Significance
Based on published chromosome conformation capture data, we investigated the effect of 3D nuclear architecture in budding yeast on repair of a broken chromosome by homologous recombination. When a nuclease-induced double-strand break (DSB) is created at one locus, efficiency of repair depends on the contact frequency of the donor locus with the DSB site. When the location of the DSB is shifted to another location, the recombination rate of a donor can change by almost 20-fold. The rate of repair of a given locus is strongly influenced by four key factors: the size of donor homology, the rate of 5′ to 3′ resection of DSB ends, and the abundance of single-strand DNA binding protein complex, replication protein factor A (RPA), as well as a cis-acting recombination enhancer.
Keywords: homologous recombination, double-strand break repair, chromosome conformation, homology search, donor location
Abstract
Repair of a chromosomal double-strand break (DSB) by gene conversion depends on the ability of the broken ends to encounter a donor sequence. To understand how chromosomal location of a target sequence affects DSB repair, we took advantage of genome-wide Hi-C analysis of yeast chromosomes to create a series of strains in which an induced site-specific DSB in budding yeast is repaired by a 2-kb donor sequence inserted at different locations. The efficiency of repair, measured by cell viability or competition between each donor and a reference site, showed a strong correlation (r = 0.85 and 0.79) with the contact frequencies of each donor with the DSB repair site. Repair efficiency depends on the distance between donor and recipient rather than any intrinsic limitation of a particular donor site. These results further demonstrate that the search for homology is the rate-limiting step in DSB repair and suggest that cells often fail to repair a DSB because they cannot locate a donor before other, apparently lethal, processes arise. The repair efficiency of a donor locus can be improved by four factors: slower 5′ to 3′ resection of the DSB ends, increased abundance of replication protein factor A (RPA), longer shared homology, or presence of a recombination enhancer element adjacent to a donor.
Homologous recombination is the predominant mechanism to repair chromosome breaks and preserve genome integrity. In eukaryotes, the broken double-strand break (DSB) ends undergo extensive 5′ to 3′ resection, promoting the binding of the Rad51 recombinase to form a nucleoprotein filament that can search the genome for a homologous sequence with which it can effect repair. Donor template sequences can be located on a sister chromatid, a homologous chromosome or an ectopic location. When ectopic sequences are used, repair results in nonallelic replacements of sequences. In budding yeast Saccharomyces cerevisiae it is possible to monitor the sequence of DSB repair events in real time by Southern blots, PCR or chromatin immunoprecipitation (1).
Haploid yeast chromosomes are arranged in a Rabl orientation, with the 16 centromeres all clustered at the spindle-pole body (SPB) whereas the telomeres are associated in loose clusters at the nuclear envelope (2). These observations have been extended by the use of chromosome conformation capture approaches (3, 4) to map the relative positions of loci along each chromosome based on their frequencies of crosslinking (contact frequencies) with many other sites in the genome. Previous studies have shown that telomere-associated sequences preferentially recombine with other telomere-associated loci whereas centromere-linked sites selectively recombine with other centromere-linked loci (5–7). However, such preferences, presumably caused by the constraints of tethering, may not reflect the general behavior of most sequences undergoing homologous recombination. It is not known how the position of the sequences, lying between the tethered ends, influences their ability to repair a DSB.
Taking advantage of the Hi-C database, we established a series of yeast strains in which a DSB, induced at a defined location, could be repaired by a short, homologous donor sequence whose location in each strain was chosen for its apparent contact frequency with the recipient. In this way we were able to learn how the relative positions of sequences influences their recombinational potential and whether, as seems to be the case for translocations arising in mammalian cells (8, 9), chromosomal proximity influences the efficiency or rate of repair. We find that there is a strong correlation between the 3D position of a donor sequence within the yeast nucleus and its efficiency in repairing a DSB and that a given locus has very different repair potential depending on the location of the chromosome break.
Results
Donor Location Determines Repair Efficiency with a Specific DSB.
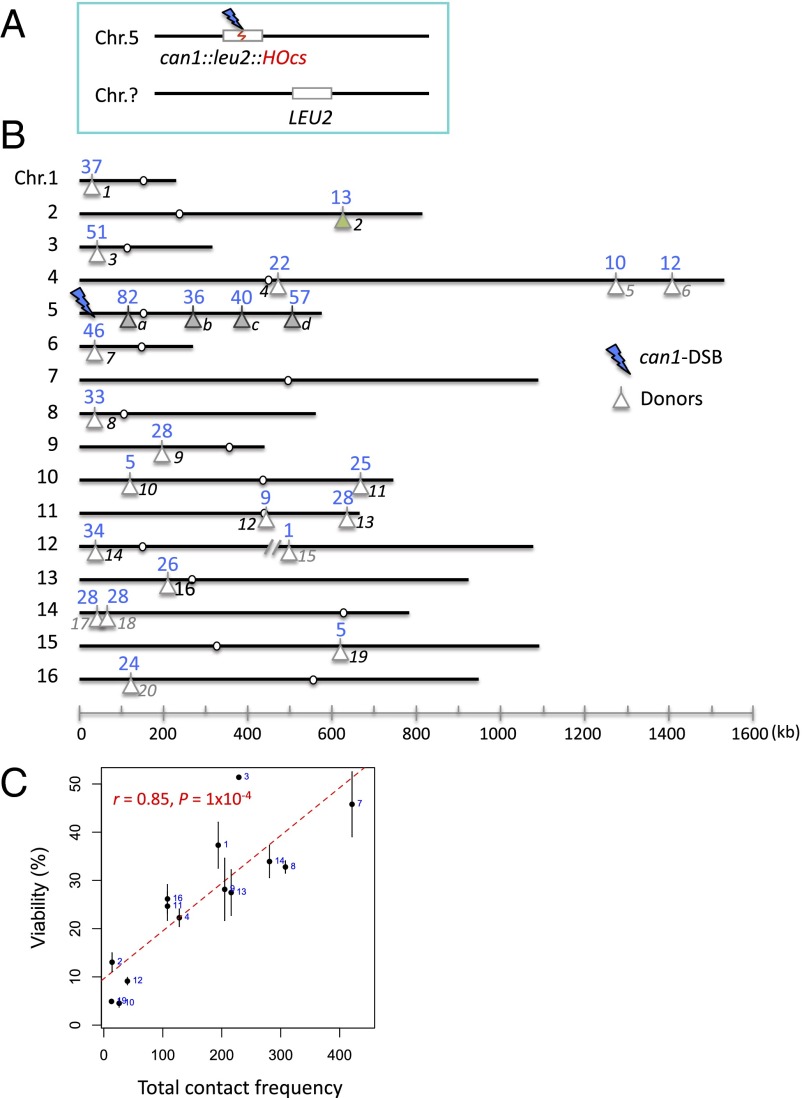
We created a series of budding yeast strains in which a galactose-inducible HO endonuclease created a DSB in the center of a leu2 gene, inserted at the can1 locus on chromosome 5 (Chr 5). In each strain, the DSB could be repaired by homologous recombination (gene conversion) using an ∼2-kb ectopic LEU2 donor sequence (Fig. 1A), integrated at one of 24 different locations (Fig. 1B). These loci were chosen to reflect a range of contact frequencies with the can1 locus (the location of the DSB target), based on the Hi-C data of Duan et al. (4). The efficiency of repair was assessed by measuring viability, which accurately reflects the amount of repair, when measured by a Southern blot (Fig. S1). Repair was generally most efficient for the four strains in which the donor was located intrachromosomally (36–82%) (Fig. 1B, loci a–d). Among the other 20 strains, in which the donor was situated on a different chromosome from the recipient (Fig. 1B, loci 1–20), viability ranged from 1% to 51%. Although cells suffering a DSB are arrested before mitosis by the DNA damage checkpoint, which should allow sufficient time for repair (10–12), half or more of the cells are unable to complete repair.
Fig. 1.
Viability assay comparing repair at 24 different donor loci. (A) The viability assay. An HO-induced DSB in a leu2 gene is repaired via an ectopic LEU2 donor. (B) Locations of the 24 donor loci and the corresponding viabilities (%, in blue). The loci a–d and the can1-DSB are on Chr 5 whereas the loci 1–20 are on different chromosomes. A 732-bp region containing the recombination enhancer was deleted in the strain having a nearby donor on Chr 3 (locus 3). Six loci (indicated in gray) are below the contact frequency threshold established by Duan et al (4) in their Hi-C analysis. (C) Correlation between the viability (%) and the total contact frequency (between regions ±30 kb around can1-DSB and ±20 kb around each donor). Error bars indicate one SD from three independent experiments. Note that (i) the four intrachromosomal loci were excluded for all of the correlation analysis and (ii) only 14 interchromosomal loci were analyzed because, for 6 loci, the contact frequency was below the threshold. Similar constraints apply to Figs. 2 C and D and 3C.
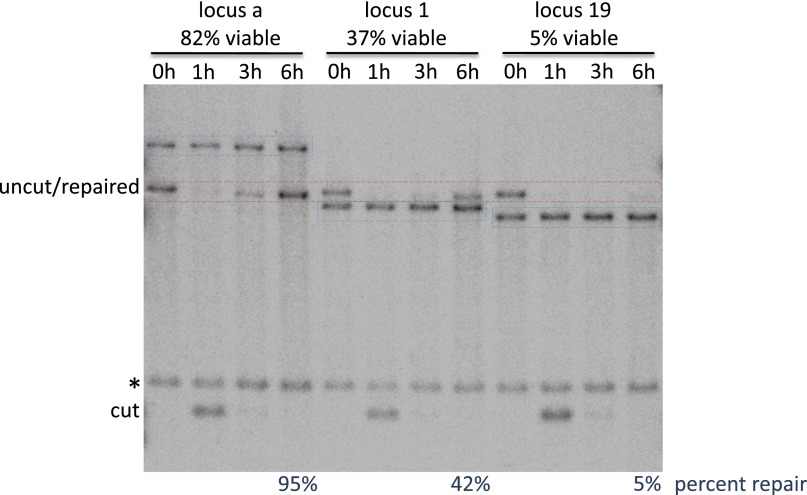
Fig. S1.
The amount of the repair product formation by Southern blot correlated with the viability measured by the viability assay in Fig. 1A. The quantification was done using the repair products at 6 h. In the red dashed square, the bands at 0 h represent the uncut restriction fragment whereas the bands at later time points are repair product. PstI-digested genomic DNAs were labeled with the U2 probe, as described by Jain et al. (29). Donor bands are indicated with blue dashed squares. The bands indicated with an asterisk (*) are regions that are also labeled by the U2 probe but could not be involved in the repair process. The location of the three tested loci are shown in Fig. 1B.
Contact Frequency Is Strongly Correlated with Repair Efficiency.
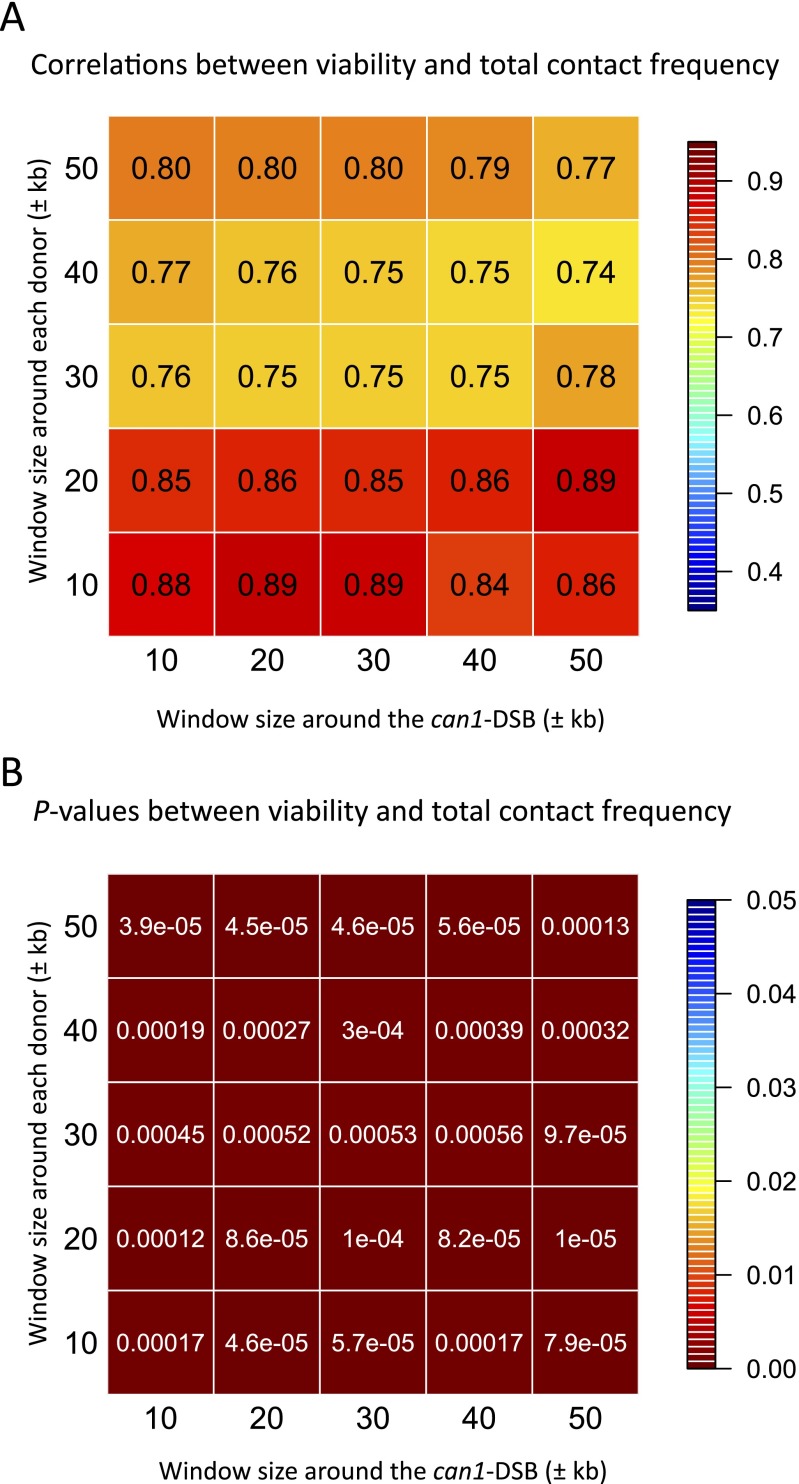
Remarkably, viability displayed a high correlation with the total contact frequency between the recombining sites. We determined contact frequency by using a ±20-kb window around the DSB site and a ±30-kb window around the donor locations; the calculated Pearson’s correlation coefficient was r = 0.85 (P = 1 × 10−4) (Fig. 1C). Similar results were observed when we used different window sizes for the loci to derive contact frequency or used the maximal contact frequency within a defined window for the correlation analysis (Figs. S2 and S3). The strong correlation demonstrated that contact frequency, and presumably distance, between a DSB and a donor largely determines the success of repair. We conclude that the Rad51-mediated homology search is subject to constraints of chromosomal organization.
Fig. S2.
Correlation analysis using different window sizes. (A) Correlations between the viability (%) and the total contact frequency, for which the window size around the can1-DSB and around each donor range from ±10 kb to ± 50kb. (B) The corresponding P values.
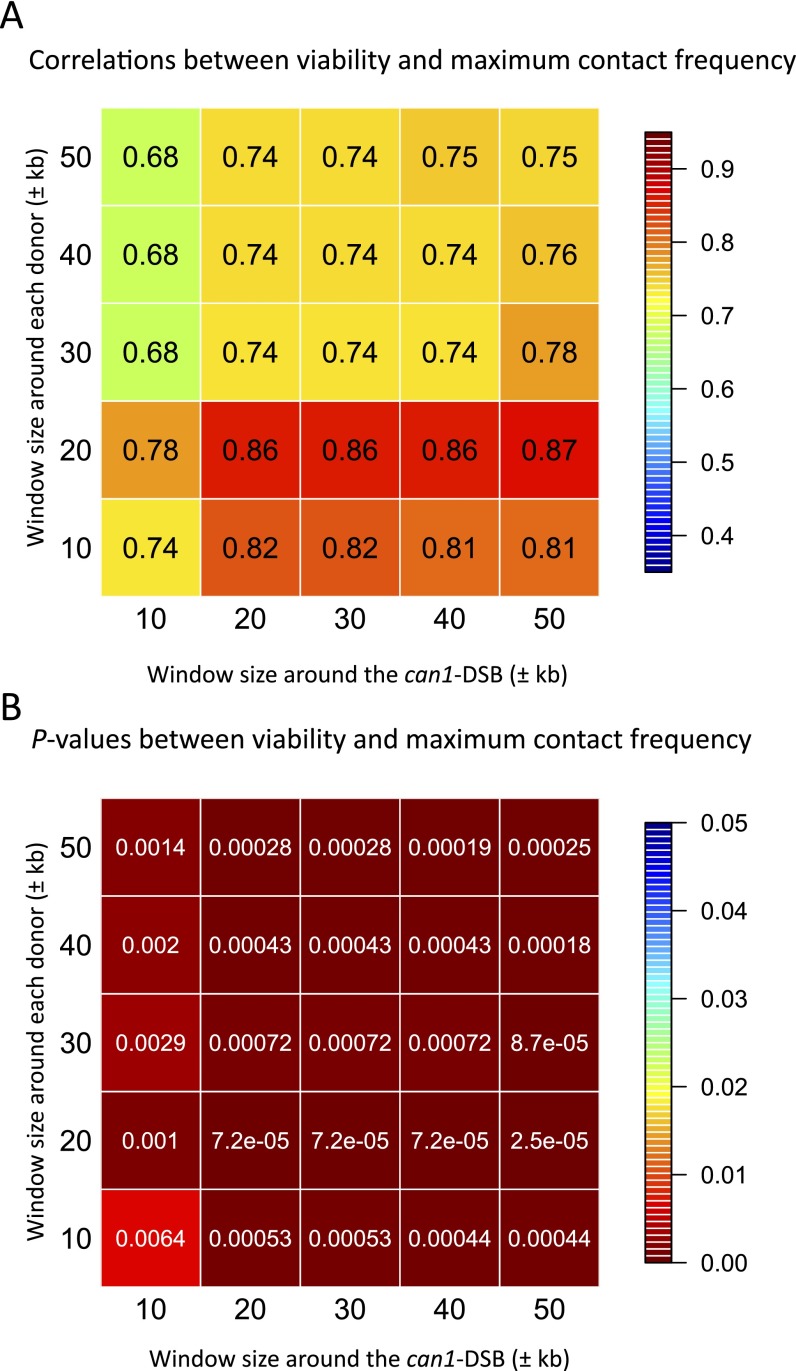
Fig. S3.
Correlation analysis using the maximum contact frequency with different window sizes. (A) Correlations between the viability (%) and the maximum contact frequency, for which the window size around the can1-DSB and around each donor range from ±10 kb to ±50 kb. (B) The corresponding P values.
Contact Frequency Is also Highly Correlated with Use in a Competition Assay.
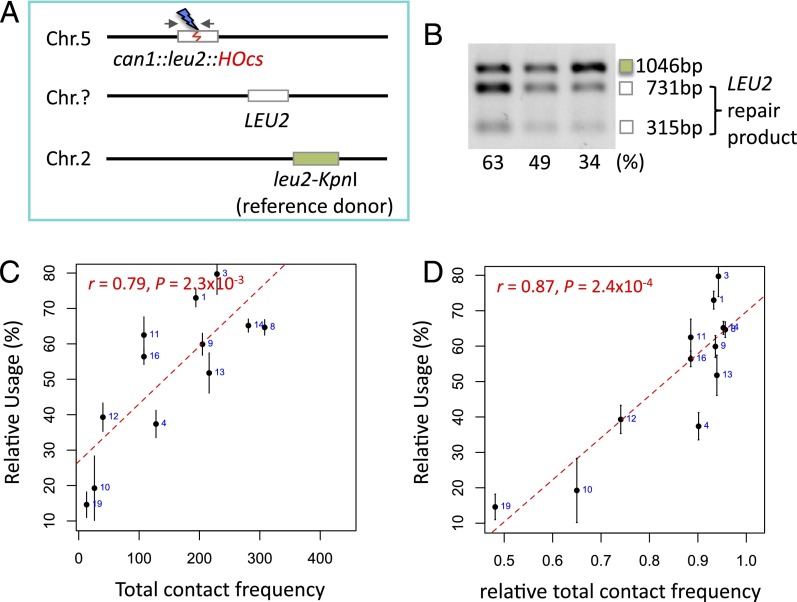
To examine more carefully the relationship between distance and donor use, we used a second approach, a competition assay in which only cells that complete repair are examined. In this assay, each LEU2 donor was forced to compete with a common second donor, leu2-K, located on Chr 2 (Fig. 2A). The relative use of the two donors was examined by using a PCR assay to measure the percentage of repaired cells inheriting a KpnI site from the LEU2 donor because the leu2-K reference donor lacks this site (Fig. 2B). The relative use of the LEU2 donor showed a strong correlation with the total contact frequency (r = 0.79, P = 2.3 × 10−3) (Fig. 2C). We also compared repair to the relative total contact frequency—the ratio of the donor site contacts divided by the total contacts of that locus and the reference donor. This comparison yielded an even stronger correlation with the relative use data (r = 0.87, P = 2.4 × 10−4) (Fig. 2D).
Fig. 2.
A competition assay showing strong correlation with donor location. (A) The competition assay differs from the viability assay in that a second copy of donor leu2-KpnI, lacking the KpnI recognition site, serves as a constant reference and only viable outcomes are scored. (B) Examples of the relative use measurement. The top band (marked with a green box) represents the leu2-KpnI repair product whereas the two lower bands (marked with white boxes) are the LEU2 repair products. The relative use of the LEU2 donor is calculated by dividing the sum of the intensity of the lower two cut bands by the total intensity. (C) Correlation between the relative use (%) and the total contact frequency (between regions ±30 kb around can1-DSB and ±20 kb around each donor). Error bars indicate one SD from three independent experiments. Note that only 12 interchromosomal loci were analyzed. (D) Correlation between the relative use (%) and the relative total contact frequency [Ci/(Ci + Cr), where Ci equals the total contact frequency between the DSB and the locus of interest and Cr equals the total contact frequency between the DSB and the reference locus]. Error bars indicate one SD from three independent experiments. Note that only 12 interchromosomal loci were analyzed.
A Similar Correlation Is Seen When the DSB Is Induced at Another Locus.
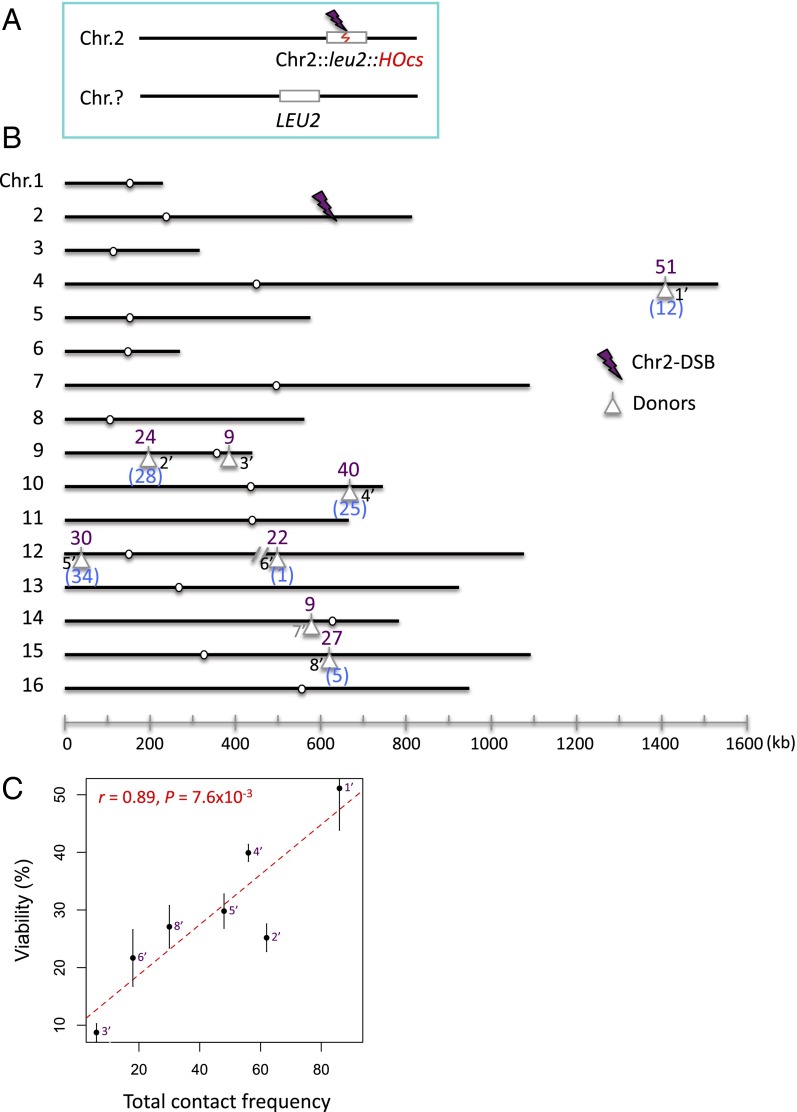
The can1 recipient locus lies only 33 kb from a telomere, and a DSB at this site might be constrained in its homology search, although we found previously that sites >20 kb from telomeres should not be confined by telomere tethering (13). To be certain that the strong correlation in repair locations was not caused by the location of the DSB at can1, we created eight additional single-donor strains in which the leu2::HOcs was inserted 200 kb from its telomere on Chr 2 (the location of the reference donor in the competition assay described above) (Fig. 3A). Six of these strains contained a donor used in the experiments involving the can1 locus, but their contact frequencies—and their repair efficiencies—were quite different with the new DSB locus (Fig. 3B). For example, a donor in the middle of the right arm of Chr 15 (Fig. 3B, locus 8′) had a viability of 5% with the DSB at can1 but 27% with the Chr 2 recipient. Using the viability assay, we found a similar correlation between repair frequency and contact frequency (r = 0.89, P = 7.6 × 10−3) (Fig. 3C), confirming that the correlations we have seen were not a consequence of some special feature of the can1 site. Moreover, these data demonstrate that the efficiencies of most donor loci are not a reflection of any inherent limitation on their use, such as their local chromatin states, but depend on their spatial distance to a particular recipient.
Fig. 3.
Effect of creating a DSB in a different location on repair efficiency with different donor locations. (A) The viability assay where the leu2::HOcs is at 625 kb on Chr 2. (B) Locations of eight donor loci and the corresponding viabilities (%, in purple) in response to the Chr2-DSB. Note that the recombination efficiencies of six loci among the eight loci were also examined in response to the can1-DSB (Fig. 1B); their viabilities are shown in blue for comparison. Locus 7′ (in gray) is below the contact frequency threshold. (C) Correlation between the viability (%) and the total contact frequency (between regions ±30 kb around the Chr2-DSB and ±20 kb around each donor). Error bars indicate one SD from three independent experiments. Seven interchromosomal loci were analyzed.
Recombination Capacity Is Inversely Correlated with Reconstituted Distance.
A number of methods have been advanced to infer 3D genome architecture from contact frequencies (14–16). Such reconstructions have the potential to refine the above contact-based analyses because they extrapolate the positions of loci whose pairwise contact frequencies were below the threshold for significance. Thus, we could infer distances between the can1 recipient and the six donor loci for which pairwise contact data were lacking. We used the reconstruction approach of Duan et al. (4) because this approach was highly customized to budding yeast via incorporation of a battery of biological constraints. Moreover, we considered several different 3D representations that arise from using differing restriction enzymes and contact quality filters (17). Interestingly, the relative use in the competition assay tends to show stronger correlations than the viability does (Table S1).
Table S1.
Correlation analysis using reconstructed distance
| DSB location | Assay | Distance | ||||
| HindIII | EcoRI | HindIII FDR < 0.01 | HindIII FDR < 0.1 | HindIII FDR < 1.0 | ||
| can1-DSB | Viability, % (20 loci) | r = −0.56 | r = −0.34 | r = −0.63 | r = −0.36 | r = −0.52 |
| P = 0.01 | P = 0.15 | P = 2.9 x 10-3 | P = 0.11 | P = 0.019 | ||
| Relative use, % (18 loci) | r = −0.73 | r = −0.5 | r = −0.8 | r = −0.47 | r = −0.66 | |
| P = 5.3 x 10-4 | P = 0.037 | P = 6.2 x 10-5 | P = 0.049 | P = 3.0 x 10-3 | ||
| Chr2-DSB | Viability, % (8 loci) | r = −0.46 | r = −0.52 | r = −0.49 | r = −0.38 | r = −0.74 |
| P = 0.25 | P = 0.18 | P = 0.22 | P = 0.36 | P = 0.035 | ||
Five reconstructed distance datasets, specified by different restriction enzymes and contact quality filters, were analyzed for the correlations with viabilities and relative use. Significant correlations (P < 0.05) are in bold.
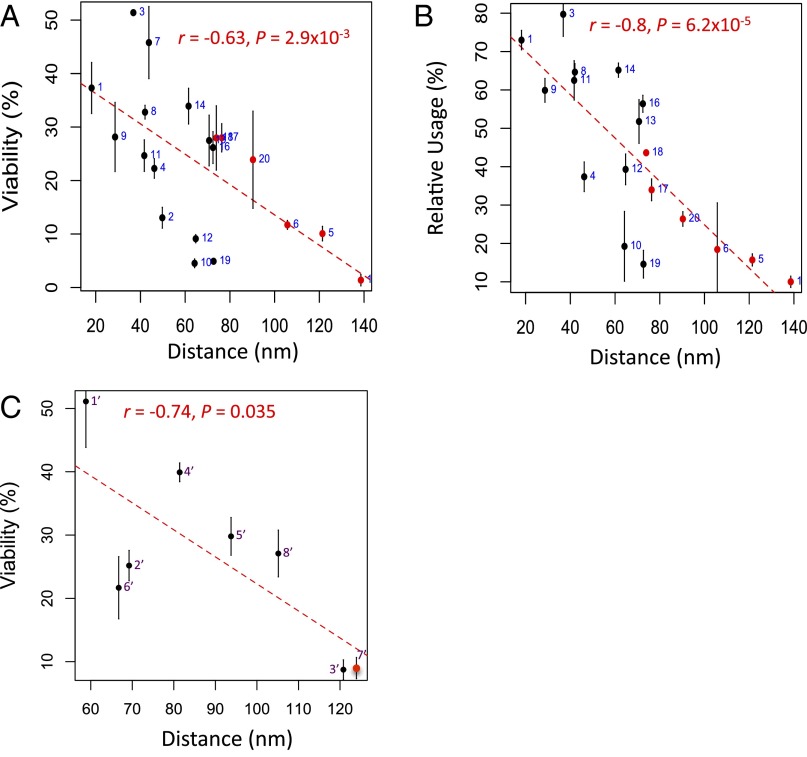
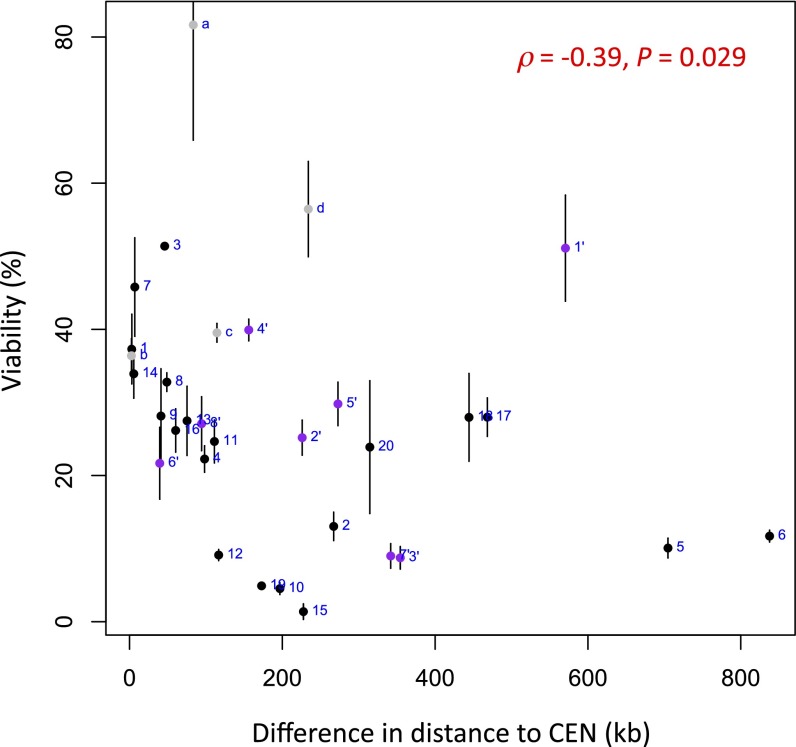
Fig. 4 A and B presents the correlations for DSBs at can1 using HindIII restriction digest data with 0.01% false discovery rate (FDR) contact filtering. The correlations are negative because they are associated with distances. Significant correlations were found for both the viability assay (r = −0.63, P = 2.9 × 10−3) and the relative use assay (r = −0.8, P = 6.2 × 10−5). When reconstructions based on EcoRI digestion were used, the correlations and P values were diminished (Table S1). However, there is appreciably lower coverage for this experiment, with fourfold fewer total contacts and threefold fewer interacting loci than for HindIII. The present DSB repair data are not sufficiently extensive to support a particular 3D representation. Similarly, the viability data for the Chr2-DSB also show a correlation with distance for the HindIII data (r = −0.74, P = 0.035) (Fig. 4C). We also asked whether some other parameters might correlate well with the results we have obtained. As shown in Fig. S4, there is a significant correlation using the difference in the genomic distances from the centromere, as also noted in the data from Agmon et al. (5), but it is much less robust than the correlations we found using contact frequency between the DSB site and the various donors.
Fig. 4.
The reconstituted 3D distance negatively correlated with repair efficiency. Error bars indicate one SD from three independent experiments. (A) Correlation between the viability (%, including all of the 20 loci, in response to the can1-DSB) and the distance in a 3D reconstruction (nm, obtained using HindIII restriction digest with 0.01% FDR contact filtering). The six loci missing in Fig. 1C are in red. (B) Correlation between the relative use (%, including all of the 18 loci, in response to the can1-DSB) and the distance in a 3D reconstruction (nm, obtained using HindIII restriction digest with 0.01% FDR contact filtering). The six loci missing in Fig. 2 C and D are in red. (C) Correlation between the viability (%, including all of the 8 loci, in response to the Chr2-DSB) and the distance in a 3D reconstruction (nm, obtained using HindIII restriction digest with 1.0% FDR contact filtering). The locus missing in Fig. 3C is in red. Note that best fits were shown for Fig. 4 A–C. Refer to Table S1 for more analysis.
Fig. S4.
Correlation between the viability (%) and the difference in the genomic distance to centromere of DSB and each donor (kb). All 20 interchromosomal loci (loci 1–20, in black), 4 intrachromosomal loci (loci a–d, in gray), and 8 loci in response to Chr2-DSB (loci 1′–8′, in purple) are included in the analysis. Spearman’s rank correlation (ρ = −0.39, P = 0.029) is used in this case because of the obviously nonnormal distribution of samples. No significant correlation is observed when using Pearson’s correlation (r = −0.30, P = 0.099).
Kinetics of Repair for both Good and Poor Donors Are Indistinguishable.
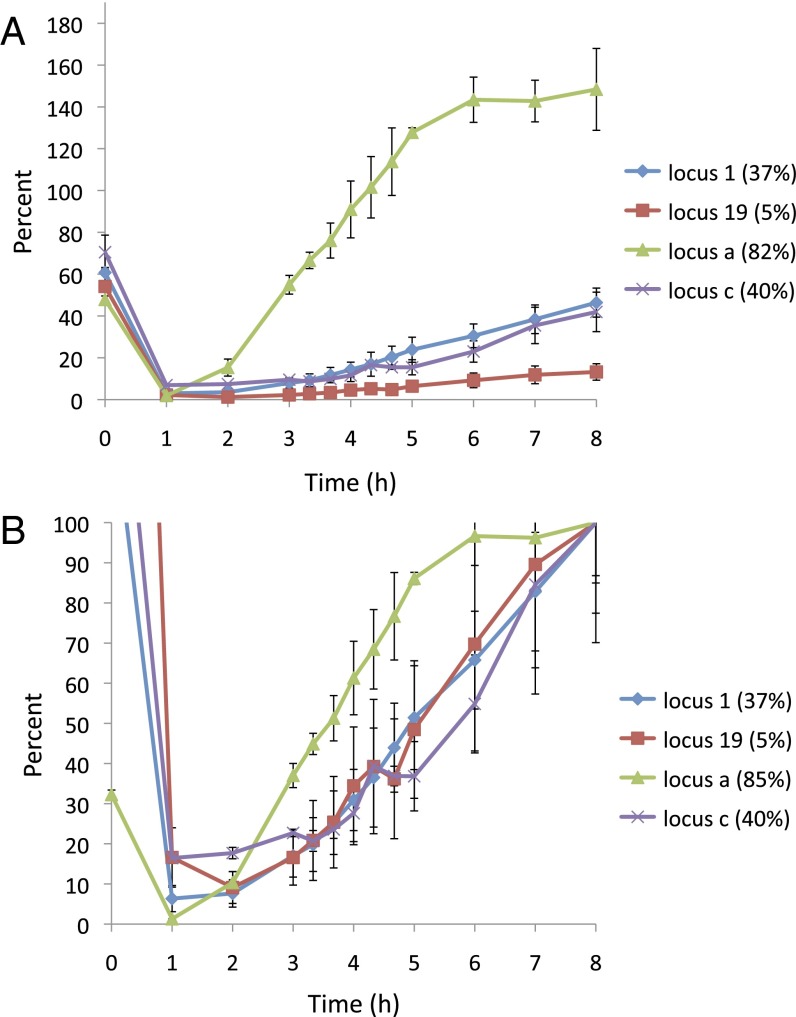
Surprisingly, poorly used donors do not have much slower kinetics of repair. To be sure that cells that had completed repair did not outgrow the unrepaired cells, log-phase cells were arrested with nocodazole before inducing a DSB and then after repair by a PCR assay that detects the restoration of the HO-cut leu2 by gene conversion. The efficiencies of the four repair events assayed are proportional to their viabilities (Fig. 5A). If we normalize the signals to that at 8 h, it is evident that the kinetics of repair for the donor with a low contact frequency are similar to those for the more frequently used donors. Consistent with previous studies (18, 19), intrachromosomal gene conversion showed faster kinetics than interchromosomal events (Fig. 5B).
Fig. 5.
Kinetics of repair for four different strains. Positions of the loci tested are shown in Fig. 1B. Error bars indicate one SE from three independent experiments. (A) PCR assay that detects the restoration of the HO-cut leu2 by gene conversion. Cells were arrested at G2/M phase by nocodazole treatment added 2 h before DSB induction. The restoration signal at each time point was normalized to the signal from a reference locus, ARG5,6, serving as input control. The signal from a derivative that had properly repaired the DSB was assigned as 100% repair. The efficiencies of the four repair events assayed are proportional to their viabilities. Note that the signals at 0 h completely disappeared 1 h after DSB was induced. (B) PCR signals were normalized to that at 8 h. The kinetics of repair for the donor with a low contact frequency (red line) are similar to those at the more frequently used donors (blue line). Intrachromosomal gene conversion (green line) showed faster kinetics than interchromosomal events.
Despite the fact that nocodazole treatment inhibits microtubule polymerization and may change chromosome configuration, this experiment, examining four loci, also leads to the important conclusion that repair in cells held in the G2/M phase of the cell cycle reflects the same contact frequency rules as we showed for cycling cells. This result is particularly interesting given the finding that mammalian metaphase chromosomes lose much of their distant 3D contacts (20). However, yeast chromosomes are likely too small to fold in the manner described for mammalian chromosomes and exhibit only modest metaphase condensation (21); thus, they may preserve their overall arrangement between different loci. At present, there is no equivalent contact frequency analysis of G2/M- or G1-arrested cells.
An Increase in the Length of Homology Improves Repair.
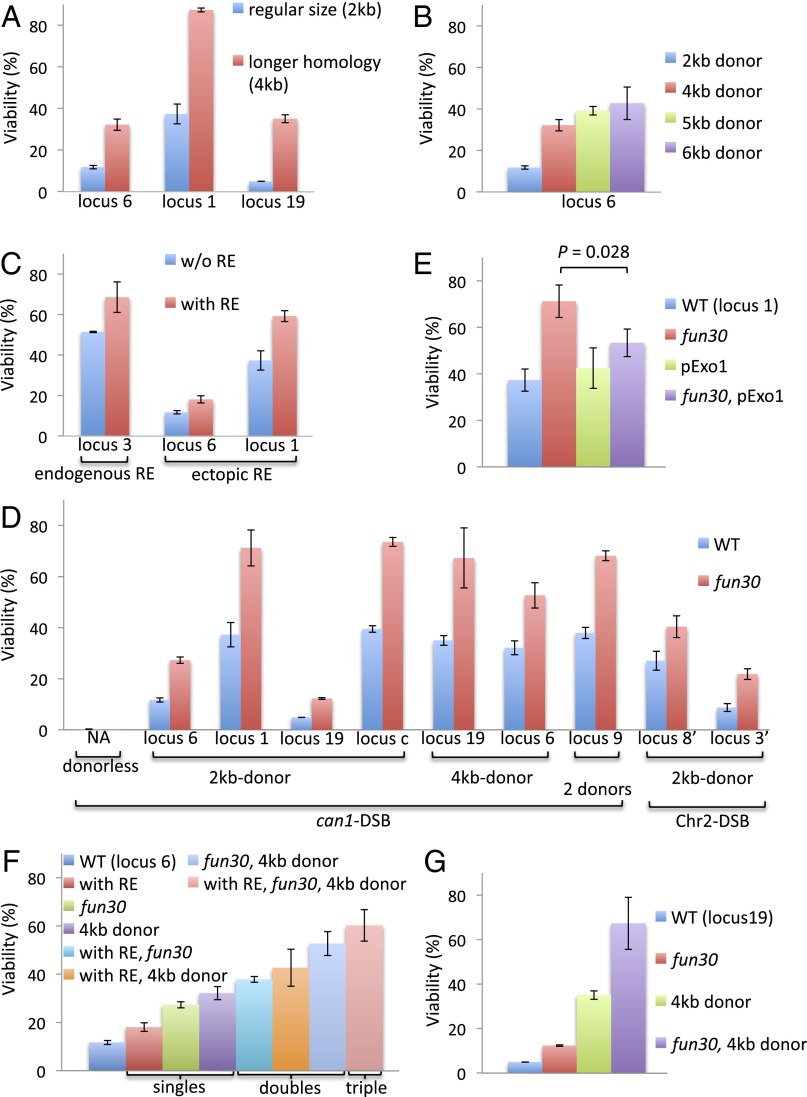
Although the relative distance between donor and recipient is the key parameter that dictates the efficiency of DSB repair, we wished to understand the reason why there was a high level of lethality even with a good donor. We asked whether increasing the length of flanking homology would increase the viability of poorly used loci, as it does in MAT switching (22). Indeed, doubling the homology on either side of the donor locus from 1 kb to 2 kb significantly increased viability in the three cases studied, both for two poorly used donors and one well-used donor (Fig. 6A). Thus, homology searching by the DSB ends is significantly improved by increasing the target size of the donor. This result agrees with previous studies that sequences more than 1 kb from the DSB end can be recognized during this search (23), but poorly used sites still showed lower viabilities than well-used sites. Interestingly, further increasing the donor size did not significantly improve repair (Fig. 6B). The use of a poor donor can also be improved by placing an ∼800-bp segment containing the cis-acting recombination enhancer (RE) sequence (24, 25) ∼5 kb from the donor (Fig. 6C).
Fig. 6.
Factors that influence repair efficiency. Positions of the loci tested are shown in Figs. 1B and 3B. Error bars indicate one SD from three independent experiments. (A) Longer homology proportionally promoted repair of both well-used donors and poorly used donors. (B) The further increase in donor size did not improve repair. (C) The presence of RE adjacent to a donor promotes repair. The endogenous RE is located at ∼11 kb to the left of the locus 3. The strain without the endogenous RE has a 732-bp deletion of the core domains of RE. The ectopic RE refers to the 798-bp RE sequence amplified with the two oligos CTTCTCAAAACCAAATTGCGCA and CTTTAGAATATAACATCTACCG and is marked with a NAT gene and inserted at ∼5 kb away from each donor. (D) Deletion of Fun30 globally and significantly improves repair (via homologous recombination). The strain indicated with “2 donors” was originally used for the competition assay; locus 9 refers to the position of the LEU2 donor in this strain whereas the leu2-KpnI donor is always at the locus 2 for all two-donor strains. (E) Increased viability in the fun30Δ strain is suppressed by expressing a second copy of Exo1. (F and G) The effects of the ectopic RE, longer homology, and fun30 on promoting viability are additive.
Extensive Resection Compromises Repair.
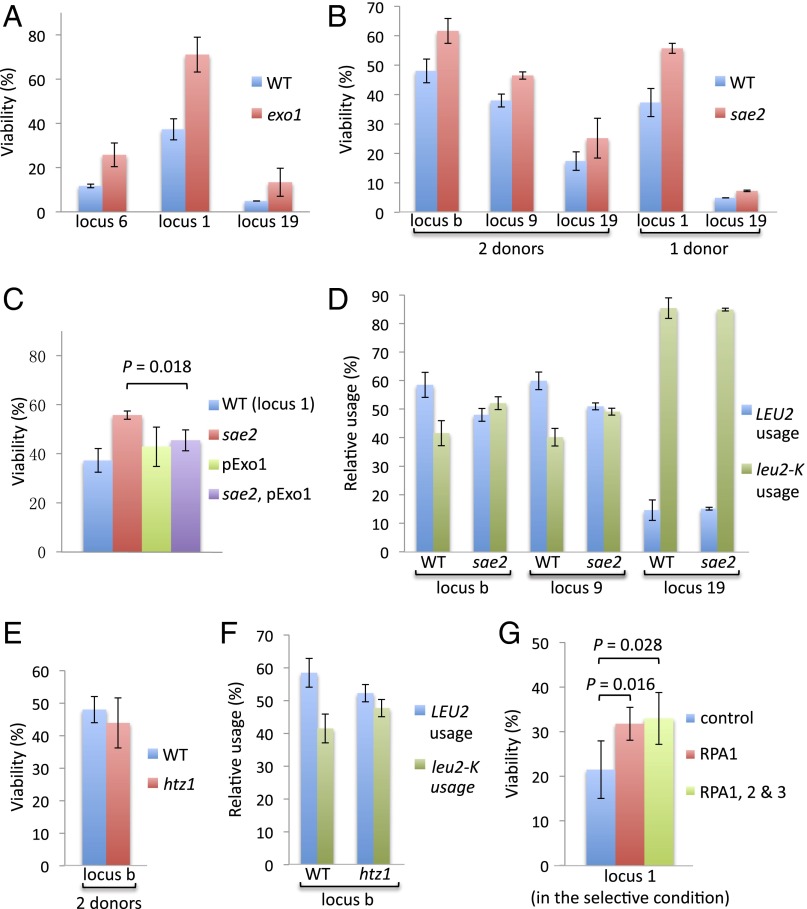
DSB ends are resected at a rate of about 4 kb/h, and, in the absence of DSB repair, this process can continue for at least 24 h (26). However, once the 1- to 2-kb sequences around the DSB that share homology with the donor are made single-stranded, it is not evident that further resection would inherently affect repair. Here, we demonstrate that viability can be dramatically improved by limiting resection. Deleting the chromatin remodeler FUN30 reduces the rate of resection from about 4 kb/h to 1.2 kb/h (27). In fun30Δ derivatives, viabilities of both good- and poor-donor strains increased, both for the can1-DSB and the Chr2-DSB, and both for 2-kb and 4-kb donors (Fig. 6D). If the effect of deleting FUN30 is attributable to slower resection, then overexpressing Exo1 should increase resection and suppress the effect of fun30Δ (27). Indeed, this prediction proved to be the case when a second copy of EXO1 was expressed (Fig. 6E). If we combine fun30Δ with a donor having larger homology, the efficiency of repair by very poor donors increased from 5–10% to 50–70% (Fig. 6 F and G). Consistent with the idea that extensive resection impedes repair, deletion of EXO1 showed a similar improvement in the efficiency of both good and poor donors (Fig. 7A).
Fig. 7.
Effect of Exo1, Sae2, and RPA on repair efficiency. Positions of the loci tested are shown in Fig. 1B. Error bars indicate one SD from three independent experiments. (A) Effect of deleting Exo1 on viability. (B) Effect of deleting Sae2 on viability. The strain indicated with “2 donors” was originally used for the competition assay; only the position of the LEU2 donor is indicated whereas the leu2-KpnI donor is always at the locus 2 for all two-donor strains. (C) Increased viability in the sae2Δ strain is suppressed by expressing a second copy of Exo1. (D) Effect of deleting Sae2 on the relative use in the competition assay. (E) Effect of deleting Htz1 on viability. The strain has two donors, but only the position of the LEU2 donor is indicated whereas the leu2-KpnI donor is always at the locus 2 for all two-donor strains. (F) Effect of deleting Htz1 on the relative use in the competition assay. (G) Expression of one additional copy of RPA1 is sufficient to improve repair whereas coexpression of all three RPA subunits doesn’t further increase viability.
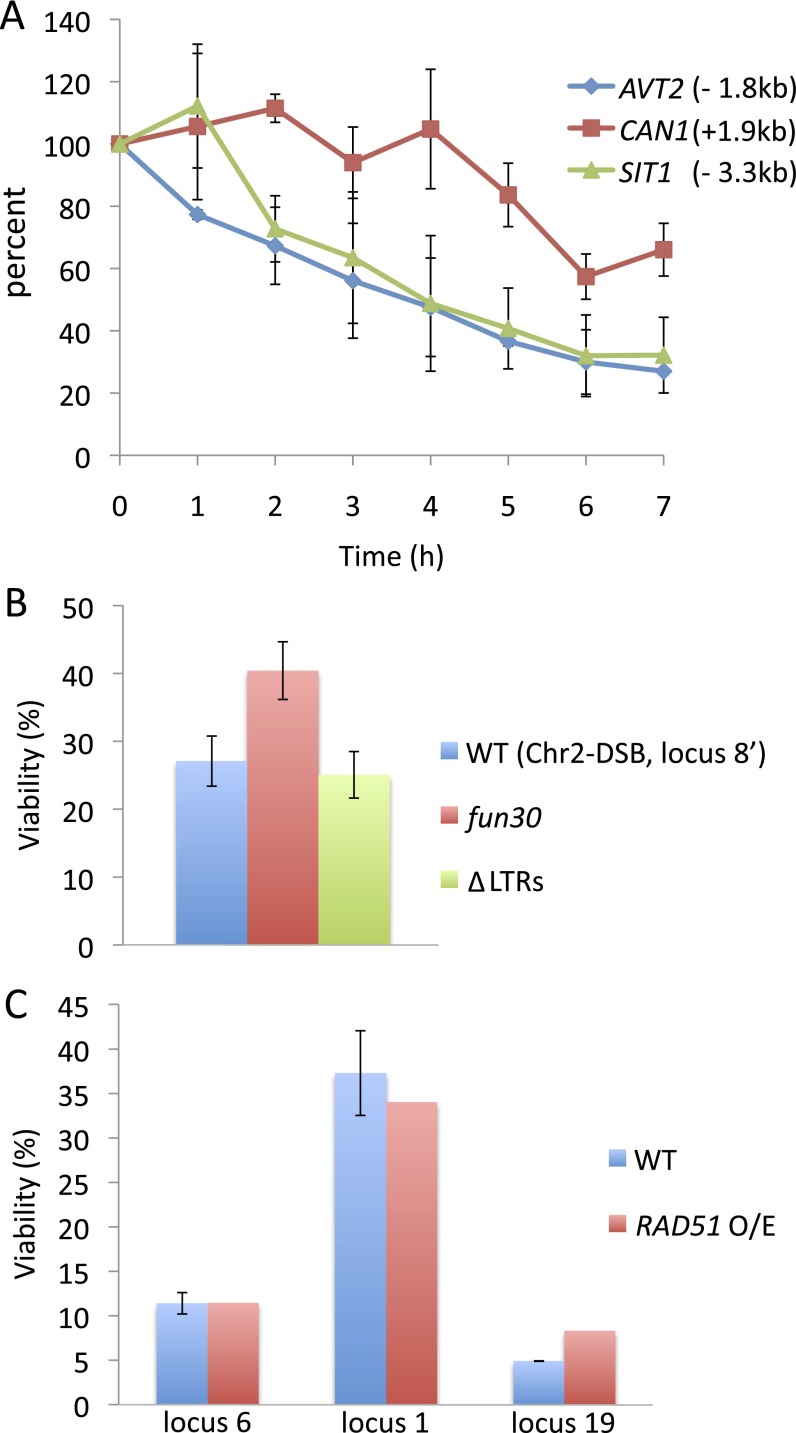
Recombination can occur only if the sequences around the DSB that share homology with a donor are not degraded. We have previously shown that the 3′-ended ssDNA tails produced by resection are quite stable (28–30), and here we directly confirmed that the 3′ end remains intact for at least 7 h. A quantitative PCR assay showed that, as expected, the signal decreased to about 50% as one strand was removed by resection (Fig. S5A), but this value persisted for at least 7 h, showing that the 3′-ended ssDNA is not lost.
Fig. S5.
(A) Resection in a donorless strain reduced the qPCR signals by about half, suggesting that the resected single-strand DNA tails were stable and available for repair (n = 3; bar stands for 1 SD). (B) Deletion of the only repeated sequences (LTR) within 50 kb of the DSB on Chr 2 did not change viability, suggesting that the increased viability in the fun30Δ mutants could not be explained by slowing down resection and thus avoiding lethal recombination events involving repeated sequences. The position of locus 8′ is shown in Fig. 3B. (C) Overexpression of Rad51 is not sufficient to promote repair via gene conversion. Rad51 was overexpressed from a yeast 2µ plasmid, pHK317, under the control of a galactose-inducible promoter. Positions of the loci tested are shown in Fig. 1B.
Failure to Repair the DSB Is Not Caused by Competing Lethal Events.
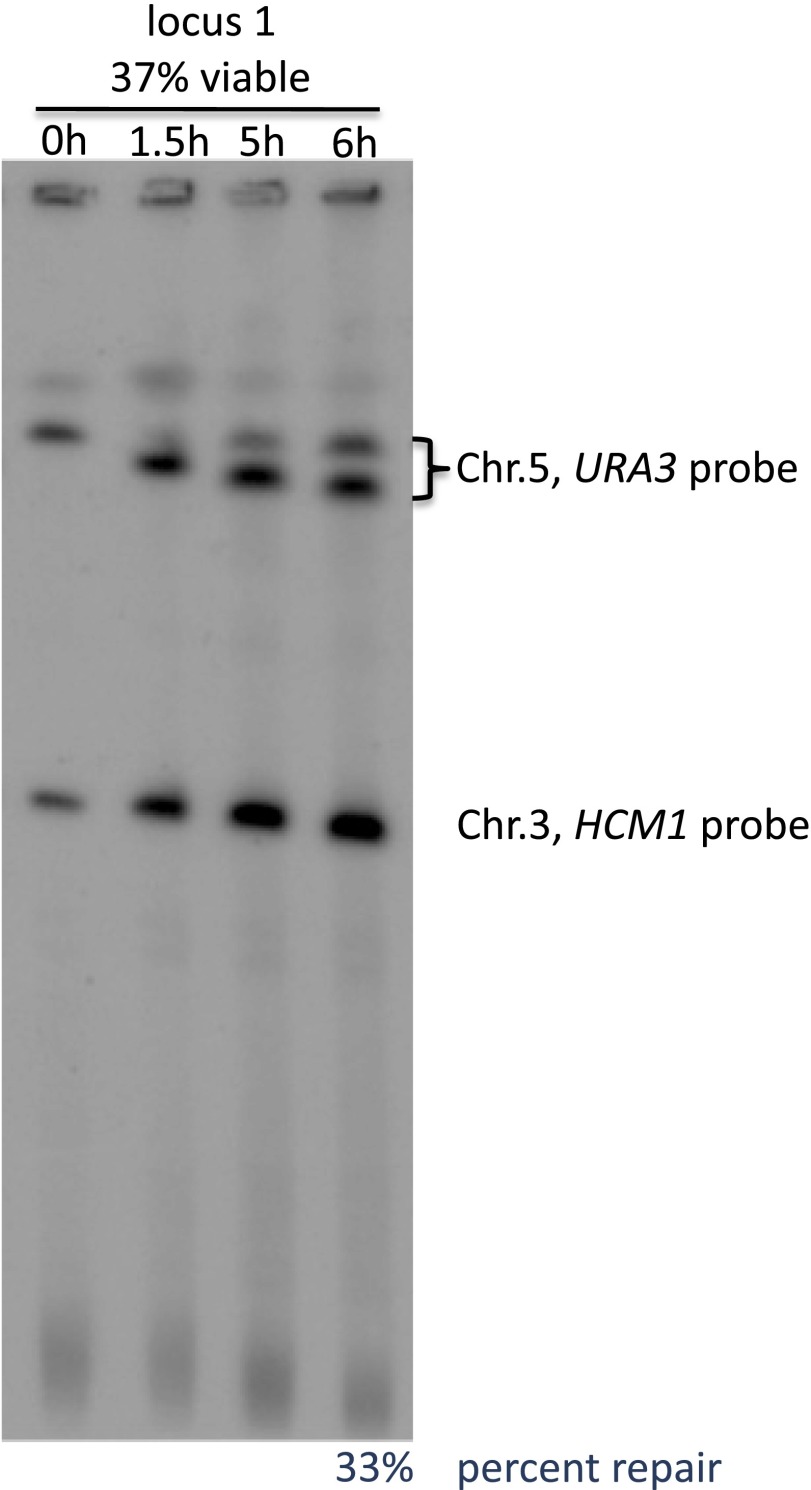
Resection activates the DNA damage checkpoint so that cells without repair arrest for more than 12 h before anaphase (31). Consequently, they should have more than enough time to repair the DSB so long as the donor and recipient can come into contact with each other; but even good ectopic donors fail to repair the DSB in half the cases. One possibility is that continuing resection exposes distal repetitive sequences, such as Ty retrotransposons, that might compete for recombination and result in unviable outcomes (32, 33). To look for such competing chromosome rearrangements, we used pulse-field gel electrophoresis (PFGE)/Southern analysis, which revealed the presence of an HO-cut band 1.5 h after HO induction and the appearance of the repaired Chr 5 product at 6 h (Fig. S6). The amount of repaired product reflected viability measurements. However, there were no additional rearranged bands representing lethal translocations, and the extensively resected broken chromosome didn’t disappear (Fig. S6). To rule out the participation of nearby repeated sequences, we deleted a cluster of three Ty LTRs (sigma sequences) that are the only repeated sequences within 50 kb of the DSB on Chr 2. The viability of a poorly used site did not change (Fig. S5B).
Fig. S6.
The majority of the unrepaired Chr 5 persist for at least 6 h after being cut. The entire chromosomes were shown by CHEF gel. URA3 and HCM1 probes were labeling Chr 5 and Chr 3, respectively. Chr 3 was used as loading control. The position of locus 1 is shown in Fig. 1B.
Deletion of Sae2 Improves Repair but Does Not Reduce Use of a Distant Donor.
Recently, Sae2 has also been implicated in regulating the way sequences near a DSB end explore the nuclear volume (34). The dramatic increase in the proportion of the nucleus that is explored after induction of a DSB is impaired by deleting SAE2 (34). Deletion of Sae2 also reduces 5′ to 3′ resection, although less severely than deleting FUN30 (35, 36). If the increased mobility after DSB induction were crucial to promoting the contact with a distant donor, viability should decrease in an sae2Δ strain even though resection is slowed down. However, in keeping with the fun30Δ results, the overall viability of four sae2Δ strains increased by 30% to ∼50% (Fig. 7B), suggesting that the effect expected from slower resection in the sae2Δ strain was most important. Similar to the fun30Δ strain, the increased viability in the sae2Δ strain is suppressed by expressing a second copy of Exo1 (Fig. 7C). Moreover, in a competition assay, deleting SAE2 did not reduce the relative use of a donor with a low contact frequency, as might be expected if the search for homology at distant locations depended on an increase in the radius of confinement (Fig. 7D). Thus, even in competition with an efficient intrachromosomal donor (locus b), the use of the reference donor on Chr 2 was increased in the absence of Sae2.
Deletion of Htz1 has also been shown to compromise the increase in the radius of confinement near a DSB (37) but has no known effect on resection. We found that deleting Htz1 did not reduce viability (Fig. 7E) or the relative use of a distant donor in the competition assay (Fig. 7F), again suggesting the DSB-induced mobility is unlikely to be an important factor for repair.
Overexpressing Replication Protein Factor A Improves Repair.
As resection proceeds, it is possible that some key protein might be titrated away, so that repair is compromised. It is unlikely that the compromised protein is solely Rad51, even though its abundance is indeed limited (38) because overexpressing Rad51 had no significant effect on repair (Fig. S5C). However, when all three subunits of the heterotrimeric replication protein factor A (RPA) complex were overexpressed, by adding an additional copy of each RFA gene, viability significantly improved (Fig. 7G). RPA is thought to stabilize the ssDNA exposed by resection (39), and it may become limited when excessive resection is proceeding. A similar result was obtained when only the largest subunit, Rfa1, was overexpressed (Fig. 7G).
Discussion
Our experiments demonstrate that the apparent proximity of chromosomal regions deduced from chromosome conformation capture provides an accurate prediction of the efficiency of repair of a DSB by ectopic recombination. Our results extend an earlier study examining primarily sites near centromeres and telomeres (5, 7); we show that sites spread along chromosome arms are strikingly nonuniform in their recombinational capacity. Our results also emphasize that the ability of a given region to participate in DSB repair is limited primarily by its proximity to the site of DNA damage rather than the underlying chromatin structure of that region. We note that the LEU2 and leu2-K donors were inserted into intergenic regions for all of the strains used in this study. Each donor has an intact LEU2 promoter and should have to have normal levels of transcription. Apparently, the presence of these transcribed sequences did not profoundly affect chromosome configuration. Also, at least in budding yeast, these correlations seem to be maintained even in G2/M-arrested cells.
The contact frequency measured from a population of cells by Hi-C reflects the average configuration. Each cell may have a different chromosome configuration (40). In this regard, the difference in the contact frequency between two pairs of loci may simply reflect the difference in the percentage of cells in which two loci are in close proximity. This assumption explains why the locus with a higher contact frequency to the DSB site did not show faster kinetics of repair than the locus with a lower contact frequency to the DSB site. The configuration within one cell is dynamic, and the change has to be continuous. Nevertheless, our data clearly demonstrate that distance is a critical factor determining the success of repair. We also show that the constraints on repair can be overcome by increasing the extent of homology between the donor and recipient, by slowing down resection, by having a recombination enhancer element adjacent to a donor, or by increasing the abundance of RPA.
The efficiency of ectopic DSB repair is strongly influenced by homology size, as shown in previous studies of how DSB ends search for a homologous sequence (22, 23, 41). Assuming that contact frequency between two loci in the genome does not change much when the extra 2 kb of homology is inserted at a donor, we postulate that extra homology facilitates recombination in two possible ways. First, the larger target provides a larger number of “minimum effective pairing segments” with which an initial collision can occur. Second, the longer homology can stabilize the initial Rad51-mediated encounter to make it more productive, consistent with our previous studies (22). It is interesting that further increases in the homology length of a donor did not further augment viability. In diploid yeast, mitotic homologous chromosomes are not paired with each other. Our result might suggest that, in diploid yeast, an ectopic homologous segment at a locus of close proximity to a DSB might effectively compete with a homologous chromosome to serve as a donor for repair.
There is apparently a limited time window in which repair must take place. This limit does not seem to reflect a loss of the 3′ end that shares homology elsewhere, nor does it seem to reflect the time at which some distant repetitive sequence is exposed and which could then participate in some competing, lethal events. Rather, the time limit is linked to the rate of resection of the DSB ends. Because HO cutting is extremely efficient and resection in a WT background occurs at a rate of ∼4 kb/h, the 1-kb flanking homologous leu2 sequence on either side of the DSB should be exposed within 1 h after DSB induction. Resection keeps exposing more ssDNA beyond the homologous leu2 sequence that is not useful for repair. Indeed, excessive resection seems to interfere with repair. Slowing down resection by deleting either FUN30 or SAE2 or EXO1 improves the efficiency of repair. Conversely, increasing resection by expressing a second copy of Exo1 (42) largely suppresses this effect. In diploid yeast cells, blocking resection in an sgs1Δ exo1Δ double mutant led to an apparently resection-independent ectopic recombination of dispersed, repeated Ty elements located 10 or more kb from a DSB (43). Were such a resection-independent mechanism operating in our system, it would be expected to enhance recombination events that would compete with, and diminish, the gene conversion scored in our assays. But we find that impairing resection has the opposite effect, enhancing recombination using the sequences close to the DSB.
Why resection is limiting is not certain. Overexpressing Rad51 does not suppress the consequences of extensive resection, but there could be other recombination factors that are titrated as longer—or additional, independent—segments of Rad51 filaments are formed. Even when Rad51 is bound to ssDNA that shares no homology elsewhere in the genome, it will still engage in a search and bind transiently to many other regions of the genome (44). As resection proceeds, there will be more and more such searches involving different segments of Rad51, and these activities may prevent efficient use of the homologous segments. A recent in vitro study showed that a minimum of 8-bp microhomology is sufficient to cause a temporary engagement of the Rad51 nucleofilament (45). As resection proceeds, more and more loci in the genome fulfill the requirement of the 8-bp microhomology. Such a small microhomology is apparently not sufficient to result in homologous recombination to repair a DSB because the viability of a strain lacking any leu2 donor sequence is about 0.1%; however, the “distraction” of microhomologies will increase and may become critical when more and more ssDNA is produced by resection, coated with Rad51, and engaged in a search for homology.
It seems that a key limiting factor in DSB repair is the abundance of RPA. Symington’s laboratory has shown that ssDNA ends become unstable when the normal level of RPA is depleted (39). Here, we find that adding an additional copy of all three subunits of RPA, or even just RFA1, leads to a significant increase in the repair efficiency.
The fact that not all interstitial chromosomal sites are equally accessible suggests that Rad51-mediated homology searching is unable to explore the entire nuclear volume efficiently enough to assure that all similarly sized donors will be used equally often. It has been suggested that such searching should be aided by an increase in the proportion of the volume that can be effectively searched (reflected in an increased radius of confinement). However, we have examined two mutations—sae2Δ and htz1Δ—that have been reported to abrogate a wider search of the nuclear volume and yet have found no change in the ability of a DSB to recombine with less efficient donors. Our data suggest that the increased mobility after DSB induction is not critical for repair. What role such apparent changes in mobility may play is not yet understood. We have suggested that DSB damage is detected by DNA damage checkpoint-mediated alterations of the kinetochore (46), which might alter the damaged chromosome’s association with the spindle pole body and thus lead to apparent changes in the radius of confinement, without necessarily changing the mobility of the DSB ends.
As noted above, our conclusions are in agreement with recent studies of nonhomologous end-joinings in mammalian cells that have concluded that proximity of interacting sites plays a key role in the rate of forming translocations (8, 9, 47). Here, we show that homologous recombination driven by a DSB on one chromosome obeys similar advantages of proximity. In this model system, we can explore how these rules are enforced and have discovered the importance of the size of the recombining sequences and the rate of resection of the DSB ends.
Materials and Methods
Strains and Plasmids.
All strains and plasmids are listed in Tables S2 and S3, respectively.
Table S2.
Strains used in these experiments
| Strains | Genotypes | Strains | Genotypes |
| YSJ278 | ho hmlΔ::ADE1 mataΔ::hisG hmrΔ::ADE1 leu2::KAN ade3::GAL::HO ade1 lys5 ura3-52 trp1 Chr3. 41.4kb::HYG can1::leu2::HOcs | ||
| YCSL064 | YSJ278, Chr1. 29kb::LEU2 | YCSL191 | YCSL064, Chr2. 625kb::URA3-leu2K |
| YCSL175 | YSJ278, Chr2. 625kb::URA3-leu2K | ||
| YCSL014 | YSJ278, Chr3. 41kb::LEU2, REΔ | YCSL214 | YCSL014, Chr2. 625kb::URA3-leu2K |
| YSJ119 | YSJ278, Chr3. 41kb::LEU2 | YCSL213 | YSJ119, Chr2. 625kb::URA3-leu2K |
| YCSL169 | YSJ278, Chr4. 471kb::LEU2 | YCSL202 | YCSL169, Chr2. 625kb::URA3-leu2K |
| YCSL065 | YSJ278, Chr4. 1274kb::LEU2 | YCSL193 | YCSL065, Chr2. 625kb::URA3-leu2K |
| YCSL011 | YSJ278, Chr4. 1407kb::LEU2 | YCSL178 | YCSL011, Chr2. 625kb::URA3-leu2K |
| YCSL015 | YSJ278, Chr5. 116kb::URA3-leu2K | ||
| YCSL006 | YSJ278, Chr5. 269kb::LEU2 | YWW002 | YCSL006, Chr2. 625kb::URA3-leu2K |
| YCSL007 | YSJ278, Chr5. 386kb::LEU2 | ||
| YCSL008 | YSJ278, Chr5. 505kb::LEU2 | ||
| YCSL483 | YSJ278, Chr6. 36kb::LEU2 | YCSL487 | YCSL483, Chr2. 625kb::URA3-leu2K |
| YCSL010 | YSJ278, Chr8. 35kb::LEU2 | YCSL162 | YCSL010, Chr2. 625kb::URA3-leu2K |
| YCSL248 | YSJ278, Chr9. 195kb::LEU2 | YWW024 | YCSL248, Chr2. 625kb::URA3-leu2K |
| YCSL250 | YSJ278, Chr10. 120kb::LEU2 | YWW026 | YCSL250, Chr2. 625kb::URA3-leu2K |
| YCSL252 | YSJ278, Chr10. 667kb::LEU2 | YWW023 | YCSL252, Chr2. 625kb::URA3-leu2K |
| YCSL254 | YSJ278, Chr11. 443kb::LEU2 | YWW019 | YCSL254, Chr2. 625kb::URA3-leu2K |
| YCSL152 | YSJ278, Chr11. 635kb::LEU2 | YWW013 | YCSL152, Chr2. 625kb::URA3-leu2K |
| YCSL256 | YSJ278, Chr12. 37kb::LEU2 | YCSL474 | YCSL256, Chr2. 625kb::URA3-leu2K |
| YCSL155 | YSJ278, Chr12. 498kb::LEU2 | YCSL181 | YCSL155, Chr2. 625kb::URA3-leu2K |
| YCSL258 | YSJ278, Chr13. 209kb::LEU2 | YWW021 | YCSL258, Chr2. 625kb::URA3-leu2K |
| YCSL158 | YSJ278, Chr14. 41kb::LEU2 | YWW016 | YCSL158, Chr2. 625kb::URA3-leu2K |
| YCSL012 | YSJ278, Chr14. 65kb::LEU2 | YCSL188 | YCSL012, Chr2. 625kb::URA3-leu2K |
| YCSL066-4 | YSJ278, Chr15. 619kb::LEU2 | YCSL197 | YCSL066-4, Chr2. 625kb::URA3-leu2K |
| YCSL160 | YSJ278, Chr16. 122kb::LEU2 | YCSL200 | YCSL160, Chr2. 625kb::URA3-leu2K |
| YCSL350 | YSJ278, Chr4. 1407kb::ca-LEU2-n1(4kb) | YCSL391 | YWW002, sae2Δ::NAT |
| YCSL348 | YSJ278, Chr1. 29kb::ca-LEU2-n1(4kb) | YCSL392 | YWW024, sae2Δ::NAT |
| YCSL354 | YSJ278, Chr15. 619kb::ca-LEU2-n1(4kb) | YWW070 | YCSL197, sae2Δ::NAT |
| YCSL541 | YSJ278, Chr4. 1407kb::ca-LEU2-n1(5kb) | YCSL438 | YCSL064, sae2Δ::NAT |
| YCSL542 | YSJ278, Chr4. 1407kb::ca-LEU2-n1(6kb) | YCSL441 | YCSL066-4, sae2Δ::NAT |
| YCSL414 | YSJ278, fun30Δ::NAT | YCSL164 | YCSL011, Chr4. 1412kb::NAT-RE |
| YCSL393 | YCSL354, fun30Δ::NAT | YCSL452 | YCSL064, Chr1. 35kb::NAT-RE |
| YCSL374 | YCSL011, fun30Δ::NAT | YCSL424 | YCSL350, Chr4. 1412kb::NAT-RE |
| YCSL376 | YCSL064, fun30Δ::NAT | YCSL464 | YCSL011, fun30Δ::URA3, Chr4. 1412kb::NAT-RE |
| YCSL386 | YCSL066-4, fun30Δ::NAT | YWW063 | YCSL011, exo1Δ::URA3 |
| YCSL408 | YCSL350, fun30Δ::NAT | YWW065 | YCSL064, exo1Δ::URA3 |
| YCSL477 | YCSL424, fun30Δ::URA3 | YWW068 | YCSL066-4, exo1Δ::URA3 |
| YCSL361 | YCSL011, +pHK317(GALp::RAD51- TRP1) | YCSL476 | YCSL007, fun30Δ::NAT |
| YCSL363 | YCSL064, +pHK317(GALp::RAD51- TRP1) | YWW104 | YWW002, htz1Δ::NAT |
| YCSL365 | YCSL066-4, +pHK317(GALp::RAD51- TRP1) | YWW157 | YCSL064, +YCP50, pWW009, pWW010 |
| YCSL484 | YCSL438, +pJH2593(EXO1-TRP1) | YWW176 | YCSL064, +pJM114(RFA1), pWW009, pWW010 |
| YCSL378 | YCSL064, +pJH2593(EXO1-TRP1) | YWW137 | YSCL064, +pJM320(RFA3), pWW001(RFA1), pCSL005(RFA2) |
| YCSL380 | YCSL376, +pJH2593(EXO1-TRP1) | ||
| YCSL305 | ho hmlΔ::ADE1 mataΔ::hisG hmrΔ::ADE1 KAN ade3::GAL::HO ade1 lys5 ura3-52 trp1 Chr2. 625kb:: leu2::HOcs | ||
| YCSL338 | YCSL305, Chr4. 1407kb::LEU2 | YCSL358 | YCSL305, Chr14. 584kb::LEU2 |
| YCSL316 | YCSL305, Chr9. 195kb::LEU2 | YCSL332 | YCSL305, Chr15. 619kb::LEU2 |
| YCSL342 | YCSL305, Chr9. 388kb::LEU2 | YCSL395 | YCSL332, fun30Δ::NAT |
| YCSL320 | YCSL305, Chr10. 667kb::LEU2 | YCSL417 | YCSL342, fun30Δ::NAT |
| YCSL324 | YCSL305, Chr12. 37kb::LEU2 | YWW055 | YCSL332, Chr2. 644kb(LTR)::NAT |
| YCSL328 | YCSL305, Chr12. 498kb::LEU2 |
Table S3.
Plasmids used in these experiments
| Plasmid name | Features |
| YCP50 | CEN4, URA3 |
| pWW009 | YCP50, ura3::NAT |
| pWW010 | YCP50, ura3::TRP1 |
| pJM114 | YCP50, RFA1 |
| pJM320 | YCP50, RFA3 |
| pWW001 | YCP50, ura3::NAT, RFA1 |
| pCSL005 | YCP50, ura3::TRP1, RFA2 |
| pHK317 | pYES, pGAL-RAD51, TRP1 |
| pJH2593 | pRS314, Exo1, TRP1 |
Viability Assay.
A single colony was inoculated into YP-Lactate medium and grew overnight. Then the culture was diluted so that 6 h later its concentration reached log phase (∼5 × 106 cells per mL). Equal amounts of cells were plated onto three yeast extract peptone dextrose (YEPD) and three YP-Galactose plates, respectively. After incubation at 30 °C for 3–4 d, the number of colonies on each plate was measured. Colonies formed on YP-Galactose plates are survivors from DSBs, mainly repaired by homologous recombination because the viability of a donorless strain is <0.1%. Viability was calculated by dividing the number of colonies on YP-Galactose plates over that on YEPD plates. The viability assay was done at least three times for each strain.
Competition Assay.
The assay was modified from the PCR-based donor preference assay described previously (25). In brief, cells were grown in YP-Lactate medium to log phase in the way described in the viability assay above. Sterile galactose solution (20% wt/vol) was added into the culture to a final concentration of 2% for DSB induction. After an overnight incubation at 30 °C, genomic DNA was extracted from the whole population of cells and used for PCR to amplify the HO targeting site using the flanking primers Can1p7 (GCCTCAATGTCTCTTCTATCGG) and Leu2p18B (CCAAATAGGCAATGGTGGCT). HO cutting is efficient and continuous; therefore, the locus would get amplified only when the DSB is repaired with the HO site being eliminated or mutated. The PCR amplicon was then digested with KpnI. The digests were separated on an agarose gel and quantified with QuantityOne. The relative use of WT LEU2 donor was calculated by dividing the sum of the intensity of the KpnI fragments by the total intensity of all amplicons. The competition assay was done three times for each strain.
Southern Blotting.
Southern blot analysis was performed as previously described (29). Briefly, genomic DNA at each time point was purified with phenol from 50 mL of log-phase culture, digested with PstI, and labeled with the U2 probe. Percent repair was estimated by normalizing the intensity of repaired product to that of the donor and using the value at 0 h as 100% repair.
Pulse-Field Gel Electrophoresis.
PFGE was performed using the Bio-Rad CHEF-DR II System, based on the manufacturer’s recommended protocol. Capillary transfer was used for the following Southern blot analysis, in which Chr 5 was labeled with a URA3 probe whereas Chr 3, used as loading control, was labeled with an HCM1 probe. Percent repair was estimated by normalizing the intensity of repaired Chr 5 to that of Chr 3 and using the value for 0 h as 100% repair.
Primer Extension Assay.
Genomic DNA at each time point was extracted with phenol and applied to qPCR using primers Can1p7 (GCCTCAATGTCTCTTCTATCGG) and CSL251-Leu2-260 (TTGTTCAGGTCTAACACTACCG). Signals were normalized to the corresponding signals acquired from a reference locus, Arg5, 6. The normalized value of a derivative that had properly repaired the DSB was set as 100% repair.
qPCR-Based Resection Assay.
The procedure was described in Eapen et al. (27). Primer sequences were as follows: CAN1 1 kb down rev (ACCATCGTTCTGGCTGAATATAG), CSL288-Avt2 p1 (AAATTGCTGATTCTATTTACAAACGGG); Can1p6 (AATGGGCCCTCTATCACTTGTTGC), CAN1-P9 (GAAAGTCGCTTCAAGCTAACCCAG); SIT1-28540 (TCGGGTGTGTATTGGTTCCATTCAC), SIT1-29265 (TGAATGTAAATGGAGAACCGTACGC).
Correlation Analysis.
Pearson's correlation test was used for testing the association between pairs of variables of interest, including contact frequency, viability, relative use, and reconstituted distance.
Acknowledgments
We thank Mirela Andronescu for providing initial information about contact probabilities and Michael Lichten for his comments on the manuscript. D.C. was supported by National Science Foundation Graduate Research Fellowship Grant 1144247 and National Institutes of Health Training Grant T32 GM007175. This work was supported by NIH Grants GM20056 (to J.E.H.) and GM109457 (to M.R.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523660113/-/DCSupplemental.
References
- 1.Hicks WM, Yamaguchi M, Haber JE. Real-time analysis of double-strand DNA break repair by homologous recombination. Proc Natl Acad Sci USA. 2011;108(8):3108–3115. doi: 10.1073/pnas.1019660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin QW, Fuchs J, Loidl J. Centromere clustering is a major determinant of yeast interphase nuclear organization. J Cell Sci. 2000;113(Pt 11):1903–1912. doi: 10.1242/jcs.113.11.1903. [DOI] [PubMed] [Google Scholar]
- 3.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 4.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465(7296):363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agmon N, Liefshitz B, Zimmer C, Fabre E, Kupiec M. Effect of nuclear architecture on the efficiency of double-strand break repair. Nat Cell Biol. 2013;15(6):694–699. doi: 10.1038/ncb2745. [DOI] [PubMed] [Google Scholar]
- 6.Therizols P, et al. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol. 2006;172(2):189–199. doi: 10.1083/jcb.200505159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong H, Arbona JM, Zimmer C. How to build a yeast nucleus. Nucleus. 2013;4(5):361–366. doi: 10.4161/nucl.26226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147(1):107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roukos V, et al. Spatial dynamics of chromosome translocations in living cells. Science. 2013;341(6146):660–664. doi: 10.1126/science.1237150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartwell LH, Weinert TA. Checkpoints: Controls that ensure the order of cell cycle events. Science. 1989;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 11.Sandell LL, Zakian VA. Loss of a yeast telomere: Arrest, recovery, and chromosome loss. Cell. 1993;75(4):729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 12.Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7(2):293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- 13.Avşaroğlu B, et al. Effect of chromosome tethering on nuclear organization in yeast. PLoS One. 2014;9(7):e102474. doi: 10.1371/journal.pone.0102474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Li G, Toh KC, Sung WK. 3D chromosome modeling with semi-definite programming and Hi-C data. J Comput Biol. 2013;20(11):831–846. doi: 10.1089/cmb.2013.0076. [DOI] [PubMed] [Google Scholar]
- 15.Varoquaux N, Ay F, Noble WS, Vert JP. A statistical approach for inferring the 3D structure of the genome. Bioinformatics. 2014;30(12):i26–i33. doi: 10.1093/bioinformatics/btu268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesne A, Riposo J, Roger P, Cournac A, Mozziconacci J. 3D genome reconstruction from chromosomal contacts. Nat Methods. 2014;11(11):1141–1143. doi: 10.1038/nmeth.3104. [DOI] [PubMed] [Google Scholar]
- 17.Segal MR, Xiong H, Capurso D, Vazquez M, Arsuaga J. Reproducibility of 3D chromatin configuration reconstructions. Biostatistics. 2014;15(3):442–456. doi: 10.1093/biostatistics/kxu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connolly B, White CI, Haber JE. Physical monitoring of mating type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8(6):2342–2349. doi: 10.1128/mcb.8.6.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keogh MC, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439(7075):497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 20.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342(6161):948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125(3):517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coïc E, et al. Dynamics of homology searching during gene conversion in Saccharomyces cerevisiae revealed by donor competition. Genetics. 2011;189(4):1225–1233. doi: 10.1534/genetics.111.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inbar O, Kupiec M. Homology search and choice of homologous partner during mitotic recombination. Mol Cell Biol. 1999;19(6):4134–4142. doi: 10.1128/mcb.19.6.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, Haber JE. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell. 1996;87(2):277–285. doi: 10.1016/s0092-8674(00)81345-8. [DOI] [PubMed] [Google Scholar]
- 25.Li J, et al. Regulation of budding yeast mating-type switching donor preference by the FHA domain of Fkh1. PLoS Genet. 2012;8(4):e1002630. doi: 10.1371/journal.pgen.1002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fishman-Lobell J, Rudin N, Haber JE. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol. 1992;12(3):1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eapen VV, Sugawara N, Tsabar M, Wu WH, Haber JE. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol Cell Biol. 2012;32(22):4727–4740. doi: 10.1128/MCB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White CI, Haber JE. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9(3):663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain S, et al. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 2009;23(3):291–303. doi: 10.1101/gad.1751209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaze MB, et al. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell. 2002;10(2):373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 31.Lee SE, et al. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94(3):399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 32.VanHulle K, et al. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol Cell Biol. 2007;27(7):2601–2614. doi: 10.1128/MCB.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Downing B, Morgan R, VanHulle K, Deem A, Malkova A. Large inverted repeats in the vicinity of a single double-strand break strongly affect repair in yeast diploids lacking Rad51. Mutat Res. 2008;645(1-2):9–18. doi: 10.1016/j.mrfmmm.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miné-Hattab J, Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol. 2012;14(5):510–517. doi: 10.1038/ncb2472. [DOI] [PubMed] [Google Scholar]
- 35.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem. 2005;280(46):38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 36.Costelloe T, et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2012;489(7417):581–584. doi: 10.1038/nature11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horigome C, et al. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol Cell. 2014;55(4):626–639. doi: 10.1016/j.molcel.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 38.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12(1):209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 39.Chen H, Lisby M, Symington LS. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol Cell. 2013;50(4):589–600. doi: 10.1016/j.molcel.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aylon Y, Kupiec M. New insights into the mechanism of homologous recombination in yeast. Mutat Res. 2004;566(3):231–248. doi: 10.1016/j.mrrev.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Amin NS, Nguyen MN, Oh S, Kolodner RD. exo1-Dependent mutator mutations: Model system for studying functional interactions in mismatch repair. Mol Cell Biol. 2001;21(15):5142–5155. doi: 10.1128/MCB.21.15.5142-5155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan FJ, Hoang ML, Koshland D. DNA resection at chromosome breaks promotes genome stability by constraining non-allelic homologous recombination. PLoS Genet. 2012;8(3):e1002633. doi: 10.1371/journal.pgen.1002633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renkawitz J, Lademann CA, Kalocsay M, Jentsch S. Monitoring homology search during DNA double-strand break repair in vivo. Mol Cell. 2013;50(2):261–272. doi: 10.1016/j.molcel.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 45.Qi Z, et al. DNA sequence alignment by microhomology sampling during homologous recombination. Cell. 2015;160(5):856–869. doi: 10.1016/j.cell.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dotiwala F, Harrison JC, Jain S, Sugawara N, Haber JE. Mad2 prolongs DNA damage checkpoint arrest caused by a double-strand break via a centromere-dependent mechanism. Curr Biol. 2010;20(4):328–332. doi: 10.1016/j.cub.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roukos V, Misteli T. The biogenesis of chromosome translocations. Nat Cell Biol. 2014;16(4):293–300. doi: 10.1038/ncb2941. [DOI] [PMC free article] [PubMed] [Google Scholar]