Significance
Many analytes in real-life samples, such as body fluids, soil contaminants, and explosives, are dispersed in liquid, solid, or air phases. However, it remains a challenge to create a platform to detect these analytes in all of these phases with high sensitivity and specificity. Here, we demonstrate a universal platform termed slippery liquid-infused porous surface-enhanced Raman scattering (SLIPSERS) that enables the enrichment and delivery of analytes originating from various phases into surface-enhanced Raman scattering (SERS)-sensitive sites and their subsequent detection down to the subfemtomolar level (<10−15 mol⋅L−1). Based on SLIPSERS, we have demonstrated detection of various chemicals, biological molecules, and environmental contaminants with high sensitivity and specificity. Our platform may lead to ultrasensitive molecular detection for applications related to analytical chemistry, diagnostics, environmental monitoring, and national security.
Keywords: spectroscopy, sensing, SERS, slippery surfaces, nanoparticles
Abstract
Detecting target analytes with high specificity and sensitivity in any fluid is of fundamental importance to analytical science and technology. Surface-enhanced Raman scattering (SERS) has proven to be capable of detecting single molecules with high specificity, but achieving single-molecule sensitivity in any highly diluted solutions remains a challenge. Here we demonstrate a universal platform that allows for the enrichment and delivery of analytes into the SERS-sensitive sites in both aqueous and nonaqueous fluids, and its subsequent quantitative detection of Rhodamine 6G (R6G) down to ∼75 fM level (10−15 mol⋅L−1). Our platform, termed slippery liquid-infused porous surface-enhanced Raman scattering (SLIPSERS), is based on a slippery, omniphobic substrate that enables the complete concentration of analytes and SERS substrates (e.g., Au nanoparticles) within an evaporating liquid droplet. Combining our SLIPSERS platform with a SERS mapping technique, we have systematically quantified the probability, p(c), of detecting R6G molecules at concentrations c ranging from 750 fM (p > 90%) down to 75 aM (10−18 mol⋅L−1) levels (p ≤ 1.4%). The ability to detect analytes down to attomolar level is the lowest limit of detection for any SERS-based detection reported thus far. We have shown that analytes present in liquid, solid, or air phases can be extracted using a suitable liquid solvent and subsequently detected through SLIPSERS. Based on this platform, we have further demonstrated ultrasensitive detection of chemical and biological molecules as well as environmental contaminants within a broad range of common fluids for potential applications related to analytical chemistry, molecular diagnostics, environmental monitoring, and national security.
Ultrasensitive detection of chemicals and biological species is important in a broad range of scientific and technological fields ranging from analytical chemistry, materials, and biomolecular diagnostics (1–5) to the inspection of pollutants, explosives, and pharmaceutical drugs (6–8). Among various analytical techniques, surface-enhanced Raman scattering (SERS) is among the most promising methods in detecting trace amounts of molecules owing to its high molecular specificity (i.e., differentiation between different types of molecules) and high sensitivity (i.e., the lowest analyte concentration from which SERS signals are distinguishable from the noise signal of a control sample) (9–17). Extensive studies have focused on the structural optimization of the SERS substrate to improve SERS sensitivity (18–22), but there are two important roadblocks that limit its practical applications. First, SERS detection in liquid media relies on highly statistical binding of analytes to the SERS-sensitive regions (or “hot spots”), a consequence of the diffusive nature of the analytes (23–27). Therefore, it is extremely challenging to achieve single-molecule detection of any SERS-active analytes in highly diluted solutions (i.e., below femtomolar concentrations). Second, many real-life analytes such as environmental contaminants and explosives may be dispersed in liquid or gas phases or may be bound to solid substrates (e.g., soil), which may require the use of nonaqueous solvents for extraction. Various approaches have been explored to improve SERS sensitivity in aqueous solvents (16, 28, 29). Among them, using superhydrophobic surfaces to overcome the “diffusion limit” of analytes in highly diluted aqueous solutions is the most successful technique (16). However, there are still no effective ways to achieve high SERS sensitivity at femtomolar concentrations or below in nonaqueous solvents. The ability to detect analytes in both aqueous and nonaqueous fluids at subfemtomolar levels using SERS would lead to many important real-world applications.
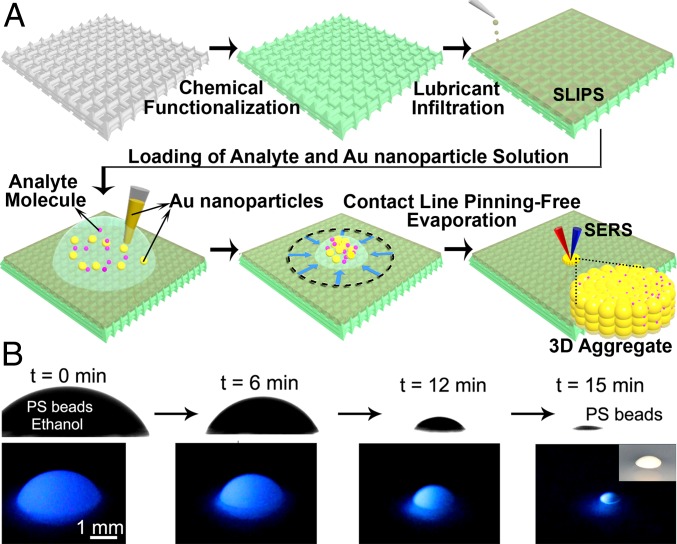
Here, we report a SERS platform based on pitcher plant-inspired slippery liquid-infused porous surfaces (SLIPS) (30), which provides a nearly pinning-free substrate for enriching and delivering analytes into a specific SERS detection area in both aqueous and, most importantly, nonaqueous liquids (Fig. 1A). A liquid droplet is nearly pinning-free when its contact line at the solid–liquid–air interface experiences small resistance to movement on a substrate. SLIPS consists of a film of lubricating fluid locked in place by a micro/nanoporous substrate, creating a smooth and stable interface that nearly eliminates pinning of the liquid contact line. Specifically, a recent study has shown that liquid pinning force on SLIPS for a low-surface-tension fluid (e.g., pentane) is an order of magnitude lower than state-of-the-art superhydrophobic surfaces (30). Based on this slippery surface, we can incorporate any SERS substrates in the form of metallic nanoparticles into either aqueous or nonaqueous fluids or their mixtures for subsequent analyte detection. This sensing platform, termed “slippery liquid-infused porous surface-enhanced Raman scattering” (SLIPSERS), is capable of ultrasensitive detection in most commonly used aqueous and nonaqueous solvents at concentrations as low as the attomolar level. Based on this platform, we have demonstrated multiphase and multiplex ultrasensitive detection of biological molecules and environmental contaminants, as well as the detection of airborne chemical molecules and soil contaminants extracted by various solvents.
Fig. 1.
Concept of SLIPSERS. (A) Schematic illustration showing the concept of SLIPSERS. Any analyte can be enriched to form a small aggregate, regardless of its interactions with the Au nanoparticles, facilitating subsequent SERS detections. (B) Visualization of the analyte enrichment process on SLIPS using luminescent PS spheres dispersed in ethanol. (Inset) Optical image of the enriched PS sphere aggregate.
Results and Discussion
Design and Characterizations of SLIPSERS.
To create SLIPSERS, we first prepared the slippery substrate by infiltrating a nano-textured surface (e.g., Teflon membranes with pore size of 200 nm) with a perfluorinated liquid (e.g., DuPont Krytox GPL 100). The perfluorinated liquid is used because it is immiscible to both the aqueous and nonaqueous phases (SI Appendix). To demonstrate that the surface is nearly free of pinning (30), we put a drop of ethanol loaded with fluorescent polystyrene (PS) beads and observed the subsequent evaporation profiles using a contact angle goniometer (SI Appendix, Fig. S1). We observed that the droplet evaporated in a constant contact angle mode without noticeable pinning at the contact line (31), until the particles clustered together to form a 3D aggregate (Fig. 1B). This mode of evaporation is consistent for various aqueous and nonaqueous colloidal solutions. The SLIPSERS platform has proven to be able to enrich particles with sizes ranging from subnanometer (e.g., small molecules and ions) to nanometer (e.g., colloids) to micrometer (e.g., PS beads) dispersed in most commonly used solvents (SI Appendix, Figs. S2–S5).
We have determined the analyte collection efficiency, defined as the percentage of total analyte in the solution that concentrates to form an aggregate, to be 97.3 ± 2.4% using copper sulfate solution (SI Appendix, Fig. S2). Owing to the high solute collection efficiency, Rhodamine 6G (R6G) molecules enriched from their ethanol solution even at subnanomolar concentrations can be observed with an unaided eye (SI Appendix, Fig. S3). To our knowledge, SLIPSERS is the only platform that can achieve nearly 100% analyte collection efficiency in most commonly used solvents. This can facilitate subsequent SERS sensing detection and significantly improve the limit of detection (LOD), which is defined as the detectable signals from the lowest analyte concentration with a signal-to-noise ratio greater than 3 in this work.
Analyte Detection in Nonaqueous Solutions by SLIPSERS.
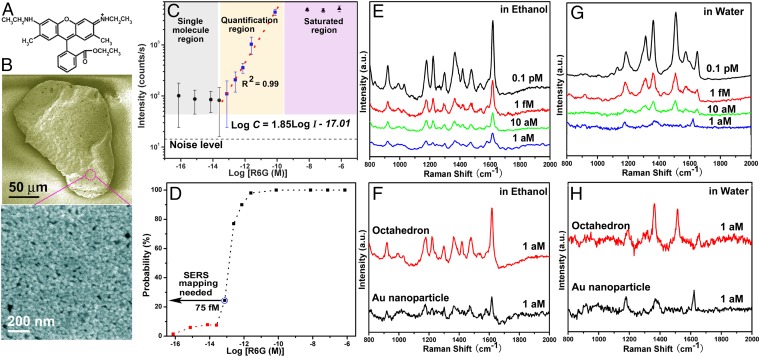
Although superhydrophobic surfaces have demonstrated ultrasensitive SERS detection in aqueous solutions (16, 32, 33), these substrates are not as effective at enriching analytes in nonaqueous solvents and biological fluids because these fluids can irreversibly pin to the micro- and nanoscale textures (30, 34–36). To demonstrate the detection capability of SLIPSERS in nonaqueous fluids, we investigate the LOD of SLIPSERS using R6G, a dye commonly used in detection tests, as a test analyte in ethanol (Fig. 2A). We chose ethanol as a representative nonaqueous liquid because of its low surface tension (∼22.3 mN/m at 20 °C)—which would cause significant pinning on superhydrophobic surfaces—and its common use as a standard solvent. In our investigation, we introduced 10 µL of ∼5 pM colloidal solution comprised of spherical Au nanoparticles (∼45 nm) (SI Appendix, Figs. S6 and S7) into 50 µL of R6G ethanol solutions of various concentrations. After the ethanol completely evaporated, a close-packed R6G/Au aggregate of ∼150 µm in diameter was formed (Fig. 2B). We estimated from our high-resolution electron micrographs that there are ∼2,000 Au nanoparticles per square micrometer (Fig. 2B). The closely packed Au nanoparticles form a large number of densely distributed hot spots for SERS detection, as supported by our finite-difference time-domain simulations (SI Appendix, Fig. S8) and SERS measurements performed on the R6G/Au aggregate.
Fig. 2.
Sensitive SERS detection of R6G molecules in aqueous and organic liquids. (A) Molecular structure of R6G molecule. (B) SEM images of the R6G/Au nanoparticle aggregate extracted from the mixture of the R6G ethanol and Au colloid solutions. (C) SERS intensity at 1,362 cm−1 as a function of R6G concentrations. Error bars indicate standard deviations from at least 20 spectra. (D) Probability of obtaining observable SERS signals at different concentrations. (E) SERS spectra of R6G molecules obtained from ethanol solutions at different concentrations. (F) Improvement of the signal intensity of 1 aM R6G in ethanol through the use of Ag octahedrons. (G) SERS spectra of R6G molecules obtained from aqueous solutions at different concentrations. (H) Improvement of the signal intensity of 1 aM R6G in aqueous solutions through the use of Ag octahedrons.
SERS measurements were performed over an area of 20 × 20 µm on the Au aggregate by performing a raster laser scan with the detection spot size of ∼1 µm in diameter. At least 400 different spectra were collected in the mapping area for quantitative and statistical SERS analysis (SI Appendix, Fig. S9). Quantitative SERS detection can be achieved when the change of the SERS signal corresponds to the change of the analyte concentration in a predictable manner. To evaluate the quantitative detection range of the SLIPSERS system, we measured R6G molecules inside ethanol solutions at concentrations ranging from 750 nM to 75 aM. Our systematic measurements showed that SLIPSERS can achieve quantitative detection in R6G concentrations ranging from ∼75 pM down to ∼75 fM (Fig. 2C). Within this concentration range, the SERS intensity at 1,362 cm−1 (C–C stretching mode of R6G) can be expressed quantitatively by an empirical equation, , where C is the R6G concentration expressed in molar concentration and I is the SERS intensity level (counts per second) (37, 38). The ability to achieve quantitative detection down to ∼75 fM is at least three orders of magnitude lower than conventional SERS substrates that do not use the analyte enrichment technique (39). When the concentration is higher than 75 pM, all of the SERS hot spots in the Au nanoparticle aggregates will be occupied by the analyte molecules. Therefore, analyte concentrations higher than 75 pM will have similar SERS signal intensity (i.e., the SERS signal intensity is saturated). However, when the concentration is lower than 75 fM, only few molecules exist within the laser spot area. The detection of molecular binding events would therefore require a mapping technique to extract the individual SERS signals (Fig. 2D).
Using the SERS mapping technique, notable SERS signals were observed at R6G concentration of 100 fM or below (Fig. 2E). As the concentration of R6G molecules further decreased down to 1 aM (10−18 mol⋅L−1), SERS signals could be detected only at random sites over a detection area of 20 × 20 µm. To investigate the probability of acquiring SERS signals of R6G at various concentrations, we performed systematic mapping measurements in the concentration range from 750 nM to 75 aM with a 10-time dilution (SI Appendix, Figs. S10–S17). When the R6G concentration is >750 fM, our measurements showed that the probability to obtain observable SERS signals is >90% (Fig. 2D). When the concentration reduces to 75 fM, the probability reduces to ∼25%. At this concentration, we also found that it is more likely to obtain SERS signals at the edge of the Au nanoparticle aggregate compared with the center region, possibly due to the coffee ring effect (40, 41). In the concentration range of subfemtomolar to ∼10 fM, the probability of observing SERS signals is <10% at the edge of the Au nanoparticle aggregate. This probability further reduced to 1.4% at a concentration of 75 aM (Fig. 2D). As a result, when the concentration of R6G molecules is <75 fM, we will need to rely on SERS mapping measurements to locate where the single-molecule binding events occur.
The LOD of SLIPSERS using organic liquids can also be further improved by simply increasing the starting solution volume (SI Appendix, Fig. S18) or by using SERS particles of different geometries. For example, detection of R6G molecules in ethanol solution using Ag octahedron particles (SI Appendix, Fig. S19) was achieved at 1 aM using SERS mapping measurement (Fig. 2F), which is at least five orders of magnitude lower than the reported LOD of conventional SERS techniques (15) (SI Appendix, Table S1). We further verified the repeatability of SLIPSERS in other commonly used organic solvents such as methanol, acetone, toluene, and dichloromethane, where an LOD of 1 fM could be achieved (SI Appendix, Fig. S20). Such a low LOD is enabled by the closely packed SERS hot spots within the Au nanoparticle aggregate, as well as the high collection efficiency of the R6G molecules on the SLIPSERS platform.
Analyte Detection in Aqueous Solutions by SLIPSERS.
In addition to the SERS detection in nonaqueous fluids, we studied detection of R6G in aqueous solutions using the SLIPSERS platform. We introduced 10 µL of ∼5 pM colloidal solution consisting of spherical Au nanoparticles (∼45 nm) into 50 µL of R6G aqueous solutions of various concentrations. Similar to the detection in nonaqueous phases, SERS mapping measurements were performed over an area of 20 × 20 µm across the R6G/Au nanoparticle aggregate. Notable SERS signals of R6G molecules could be observed even for aggregates formed using 1 aM R6G solutions (Fig. 2G), which is at least one order of magnitude improvement compared with the demonstrated detection limit based on superhydrophobic surfaces (16). This LOD can be further improved to 0.1 aM by simply increasing the volume of the starting solution (SI Appendix, Fig. S18) or by using SERS substrates of different geometries, such as Ag octahedrons (Fig. 2H and SI Appendix, Fig. S19).
In any of these analyte enrichment processes, we can further accelerate the solvent evaporation (e.g., <5 min for 50 µL of solution) by thermal treatment to achieve rapid analysis without influencing the LOD (SI Appendix, Figs. S21 and S22). In contrast, evaporation of 50-µL aqueous solutions on superhydrophobic surfaces will take several hours and the heat treatment will negatively affect the analyte enrichment efficiency (SI Appendix, Figs. S24–S26 and section S3). Our experimental characterizations demonstrated that SLIPSERS can reach subfemtomolar detection limits of analytes dispersed in nonaqueous and aqueous fluids, showing that the SLIPSERS platform outperforms other state-of-the-art SERS detection methods (21, 22, 42, 43) (SI Appendix, Table S1).
SLIPSERS in Liquid-Phase Detection.
To demonstrate the potential of SLIPSERS as a practical sensing platform, it is important to investigate its ability to detect technologically significant molecules in realistic conditions. For real-life samples, many of the analytes can be suspended in a liquid phase (e.g., biological fluids), dispersed in gas medium (e.g., explosives), or bound to solid substrates (e.g., soil). We have conducted experiments to investigate each of these three cases.
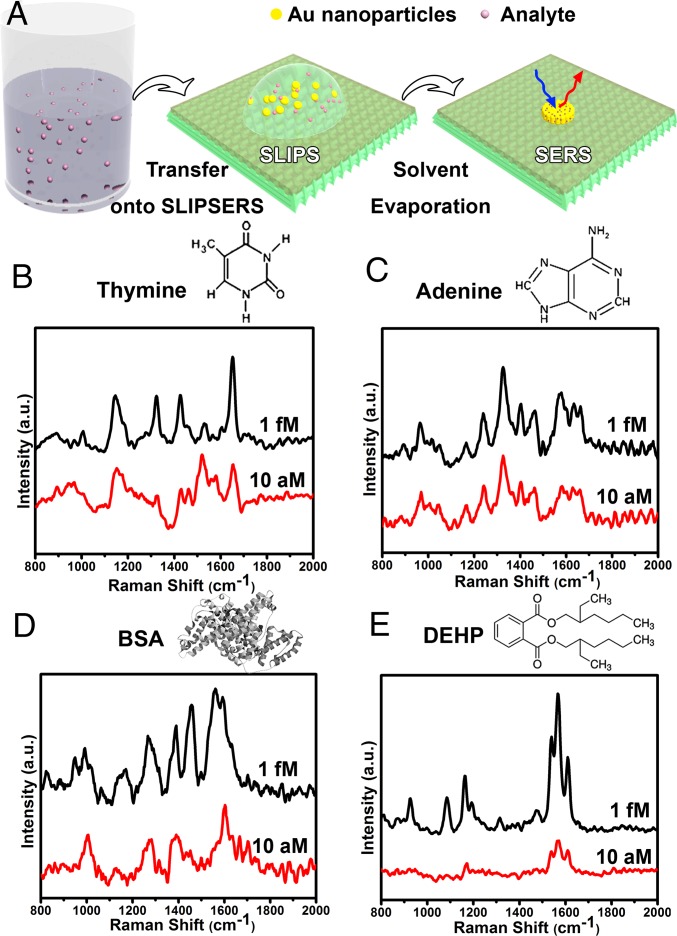
First, we demonstrated the ultrasensitive detection of biological species and environmental pollutants using various liquid media (Fig. 3A). Sensitive detection of biological species (i.e., biomarkers, antigens, etc.) is crucial in life sciences and medical diagnostics (1–5, 44, 45). Over the years, SERS has already proven to be a powerful platform to detect and analyze various biological entities such as proteins (46, 47), viruses (48), and cells (49) as well as DNA (e.g., discriminate mutations) (50, 51) and RNA (11). As proof-of-concept examples, we have shown that DNA bases of thymine and adenine as well as proteins such as bovine serum albumin (BSA) can be detected in water at subfemtomolar concentrations without additional labeling processes using SLIPSERS (Fig. 3 B–D). In addition to biological molecular detection, we have demonstrated the detection of environmental contaminants, such as bis(2-ethylhexyl) phthalate (DEHP). DEHP is an organic plasticizer commonly absorbed into food and water due to its low vapor pressure, and is only soluble in nonaqueous solvents. Using ethanol as the dispersion medium, we have shown that SLIPSERS is capable of detecting DEHP at subfemtomolar concentrations (Fig. 3E). Furthermore, multiplex molecular sensing of R6G (75 fM in ethanol) and adenine (37 fM in water) in multiple liquid phases can be achieved using the SLIPSERS platform (SI Appendix, Fig. S27). All of these demonstrations have shown that SLIPSERS is capable of detecting these molecules at ultralow concentrations with high specificity in either aqueous or nonaqueous solutions or their mixtures.
Fig. 3.
Liquid-phase detection of biological species and environmental pollutants using SLIPSERS. (A) Schematic illustration of the liquid phase detection using SLIPSERS. (B) Thymine in water. (C) Adenine in water. (D) BSA in water. (E) DEHP in ethanol. An initial volume of 50 µL of analyte solution was used here.
SLIPSERS in Gas-Phase Extraction and Detection.
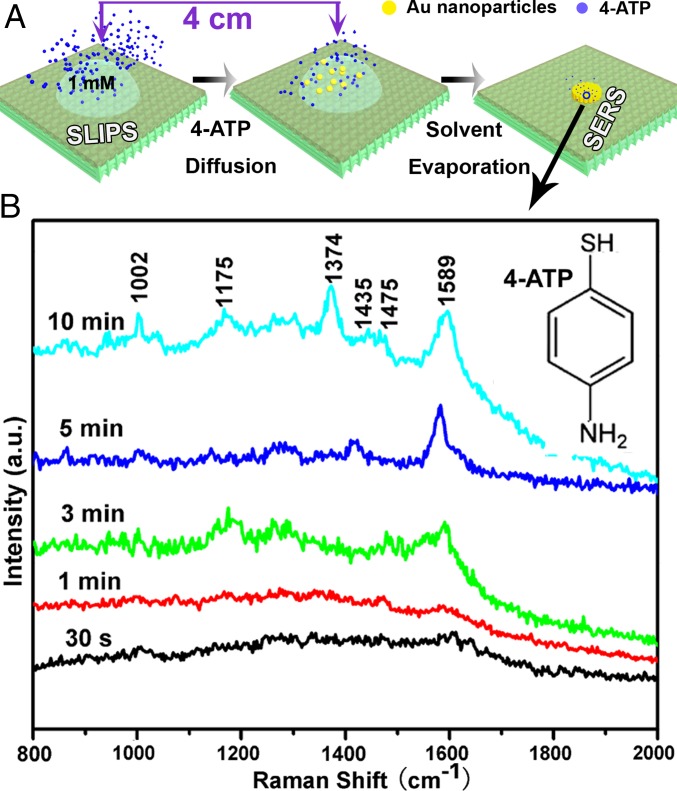
Next, we demonstrated that SLIPSERS is capable of detecting analytes in the gas phase using a suitable liquid solvent of high analyte solubility. Despite the significance of detecting explosives, poisonous airborne compounds, chemical warfare agents, and environmental pollutants dispersed in air, gas-phase SERS detection remains extremely challenging. Detection of these species generally needs specially designed molecule collection systems [e.g., electrodynamic precipitation (17)] and is limited to specific airborne species (52). We used our SLIPSERS for sensitive detection of airborne species without any periphery molecule collection equipment (Fig. 4).
Fig. 4.
Gas-phase detection using SLIPSERS. (A) Schematic showing the experimental procedures for airborne species detection. (B) SERS spectra of 4-ATP captured on the Au nanoparticle aggregate exposed to the 4-ATP environment at different amounts of time. After exposing for 10 min, most of the Raman peaks of 4-ATP molecules are distinguishable.
To demonstrate the ability of SLIPSERS to detect airborne analytes, 100 µL of 1-mM solution of 4-aminothiophenol (4-ATP) in ethanol was heated at 60 °C inside a Petri dish (14 cm in diameter and 1.4 cm in height, with a total volume of 215 cm3) to create 4-ATP molecules in the gas phase (Fig. 4A). Near the solution, we placed a 10-µL droplet of Au colloid solution diluted with 50 µL of ethanol to capture the 4-ATP molecules in the gas phase for about 10 min. After that, the ethanol droplet was allowed to dry up completely, and the Au–4-ATP aggregate was collected for further SERS analysis. If we assume that all of the analyte molecules (∼6 × 1016 molecules) are evenly distributed inside the enclosed container, the number of molecules attached onto the surface of the Au nanoparticle aggregate can be estimated to be ∼6 × 105 (i.e., a number density of ∼35 molecules per square micrometer). Note that this is an extreme case because there must be substantial 4-ATP residue in the solid state. However, the estimation provides us with the maximum number of molecules that would be available for surface adsorption from the vapor phase.
To compare with this estimated value, we have performed additional experiments to quantify the amount of 4-ATP molecules that were captured by the ethanol solutions. To achieve this, we compared the intensities of the Raman peaks of 4-ATP molecules from the aggregate sample (Fig. 4B) and a control sample obtained by drying 50 µL of 4-ATP ethanol solutions at various concentrations. We found that the intensity of the Raman peaks of the 4-ATP molecules of the aggregate sample (about 45 counts per second at 1,589 cm−1) is very close to that of the control sample at >180 fM (SI Appendix, Fig. S28). Thus, the number density of 4-ATP molecules on the Au nanoparticle aggregate is estimated to be >240 per square micrometer, which is much higher than the upper-limit estimation of ∼35 molecules per square micrometer. This indicates that using a suitable solvent can allow for efficient capturing of target analytes in air.
This proof-of-concept experiment demonstrates that it is possible to use SLIPSERS to detect gas-phase molecules given a suitable solvent. Because a broad range of liquids can be used in the SLIPSERS system, one could achieve gas-phase detection of different SERS-active airborne species by using appropriate solvents. This capability may open up a new opportunity toward practical applications of SLIPSERS in gas-phase detection, because field tests typically involve several airborne species that may need to be monitored at the same time.
SLIPSERS in Solid-Phase Extraction and Detection.
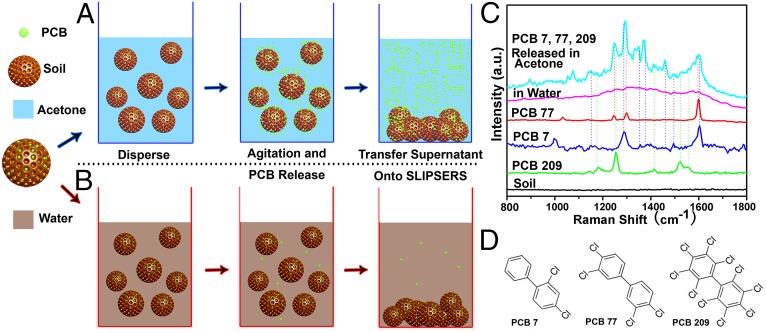
Finally, we demonstrated that SLIPSERS is capable of detecting analytes that are initially bound to solid substrates. We illustrated this concept by detecting soil contaminants. Real soil samples typically contain a complex mixture of contaminants, many of which need to be identified simultaneously. Among the soil contaminants, polychlorinated biphenyls (PCBs) can cause a variety of adverse health effects even at trace levels (53). Soil contamination by PCBs can be found all over the world and has become a serious worldwide problem (6). The maximum allowable contamination level in drinking water is 0.5 part per billion (ppb) set by the Environmental Protection Agency in the United States (54).
It is well known that the solubility of a chemical depends on the liquid in which it is dissolved. Therefore, proper solvents have to be used to release contaminants from the contaminated soil. As a proof-of-concept demonstration, we first contaminated soil with PCB 7 and PCB 209, each at a concentration of 1 ppb (SI Appendix). Then, the PCB-contaminated soil was dispersed into different liquids to release the PCB molecules (Fig. 5 A and B). After the soil precipitated, the supernatant was loaded onto our SLIPSERS platform to be analyzed (Fig. 1A). After obtaining a SERS spectrum from a sample, we can analyze the possible compositions of the analytes, as well as determine the quantity of each analyte in the sample. Through the use of commercially available software, we can calculate the location of Raman vibration modes of different molecules, facilitating the analysis of the samples with unknown or unspecified analytes.
Fig. 5.
Solid-phase detection of PCBs in contaminated soil using SLIPSERS. (A) Using acetone as dispersion liquid. PCB molecules can be released from the soil particulates. (B) Using water as dispersion liquid. No detectable PCB molecules are released to the liquid medium. (C) Multiplex SERS detection of PCB 7, 77, and 209 released from contaminated soil. (D) Molecular structures of PCB 7, 77, and 209.
It was found that when acetone was chosen as the dispersion liquid, strong SERS peaks of PCB 7 and PCB 209 could be observed even at such a low contamination level (i.e., 1 ppb) (SI Appendix, Fig. S29). We have further demonstrated multiplex detection of PCB 7, PCB 77, and PCB 209 released from the contaminated soil, each at a concentration of 1 ppb (Fig. 5C and SI Appendix, Fig. S30). As a control, no SERS peaks could be observed when water was used as the dispersion medium (Fig. 5C). To our knowledge, this is the first experimental demonstration of multiplex detection of PCBs from soil samples at a concentration on the order of 1 ppb despite their similar molecular structures (Fig. 5D).
The LOD of the SLIPSERS system is currently limited by the smallest size of the Au nanoparticle aggregate that can be formed on the platform. By further reducing the size of the Au nanoparticle aggregate, the analyte packing density will increase and the analyte distribution will become more uniform on the nanoparticle aggregate (i.e., more analytes can be packed within smaller surface area). This will in turn increase the probability of detecting analytes at very low concentrations and hence increase the accuracy of determining the analyte concentration. One method to reduce the overall size of the aggregate is to reduce the amount of Au nanoparticles used in the solution. However, using low concentrations of Au nanoparticles with our current particle enrichment method would lead to the formation of a very thin Au nanoparticle aggregate. This aggregate could be damaged by the laser beam used during the SERS measurements (SI Appendix, Fig. S7). Future studies should focus on the use of SLIPSERS in conjunction with supraparticles, which are preassembled, highly stable forms of Au nanoparticle aggregates (55). By using supraparticles of micrometer size or smaller, one may be able to further enhance the LOD, as well as the accuracy for determining the analyte concentration for extremely diluted solutions (i.e., <1 fM concentrations). In addition, although we demonstrated multiplex detection of three different molecules of similar chemical structures, the maximum number of molecules one could distinguish using our platform remains uncertain. Therefore, multiplex detection of various molecules in highly complex chemical and biological environments using SLIPSERS should be a subject of systematic investigation in the future.
Conclusion
In summary, SLIPSERS overcomes the long-standing limitations of SERS regarding collection and precise delivery of analytes to SERS hot spots in both aqueous and, most importantly, nonaqueous fluids. We have demonstrated that SLIPSERS is capable of detecting chemicals and biological species that are either dispersed in liquid or gas phases or bound to solid substrates. Furthermore, we have demonstrated that SLIPSERS is capable of quantitative detection down to ∼75 fM level. When combined with our SERS mapping technique, we have shown that it is possible to reach attomolar detection limits, which is the lowest LOD for any SERS-based detection reported in the literature. The detection limit could be lowered further by simply increasing the starting sample volume, by using engineered nanoparticles such as core-shell (14), core-gap-shell (20), or dumbbell structures (56), or by further reducing the size of Au nanoparticle aggregate (55). SLIPSERS could be integrated with other analytical techniques such as infrared spectroscopy, fluorescence, absorption spectroscopy, and photonic crystals (57) to further expand its capabilities for chemical, biological, and materials analysis. With all of the capabilities of SLIPSERS, we anticipate that it could be developed to meet the emerging needs in ultrasensitive biological sensing, environmental pollution monitoring, food safety evaluation, and defense applications.
Materials and Methods
SLIPS Preparation.
Teflon membranes (200-nm pore size and ∼70 µm in thickness) were attached on a flat glass slide or a concave glass bowl. Perfluorinated fluids (Dupont Krytox GPL 100) serving as lubricating liquids were then sprayed onto the Teflon membranes or the silanized silica bowl arrays to form an overcoat. The lubricated sample was then spun at 1,000 rpm for 1 min to remove the excess lubricant.
SERS Measurement.
SERS measurement was performed on a WITec confocal Raman instrument equipped with a 633-nm wavelength laser. We have performed SERS mapping measurements over an area of 20 × 20 µm on the Au aggregate by performing a raster laser scan with a detection spot size of ∼1 µm in diameter (SI Appendix, Figs. S9–S16). SERS spectra were extracted from sample locations with high SERS intensity (i.e., bright spots on the mapping results). The reproducibility of the SERS measurements was evaluated based on more than 20 independent SERS spectra at different analyte concentrations (SI Appendix, Fig. S9).
Analyte Detection on SLIPSERS.
Organic or aqueous solutions (50 µL, unless otherwise specified) containing analyte molecules at different concentrations were loaded onto the SLIPSERS platform with a pipette. Ten microliters of 5 pM Au colloidal solution was injected into a droplet of the analyte solution and mixed thoroughly.
After all of the solvent evaporated, the analyte and Au particles formed an aggregate that appeared as a small black dot visible to an unaided eye. SERS measurements were conducted on this aggregate, which consisted of closely packed Au nanoparticles and analyte molecules.
Detailed information about fabrication of SLIPS, Au nanoparticles, Ag octahedrons, and black Si as well as parameters used for SERS measurements and analyte detection on SLIPSERS can be found in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation (NSF) CAREER Award 1351462 and the Start-Up Fund from The Pennsylvania State University and by NSF Graduate Research Fellowship Grant DGE1255832 (to B.B.S.). Part of the work was conducted at the Pennsylvania State University node of the NSF-funded National Nanotechnology of Infrastructure Network.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518980113/-/DCSupplemental.
References
- 1.Homola J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem Rev. 2008;108(2):462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 2.Reiner JE, et al. Disease detection and management via single nanopore-based sensors. Chem Rev. 2012;112(12):6431–6451. doi: 10.1021/cr300381m. [DOI] [PubMed] [Google Scholar]
- 3.Zhang C-Y, Yeh H-C, Kuroki MT, Wang T-H. Single-quantum-dot-based DNA nanosensor. Nat Mater. 2005;4(11):826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 4.Kelley SO, et al. Advancing the speed, sensitivity and accuracy of biomolecular detection using multi-length-scale engineering. Nat Nanotechnol. 2014;9(12):969–980. doi: 10.1038/nnano.2014.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosaka PM, et al. Detection of cancer biomarkers in serum using a hybrid mechanical and optoplasmonic nanosensor. Nat Nanotechnol. 2014;9(12):1047–1053. doi: 10.1038/nnano.2014.250. [DOI] [PubMed] [Google Scholar]
- 6.Reiner EJ, Clement RE, Okey AB, Marvin CH. Advances in analytical techniques for polychlorinated dibenzo-p-dioxins, polychlorinated dibenzofurans and dioxin-like PCBs. Anal Bioanal Chem. 2006;386(4):791–806. doi: 10.1007/s00216-006-0479-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pushkarsky MB, et al. High-sensitivity detection of TNT. Proc Natl Acad Sci USA. 2006;103(52):19630–19634. doi: 10.1073/pnas.0609789104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker BR, et al. An electronic, aptamer-based small-molecule sensor for the rapid, label-free detection of cocaine in adulterated samples and biological fluids. J Am Chem Soc. 2006;128(10):3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 9.Nie S, Emory SR. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275(5303):1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 10.Kneipp K, et al. Single molecule detection using surface-enhanced Raman scattering (SERS) Phys Rev Lett. 1997;78(9):1667–1670. [Google Scholar]
- 11.Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297(5586):1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 12.Moskovits M. Surface-enhanced spectroscopy. Rev Mod Phys. 1985;57(3):783–826. [Google Scholar]
- 13.Anker JN, et al. Biosensing with plasmonic nanosensors. Nat Mater. 2008;7(6):442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 14.Li JF, et al. Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nature. 2010;464(7287):392–395. doi: 10.1038/nature08907. [DOI] [PubMed] [Google Scholar]
- 15.Cecchini MP, Turek VA, Paget J, Kornyshev AA, Edel JB. Self-assembled nanoparticle arrays for multiphase trace analyte detection. Nat Mater. 2013;12(2):165–171. doi: 10.1038/nmat3488. [DOI] [PubMed] [Google Scholar]
- 16.De Angelis F, et al. Breaking the diffusion limit with super-hydrophobic delivery of molecules to plasmonic nanofocusing SERS structures. Nat Photonics. 2011;5(11):682–687. [Google Scholar]
- 17.Lin E-C, Fang J, Park S-C, Johnson FW, Jacobs HO. Effective localized collection and identification of airborne species through electrodynamic precipitation and SERS-based detection. Nat Commun. 2013;4:1636. doi: 10.1038/ncomms2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson JB, Halas NJ. Surface-enhanced Raman scattering on tunable plasmonic nanoparticle substrates. Proc Natl Acad Sci USA. 2004;101(52):17930–17935. doi: 10.1073/pnas.0408319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham D, Thompson DG, Smith WE, Faulds K. Control of enhanced Raman scattering using a DNA-based assembly process of dye-coded nanoparticles. Nat Nanotechnol. 2008;3(9):548–551. doi: 10.1038/nnano.2008.189. [DOI] [PubMed] [Google Scholar]
- 20.Lim D-K, et al. Highly uniform and reproducible surface-enhanced Raman scattering from DNA-tailorable nanoparticles with 1-nm interior gap. Nat Nanotechnol. 2011;6(7):452–460. doi: 10.1038/nnano.2011.79. [DOI] [PubMed] [Google Scholar]
- 21.Yang S, Cai W, Kong L, Lei Y. Surface nanometer-scale patterning in realizing large-scale ordered arrays of metallic nanoshells with well-defined structures and controllable properties. Adv Funct Mater. 2010;20(15):2527–2533. [Google Scholar]
- 22.Yang S, et al. Large-scale fabrication of three-dimensional surface patterns using template-defined electrochemical deposition. Adv Funct Mater. 2013;23(6):720–730. doi: 10.1002/adfm.201201466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Squires TM, Messinger RJ, Manalis SR. Making it stick: Convection, reaction and diffusion in surface-based biosensors. Nat Biotechnol. 2008;26(4):417–426. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 24.Fang Y, Seong N-H, Dlott DD. Measurement of the distribution of site enhancements in surface-enhanced Raman scattering. Science. 2008;321(5887):388–392. doi: 10.1126/science.1159499. [DOI] [PubMed] [Google Scholar]
- 25.Michaels AM, Jiang, Brus L. Ag nanocrystal junctions as the site for surface-enhanced Raman scattering of single Rhodamine 6G molecules. J Phys Chem B. 2000;104(50):11965–11971. [Google Scholar]
- 26.Bosnick KA, Jiang, Brus LE. Fluctuations and local symmetry in single-molecule Rhodamine 6G Raman scattering on silver nanocrystal aggregates. J Phys Chem B. 2002;106(33):8096–8099. [Google Scholar]
- 27.Dieringer JA, Lettan RB, 2nd, Scheidt KA, Van Duyne RP. A frequency domain existence proof of single-molecule surface-enhanced Raman spectroscopy. J Am Chem Soc. 2007;129(51):16249–16256. doi: 10.1021/ja077243c. [DOI] [PubMed] [Google Scholar]
- 28.Escobedo C, Brolo AG, Gordon R, Sinton D. Optofluidic concentration: plasmonic nanostructure as concentrator and sensor. Nano Lett. 2012;12(3):1592–1596. doi: 10.1021/nl204504s. [DOI] [PubMed] [Google Scholar]
- 29.Eftekhari F, et al. Nanoholes as nanochannels: Flow-through plasmonic sensing. Anal Chem. 2009;81(11):4308–4311. doi: 10.1021/ac900221y. [DOI] [PubMed] [Google Scholar]
- 30.Wong T-S, et al. Bioinspired self-repairing slippery surfaces with pressure-stable omniphobicity. Nature. 2011;477(7365):443–447. doi: 10.1038/nature10447. [DOI] [PubMed] [Google Scholar]
- 31.Erbil HY, McHale G, Newton MI. Drop evaporation on solid surfaces: Constant contact angle mode. Langmuir. 2002;18(7):2636–2641. [Google Scholar]
- 32.Yang S, et al. Superhydrophobic surface enhanced Raman scattering sensing using Janus particle arrays realized by site-specific electrochemical growth. J Mater Chem C Mater Opt Electron Devices. 2014;2014(3):542–547. doi: 10.1039/C3TC31635A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallace RA, et al. Superhydrophobic analyte concentration utilizing colloid-pillar array SERS substrates. Anal Chem. 2014;86(23):11819–11825. doi: 10.1021/ac5033947. [DOI] [PubMed] [Google Scholar]
- 34.Epstein AK, Wong T-S, Belisle RA, Boggs EM, Aizenberg J. Liquid-infused structured surfaces with exceptional anti-biofouling performance. Proc Natl Acad Sci USA. 2012;109(33):13182–13187. doi: 10.1073/pnas.1201973109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao X, Song Y, Jiang L. Applications of bio-inspired special wettable surfaces. Adv Mater. 2011;23(6):719–734. doi: 10.1002/adma.201002689. [DOI] [PubMed] [Google Scholar]
- 36.Liu TL, Kim C-J. Repellent surfaces. Turning a surface superrepellent even to completely wetting liquids. Science. 2014;346(6213):1096–1100. doi: 10.1126/science.1254787. [DOI] [PubMed] [Google Scholar]
- 37.Liu W, et al. Superhydrophobic Ag nanostructures on polyaniline membranes with strong SERS enhancement. Phys Chem Chem Phys. 2014;16(41):22867–22873. doi: 10.1039/c4cp02677b. [DOI] [PubMed] [Google Scholar]
- 38.Hill AV. The combinations of haemoglobin with oxygen and with carbon monoxide. I. Biochem J. 1913;7(5):471–480. doi: 10.1042/bj0070471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phan-Quang GC, Lee HK, Phang IY, Ling XY. Plasmonic colloidosomes as three-dimensional SERS platforms with enhanced surface area for multiphase sub-microliter toxin sensing. Angew Chem Int Ed Engl. 2015;54(33):9691–9695. doi: 10.1002/anie.201504027. [DOI] [PubMed] [Google Scholar]
- 40.Deegan RD, et al. Capillary flow as the cause of ring stains from dried liquid drops. Nature. 1997;389(6653):827–829. [Google Scholar]
- 41.Shen X, Ho C-M, Wong T-S. Minimal size of coffee ring structure. J Phys Chem B. 2010;114(16):5269–5274. doi: 10.1021/jp912190v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen B, et al. Green synthesis of large-scale highly ordered core@shell nanoporous Au@Ag nanorod arrays as sensitive and reproducible 3D SERS substrates. ACS Appl Mater Interfaces. 2014;6(18):15667–15675. doi: 10.1021/am505474n. [DOI] [PubMed] [Google Scholar]
- 43.Wang W, Li Z, Gu B, Zhang Z, Xu H. Ag@SiO2 core-shell nanoparticles for probing spatial distribution of electromagnetic field enhancement via surface-enhanced Raman scattering. ACS Nano. 2009;3(11):3493–3496. doi: 10.1021/nn9009533. [DOI] [PubMed] [Google Scholar]
- 44.Vollmer F, Arnold S. Whispering-gallery-mode biosensing: Label-free detection down to single molecules. Nat Methods. 2008;5(7):591–596. doi: 10.1038/nmeth.1221. [DOI] [PubMed] [Google Scholar]
- 45.Arnold S, et al. Whispering Gallery Mode Carousel--a photonic mechanism for enhanced nanoparticle detection in biosensing. Opt Express. 2009;17(8):6230–6238. doi: 10.1364/oe.17.006230. [DOI] [PubMed] [Google Scholar]
- 46.Kahraman M, Sur I, Çulha M. Label-free detection of proteins from self-assembled protein-silver nanoparticle structures using surface-enhanced Raman scattering. Anal Chem. 2010;82(18):7596–7602. doi: 10.1021/ac101720s. [DOI] [PubMed] [Google Scholar]
- 47.Costas C, et al. Using surface enhanced Raman scattering to analyze the interactions of protein receptors with bacterial quorum sensing modulators. ACS Nano. 2015;9(5):5567–5576. doi: 10.1021/acsnano.5b01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shanmukh S, et al. Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Lett. 2006;6(11):2630–2636. doi: 10.1021/nl061666f. [DOI] [PubMed] [Google Scholar]
- 49.Pallaoro A, Hoonejani MR, Braun GB, Meinhart CD, Moskovits M. Rapid identification by surface-enhanced Raman spectroscopy of cancer cells at low concentrations flowing in a microfluidic channel. ACS Nano. 2015;9(4):4328–4336. doi: 10.1021/acsnano.5b00750. [DOI] [PubMed] [Google Scholar]
- 50.Johnson RP, Richardson JA, Brown T, Bartlett PN. A label-free, electrochemical SERS-based assay for detection of DNA hybridization and discrimination of mutations. J Am Chem Soc. 2012;134(34):14099–14107. doi: 10.1021/ja304663t. [DOI] [PubMed] [Google Scholar]
- 51.Barhoumi A, Zhang D, Tam F, Halas NJ. Surface-enhanced Raman spectroscopy of DNA. J Am Chem Soc. 2008;130(16):5523–5529. doi: 10.1021/ja800023j. [DOI] [PubMed] [Google Scholar]
- 52.Piorek BD, et al. Free-surface microfluidic control of surface-enhanced Raman spectroscopy for the optimized detection of airborne molecules. Proc Natl Acad Sci USA. 2007;104(48):18898–18901. doi: 10.1073/pnas.0708596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van den Berg M, et al. Toxic equivalency factors (TEFs) for PCBs, PCDDs, PCDFs for humans and wildlife. Environ Health Perspect. 1998;106(12):775–792. doi: 10.1289/ehp.98106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. US Environmental Protection Agency (2015) Drinking Water Contaminants – Standards and Regulations (US Environmental Protection Agency, Washington, DC). Available at www.epa.gov/dwstandardsregulations.
- 55.Thai T, et al. Self-assembly of vertically aligned gold nanorod arrays on patterned substrates. Angew Chem Int Ed Engl. 2012;51(35):8732–8735. doi: 10.1002/anie.201204609. [DOI] [PubMed] [Google Scholar]
- 56.Lim D-K, Jeon K-S, Kim HM, Nam J-M, Suh YD. Nanogap-engineerable Raman-active nanodumbbells for single-molecule detection. Nat Mater. 2010;9(1):60–67. doi: 10.1038/nmat2596. [DOI] [PubMed] [Google Scholar]
- 57.Kolle M, et al. Mimicking the colourful wing scale structure of the Papilio blumei butterfly. Nat Nanotechnol. 2010;5(7):511–515. doi: 10.1038/nnano.2010.101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.