Significance
Modern Europe has been shaped by two episodes in prehistory, the advent of agriculture and later metallurgy. These innovations brought not only massive cultural change but also, in certain parts of the continent, a change in genetic structure. The manner in which these transitions affected the islands of Ireland and Britain on the northwestern edge of the continent remains the subject of debate. The first ancient whole genomes from Ireland, including two at high coverage, demonstrate that large-scale genetic shifts accompanied both transitions. We also observe a strong signal of continuity between modern day Irish populations and the Bronze Age individuals, one of whom is a carrier for the C282Y hemochromatosis mutation, which has its highest frequencies in Ireland today.
Keywords: ancient DNA, genomics, population genetics
Abstract
The Neolithic and Bronze Age transitions were profound cultural shifts catalyzed in parts of Europe by migrations, first of early farmers from the Near East and then Bronze Age herders from the Pontic Steppe. However, a decades-long, unresolved controversy is whether population change or cultural adoption occurred at the Atlantic edge, within the British Isles. We address this issue by using the first whole genome data from prehistoric Irish individuals. A Neolithic woman (3343–3020 cal BC) from a megalithic burial (10.3× coverage) possessed a genome of predominantly Near Eastern origin. She had some hunter–gatherer ancestry but belonged to a population of large effective size, suggesting a substantial influx of early farmers to the island. Three Bronze Age individuals from Rathlin Island (2026–1534 cal BC), including one high coverage (10.5×) genome, showed substantial Steppe genetic heritage indicating that the European population upheavals of the third millennium manifested all of the way from southern Siberia to the western ocean. This turnover invites the possibility of accompanying introduction of Indo-European, perhaps early Celtic, language. Irish Bronze Age haplotypic similarity is strongest within modern Irish, Scottish, and Welsh populations, and several important genetic variants that today show maximal or very high frequencies in Ireland appear at this horizon. These include those coding for lactase persistence, blue eye color, Y chromosome R1b haplotypes, and the hemochromatosis C282Y allele; to our knowledge, the first detection of a known Mendelian disease variant in prehistory. These findings together suggest the establishment of central attributes of the Irish genome 4,000 y ago.
The oldest Gaelic literature describes the origins of the Irish people as a series of ancient invasions, and the archaeological record in Ireland, as elsewhere in Europe, exhibits several horizons where major cultural shifts are apparent (1). The two most transformative are the arrival of agriculture (∼3750 BC) followed by the onset of metallurgy (∼2300 BC). The Neolithic package characterized by animal husbandry, cereal crops, ceramics, and timber houses reached the shores of Ireland some 5,000 years after its beginnings in the Near East. The second great wave of change starts with the appearance of copper mines, associated with Bell Beaker pottery, which are quickly followed by Bronze tool-making, weaponry, and gold-working, with distinct Food Vessel pottery succeeding from the earlier beakers (2). This period coincides with the end of the large passage graves of Neolithic Ireland in favor of single burials and smaller wedge tombs.
Twentieth-century archaeology was dominated by two non-mutually-exclusive paradigms for how such large scale social change occurs (3). The first, demic diffusion, linked archaeological change with the displacement and disruption of local populations by inward migrations. However, from the 1960s onwards, this assertion was challenged by a paradigm of cultural diffusion whereby social change happened largely through indigenous processes.
High-throughput sequencing has opened the possibility for genome-wide comparisons of genetic variation in ancient populations, which may be informatively set in the context of extensive modern data (4–10). In Europe, these clearly show population replacement by migrating farmers from southwest Asia at the onset of the Neolithic with some retrenchment of the earlier Mesolithic genome at later stages (5–9, 11, 12). Three longitudinal genome studies have also shown later genome-wide shifts around the beginnings of the Bronze Age in central Europe with substantial introgression originating with the Yamnaya steppe herders (7, 9, 10). However, replacement coupled to archaeological horizons is unlikely to be a universal phenomenon, and whether the islands of Britain and Ireland, residing at the temporal and geographical edges of both the Neolithic and steppe migrations, were subject to successive substantial population influxes remains an open and debated question. For example, a recent survey of archaeological opinion on the origins of agriculture in Ireland showed an even split between adoption and colonization as explanatory processes (13). Recent archaeological literature is also divided on the origins of the insular Bronze Age, with most opinion favoring incursion of only small numbers of technical specialists (1, 2, 14, 15).
To address this controversy, we present here the first, to our knowledge, genome-wide data from four ancient Irish individuals, a Neolithic woman (3343–3020 cal BC) from Ballynahatty, Co. Down, found in the context of an early megalithic passage-like grave, and three Early Bronze Age men from a cist burial in Rathlin Island, Co. Antrim (2026–1534 cal BC) with associated Food Vessel pottery (16) (SI Appendix, Section S1).
Results
DNA Extraction, Sequencing, and Uniparental Haplotypes.
DNA was extracted (SI Appendix, Section S2) from the petrous portion of the temporal bone (7) yielding high levels of endogenous content (38–68%) for all samples (Table 1). Two individuals, Ballynahatty and Rathlin1, were shotgun sequenced to high coverage (10–11×), whereas the two remaining Bronze Age samples, Rathlin2 and Rathlin3, were sequenced to 1.5× and 0.75×, respectively (SI Appendix, Section S3). Data authenticity was assessed by using several approaches (SI Appendix, Section S5). Molecular damage patterns were found to be consistent with those of ancient DNA (SI Appendix, Fig. S5.1). Contamination estimates were found to be low from negative controls (0.07–0.16%), as well as heterozygosity of the mitochondrial genome (0.36–0.50%) and the X chromosome in males (0.688–1.646%).
Table 1.
Sequencing results from four ancient Irish individuals
| Sample | Date, cal BC | Archaeological context | Final endogenous DNA content, % | Mean genome coverage, X | Genetic sex | mtDNA haplogroup | Y chromosome haplogroup |
| Ballynahatty | 3343–3020 | Middle to Late Neolithic | 67.86 | 10.27 | Female | HV0 +195 | — |
| Rathlin1 | 2026–1885 | Early Bronze Age | 60.74 | 10.50 | Male | U5a1b1e | R1b1a2a1a2c1g |
| Rathlin2 | 2024–1741 | Early Bronze Age | 38.15 | 1.49 | Male | U5b2a2 | R1b1a2a1a2c1 |
| Rathlin3 | 1736–1534 | Early Bronze Age | 61.42 | 0.75 | Male | J2b1a | R1b1a2a1a2c |
Molecular sex determination agreed with archaeological results (SI Appendix, Section S7), and whole mitochondrial chromosome haplogroups were assigned (Table 1 and SI Appendix, Section S7). The Neolithic individual, Ballynahatty, belongs to HV0, a clade with highest frequencies seen in Early and Middle Neolithic (MN) groups from Germany and France (9, 17, 18) and which has not been identified in any Mesolithic Europeans. The Bronze Age individuals belonged to the haplogroups U5 and J2b. Notably, the three Bronze Age males each belong to the Y chromosome haplogroup R1b1a2a1a2c (SI Appendix, Section S8). This lineage is characterized by a modern east–west frequency gradient in the British Isles, and is almost fixed (94%) in the West of Ireland (19, 20). R1b haplotypes have been argued as first appearing with steppe incursion in the central European Bronze Age (9).
Principal Component Analysis and ADMIXTURE.
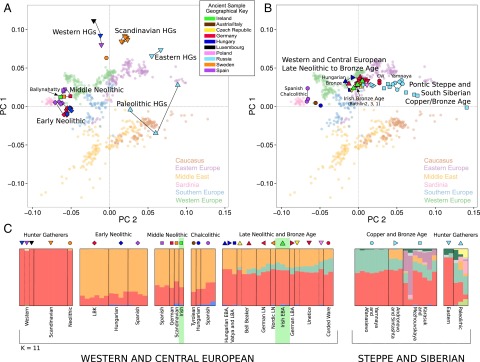
A projection principal component analysis (PCA) was used to investigate the genetic affinities of our four ancient Irish genomes, alongside 78 published ancient samples (SI Appendix, Table S9.1; selection criteria detailed in SI Appendix, Section S4) (5–10, 21–26) using 677 modern individuals from Western Eurasia (8) as a reference population (Fig. 1 A and B and SI Appendix, Sections S9.1 and S10). In Fig. 1A, we see a clear division between hunter–gatherer and early farmer individuals, with the Ballynahatty female plotting with five other MN samples from Germany, Spain, and Scandinavia. However, a large shift in genetic variation is seen between Ballynahatty and the three Irish Early Bronze Age samples, Rathlin1, Rathlin2, and Rathlin3, who fall in a separate central region of the graph along with Unetice and other Early Bronze Age genomes from Central and North Europe. These plots imply that ancient Irish genetic affinities segregate within European archaeological horizons rather than clustering geographically within the island.
Fig. 1.
Genetic affinities of ancient Irish individuals. (A and B) Genotypes from 82 ancient samples are projected onto the first two principal components defined by a set of 354,212 SNPs from Eurasian populations in the Human Origins dataset (29) (SI Appendix, Section S9.1 and S10). (A) This PCA projects ancient Eurasian Hunter–Gatherers and Neolithic Farmers, where they separate clearly into Early Neolithic, MN (including the Irish Ballynahatty genome), and several hunter–gatherer groups. (B) PCA projection of Late Neolithic, Copper, and Bronze Age individuals where the three Rathlin genomes adopt a central position within a large clustering of European Bronze Age individuals. (C) A plot of ADMIXTURE ancestry components (K = 11) of these same ancient genomes. In West and Central Europe, ancient individuals are composed almost entirely of two dominant strands of ancestry, linked to hunter–gatherer (red) and early farmer (orange) populations, until the Late Neolithic. At this point, a third (green) Caucasus component features. Previously, this component was only seen in ancient Steppe and Siberian populations such as the Yamnaya. The three Rathlin genomes each display this Caucasus strand of ancestry whereas the Irish Neolithic does not.
Model-based approaches allow the decomposition of individuals into coefficients contributed by a set number (K) of ancestral populations. We investigated this by using ADMIXTURE (Version 1.23) (27) with ancient individuals included in the analysis alongside 1,941 modern samples from diverse worldwide populations (8) (SI Appendix, Section S11). Fig. 1C shows the plot of estimated ancestry proportions for each ancient genome at a value of K = 11. These partition similarly to previous analyses (7, 9, 10), with three major ancestral coefficients manifesting in west and central Europe. The first (colored red) forms the near totality of ancestry in hunter–gatherer samples, and admixes with a second (orange) component found at high levels in Neolithic and also modern Near Eastern populations (SI Appendix, Section S11.1). Ballynahatty is similar to other MN samples with a majority orange “early farmer” component but with an elevated level of the red “hunter–gatherer” component compared with Early Neolithic genomes. The ancestry of Yamnaya, Early Bronze Age herders from the Pontic Steppe, is evenly divided between this same red component and the third major European coefficient, colored in green, which has been identified with a Caucasus origin (28). This Caucasus component is encountered subsequently, introduced via Yamnaya, in Late Neolithic and Bronze Age samples in central Europe (9, 10) and features in the three Irish Early Bronze Age samples within profiles similar to continental Bronze Age genomes.
Neolithic Origins.
D and f statistics (29, 30) (Dataset S1 and SI Appendix, Section S12) support identification of Ballynahatty with other MN samples who show a majority ancestry from Near Eastern migration but with some western Mesolithic introgression. Outgroup f3 statistics indicated that Ballynahatty shared most genetic drift with other Neolithic samples with maximum scores observed for Spanish Middle, Epicardial, and Cardial Neolithic populations, and the Scandinavian individual Gok2 (SI Appendix, Section S12.1). D statistics confirmed these affinities. Interestingly, both the Spanish MN individuals and Gok2 also belong to Megalithic passage tomb cultures. Along with other MNs, Ballynahatty displays increased levels of hunter–gatherer introgression compared with earlier farming populations. D statistics revealed that, out of the three Mesolithic hunter–gatherer groupings from Haak et al. (9), eastern hunter–gatherer (EHG), Scandinavian hunter–gatherer (SHG), and western hunter–gatherer (WHG), Ballynahatty shares the most alleles with WHG and furthermore has the strongest preference for WHG over EHG and SHG out of all contemporaneous Neolithic individuals so far sampled. However, Ballynahatty forms a clade with other MN genomes to the exclusion of WHG and, symmetrically, WHG genomes form a clade to the exclusion of Ballynahatty. Of the three WHG individuals, Ballynahatty appeared to share the least amount of affinity with LaBrana (Spain) and the largest amount of affinity with Loschbour (Luxembourg). To gauge the proportion of Ballynahatty’s ancestry derived from WHG, we used the f4 ratio, f4(Mbuti, Loschbour; Ballynahatty, Dai)/f4(Mbuti, Loschbour; KO1, Dai) (29), to estimate a hunter–gatherer component of 42 ± 2% within a predominantly early farmer genome. Thus, we deduce migration has a primary role in Irish agricultural origins.
Bronze Age Replacement.
Prior studies (7, 9, 10) convincingly demonstrate that Central European genomes from the late Neolithic and Early Bronze Age differ from the preceding MN due to a substantial introgression originating with Steppe herders linked to cultures such as the Yamnaya. Accordingly, we used a series of tests to gauge whether the ancestries of the Rathlin Early Bronze Age genomes were subject to this influence. D statistics confirmed that Ballynahatty and other MN individuals form clades with each other to the exclusion of these Irish Bronze Age samples. Specific disruption of continuity between the Irish Neolithic and Bronze Age is clear with significant evidence of both Yamnaya and EHG introgression into Irish Bronze Age samples when placed in a clade with any MN. However, like other European Bronze Age samples, this introgression is incomplete, as they also show significant MN ancestry when placed in a clade with Yamnaya. The highest levels of MN ancestry were observed when either Ballynahatty or Gok2 (Scandinavian) was the sample under study. However, when paired with central European Bronze Age populations, the Rathlin samples show no trace of significant introgression from Ballynahatty, suggesting that earlier Irish populations may not have been a source of their partial MN ancestry.
These analyses, taken with the PCA and ADMIXTURE results, indicate that the Irish Bronze Age is composed of a mixture of European MN and introgressing Steppe ancestry (9, 10). To estimate the proportion of Yamnaya to MN ancestry in each Irish Bronze Age sample, we took three approaches. First, from ADMIXTURE analysis (Fig. 1), we examined the green Caucasus ancestry component. We presume an ultimate source of this as the Yamnaya where it features at a proportion of 40% of their total ancestry. In our three Irish Bronze Age samples, it is present at levels between 6–13%, which, when scaled up to include the remaining 60% of Yamnaya ancestry, imply a total of 14–33% Yamnaya ancestry and therefore 67–86% MN in the Irish Bronze Age. Second, for each Bronze Age Irish individual, we calculated the proportion of MN ancestry by using the ratio f4(Mbuti, Ballynahatty; X, Dai)/f4(Mbuti, Ballynahatty; Gok2, Dai), which gave estimates between 72 ± 4% to 74 ± 5%, implying again a substantial Yamnaya remainder. Third, we followed the methods described in Haak et al. (9), which use a collection of outgroup populations, to estimate the mixture proportions of three different sources, Linearbandkeramik (Early Neolithic; 35 ± 6%), Loschbour (WHG; 26 ± 12%), and Yamnaya (39 ± 8%), in the total Irish Bronze Age group. These three approaches give an overlapping estimate of ∼32% Yamnaya ancestry.
Runs of Homozygosity.
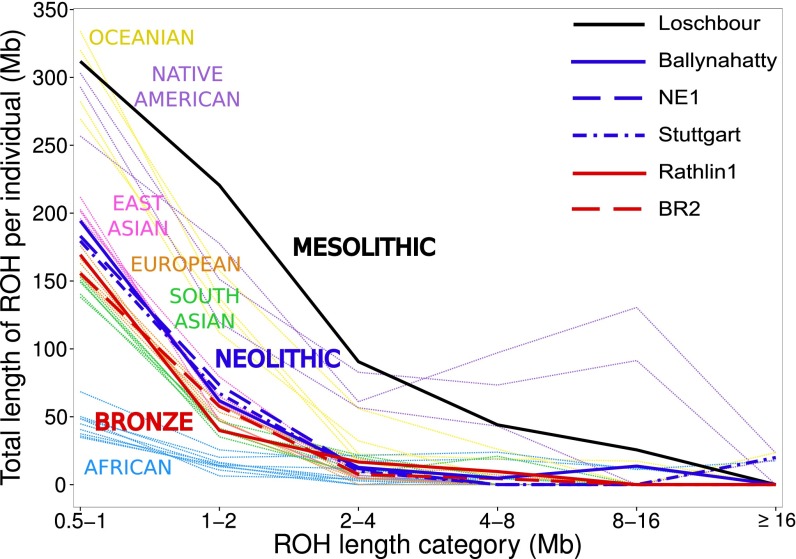
We coanalyzed Ballynahatty, Rathlin1, and the four other ancient Europeans sequenced to high coverage (7, 8) to estimate the fraction of each genome under runs of homozygosity (ROH) (SI Appendix, Section S13). This approach may be useful to inform on past demography; long contiguous homozygous segments indicate recent endogamy, whereas shorter runs result from older restrictions in ancestral population size (31, 32). Fig. 2 plots these ROH estimates in a background of data from modern genomes of diverse origins (33, 34). The single ancient hunter–gatherer genome in this analysis, Loschbour, shows extremely high levels of ROH, indicative of a restricted ancestral population size. However, Ballynahatty and the two early Neolithic genomes, Stuttgart and NE1, each show levels of ROH comparable to those seen in modern East Asians. This pattern argues against a genetic bottleneck, such as that due to a narrow pioneer colonization, in the ancestry of this Irish Neolithic genome and supports the idea of large-scale Neolithic migration rather than assimilation of farming by local hunter–gatherers. The Bronze Age genomes, Rathlin1 and BR2, both show further reductions of ROH, producing distributions similar to each other and to that of modern Europeans.
Fig. 2.
Estimated distribution of ROH for ancient samples, placed in the context of values from modern populations. This plot shows total length of ROH for a series of length categories in 32 modern and 6 ancient individuals. Of these modern individuals, 18 were selected from a subset of 1000 Genomes populations (33) with no evidence of recent historical admixture. These individuals represented the median value of total length of ROH for their population. Thirteen modern individuals from the Simons Genome Diversity Project (34) were also included. ROH calls for each sample were based on 1.6M autosomal transversions with minor allele frequencies above 0.5%.
Haplotype-Based Resolution of Continuity.
Haplotype-based approaches are more powerful than those using unlinked genetic loci in identifying fine genetic structure, such as that displayed among Europeans, and are relatively robust to bias from marker ascertainment (35, 36). We ran ChromoPainter in fineSTRUCTURE (Version 2) (35) to decompose each ancient genome into a series of haplotypic chunks, and identified which modern individuals from a diverse set of Eurasian populations (37) shared the same, or most similar, haplotype at each given chunk. We then considered the pattern of chunk donation between each ancient genome and modern populations.
Unsurprisingly, the pattern of haplotypic affinity of Ballynahatty among modern European populations is strongly correlated to that of the earlier Neolithic samples (SI Appendix, Fig. S14.2; r > 0.74, P < 10−7), with southern Mediterranean samples in each analysis showing highest levels of chunk copying. However, some differences are discernable; the Hungarian and Stuttgart Neolithic genomes tend toward higher values in eastern Mediterranean (Sicilian, Italian, and Greek samples; Fig. 3 and SI Appendix, Fig. S14.1), and the Irish Neolithic has highest values in the west (Sardinian and Spanish). A further difference lies in the comparison of each to the affinities shown by the Luxembourg WHG, Loschbour, which shows no correlation in its modern affinities with the earlier continental Neolithics but does show a significant relationship (P = 5.4 × 10−4) with those of Ballynahatty, undoubtedly because of greater WHG admixture in her ancestry.
Fig. 3.
Comparison of Irish and Central European ancient genomes for haplotype-based affinity to modern populations. Interpolated heatmaps comparing relative haplotype donations by two Irish (Ballynahatty, Rathlin1) and two Hungarian (NE1, BR2) ancient genomes.
The most striking feature of the haplotype sharing by the Irish Bronze Age genome is its high median donation levels to Irish, Scottish, and Welsh populations (Fig. 3). In regression with results from the other ancient genomes, these insular Celtic populations, and to a lesser degree the English, show an excess of sharing with Rathlin1, suggesting some level of local continuity at the edge of Europe persisting over 4,000 y. The Hungarian Bronze Age genome shows more affinity with central European populations. Interestingly, for both Bronze Age genomes, the modern Basque population displays outlying low-affinity scores compared with neighboring western European samples, supporting recent findings that suggest a continuity between the Basques and Iberian Chalcolithic groups (25).
Phenotypic Analysis.
Ireland is unusual in displaying world maximum frequencies of a number of important genetic variants, particularly those involved in lactase persistence and two recessive diseases: cystic fibrosis and hemochromatosis (38–41, 42, 43). Interestingly, both the high coverage Neolithic and Bronze Age individuals were heterozygous for the hemochromatosis alleles H63D and C282Y, respectively. Modern Irish allele frequencies are 15% and 11% for these variants, with the latter, more penetrant variant responsible for a world maximum of this disease in Ireland (44, 45). Additionally and in accordance with other data suggesting a late spread of the lactase persistence phenotype now prevalent in western Europe (7, 10), the Neolithic Ballynahatty was homozygous for the nonpersistent genotype and Rathlin1 was heterozygous and thus tolerant of drinking raw milk into adulthood.
We were able to deduce that Neolithic Ballynahatty had a dark hair shade (99.5% probability), most likely black (86.1% probability), and brown eyes (97.3% probability) (46). Bronze Age Rathlin1 probably had a light hair shade (61.4%) and brown eyes (64.3%). However, each Rathlin genome possessed indication of at least one copy of a haplotype associated with blue eye color in the HERC2/OCA2 region.
Discussion
The advent of farming in Ireland lies at the temporal and geographical terminus of several thousand years of transition across Europe and seems to occur rapidly ∼ 3750 BC (47). The nature of this transition remains a long-standing archaeological controversy between proposals of migration by incoming farmers versus those of adoption of agriculture by indigenous Mesolithic populations (48–51). The Irish MN female farmer (3343–3020 cal. BC) from a Megalithic tomb in Ballynahatty near Belfast affords, to our knowledge, a first direct genetic view of the transition at Europe’s western edge. She displays predominant ancestry from early farmers that ultimately originated in migrating agriculturists from the Near East. This derivation is attested by her PCA and ADMIXTURE profile, her correlated allele frequencies with other Neolithic genomes, reflected in D statistics, and by her haplotypic affinity with modern southern Mediterranean populations such as Sardinians. Her early European farmer coefficient is estimated at ∼60%; an ancestry which is difficult to reconcile with extensive indigenous adoption of agriculture in Ireland only several hundred years earlier. She shares higher levels of genetic drift with Early and MN samples from Spain rather than those from Germany, supporting a link between the early farming cultures of Atlantic Europe and arguing for the possible passage of farming to Ireland via a southern coastal route rather than via the migrations through central Europe (2). A high affinity to Scandinavian farmer, Gok2 is more difficult to interpret as it is associated with the TRB (Funnelbeaker) culture whose origins are generally derived from Central European farming cultures but perhaps it is noteworthy that this later sample is also recovered from a Megalithic context.
From examination of the fraction of her genome, which is under ROH, she seems similar to other ancient Neolithics, suggesting that she belonged to a large outbreeding population. This analysis argues against a marked population bottleneck in her ancestry, such as might have occurred had she been descended from a small pioneering group of migrating farmers. Either a restricted colonization does not reflect the nature of the first Irish farmers or her ancestry was augmented by substantial additional Neolithic communication from elsewhere in intervening centuries.
However, like other MN genomes, Ballynahatty shows some elevation of hunter–gatherer ancestry relative to earlier Neolithic samples, suggesting admixture with surviving Mesolithic populations somewhere within the temporal and spatial trajectory between the Neolithic heartland and north east Ireland ∼3300 BC (5, 9). Further sampling will be needed to determine the local or continental origins of these hunter–gatherer ancestral components but it is interesting that she shows slightly higher affinity to a Luxembourger Mesolithic genome, than to Hungarian and Spanish equivalents.
There are several lines of evidence for discontinuity between the MN and Early Bronze Age within our genome samples from northeast Ireland. First, they occupy different regions in the PCA plot; second, there is a signal from the ADMIXTURE analysis of a Caucasus ancestral component within the Rathlin genomes, which is absent in Ballynahatty and other western MN genomes. Third, D statistics also show highly significant evidence for this eastern introgression. Finally, each Bronze Age sample (all male) exhibits the Bronze Age-associated Y chromosome lineage R1b-M269, the appearance of which has been strongly linked with Steppe incursion into Central Europe (9). Thus, it is clear that the great wave of genomic change which swept from above the Black Sea into Europe around 3000 BC washed all of the way to the northeast shore of its most westerly island. At present, the Beaker culture is the most probable archaeological vector of this Steppe ancestry into Ireland from the continent, although further sampling from Beaker burials across western Europe will be necessary to confirm this. The extent of this change, which we estimate at roughly a third of Irish Bronze Age ancestry, opens the possibility of accompanying language change, perhaps the first introduction of Indo-European language ancestral to Irish. This assertion gains some support by the relative lack of affinity of non-Indo-European speakers, Basques, to the ancient Bronze Age genomes (Fig. 3).
The high coverage of both Ballynahatty and Rathlin1 allows a sensitive test of haplotype donation to modern populations, with interesting and contrasting results. Whereas Ballynahatty shows closest affinity with the southwest Mediterranean, Rathlin1 has highest sharing with the geographically closest modern populations, a trend not seen with the other high coverage ancient genome samples. This affinity with Irish, Scottish, and Welsh (a weaker signal from modern English populations is undoubtedly due to the effects of Anglo-Saxon migrations; ref. 36) suggests a degree of continuity stretching over 4,000 y at the insular Celtic edge of Europe. Ireland shows the global maxima for frequencies of the R1b1a2a1a2c (M529) Y chromosome haplotype, lactase persistence, and the C282Y hemochromatosis allele. Our data show that these genotypes had arrived by the time of the Irish Bronze Age. Hemochromatosis, known as the “Celtic” disease (44), is a variably penetrant recessive disorder, potentially fatal, due to excessive retention of dietary iron. It has been suggested that the high frequencies of both C282Y and H63D are due to heterozygote advantage related to nutritional advantage in Neolithic iron-poor diets (52), mitigation of celiac disease (53), and increased resistance to parasitic infection (54). To our knowledge, our data provide the first evidence for known Mendelian disease alleles in ancient genomes and mark the associations of both hemochromatosis alleles with the island of Ireland as ancient.
Materials and Methods
A full description of the materials and methods is provided in SI Appendix. Briefly, we extracted DNA by using a modified silica column based method (55, 56) from four petrous bones (7) belonging to one Neolithic (Ballynahatty) and three Bronze Age (Rathlin1–3) individuals (SI Appendix, Section S2). DNA fragments were then incorporated into next-generation sequencing libraries (7, 57), amplified with distinct indexing oligos, purified (MinElute PCR Purification kit, Qiagen), pooled, and sequenced with Illumina technology (SI Appendix, Section S3). Sample reads were subsequently trimmed of adapter sequences by using cutadapt (Version. 1.2.1; ref. 58) and aligned to the human reference genome (hg19/GRCh37), with the mitochondrial genome replaced by the revised Cambridge reference sequence using BWA (Version 0.7.5; ref. 59). Reads were filtered for mapping quality, and clonal amplification products were removed with SAMtools (60). Base quality scores were recalibrated by using mapDamage (Version 2.0; ref. 61) (SI Appendix, Section S4).
Data authenticity was established by examination of molecular damage patterns and levels of mtDNA and X chromosome contamination (SI Appendix, Section S5). Molecular sex determination, mtDNA, and Y chromosome analysis were carried out following established methods (SI Appendix, Sections S6–S8). Sample genotypes at known SNP positions were called by using GATK (Version 2.4; ref. 62), and merged with modern data from the Human Origins array (8), Hellenthal et al. (37), 1000 Genomes Phase 3 (Version 5; ref. 33), and the Simons Genome Diversity Project (34), as well as published ancient data (5–10, 21–26) (SI Appendix, Section S9). Relationships between the ancient Irish samples and other ancient and modern individuals were investigated by using PCA (SI Appendix, Section S10) and ADMIXTURE (27) (SI Appendix, Section S11), with formal confirmation of these relationships assessed by using D and f statistics (29, 30, 63) (SI Appendix, Section S12). For our high-coverage samples, Ballynahatty and Rathlin1, we performed haplotype-based analysis by using FineSTRUCTURE (Version 2; ref. 35) (SI Appendix, Section S14) and investigated the proportion of their genomes under ROH with PLINK (Version 1.90; ref. 64) (SI Appendix, Section S13). Finally, we investigated, by direct observation, a panel of SNPs, small indels, and multiallelic markers known to have well established phenotypic associations with traits such as pigmentation, immunity, disease susceptibility, diet, and blood group (SI Appendix, Section S15).
Supplementary Material
Acknowledgments
We thank Hellenthal et al. (37) for sharing the genotype data in PLINK format and David McConnell for a critical reading of the manuscript. L.M.C. is funded by the Irish Research Council Government of Ireland Scholarship Scheme (GOIPG/2013/1219). R.M. was financed by the Bridging the European and Anatolian Neolithic project of the Marie Curie Innovative Training Networks (Grant 289966). M.D.T. was supported by European Research Council (ERC) Investigator Grant 295729-CodeX. We acknowledge Science Foundation Ireland (SFI) ERC Support Award 12/ERC/B2227, Valeria Mattiangeli, and TrinSeq for MiSeq support. We acknowledge the DJEI/DES/SFI/HEA Irish Centre for High-End Computing for the provision of computational facilities and support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the European Nucleotide Archive (project accession no. PRJEB11995).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518445113/-/DCSupplemental.
References
- 1.Waddell J. The Prehistoric Archaeology of Ireland. Galway Univ Press; Galway, Ireland: 1998. [Google Scholar]
- 2.Mallory JP. The Origins of the Irish. Thames & Hudson; London: 2013. [Google Scholar]
- 3.Hakenbeck S. Migration in archaeology: Are we nearly there yet? Archaeol Rev Camb. 2008;23:9–26. [Google Scholar]
- 4.Skoglund P, et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336(6080):466–469. doi: 10.1126/science.1216304. [DOI] [PubMed] [Google Scholar]
- 5.Skoglund P, et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science. 2014;344(6185):747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- 6.Keller A, et al. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat Commun. 2012;3:698. doi: 10.1038/ncomms1701. [DOI] [PubMed] [Google Scholar]
- 7.Gamba C, et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat Commun. 2014;5:5257. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513(7518):409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522(7555):207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522(7555):167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 11.Haak W, et al. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310(5750):1016–1018. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- 12.Bramanti B, et al. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science. 2009;326(5949):137–140. doi: 10.1126/science.1176869. [DOI] [PubMed] [Google Scholar]
- 13.Warren G. The adoption of agriculture in Ireland: Perceptions of key research challenges. J Archaeol Method Theory. 2013;20(4):525–551. [Google Scholar]
- 14.Cooney G, Grogan E. Irish Prehistory: A Social Perspective. Wordwell; Dublin: 1994. [Google Scholar]
- 15.Jones C, Mcveigh TO, Maolduin R. Monuments landscape and identity in Chalcolithic Ireland. In: Springs KD, editor. Landscape and Identity; Archaeology and Human Geography. Archaeopress; Oxford: 2015. pp. 3–25. [Google Scholar]
- 16.Sloan B, Carver N, Murphy E, McGranaghan C. A Bronze Age cist burial at Glebe, Rathlin Island, County Antrim. Ulster J Archaeology. 2008;67:60–83. [Google Scholar]
- 17.Brandt G, et al. Genographic Consortium Ancient DNA reveals key stages in the formation of central European mitochondrial genetic diversity. Science. 2013;342(6155):257–261. doi: 10.1126/science.1241844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacan M, et al. Ancient DNA reveals male diffusion through the Neolithic Mediterranean route. Proc Natl Acad Sci USA. 2011;108(24):9788–9791. doi: 10.1073/pnas.1100723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore LT, McEvoy B, Cape E, Simms K, Bradley DG. A Y-chromosome signature of hegemony in Gaelic Ireland. Am J Hum Genet. 2006;78(2):334–338. doi: 10.1086/500055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myres NM, et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur J Hum Genet. 2011;19(1):95–101. doi: 10.1038/ejhg.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olalde I, et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature. 2014;507(7491):225–228. doi: 10.1038/nature12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Q, et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514(7523):445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seguin-Orlando A, et al. Paleogenomics. Genomic structure in Europeans dating back at least 36,200 years. Science. 2014;346(6213):1113–1118. doi: 10.1126/science.aaa0114. [DOI] [PubMed] [Google Scholar]
- 24.Raghavan M, et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014;505(7481):87–91. doi: 10.1038/nature12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Günther T, et al. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc Natl Acad Sci USA. 2015;112(38):11917–11922. doi: 10.1073/pnas.1509851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olalde I, et al. A common genetic origin for early farmers from Mediterranean Cardial and Central European LBK cultures. Mol Biol Evol. 2015 doi: 10.1093/molbev/msv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones ER, et al. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat Commun. 2015;6:8912. doi: 10.1038/ncomms9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192(3):1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461(7263):489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacLeod IM, Larkin DM, Lewin HA, Hayes BJ, Goddard ME. Inferring demography from runs of homozygosity in whole-genome sequence, with correction for sequence errors. Mol Biol Evol. 2013;30(9):2209–2223. doi: 10.1093/molbev/mst125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pemberton TJ, et al. Genomic patterns of homozygosity in worldwide human populations. Am J Hum Genet. 2012;91(2):275–292. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505(7481):43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8(1):e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leslie S, et al. Wellcome Trust Case Control Consortium 2; International Multiple Sclerosis Genetics Consortium The fine-scale genetic structure of the British population. Nature. 2015;519(7543):309–314. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hellenthal G, et al. A genetic atlas of human admixture history. Science. 2014;343(6172):747–751. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devaney J, et al. Cystic fibrosis mutation frequencies in an Irish population. Clin Genet. 2003;63(2):121–125. doi: 10.1034/j.1399-0004.2003.00017.x. [DOI] [PubMed] [Google Scholar]
- 39.Distante S, et al. The origin and spread of the HFE-C282Y haemochromatosis mutation. Hum Genet. 2004;115(4):269–279. doi: 10.1007/s00439-004-1152-4. [DOI] [PubMed] [Google Scholar]
- 40.Murphy M, et al. Genetic basis of transferase-deficient galactosaemia in Ireland and the population history of the Irish Travellers. Eur J Hum Genet. 1999;7(5):549–554. doi: 10.1038/sj.ejhg.5200327. [DOI] [PubMed] [Google Scholar]
- 41.Blanco I, Fernández E, Bustillo EF. Alpha-1-antitrypsin PI phenotypes S and Z in Europe: An analysis of the published surveys. Clin Genet. 2001;60(1):31–41. doi: 10.1034/j.1399-0004.2001.600105.x. [DOI] [PubMed] [Google Scholar]
- 42.Itan Y, Jones BL, Ingram CJE, Swallow DM, Thomas MG. A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol Biol. 2010;10:36. doi: 10.1186/1471-2148-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fielding JF, Harrington MG, Fottrell PF. The incidence of primary hypolactasia amongst native Irish. Ir J Med Sci. 1981;150(9):276–277. doi: 10.1007/BF02938254. [DOI] [PubMed] [Google Scholar]
- 44.Byrnes V, et al. Genetic hemochromatosis, a Celtic disease: Is it now time for population screening? Genet Test. 2001;5(2):127–130. doi: 10.1089/109065701753145583. [DOI] [PubMed] [Google Scholar]
- 45.Lucotte G, Dieterlen F. A European allele map of the C282Y mutation of hemochromatosis: Celtic versus Viking origin of the mutation? Blood Cells Mol Dis. 2003;31(2):262–267. doi: 10.1016/s1079-9796(03)00133-5. [DOI] [PubMed] [Google Scholar]
- 46.Walsh S, et al. The HIrisPlex system for simultaneous prediction of hair and eye colour from DNA. Forensic Sci Int Genet. 2013;7(1):98–115. doi: 10.1016/j.fsigen.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 47.Whitehouse NJ, et al. Neolithic agriculture on the European western frontier: The boom and bust of early farming in Ireland. J Archaeol Sci. 2014;51(0):181–205. [Google Scholar]
- 48.Thomas J. 2004 Current debates on the Mesolithic-Neolithic transition in Britain and Ireland. Documenta Praehistorica. Available at: arheologija.ff.uni-lj.si/documenta/pdf31/31thomas.pdf.
- 49.Bonsall C, Macklin MG, Anderson DE, Payton RW. Climate change and the adoption of agriculture in north-west Europe. Eur J Archaeol. 2002;5(1):9–23. [Google Scholar]
- 50.Sheridan JA. The Neolithization of Britain and Ireland: The big picture. In: Finlayson B, Warren G, editors. Landscapes in Transition. Oxbow; Oxford: 2010. pp. 89–105. [Google Scholar]
- 51.Collard M, Edinborough K, Shennan S, Thomas MG. Radiocarbon evidence indicates that migrants introduced farming to Britain. J Archaeol Sci. 2010;37(4):866–870. [Google Scholar]
- 52.Naugler C. Hemochromatosis: A Neolithic adaptation to cereal grain diets. Med Hypotheses. 2008;70(3):691–692. doi: 10.1016/j.mehy.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 53.Whittington C. The C282Y mutation may have been positively selected as it mitigates the infertility of celiac disease. Med Hypotheses. 2006;66(4):769–772. doi: 10.1016/j.mehy.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 54.Uzoigwe OF. The distribution of the parasitic fauna dictates the distribution of the haemochromatosis genes. Med Hypotheses. 2010;75(5):415–417. doi: 10.1016/j.mehy.2008.12.057. [DOI] [PubMed] [Google Scholar]
- 55.Yang DY, Eng B, Waye JS, Dudar JC, Saunders SR. Technical note: Improved DNA extraction from ancient bones using silica-based spin columns. Am J Phys Anthropol. 1998;105(4):539–543. doi: 10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 56.MacHugh DE, Edwards CJ, Bailey JF, Bancroft DR, Bradley DG. The extraction and analysis of ancient DNA from bone and teeth: A survey of current methodologies. Anc Biomol. 2000;3:81. [Google Scholar]
- 57.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010(6):db.prot5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 58.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17(1):10–12. [Google Scholar]
- 59.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29(13):1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKenna A, et al. The Genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.