA. 2012;109(20):7717–7722. doi: 10.1073/pnas.1203193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupuis NF, Wu C, Shea JE, Bowers MT. The amyloid formation mechanism in human IAPP: Dimers have β-strand monomer-monomer interfaces. J Am Chem Soc. 2011;133(19):7240–7243. doi: 10.1021/ja1081537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armstrong BD, Han S. Overhauser dynamic nuclear polarization to study local water dynamics. J Am Chem Soc. 2009;131(13):4641–4647. doi: 10.1021/ja809259q. [DOI] [PubMed] [Google Scholar]
- 17.Franck JM, Pavlova A, Scott JA, Han S. Quantitative cw Overhauser effect dynamic nuclear polarization for the analysis of local water dynamics. Prog Nucl Magn Reson Spectrosc. 2013;74:33–56. doi: 10.1016/j.pnmrs.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavlova A, et al. Site-specific dynamic nuclear polarization of hydration water as a generally applicable approach to monitor protein aggregation. Phys Chem Chem Phys. 2009;11(31):6833–6839. doi: 10.1039/b906101k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussain S, Franck JM, Han S. Transmembrane protein activation refined by site-specific hydration dynamics. Angew Chem Int Ed Engl. 2013;52(7):1953–1958. doi: 10.1002/anie.201206147. [DOI] [PubMed] [Google Scholar]
- 20.Armstrong BD, et al. Site-specific hydration dynamics in the nonpolar core of a molten globule by dynamic nuclear polarization of water. J Am Chem Soc. 2011;133(15):5987–5995. doi: 10.1021/ja111515s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franck JM, Ding Y, Stone K, Qin PZ, Han S. Anomalously rapid hydration water diffusion dynamics near DNA surfaces. J Am Chem Soc. 2015;137(37):12013–12023. doi: 10.1021/jacs.5b05813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nucci NV, Pometun MS, Wand AJ. Mapping the hydration dynamics of ubiquitin. J Am Chem Soc. 2011;133(32):12326–12329. doi: 10.1021/ja202033k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebbinghaus S, et al. An extended dynamical hydration shell around proteins. Proc Natl Acad Sci USA. 2007;104(52):20749–20752. doi: 10.1073/pnas.0709207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barghorn S, Mandelkow E. Toward a unified scheme for the aggregation of tau into Alzheimer paired helical filaments. Biochemistry. 2002;41(50):14885–14896. doi: 10.1021/bi026469j. [DOI] [PubMed] [Google Scholar]
- 25.Peterson DW, Zhou H, Dahlquist FW, Lew J. A soluble oligomer of tau associated with fiber formation analyzed by NMR. Biochemistry. 2008;47(28):7393–7404. doi: 10.1021/bi702466a. [DOI] [PubMed] [Google Scholar]
- 26.Gustke N, Trinczek B, Biernat J, Mandelkow EM, Mandelkow E. Domains of tau protein and interactions with microtubules. Biochemistry. 1994;33(32):9511–9522. doi: 10.1021/bi00198a017. [DOI] [PubMed] [Google Scholar]
- 27.von Bergen M, et al. Assembly of τ protein into Alzheimer paired helical filaments depends on a local sequence motif (306VQIVYK311) forming β structure. Proc Natl Acad Sci USA. 2000;97(10):5129–5134. doi: 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Bergen M, et al. Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local beta-structure. J Biol Chem. 2001;276(51):48165–48174. doi: 10.1074/jbc.M105196200. [DOI] [PubMed] [Google Scholar]
- 29.Margittai M, Langen R. Template-assisted filament growth by parallel stacking of tau. Proc Natl Acad Sci USA. 2004;101(28):10278–10283. doi: 10.1073/pnas.0401911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sibille N, et al. Structural impact of heparin binding to full-length tau as studied by NMR spectroscopy. Biochemistry. 2006;45(41):12560–12572. doi: 10.1021/bi060964o. [DOI] [PubMed] [Google Scholar]
- 31.Goedert M, et al. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature. 1996;383(6600):550–553. doi: 10.1038/383550a0. [DOI] [PubMed] [Google Scholar]
- 32.von Bergen M, et al. The core of tau-paired helical filaments studied by scanning transmission electron microscopy and limited proteolysis. Biochemistry. 2006;45(20):6446–6457. doi: 10.1021/bi052530j. [DOI] [PubMed] [Google Scholar]
- 33.Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. J Magn Reson A. 1996;120(2):155–189. [Google Scholar]
- 34.Hwang JS, Mason RP, Hwang LP, Freed JH. Electron-spin resonance studies of anisotropic rotational reorientation and slow tumbling in liquid and frozen media. 3. Perdeuterated 2,2,6,6-tetramethyl-4-piperidone N-oxide and an analysis of fluctuating torques. J Phys Chem. 1975;79(5):489–511. [Google Scholar]
- 35.Columbus L, Kálai T, Jekö J, Hideg K, Hubbell WL. Molecular motion of spin labeled side chains in alpha-helices: Analysis by variation of side chain structure. Biochemistry. 2001;40(13):3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 36.Serag AA, Altenbach C, Gingery M, Hubbell WL, Yeates TO. Arrangement of subunits and ordering of beta-strands in an amyloid sheet. Nat Struct Biol. 2002;9(10):734–739. doi: 10.1038/nsb838. [DOI] [PubMed] [Google Scholar]
- 37.Cobb NJ, Sönnichsen FD, McHaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: A parallel, in-register β-structure. Proc Natl Acad Sci USA. 2007;104(48):18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jayasinghe SA, Langen R. Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J Biol Chem. 2004;279(46):48420–48425. doi: 10.1074/jbc.M406853200. [DOI] [PubMed] [Google Scholar]
- 39.Chen M, Margittai M, Chen J, Langen R. Investigation of alpha-synuclein fibril structure by site-directed spin labeling. J Biol Chem. 2007;282(34):24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- 40.Gu L, et al. Antiparallel triple-strand architecture for prefibrillar Aβ42 oligomers. J Biol Chem. 2014;289(39):27300–27313. doi: 10.1074/jbc.M114.569004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Török M, et al. Structural and dynamic features of Alzheimer’s Aβ peptide in amyloid fibrils studied by site-directed spin labeling. J Biol Chem. 2002;277(43):40810–40815. doi: 10.1074/jbc.M205659200. [DOI] [PubMed] [Google Scholar]
- 42.Bibow S, et al. The dynamic structure of filamentous tau. Angew Chem Int Ed Engl. 2011;50(48):11520–11524. doi: 10.1002/anie.201105493. [DOI] [PubMed] [Google Scholar]
- 43.Bitan G, Lomakin A, Teplow DB. Amyloid beta-protein oligomerization: Prenucleation interactions revealed by photo-induced cross-linking of unmodified proteins. J Biol Chem. 2001;276(37):35176–35184. doi: 10.1074/jbc.M102223200. [DOI] [PubMed] [Google Scholar]
- 44.Daidone I, Ulmschneider MB, Di Nola A, Amadei A, Smith JC. Dehydration-driven solvent exposure of hydrophobic surfaces as a driving force in peptide folding. Proc Natl Acad Sci USA. 2007;104(39):15230–15235. doi: 10.1073/pnas.0701401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheon M, et al. Structural reorganisation and potential toxicity of oligomeric species formed during the assembly of amyloid fibrils. PLOS Comput Biol. 2007;3(9):1727–1738. doi: 10.1371/journal.pcbi.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]