Significance
The microbial Wood−Ljungdahl pathway is the biological equivalent of the Monsanto process, responsible for converting greenhouse gas CO2 into acetate. In addition to CO2, this microbial pathway requires low-potential electrons. The recently discovered oxalate oxidoreductase produces both low-potential electrons and CO2 through the oxidation of oxalate. Here our structural data allow us to visualize intermediates along the reaction cycle that have not been previously described, providing insight into the molecular mechanism by which oxalate is metabolized.
Keywords: oxalate, oxidoreductase, carbon dioxide, thiamine pyrophosphate
Abstract
Thiamine pyrophosphate (TPP)-dependent oxalate oxidoreductase (OOR) metabolizes oxalate, generating two molecules of CO2 and two low-potential electrons, thus providing both the carbon and reducing equivalents for operation of the Wood−Ljungdahl pathway of acetogenesis. Here we present structures of OOR in which two different reaction intermediate bound states have been trapped: the covalent adducts between TPP and oxalate and between TPP and CO2. These structures, along with the previously determined structure of substrate-free OOR, allow us to visualize how active site rearrangements can drive catalysis. Our results suggest that OOR operates via a bait-and-switch mechanism, attracting substrate into the active site through the presence of positively charged and polar residues, and then altering the electrostatic environment through loop and side chain movements to drive catalysis. This simple but elegant mechanism explains how oxalate, a molecule that humans and most animals cannot break down, can be used for growth by acetogenic bacteria.
Oxalate is a difficult molecule to metabolize, so much so that humans and most animals lack the capacity to break it down. This inability to metabolize oxalate is associated with chronic renal disease in humans, some of the manifestations of which are painful kidney stones, crystalline arthritis, and total kidney failure (1–3). Plants, fungi, and some Gram-positive bacteria have evolved oxygen-dependent pathways for metabolizing oxalate with the help of manganese-dependent enzymes in the cupin family of proteins (4, 5). These aerobic enzymes, oxalate oxidase and oxalate decarboxylase, take advantage of the propensity of oxalate to ligate metal ions to form a Mn oxalate complex, following which dioxygen is used to generate an oxalate-based radical species that causes rapid decarboxylation. The reduced products of these enzymes are hydrogen peroxide in the case of the oxidase and formate in the case of the decarboxylase.
Before 2010, only one enzyme pathway was known to catalyze the degradation of oxalic acid in the absence of dioxygen: oxalyl-CoA decarboxylase (6). This enzyme is best known for its role in the metabolism of Oxalobacter formigenes, a bacterium that colonizes the human gut and has been the subject of research into the role of the human gut microbiome in preventing, and possibly treating, human renal disease (7–9). As its name implies, however, oxalyl-CoA decarboxylase first requires the activation of oxalate by CoA, a feat that is accomplished by a formyl-CoA transferase. This activated oxalate, featuring a thioester in place of a carboxylate, is susceptible to nucleophilic attack by thiamin pyrophosphate (TPP; SI Appendix, Fig. S1A), which is the catalytic cofactor in this anaerobic enzyme. The end product of this reaction is formate, which can either enter cellular metabolism or be pumped out of the cell by a formate:oxalate antiporter to generate ATP (8).
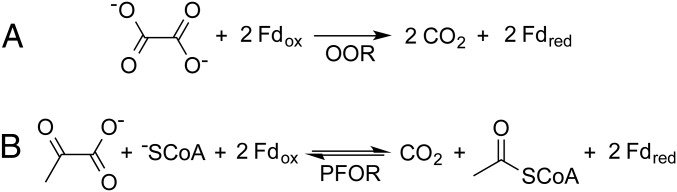
Recently, an oxalate oxidoreductase (OOR) was reported that performs a previously unknown metabolic transformation of oxalate by splitting it into two molecules of carbon dioxide and recovering two reducing equivalents for use in cellular processes (Fig. 1A) (10). This OOR, discovered in Moorella thermoacetica (11, 12), feeds both carbon dioxide and low-potential electrons into the Wood−Ljungdahl pathway for acetogenesis, allowing M. thermoacetica to grow exclusively on oxalate—a boon to this anaerobic bacterium commonly found in soil, which is often rich in oxalate (13). Like oxalyl-CoA decarboxylase, OOR uses TPP as its catalytic cofactor, but differs in that it does not require CoA to activate oxalate, and it uses a chain of [4Fe-4S] clusters to store and transfer the electrons captured from the C−C bond.
Fig. 1.
OOR and PFOR catalyze the oxidation of oxalate and pyruvate, respectively, reducing two ferredoxin equivalents in the process. (A) The reaction scheme for OOR. (B) The reaction scheme for PFOR.
OOR belongs to a larger superfamily of microbial enzymes, known as 2-oxoacid:ferredoxin oxidoreductases (OFORs), which use TPP and [4Fe-4S] clusters to oxidize pyruvate and other cellular building blocks (14, 15). The best characterized family member is pyruvate:ferredoxin oxidoreductase (PFOR), for which X-ray structures of substrate-free and substrate-bound forms (SI Appendix, Fig. S1B) as well as some biochemical data are available for the Desulfovibrio africanus enzyme (Da PFOR) (16–19), and more biochemical and spectroscopic data are available for the M. thermoacetica enzyme (20–23). PFOR catalyzes the reversible cleavage of pyruvate to form CO2, acetyl-CoA, and two electrons that are capable of reducing low-potential ferredoxins (Fig. 1B). In this reaction, TPP acts as a nucleophile and forms a covalent adduct with pyruvate to initiate the subsequent decarboxylation. Although we assume the same will be true for TPP-dependent OOR, oxalate is a very different substrate than pyruvate and the other 2-oxoacid substrates of the OFOR superfamily, all of which are characterized by a ketone immediately adjacent to a carboxylic acid. Oxalate, on the other hand, is a diprotic acid and carries two negative charges at neutral pH (24), and is therefore less amenable to nucleophilic attack by a TPP cofactor.
To probe how OOR was adapted to act on oxalate, we previously solved the structure of OOR from M. thermoacetica in its resting state (25). Structural analysis revealed that OOR is a dimer of trimers, (αγβ)2, with each αγβ forming a catalytic unit that is composed of five domains (SI Appendix, Fig. S1 C and D). The active site, which houses the TPP and one of the three [4Fe-4S] clusters, appears to be more polar than that found in PFOR, consistent with OOR’s unique substrate (25). Here, we further interrogate substrate binding to OOR through soaking and cocrystallization experiments with oxalate. In addition to identifying residues responsible for activating oxalate, these structures allow us to consider the molecular mechanism responsible for conversion of oxalate to CO2 and release of oxalate-derived CO2 from TPP. Also, the previously determined OOR structure showed a closed active site (25), raising the question of how oxalate gains access to the active site and how CO2 dissociates from the enzyme. Here, structures reveal conformational changes that appear to gate substrate and product access.
Results
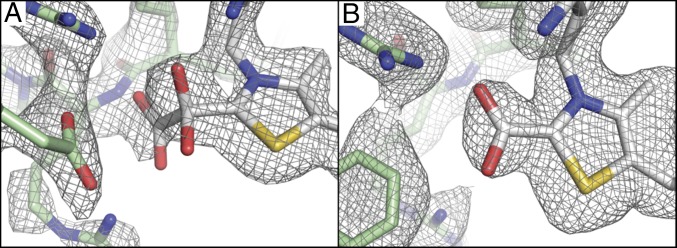
A crystal incubated with oxalate, which diffracted to 2.50-Å resolution, was found to have two OOR dimers in the asymmetric unit (see SI Appendix, Tables S1 and S3 for refinement statistics and a summary of all four monomeric units, respectively). In the active site of monomer 2, there was positive difference density adjacent to the C2 carbon of the TPP thiazole ring. A model for a carboxy-di-oxido-methyl-TPP (COOM-TPP) covalent adduct was refined in this active site, and was found to account well for the electron density (Fig. 2A and SI Appendix, Fig. S2A). The other three active sites of the oxalate-soaked crystal, monomers 1, 3, and 4, also had positive difference density adjacent to C2 of TPP, but the electron density was not consistent with a specific adduct and likely represents a mixture of states (SI Appendix, Table S3). This variation in the active sites of the four monomers appears to be due to differential lattice contacts, which alternatively serve to stabilize a closed active site in molecule 2 (the molecule with oxalate bound), and open active site states in the other molecules (see Discussion and SI Appendix, Fig. S11). To obtain support for the electron density at C2 of TPP being due to enzymatic turnover, methyl viologen was used as an indicator of the oxidation of oxalate to show that crystallized OOR retains activity (SI Appendix, Fig. S3).
Fig. 2.
Substrate-bound intermediates in OOR and corresponding composite-omit 2Fo-Fc (gray mesh) electron density maps contoured at 1 σ (see SI Appendix, Fig. S2 for stereoviews). (A) Composite-omit electron density of the COOM-TPP intermediate (white) and the side chains of nearby residues including Arg31α, Arg109α, and Asp116α (green) are shown as sticks. (B) Composite-omit electron density of the carboxy-TPP intermediate from monomer 1 of the oxalate cocrystal is shown with the same representation as in A, but, due to a conformational change of the Switch loop, Phe117α is now shown instead of Asp116α.
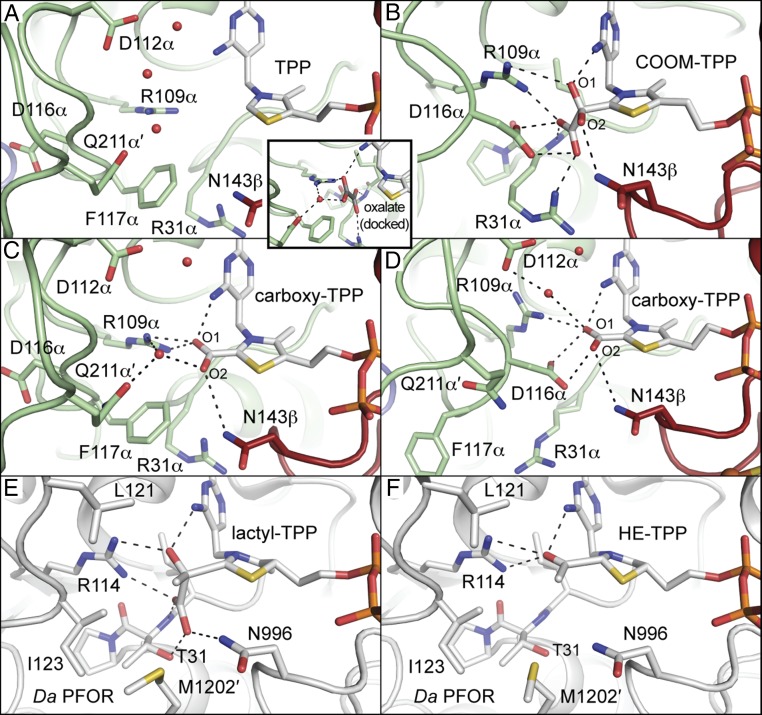
The structure of OOR with the COOM-TPP intermediate has allowed us to evaluate the previous proposal for how oxalate would bind to OOR. In particular, it was previously hypothesized (10, 25) that oxalate would bind in a similar fashion as pyruvate binds to PFOR, with the exception that replacement of PFOR’s 28YPITP32 motif with OOR’s 28YPIRP32 motif would provide oxalate with a hydrogen bond to the positively charged Arg31α instead of the neutral Thr31, countering the additional negative charge that oxalate has compared with pyruvate (Fig. 3A and SI Appendix, Fig. S4A) (25). Note that the Greek symbols after the residue number indicate the OOR chain ID (see SI Appendix, Fig. S1D). The structure presented here of OOR with oxalate covalently attached to TPP is consistent with this hypothesis. Arg31α occupies an analogous position to Thr31, and substrate interactions with Arg109α (OOR)/Arg114 (PFOR) and the imine group of the TPP iminopyrimidine are also conserved (Fig. 3 B and E). Additionally, Asn143β (OOR)/Asn996 (PFOR) is in a similar position to hydrogen bond with the bound substrate, although this 3.1-Å interaction appears to be with O2 of the bound oxalate, whereas, in PFOR, Asn996 interacts with the carboxylate group of the substrate (Fig. 3 B and E). To facilitate hydrogen-bonding interactions, rotation of the ethyl linker of TPP (that links the thiazole ring to the pyrophosphate moiety) changes the angle of the thiazole plane, tilting the bound substrate toward the various hydrogen bond donors. This ethyl linker rotation is akin to previously described TPP flexibility in PFOR (19), and it is not observed in any active sites in which oxalate is not bound.
Fig. 3.
Snapshots of OOR catalysis compared with PFOR with close interactions, <4 Å, shown as dashed lines. (A) The native structure of OOR (PDB ID: 5C4I) revealed a substrate-binding pocket formed by hydrophilic and positively charged residues. Select active site residues are also shown as stick models. Carbon atoms from domain I are colored green, whereas those from domain VI are colored dark red. Oxygen atoms are colored red, nitrogen atoms are colored blue, sulfur atoms are colored yellow, and phosphorous atoms are colored dark orange. Active site water molecules are shown as red spheres. For distances, see SI Appendix, Fig. S4. Inset shows a model of oxalate (with carbons colored dark gray) docked into the substrate-binding pocket based on a structure of PFOR with pyruvate (19). Proposed interactions between oxalate and surrounding protein side chains and water molecules are indicated with dashed lines. (B) The COOM-TPP intermediate from monomer 2 of the oxalate-soaked crystal is shown as in A. (C) The carboxy-TPP intermediate from monomer 1 of the oxalate cocrystal is shown as in A. (D) The carboxy-TPP intermediate from monomer 2 of the oxalate cocrystal is shown as above. (E) The lactyl-TPP intermediate observed in Da PFOR (19) (PDB ID: 2C3P) is shown with a similar representation as A, but with all carbons colored white. Substrate-binding residues are shown as sticks. (F) The HE-TPP intermediate observed in Da PFOR (18) (PDB ID: 1KEK) is shown as in E.
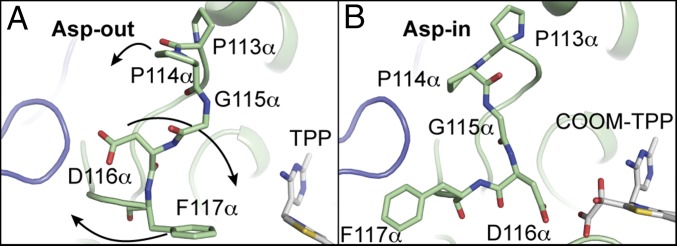
Unexpectedly, a close contact with the COOM intermediate is also made by Asp116α, which is part of a loop from residues 107–121 of chain α that helps form the substrate-binding pocket. In the previously reported structure (Fig. 4A), Asp116α points away from the active site and Phe117α points toward where substrate would bind (25) (an Asp-out conformation). However, in each of the four active sites of the oxalate-soaked structure, we see this situation reversed, with Asp116α rotated about 150° to point toward the carboxylate group of oxalate, Phe117α rotated about 90°, and the phenyl group swung away from the active site (Asp-in conformation) (Fig. 4B). To enable this rearrangement, a hairpin turn with sequence PPG (residues 113–115 of chain α) that immediately precedes Asp116α folds back away from the active site, allowing rotation of the peptide bonds of Asp116α and Phe117α. Together, these residues form what we will call the “Switch loop.” PFOR does not have an equivalent residue to Asp116α, nor does it have an alternate conformation of the loop that occupies the position equivalent to OOR’s Switch loop (17–19). Instead, the loop in PFOR, which resembles the Asp-out conformation of OOR’s Switch loop, contains Leu121 and Ile123, which are observed providing hydrophobic contacts for the methyl group of pyruvate (Fig. 3E and SI Appendix, Fig. S5).
Fig. 4.
Residues 113–117 of chain α in OOR form a Switch loop that has two alternate conformations. (A) The Asp-out conformation of the native OOR structure (PDB ID: 5C4I) is shown with the Switch residues as stick models. In this conformation, Phe117α is oriented toward the substrate-binding pocket. Arrows from Pro113α, Asp116α, and Phe117α indicate the direction of movement of these residues to go from Asp-out to Asp-in. (B) The Asp-in conformation from the oxalate-soaked structure is shown as in A. In this conformation, Asp116α is oriented toward the substrate-binding pocket. Structure is shown with the COOM-TPP intermediate in the active site.
Cocrystallization and Soaking Yield Different Intermediate Bound Structures.
The structure of OOR incubated with oxalate during the crystallization process was solved to 1.88-Å resolution and found to contain one dimer in the asymmetric unit (SI Appendix, Tables S1 and S4). In both active sites, electron density was observed adjacent to the C2 carbon of the TPP thiazole ring that was best modeled as the CO2 product bound to TPP (Fig. 2B and SI Appendix, Fig. S2B). We will refer to this intermediate as the fully oxidized carboxy-TPP intermediate, although it may also be the once-reduced carboxy-TPP radical intermediate, or the twice-reduced hydroxy- or dihydroxy(oxido)methylidene-TPP intermediate. These intermediates are indistinguishable at the resolution of the structure. In each case, the carboxy group was refined at partial occupancy (60–70%) (SI Appendix, Table S4). Difference density also indicates two distinct conformations of the Switch loop in both active sites. Both conformations of this loop (residues 108–119 of chains A and D) were modeled and refined with split occupancy (SI Appendix, Fig. S6). In the active site of monomer 1, the Asp-out conformation is predominant, whereas, in the other active site, monomer 2, the Asp-in conformation is more prevalent. Therefore, two active sites provide two different sets of environmental interactions with the carboxy-TPP intermediate. In monomer 1 (Fig. 3C and SI Appendix, Figs. S4C, S7A, and S8A), a number of potential hydrogen bond donors are making interactions with the bound CO2, including Arg109α, Asn143β, and Gln211α′ through an ordered water molecule (the prime indicates that residues come from the opposite monomeric unit of the OOR dimer). In monomer 2, the ordered water molecule has moved up in the active site to interact with Asp112α and, due to the hydrogen-bonding bond angle (Asp-H2O-[carboxy-TPP] = 143°), is likely not able to donate a hydrogen bond to the bound intermediate (Fig. 3D and SI Appendix, Figs. S4D, S7B, and S8B). Asp116α from the Switch loop is also nearby the bound intermediate, but is not positioned at an angle that suggests that it makes a hydrogen-bonding interaction. Instead, Asp116α may engage in a charge repulsion with the bound intermediate.
In addition to direct interactions with the active site, the Switch loop interacts with residues 211α′−214α′. In the Asp-in conformation, Pro214α′ is propped up by Phe117α through a stacking interaction, whereas, in the Asp-out conformation, Pro214α′ has moved down by 2.5 Å into the space left empty by Phe117α, which is now flipped almost 180° into the active site (SI Appendix, Fig. S9). The movement of Pro214α′ affects the other residues in its loop, 210α′ −220α′, resulting in two different loop conformations, which, like the Switch loop, were modeled with split occupancy. These distinct loop conformations for 210α′−220α′ alternatively position the side chain of Gln211α′ toward and away from the TPP-bound intermediate (Fig. 3 C and D and SI Appendix, Fig. S9), which in turn alters the positions of bound water molecules in the active site, as shown in SI Appendix, Figs. S7 and S8 and described above.
We can also compare these OOR structures with that of PFOR following its first decarboxylation (the so-called hydroxyethyl-TPP, or HE-TPP, species). Overall, the structures look similar (Fig. 3 C, D, and F). Key differences include the two positions of OOR’s Switch loop, and the fact that the side chain of Arg31α of the YPIRP motif (OOR) can have two positions (see below), whereas Thr31 of the YPITP motif (PFOR) cannot. Thus, OOR, which cannot use CoA, seems to make use of a protein conformational change to generate a number of interactions that are responsible for substrate activation, decarboxylation, and conversion to the carboxy-TPP intermediate.
Arg31α and Domain III Appear to Play a Role in Active Site Access.
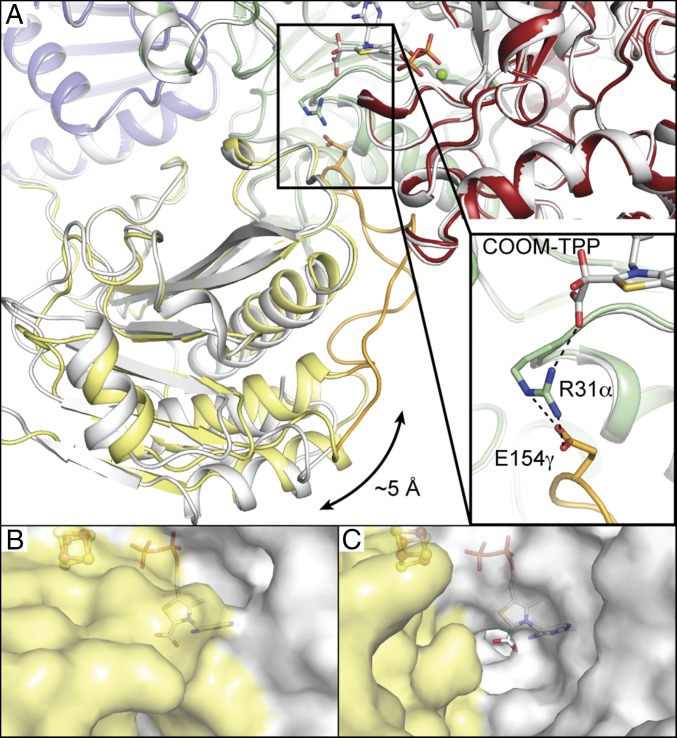
Arg31α is observed in two distinct conformations relative to the substrate-binding pocket (SI Appendix, Tables S3 and S4). Before and after oxalate is bound to TPP, Arg31α is positioned to make hydrogen-bonding interactions with oxalate (Fig. 3 A and B). In active sites modeled with carboxy-TPP, however, Arg31α is in two conformations: one where it is positioned at the bottom of the substrate-binding pocket as described above and one where it has swung away from the active site such that it is well away from the carboxy-TPP intermediate (Fig. 3 C and D). This swung-out conformation of Arg31α is also seen in other monomers of the soaked crystal, in which TPP was modeled without a bound adduct. As was noted previously (25), the swung-in conformation of Arg31α is stabilized by a salt bridge interaction with Glu154γ off of a domain III loop (Fig. 5A). This domain III loop, comprising residues 146γ−163γ, occupies a channel that leads directly to the active site and appears to regulate active site access (Fig. 5 B and C). Because this loop can seal the active site, we will refer to this loop as the “plug loop.”
Fig. 5.
Solvent access to the OOR active site is gated by the domain III plug loop. (A) Monomer 2 of the oxalate-soaked crystal (shown colored by domain) is superimposed upon monomer 4 from the same crystal (colored white). Domain III in monomer 2, colored yellow, is up to 5 Å closer to the active site than in monomer 4, as indicated by the arrow. The domain III plug loop of monomer 2, colored orange, is ordered and forms a salt bridge through Arg31α to the COOM-TPP intermediate, shown in Inset. (B) The plug loop is ordered in monomer 1 of the oxalate cocrystal. Carboxy-TPP and the proximal [4Fe-4S] cluster are shown buried underneath the protein surface. Domain III is colored yellow. (C) Domain III is rotated away in monomer 2 of the oxalate cocrystal, causing the plug loop to be disordered. Together with Arg31α being rotated away from the active site, a channel to the carboxy-TPP intermediate is visible.
When Arg31α is observed swung away from the active site, Glu154γ and the plug loop are no longer visible in the electron density, suggesting that they have enhanced flexibility. Further, these movements are accompanied by a large movement of domain III, which rotates outward such that the terminal connectors of the plug loop are displaced from the active site by up to 5 Å (Fig. 5A). Together, the movements of domain III and Arg31α appear to both open and close the active site (Fig. 5). Interestingly, Da PFOR has no plug loop off of domain III, instead using a unique domain VII to occupy this active site cavity. M1202′ from domain VII occupies an equivalent position to Arg31α (Fig. 3 E and F). Mutation of Da PFOR to remove domain VII results in enhanced activity, which is believed to be linked to the easier flow of substrates and products to and from the active site (26).
Discussion
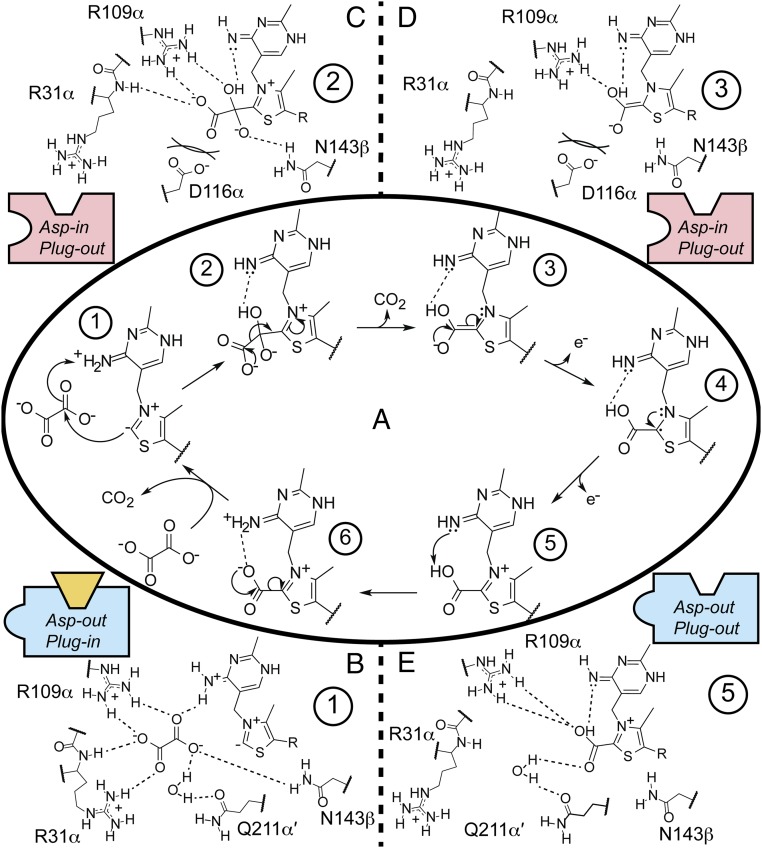
These structural snapshots of OOR inform our understanding of how the common soil component oxalate can be metabolized to provide both the carbon and the reducing equivalents to fuel the Wood−Ljungdahl pathway in M. thermoacetica (11, 12). Taking into account what is known about TPP reaction intermediates in PFOR (SI Appendix, Fig. S10) (10, 27) as well as our structural snapshots of OOR, we can propose a mechanism for OOR catalysis (Fig. 6).
Fig. 6.
Proposed mechanism for oxalate oxidation by OOR. (A) The catalytic cycle showing just TPP and substrate with intermediates numbered. Surrounding panels show proposed interactions of the surrounding protein residues with intermediates (B) 1, (C) 2, (D) 3, and (E) 5. Cartoon boxes indicate the position of the Switch and plug loops, with blue boxes indicating a more positively charged active site (Switch loop in Asp-out conformation), red boxes indicating a more negatively charged active site (Switch loop in Asp-in conformation), and yellow trapezoid indicating plug loop in plug-in conformation. Oxalate binding to the OOR active site and subsequent nucleophilic attack is facilitated by a number of hydrogen bond donating and positively charged residues. After the COOM-TPP intermediate is formed, movement of domain III (plug-in to plug-out) allows Arg31α to swing away from the active site, which, coupled with the Switch loop flipping to the Asp-in conformation, causes the net charge of the active site to become more negative. This change, and especially the interaction between Asp116α and COOM-TPP, may facilitate the first decarboxylation and two subsequent one-electron oxidations. The final deprotonation before the second decarboxylation may be facilitated by Asp116α flipping back out, allowing Gln211α′ to organize a water for hydrogen bonding to the TPP-bound acid.
The first step in the mechanism is the binding and activation of oxalate for nucleophilic attack by TPP (Fig. 6, state 1). Because neither carbon of oxalate is particularly electrophilic, it was expected that OOR would be designed such that it was capable of activating this dianion (10). Previously, we saw that the OOR active site is tailored to provide a number of positive charges and hydrogen bond donors (25). This tailoring includes substitution of PFOR’s 28YPITP32 motif with a 28YPIRP32 motif, which places Arg31α (instead of Thr31) in the active site (Fig. 3A). Also, the plug loop from domain III, which is not present in PFOR, provides Glu154γ, which can form a salt bridge with Arg31α and stabilize this Arg pointing in toward the active site.
The structure of COOM-TPP presented in this work agrees with the previous predictions about oxalate activation by OOR. Additionally, this structure provides unexpected insight into the next step in the reaction mechanism, i.e., the first decarboxylation to generate carboxy-TPP (Fig. 6, state 2 to state 3). Here, our structural data suggest that a flip of the Switch loop from the Asp-out to the Asp-in conformation could represent part of the driving force behind this first decarboxylation, because this loop flip results in a negative charge being in direct contact with the COOM-TPP intermediate (Fig. 6, state 2). Although we see oxalate (and not CO2) bound in the structure that has the Asp-in conformation, we believe that decarboxylation cannot occur within the active site of this molecule (molecule 2), due to lattice contacts. In particular, lattice contacts made with molecule 2 favor a conformation of domain III and its plug loop, which position Arg31α such that the positively charged side chain is interacting with the negatively charged oxalate (SI Appendix, Fig. S11). On the other hand, when lattice contacts stabilize domain III away from the active site (molecules 1, 3, and 4), the plug loop is disordered, Arg31α is swung out of the active site, and no density for oxalate is present. Thus, we propose that the first decarboxylation involves both the Switch loop adopting the Asp-in conformation and the Arg31α moving out of the substrate-binding pocket, which together create a net −2 charge difference between the state that binds and activates oxalate for TPP attack and the state that is responsible for the first decarboxylation (Fig. 6, state 1 to state 2).
In other words, we are proposing a “bait-and-switch” mechanism, in which the positively charged environment of the active site that includes Arg31α is the bait, and the introduction of the negatively charged Asp116α, and removal of the positively charged Arg31α, is the switch. The use of this Asp as part of the driving force for decarboxylation is consistent with the observation that the active sites of decarboxylases are either hydrophobic (stabilizing the production of hydrophobic CO2) (28) or negatively charged [destabilizing the negatively charged carboxylate (29, 30)]. Additionally, computational studies have shown that adjacent negative charges in protein environments can accelerate decarboxylation reactions by up to 108 (31). An example of this type of mechanism was described in indolocarbazole biosynthesis, in which substrate binds to a StaC enzyme variant before a mobile, negatively charged FAD swings into the active site to drive decarboxylation (32). Decarboxylation by adjacent negative charge is a simple mechanism that relies on the fundamental principle of charge−charge repulsion, and it is no surprise that enzymes make use of this principle to accomplish their goals.
Protonation of the COOM-TPP intermediate, which would reduce the overall negative charge of the TPP-bound adduct, is likely to be important in the first decarboxylation. By analogy to PFOR, we expect the imino group of the iminopyrimidine moiety of TPP to have an acidic proton and, in particular, to have the acidic proton that was removed from C2 of the TPP thiazole to activate the cofactor for catalysis (33) (Fig. 6, state 1). The oxygen atom of oxalate closest to the imino group (O1 in Fig. 3B) will likely pick up this acidic proton (Fig. 6, state 2). The other oxygen atom of oxalate (O2 in Fig. 3B), however, is not in close contact with any general acids, and so it may remain deprotonated. Asn143β, which is within hydrogen-bonding distance of O2, might stabilize a deprotonated O2 by donating a hydrogen bond. We expect that even monoprotonation will facilitate decarboxylation to form a hydroxy(oxido)methylidene-TPP (Fig. 6, state 2 to state 3). In order for this species to subsequently lose an electron to generate a carboxy-TPP radical intermediate (Fig. 6, state 4), charge−charge repulsion from Asp116α in the Asp-in conformation may once again help to overcome the barrier for this first oxidation.
A possible structure for the carboxy-TPP radical intermediate is shown in Fig. 6, state 4. This intermediate is analogous to the proposed state of the HE-TPP adduct in PFOR (SI Appendix, Fig. S10) (23, 27). Such neutral species are thought to have a high barrier for electron transfer (34). In particular, this radical species in PFOR is long-lived in the absence of CoA, and electron transfer to the [4Fe-4S] clusters happens slowly. Upon CoA binding to PFOR, however, the second electron transfer is accelerated 100,000-fold, and acetyl-CoA is eliminated as the reaction product (21). One explanation for how CoA binding to PFOR accelerates the second electron transfer step is that the additional negative charge brought to the active site by the deprotonated CoA thiol drives down the reduction potential of the HE-TPP radical, allowing the electron to move into the low-potential [4Fe-4S] clusters (21). Whether this happens before or after nucleophilic attack by CoA, however, is still a matter of investigation (27).
In contrast to the slow rate of oxidation of the neutral intermediate in PFOR in the absence of CoA, the electron transfer from the analogous intermediate state in OOR appears facile, as judged in vitro by the fact that OOR’s [4Fe-4S] clusters are reduced with no TPP-based radical observable within 30 s of initiating OOR catalysis (10). Thus, it is tempting to propose that oxidation of this neutral species in OOR could be driven by charge−charge repulsion from Asp116α in a manner that may be analogous to the binding of a negatively charged CoA in PFOR, although without the formation of a covalent adduct. This mechanistic idea is also consistent with the finding that OOR is capable of oxidizing glyoxylate in a CoA-independent fashion (10), although we do not have data that indicate the precise mechanism by which glyoxylate is oxidized.
Although Asp116α, in the Asp-in conformation, may facilitate the first decarboxylation step and the subsequent electron transfers, once the protonated TPP-carboxylic acid intermediate is reached (Fig. 6, state 5), interaction with Asp116α will likely raise the pKa of the bound carboxylic acid, hindering deprotonation and subsequent decarboxylation. Therefore, we propose a second rearrangement of the Switch loop back to the Asp-out conformation as observed in one of the two active sites with a carboxy-TPP intermediate (Fig. 3C). This movement not only removes the negatively charged Asp116α, but allows for Gln211α′ to return to the active site and for water molecules to rearrange such that they are in position to H-bond to the TPP-bound carboxylic acid. These H bonds could lower the pKa of the carboxy-TPP intermediate, allowing the imino group from the pyrimidine moiety to remove the remaining proton, facilitating the second and final decarboxylation (Fig. 6, state 6 to state 1).
Thus, through a series of structures of OOR from M. thermoacetica, we have gained insight into substrate access to the active site, substrate binding and activation by residues in the active site, and substantial protein movements that change the charge profile of the active site and drive catalysis. These protein motions, which involve a nearly 180° flip of an active site loop, the rotation of an entire 200-amino acid domain, and the disordering of a previously unknown 20-residue loop that plugs a channel to the active site, are without precedent in the OFOR superfamily and, as far as we know, in TPP-using enzymes at large. This mechanism is particularly of interest because of the difficulty with which oxalate is metabolized in nature. OOR is the only oxalate-degrading enzyme that does not require either the oxidizing power of dioxygen or the carbonyl-activating capabilities of CoA, instead making use of active-site residues to both bind and activate oxalate.
Methods
OOR was purified from M. thermoacetica by the methods described (10) and crystallized by hanging-drop vapor diffusion in an anoxic atmosphere. Native OOR crystals obtained in the previously described crystallization condition (25) were soaked in a 10-mM oxalate solution before cryocooling in liquid nitrogen to yield the oxalate-soaked structure. OOR-oxalate cocrystals were obtained by the addition of 10-mM oxalate to the previously described crystallization drop (25) before crystal formation. Both structures were solved by molecular replacement using the native structure of OOR (PDB ID 5C4I) (25) and refined to 2.50-Å and 1.88-Å resolution, respectively. Final R-work and R-free values for the OOR-oxalate-soaked structure were 19.6% and 22.9%, respectively, whereas those for the OOR-oxalate-cocrystal structure were 17.7% and 20.7%, respectively. Detailed protocols can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health (NIH) Grants GM069857 (to C.L.D.) and GM39451 (to S.W.R.), and by the National Science Foundation Graduate Research Fellowship under Grant 1122374 (to M.I.G.). This research was also made possible by the support of the Martin Family Society of Fellows for Sustainability (M.I.G.). C.L.D. is a Howard Hughes Medical Institute Investigator. This work is also based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by a grant from the National Institute of General Medical Sciences (P41 GM103403) from NIH. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5EXD and 5EXE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518537113/-/DCSupplemental.
References
- 1.Hodgkinson A. Oxalic Acid in Biology and Medicine. Academic; London: 1977. [Google Scholar]
- 2.Hoffman GS, et al. Calcium oxalate microcrystalline-associated arthritis in end-stage renal disease. Ann Intern Med. 1982;97(1):36–42. doi: 10.7326/0003-4819-97-1-36. [DOI] [PubMed] [Google Scholar]
- 3.Marengo SR, Romani AMP. Oxalate in renal stone disease: The terminal metabolite that just won’t go away. Nat Clin Pract Nephrol. 2008;4(7):368–377. doi: 10.1038/ncpneph0845. [DOI] [PubMed] [Google Scholar]
- 4.Kotsira VP, Clonis YD. Oxalate oxidase from barley roots: Purification to homogeneity and study of some molecular, catalytic, and binding properties. Arch Biochem Biophys. 1997;340(2):239–249. doi: 10.1006/abbi.1997.9896. [DOI] [PubMed] [Google Scholar]
- 5.Tanner A, Bornemann S. Bacillus subtilis YvrK is an acid-induced oxalate decarboxylase. J Bacteriol. 2000;182(18):5271–5273. doi: 10.1128/jb.182.18.5271-5273.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quayle JR. Carbon assimilation by Pseudomonas oxalaticus (Ox 1). 7. Decarboxylation of oxalyl-coenzyme A to formyl-coenzyme A. Biochem J. 1963;89:492–503. doi: 10.1042/bj0890492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allison MJ, Dawson KA, Mayberry WR, Foss JG. Oxalobacter formigenes gen. nov., sp. nov.: Oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol. 1985;141(1):1–7. doi: 10.1007/BF00446731. [DOI] [PubMed] [Google Scholar]
- 8.Anantharam V, Allison MJ, Maloney PC. Oxalate:formate exchange. The basis for energy coupling in Oxalobacter. J Biol Chem. 1989;264(13):7244–7250. [PubMed] [Google Scholar]
- 9.Sidhu H, et al. Direct correlation between hyperoxaluria/oxalate stone disease and the absence of the gastrointestinal tract-dwelling bacterium Oxalobacter formigenes: Possible prevention by gut recolonization or enzyme replacement therapy. J Am Soc Nephrol. 1999;10(Suppl 14):S334–S340. [PubMed] [Google Scholar]
- 10.Pierce E, Becker DF, Ragsdale SW. Identification and characterization of oxalate oxidoreductase, a novel thiamine pyrophosphate-dependent 2-oxoacid oxidoreductase that enables anaerobic growth on oxalate. J Biol Chem. 2010;285(52):40515–40524. doi: 10.1074/jbc.M110.155739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel SL, Drake HL. Oxalate- and glyoxylate-dependent growth and acetogenesis by Clostridium thermoaceticum. Appl Environ Microbiol. 1993;59(9):3062–3069. doi: 10.1128/aem.59.9.3062-3069.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel SL, Pilsl C, Drake HL. Oxalate metabolism by the acetogenic bacterium Moorella thermoacetica. FEMS Microbiol Lett. 2004;231(1):39–43. doi: 10.1016/S0378-1097(03)00924-8. [DOI] [PubMed] [Google Scholar]
- 13.Dutton MV, Evans CS. Oxalate production by fungi: Its role in pathogenicity and ecology in the soil environment. Can J Microbiol. 1996;42(9):881–895. [Google Scholar]
- 14.Kletzin A, Adams MW. Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J Bacteriol. 1996;178(1):248–257. doi: 10.1128/jb.178.1.248-257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mai X, Adams MW. Characterization of a fourth type of 2-keto acid-oxidizing enzyme from a hyperthermophilic archaeon: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J Bacteriol. 1996;178(20):5890–5896. doi: 10.1128/jb.178.20.5890-5896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pieulle L, et al. 1995. Isolation and characterization of the pyruvate-ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus. Biochim Biophys Acta Protein Struct Mol Enzymol 1250(1):49–59.
- 17.Chabrière E, et al. Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat Struct Biol. 1999;6(2):182–190. doi: 10.1038/5870. [DOI] [PubMed] [Google Scholar]
- 18.Chabrière E, et al. Crystal structure of the free radical intermediate of pyruvate:ferredoxin oxidoreductase. Science. 2001;294(5551):2559–2563. doi: 10.1126/science.1066198. [DOI] [PubMed] [Google Scholar]
- 19.Cavazza C, et al. Flexibility of thiamine diphosphate revealed by kinetic crystallographic studies of the reaction of pyruvate-ferredoxin oxidoreductase with pyruvate. Structure. 2006;14(2):217–224. doi: 10.1016/j.str.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Furdui C, Ragsdale SW. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J Biol Chem. 2000;275(37):28494–28499. doi: 10.1074/jbc.M003291200. [DOI] [PubMed] [Google Scholar]
- 21.Furdui C, Ragsdale SW. The roles of coenzyme A in the pyruvate:ferredoxin oxidoreductase reaction mechanism: Rate enhancement of electron transfer from a radical intermediate to an iron-sulfur cluster. Biochemistry. 2002;41(31):9921–9937. doi: 10.1021/bi0257641. [DOI] [PubMed] [Google Scholar]
- 22.Astashkin AV, Seravalli J, Mansoorabadi SO, Reed GH, Ragsdale SW. Pulsed electron paramagnetic resonance experiments identify the paramagnetic intermediates in the pyruvate ferredoxin oxidoreductase catalytic cycle. J Am Chem Soc. 2006;128(12):3888–3889. doi: 10.1021/ja0585275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansoorabadi SO, et al. EPR spectroscopic and computational characterization of the hydroxyethylidene-thiamine pyrophosphate radical intermediate of pyruvate:ferredoxin oxidoreductase. Biochemistry. 2006;45(23):7122–7131. doi: 10.1021/bi0602516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haynes WM, editor. CRC Handbook of Chemistry and Physics. 95th Ed CRC; Boca Raton, FL: 2015. [Google Scholar]
- 25.Gibson MI, et al. The structure of an oxalate oxidoreductase provides insight into microbial 2-oxoacid metabolism. Biochemistry. 2015;54(26):4112–4120. doi: 10.1021/acs.biochem.5b00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pieulle L, Magro V, Hatchikian EC. Isolation and analysis of the gene encoding the pyruvate-ferredoxin oxidoreductase of Desulfovibrio africanus, production of the recombinant enzyme in Escherichia coli, and effect of carboxy-terminal deletions on its stability. J Bacteriol. 1997;179(18):5684–5692. doi: 10.1128/jb.179.18.5684-5692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed GH, Ragsdale SW, Mansoorabadi SO. Radical reactions of thiamin pyrophosphate in 2-oxoacid oxidoreductases. Biochim Biophys Acta. 2012;1824(11):1291–1298. doi: 10.1016/j.bbapap.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemp DS, Cox DD, Paul KG. Physical organic chemistry of benzisoxazoles. IV. Origins and catalytic nature of the solvent rate acceleration for the decarboxylation of 3-carboxybenzisoxazoles. J Am Chem Soc. 1975;97(25):7312–7318. [Google Scholar]
- 29.Gallagher T, Snell EE, Hackert ML. Pyruvoyl-dependent histidine decarboxylase. Active site structure and mechanistic analysis. J Biol Chem. 1989;264(21):12737–12743. [PubMed] [Google Scholar]
- 30.Appleby TC, Kinsland C, Begley TP, Ealick SE. The crystal structure and mechanism of orotidine 5′-monophosphate decarboxylase. Proc Natl Acad Sci USA. 2000;97(5):2005–2010. doi: 10.1073/pnas.259441296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran NL, Colvin ME, Gronert S, Wu W. Catalysis of decarboxylation by an adjacent negative charge: A theoretical approach. Bioorg Chem. 2003;31(4):271–277. doi: 10.1016/s0045-2068(03)00028-2. [DOI] [PubMed] [Google Scholar]
- 32.Goldman PJ, et al. An unusual role for a mobile flavin in StaC-like indolocarbazole biosynthetic enzymes. Chem Biol. 2012;19(7):855–865. doi: 10.1016/j.chembiol.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breslow R. On the mechanism of thiamine action. IV.1 Evidence from studies on model systems. J Am Chem Soc. 1958;80(14):3719–3726. [Google Scholar]
- 34.Ragsdale SW. Pyruvate ferredoxin oxidoreductase and its radical intermediate. Chem Rev. 2003;103(6):2333–2346. doi: 10.1021/cr020423e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.