Significance
Using computational methodologies, we refined the binding modes of the transient receptor potential cation channel subfamily V member 1 (TRPV1) modulators, capsaicin and resiniferatoxin, provided by the recent experimental cryo-electron microscopy electron density. The resulting insights enable us to predict the binding pose of 96 additional TRPV1 agonists, which we compare with reported mutagenesis studies. Specifically, we characterize the response of five previously unidentified mutants to capsaicin and resiniferatoxin. Analysis of the amino acids engaged in favorable ligand–channel interactions defines the key structural determinants of the TRPV1 vanilloid binding site.
Keywords: nociception, vanilloid, ligand-gated, docking, heat-sensitive
Abstract
The transient receptor potential cation channel subfamily V member 1 (TRPV1) or vanilloid receptor 1 is a nonselective cation channel that is involved in the detection and transduction of nociceptive stimuli. Inflammation and nerve damage result in the up-regulation of TRPV1 transcription, and, therefore, modulators of TRPV1 channels are potentially useful in the treatment of inflammatory and neuropathic pain. Understanding the binding modes of known ligands would significantly contribute to the success of TRPV1 modulator drug design programs. The recent cryo-electron microscopy structure of TRPV1 only provides a coarse characterization of the location of capsaicin (CAPS) and resiniferatoxin (RTX). Herein, we use the information contained in the experimental electron density maps to accurately determine the binding mode of CAPS and RTX and experimentally validate the computational results by mutagenesis. On the basis of these results, we perform a detailed analysis of TRPV1–ligand interactions, characterizing the protein ligand contacts and the role of individual water molecules. Importantly, our results provide a rational explanation and suggestion of TRPV1 ligand modifications that should improve binding affinity.
Advances in molecular genetics have allowed the identification of a set of ion channels that are expressed in primary afferent neurons and play an important role in the detection and transduction of nociceptive stimuli (1). Among them, transient receptor potential (TRP) channels form a large family. Mammalian TRPs are classified in six subfamilies (2, 3): TRPC (canonical), TRPV (vanilloid), TRPM (melastatin), TRPA (ankyrin), TRPML (mucolipin), and TRPP (polycysteine). TRP channels are nonselective cationic channels, distributed in a diverse range of tissues, with local expression in the free terminals of nociceptive nerve fibers and the skin (4). They are involved in the direct detection of stimuli associated with senses and maintenance of ionic homeostasis (5). The transient receptor potential cation channel subfamily V member 1 (TRPV1) or vanilloid receptor 1 is a polymodal mammalian nociceptive integrator (6) abundantly expressed in the free nerve endings of primary pain sensing afferent Aδ and C fibers (7, 8). Structurally, the TRPV1 channel is a homotetramer, symmetrically organized around a solvent exposed central pore (9, 10). Each subunit is formed by six transmembrane helices (S1–S6) with the channel's N and C termini located in the intracellular medium (11).
TRPV1 is activated by a wide range of proinflammatory and proalgesic mediators (12), including temperatures above 43 °C, external pH, bradykinin, anandamide, arachidonic acid metabolites, jellyfish and spider toxins, and vanilloid. The scope of the TRPV1 pharmacological spectrum (13–15) is mainly in the area of analgesics: novel painkillers could be either TRPV1 agonists or antagonists (16, 17). Moving forward toward the rational drug design of TRPV1 modulators requires a basic understanding of how known ligands interact with TRPV1.
TRPV1 is known to be the target of capsaicin (CAPS), the active component of chili peppers, and it can also be referred to as the capsaicin receptor (18). Resiniferatoxin (RTX), a phorbol ester isolated from the irritant lattices of the Moroccan cactus, shows a much higher affinity for TRPV1 than CAPS (19). Both compounds activate TRPV1, causing the channel to be more permeable to cations, ultimately resulting in an analgesic effect due to channel desensitization. CAPS can be subdivided into three structural regions (20) (Fig. S1): A (aromatic ring), B (amide bond), and C (hydrophobic side chain). RTX can be analogously subdivided into three similar regions: A (aromatic ring), B (ester bond), and C (polyring group) (21) (Fig. S1). Structure–activity relationship studies have provided information about the acceptable structural modifications for CAPS and RTX (22). For example the three- and four-position aryl substituents in the A region were found to be required for activity in CAPS analogs but not that important in RTX ones (23, 24). Replacement of the homovanillyl amide group by an ester in CAPS decreased its activity while increasing the potency of RTX. Additionally, the functionalized five-membered diterpene ring was found to be important for the activity of RTX (24). These studies provided abundant information concerning the structural requirements for CAPS and RTX analog binding. Importantly, organizing such information into a coherent framework can help formulate predictive models about putative TRPV1 binders. To this end, a molecular picture of ligand channel interactions is highly desirable.
Fig. S1.
Chemical structures of CAPS (Upper) and RTX (Lower). The three similar structural regions are described as (A) aromatic, (B) amide, ester, and (C) hydrophobic and polyring.
Advanced single particle electron cryo-electron microscopy (cryoEM) techniques were recently used to obtain the structures of TRPV1 in the apo form and in complex with CAPS and RTX bound in the vanilloid pocket at a resolution of 3.4, 4.2, and 3.8 Å, respectively (9, 10). Despite the crucial insight provided by a recent investigation of CAPS binding mode (25), the binding modes of RTX are still largely uncharacterized. Indeed, the electron density maps of the TRPV1–CAPS and TRPV1–RTX complexes do not carry enough information to confidently infer the location and conformation of the ligands. In this manuscript, we report an investigation of the binding mode of CAPS and RTX based on a pose-directed extraction of the ligand electron density, followed by mutagenesis study to validate our predictions.
Results
Vanilloid Pocket.
We determined four identical vanilloid pockets in the TRPV1 protein. Several other cavities were detected mostly connected among themselves through narrow enclosures or tunnels (Fig. 1). TRPV1 is a homotetramer, and the vanilloid pocket is found between two adjacent chains. The structure of the vanilloid pocket (Fig. 1) is essentially different in the three structures (Fig. S2). In the apo protein, it has a molecular surface of about 9,456 Å2, which is more extended than in the TRPV1–CAPS (9,211 Å2) and in the TRPV1–RTX complexes (8,527 Å2). The vanilloid pocket in the TRPV1–CAPS complex is more accessible to water than in the other two structures, showing solvent accessible surface area of 2,810 Å2, whereas in the TRPV1–RTX complex, it is 2,665 Å2, and in the apo protein, it is 2,291 Å2. The apo protein showed a wider vanilloid pocket (Fig. S2) than those of TRPV1–ligand complexes due to the orientation of Tyr511 outside the pocket and therefore it is not expected to promote tight binding of any ligand. Accordingly, the cryoEM density map of the apo structure shows low values of the density in the region of the cavity, suggestive of occasional binding events of lipid molecules. Tyr511 is oriented upward in the TRPV1–CAPS complex (Fig. S2), thereby closing the pocket and providing a source of hydrophobic, electrostatic and hydrogen bonding contacts with CAPS. Glu570 is about 2 Å apart from Tyr511 in the TRPV1–RTX complex, narrowing the vanilloid pocket in this region. As well, the pocket is wide toward Leu669, Ala665, and Phe591 and narrow in the cleft between S3 and S4, toward the Met547. The features of the pocket of TRPV1–RTX allow for fitting of large structures such as RTX. In conclusion, owing to the intrinsic structural flexibility of the vanilloid pocket, a detailed analysis of the two distinct structures is crucial to study the determinants of the binding affinity of CAPS and RTX for TRPV1.
Fig. 1.
Molecular cavities of TRPV1. (Left) Connolly surface of the molecular cavities; the vanilloid pocket is highlighted by the black box. (Right) The cavity corresponding to the vanilloid pocket is represented as a set of space-filling spheres for the three distinct structures. The different shades of surface coloring represent distinct subpockets. Note how the shape and the volume of the pocket are significantly different in the three cases, suggesting that significant structural rearrangements of the target (induced-fit) take place on binding of the ligand.
Fig. S2.
The vanilloid pocket of the apo protein (cyan, A), the TRPV1–CAPS complex (yellow, B), and the TRPV1–RTX complex (pink, C). (A) Tyr511 is oriented downward in the apo and upward in the protein–ligand complex structures. In the apo structure, the pocket is wide in all directions, facilitating ligand to get in. Tyr511 and Arg557 are oriented in such a way allowing pocket extension. Leu573 and Leu577 are oriented away from each other, leading to a wide subpocket in this direction. This wide pocket does not support tight ligand binding. (B) In the TRPV1–CAPS complex, the pocket is smaller in size compared with the one of the apo protein due to the upward orientation of Tyr511. A small volume is observed between Met547, Asn551, and Leu515. The bulk of the cavity is close to Tyr511 to accommodate the vanillyl group of CAPS and leading to hydrophobic contacts with between with Tyr511. (C) In the TRPV1–RTX complex, the pocket volume is smaller than the one of the apo protein due to upward orientation of Tyr511. Tyr511 and Glu570 are very close in TRPV1–RTX (∼2 Å apart). A wide volume is observed between Ala665, Leu669, and Leu674 and a small volume between Met547, Asn551, and Leu515. The bulk of the cavity is close to Leu669 to accommodate the large diterpene group of RTX.
Binding Pose Prediction.
We docked CAPS and RTX to provide a starting point for a more precise atomic fit. The orientations of key amino acid residues in the vanilloid pocket (Fig. S2) provide a tentative explanation for the differences in binding affinity of ligand in each Protein Data Bank (PDB) structure. CAPS fits well inside the vanilloid pocket of TRPV1–CAPS (Fig. 2B). The subpocket formed between Tyr511, Glu570, and Ile569 is deep enough in the apo and TRPV1–CAPS complex structures to accommodate the vanilloid group. Although the cryoEM experiment was not able to define unambiguously the conformation of all of the sidechains of the binding site, the rotameric state of only two residues (Met547 and Leu669) appear to be only weakly restrained by the electron density (Fig. S3). The flexible aliphatic moiety occupies alternatively distinct cavities in the upper part of the vanilloid pocket. The best docking pose is characterized by a Chemgauss4 score of about −8 kcal/mol. The subpocket close to Tyr511 is shallow in TRPV1–RTX due to the Tyr511-Glu570 proximity and the orientation of Ile-569 toward the vanilloid pocket. Indeed, if CAPS is placed into the vanilloid pocket of TRPV1–RTX using the docking pose determined for TRPV1–CAPS, its methoxy group overlaps with several side chains (Fig. 2C). The apo protein has a wider and deeper subpocket than the CAPS complex, preventing CAPS to achieve tight binding in this region (Fig. 2A).
Fig. 2.
The docking poses of CAPS and RTX, determined in the TRPV1–CAPS and TRPV1–RTX complexes, respectively, and then pasted in the other two structures. (A) CAPS in the apo potein. A loose binding is observed inside the shallow binding pocket. (B) CAPS in the TRPV1–CAPS complex. (C) CAPS in TRPV1–RTX. The methoxy group of CAPS clashes with the deep pocket. (D) RTX in the apo protein. The aromatic head group of RTX clashes with the deep subpocket. (E) RTX in the TRPV1–CAPS complex. The aromatic head group and the diterpene moiety clash with the vanilloid pocket. (F) RTX in the TRPV1–RTX complex. RTX fits well inside the binding pocket.
Fig. S3.
Rotameric state of binding site residues as determined from cryoEM experiments. Shown is the experimentally determined electron density (gray shading, arbitrary isovalue) superimposed to the structure of the protein (atomic coordinates as deposited in the protein databank, gray sticks) and to our docking pose of capsaicin (orange sticks). Purple color highlights the side-chains of residues Y511, L515, F522, F543, T550, F591, I661, and A665, whose conformation is highly restrained by the density, whereas cyan indicates those (M547 and L669) for which the experimental information is less abundant.
The docking (Chemgauss4) score of RTX in the TRPV1–RTX complex is about −6.0 kcal/mol. The subpocket close to Leu669, Val583, and Phe587 is wide due to the projection of these amino acids out of the vanilloid pocket. This subpocket allows accommodation of the diterpene group of RTX without any clashes (Fig. 2F). However, the orientation of Leu515 and Met547 makes this region of the vanilloid pocket narrow, thereby greatly constraining the nature of the tolerated fragments. On the other hand, the diterepene group of RTX would cause clashes with several protein residues in the apo and TRPV1–CAPS structures because the subpocket close to Leu669 is narrow (Fig. 2 C and D).
To extract the ligand electron density, to find the best ligand fit, and to restrict the scope of conformational exploration, we used the information obtained from the docking step. The approximate binding mode determined by docking enabled the extraction of the low-resolution ligand electron density. The pose of CAPS was tuned to match the extracted ligand electron density (Fig. 3A). The density fit binding mode (Dataset S1) is close to the docked one, only with a flipping of the vanillyl group, which leads to an RMSD of ∼4.05 Å (Fig. 3C). CAPS was placed in the ligand electron density with a real space correlation coefficient (RSCC) of 0.41. The accompanying pairwise linear potential (PLP) and Chemscore values are −31.5 and −9.9 kcal/mol, respectively, characteristic of a good fit. The electron density representing CAPS shows three lobes, suggesting a dynamic binding of the aliphatic side chain. The electron density is pointing toward Tyr511 to accommodate the methoxy group of the vanillyl ring. The hydroxyl group and amide nitrogen of CAPS are hydrogen bonded to Glu570 and Tyr511, and to Thr550 (Fig. 3D). The rest of the molecule is involved in hydrophobic interactions with several amino acids of the pocket (Fig. 3D). Our predicted binding mode for CAPS is in a tail-up, head-down configuration; interestingly, the vanillyl group is rotated by 180° compared with the most accurate prediction available thus far (25). Further studies, based on molecular dynamics simulations, will likely provide insight on which of these two viable conformations is the most populated one; however, it is worth noting that the fact that no electron density was detected via cryoEM in the region of the head group suggests a certain degree of structural heterogeneity. RTX displayed better fitness inside the vanilloid pocket than CAPS.
Fig. 3.
Binding mode of CAPS based on the atomic fitting with the electron density map. (A) CAPS fit within the electron density. (B) CAPS binding mode as balls and sticks, and the surrounding amino acids as surface. (C) The docked and density-fit binding modes of CAPS are very similar and involve merely a rotation along its long axis, with little penetration of the pocket. (D) The ligand interaction model of the binding modes of CAPS. Atoms of the amino acid residues in hydrophobic contacts with the ligand are shown as spheres. Hydrogen bonds are shown as green lines. The interatomic distances between the hydrogen bond donor and acceptor are shown up to 4.0 Å.
RTX fits well in the electron density (Fig. 4A) with an RSCC of 0.36, PLP of −43.8 kcal/mol, and Chemscore of −16.5 kcal/mol. The blob has the shape of RTX, with the upper section able to fit the diterpene group, and the lower one of the size of the aromatic group. The density fit and docking binding modes aligned with an RMSD of ∼2.35 Å (Fig. 4C and Dataset S2), highlighting the good performance of the docking with fast rigid exhaustive docking (FRED). Compared with the docking pose, the aromatic group is located deeper inside the subpocket close to Tyr511 and is oriented almost parallel to the aromatic side chain of Tyr511, so that it establishes a strong π–π interaction (Fig. 4D). The aromatic hydroxyl and methoxy groups of RTX form strong hydrogen bonds with Glu570, Arg557, and Ser512 (Fig. 4D). The ester group is also in the appropriate position to be hydrogen bonded to Tyr511 and Thr550 (Fig. 4D). Taken together, these results rationalize the greater potency of RTX compared with CAPS.
Fig. 4.
Binding mode of RTX based on the atomic fitting with the electron density map. (A) RTX fit within the electron density. (B) RTX binding mode as balls and sticks, and the surrounding amino acids as surface. (C) The docked and density-fit binding modes of RTX are very similar. RTX is placed slightly more downward in the density-fit binding mode. (D) The ligand interaction model of the binding modes of RTX. Atoms of the amino acid residues in hydrophobic contacts with the ligand are shown as spheres. Hydrogen bonds are shown as green lines. The interatomic distances between the hydrogen bond donor and acceptor are shown up to 4.0 Å.
Protein–Ligand Interaction Profiles.
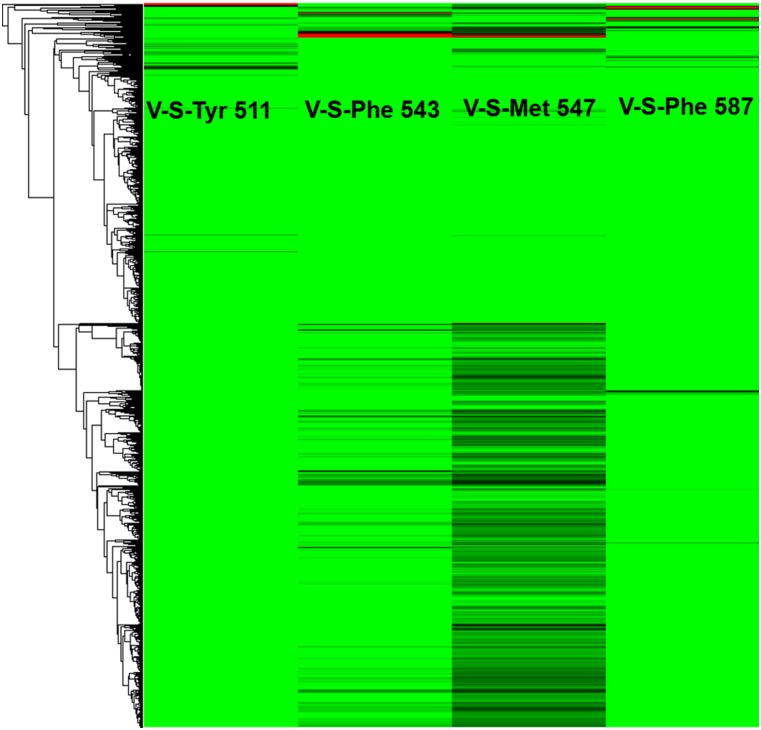
To identify the pharmacological preferences of the amino acid residues in the vanilloid pocket, we generated protein–ligand interaction profiles P(I) by docking a library of CAPS analogs to the TRPV1 structures (21, 26, 27). The profiles are divided according to interaction type (28), electrostatic (E), hydrogen bonding (H), and van der Waals (V) (Fig. S4), and to interaction site, main chain (M) or side chain (S). To do this, we used both the apo protein and TRPV1–CAPS complex. From the apo protein results, the amino acids that are found to be important for ligand binding are Thr550 (H-S, 1.00 and V-S, 0.52), Leu553 (V-M, 0.95), Tyr554 (V-M, 0.56), Arg557 (H-S, 0.53), Glu570 (V-S, 1.00), Leu-669 (V-S, 0.56), and Ile573 (V-S, 0.62) (Fig. S4). Carrying out the same analysis in the TRPV1–CAPS complex, we add as important residues Tyr511 (V-S, 1.00), Phe543 (V-S, 0.43), Met547 (V-S, 0.91), and Phe587 (V-S, 0.69) (Fig. S5). More than 80% of the docked poses are forming hydrogen bonding and van der Waals interactions with the side chains of Thr550 (H-S) and Tyr511 (V-S). This observation indicates that these interactions are essential for ligand binding and TRPV1 activity. Our results are consistent with the mutation studies of Tyr511, Met547, Thr550, Arg557, and Glu570, which were reported to be important for CAPS and RTX binding (29). We suggest that other residues, whose role has not yet been investigated experimentally, might be crucial for binding to the vanilloid pocket, including Leu515, Leu553, Tyr-554, Ile573, and Phe-587. In this list, Leu553 and Tyr554 interact mostly through their main chain groups. Despite the fact that single residue mutation at these positions cannot change the chemical nature of the main chain, it is reasonable to assume that changing the side chains of these amino acids may perturb the local geometry and thus the favorable energetic interactions between CAPS and TRPV1.
Fig. S4.
The interaction profile of TRPV1 modulators with the TRPV1 apo structure. Favorable interactions are shown in green and unfavorable ones in red. The docked poses (y axis) are clustered based on their contacts with the commonly interacting amino acids (x axis). (Left) Clustered interaction profiles of the ensemble of ligands. (Right) Zoom on a small cluster of six compounds, and their docking poses in the vanilloid pocket.
Fig. S5.
We are showing only the new commonly interacting amino acid residues from the interaction profile of TRPV1 modulators with TRPV1–CAPS. Favorable interactions are shown in green and unfavorable ones in red. The docked poses (y axis) are clustered based on their contacts with the commonly interacting amino acids (x axis).
Mutagenesis Study.
We generated individual alanine mutants of residues that our computational modeling predicted to be involved in channel activation by CAPS and RTX. To test their sensitivity to CAPS and RTX, we expressed WT and mutant channels in Xenopus laevis oocytes and measured currents induced by CAPS (1 and 100 μM) and RTX (10 and 100 nM). Low extracellular pH (pH = 4) was used as a control stimulus to test for functionality of the mutant channels. All mutants displayed clear responses to low pH, even though there was a variable extent of delay in the responses in some mutants, as shown in Fig. 5 A and B. All five mutants had somewhat smaller, but comparable current amplitudes induced by pH 4 to those in the WT TRPV1 (Fig. S6). Noninjected oocytes showed no current responses to pH 4. CAPS- and RTX-induced currents in the WT TRPV1 that were similar to those in earlier publications (30, 31). CAPS at 1 μM induced a somewhat larger current than pH 4 and somewhat smaller than 100 μM CAPS (Fig. 5 A and C), whereas RTX at 10 nM induced currents that were similar amplitude to those induced by pH 4 and substantially smaller than those induced by 100 nM RTX (Fig. 5 B and C). The effect of RTX developed much slower than the effect of either pH 4 or CAPS. All of the alanine point mutants showed impaired responses to both CAPS and RTX; results are summarized in Fig. 5C, where amplitudes were compared with those induced by pH 4. Leu553, Tyr554, and Ile573 mutants showed no, or minimal, responses to both CAPS and RTX, even at high concentrations, whereas the Leu515 and Phe587 mutants showed clear, yet somewhat smaller, responses than WT, to the higher concentrations of CAPS (100 μM) and RTX (100 nM), but did not respond to the lower concentrations: 1 μM and 10 nM, respectively. These data confirm our computational predictions.
Fig. 5.
Altered CAPS and RTX responses in TRPV1 mutants. TEVC oocyte electrophysiology was performed as described in Materials and Methods. (A) Representative traces, currents from repeated ramp protocols are plotted for −100, 0, and 100 mV; bottom, middle, and top traces, respectively. The effects of pH 4 (green), 1 μM CAPS (pink), and 100 μM CAPS (red) are indicated with the horizontal lines. (B) Similar representative measurements with 10 nM RTX (cyan) and 100 nM RTX (blue). (C) Summary of current amplitudes at 100 mV (mean ± SE); data were normalized to the current evoked by pH 4 at the beginning of each measurement (n = 4 for each construct both for CAPS and for RTX).
Fig. S6.
Current amplitudes induced by pH 4 in WT TRPV1 and five mutants. Data are summarized from n = 8 measurements for all channels; mean current amplitudes ± SE are plotted.
Water Mapping of the Vanilloid Pocket.
We analyzed the active site of TRPV1–ligand complexes to find the most favorable regions for binding polar or nonpolar moieties using a fragment-based–like approach. To this end, we calculated the binding free energy of explicit water molecules placed at different positions within the binding site. To differentiate locations with large affinity for polar or nonpolar moieties, we used, in addition to the canonical water model (Fig. 6A), a modified one lacking any electrostatic interaction with the protein (uncharged water). These results are summarized in the 3D maps shown in Fig. 6 B and C. The red map shows the regions of space with large putative occupancy of uncharged water, whereas the yellow one shows the same property for the canonical water model. Finally, the green map shows the regions in which uncharged waters have the largest van der Waals interactions with the protein. Inspection of these maps is, in general, informative about the spatial distribution of chemical groups that gives rise to large ligand-target affinities. Regions deemed favorable for binding uncharged waters are putative binding spots of nonpolar moieties. Conversely, regions showing large propensity for canonical water are probable binding spots for polar and charged groups. More detailed information can be extracted from the green map: the regions showing the largest van der Waals interaction energies with the uncharged water molecules are those favoring strongly van der Waals-interacting groups. The analysis of these maps calculated for TRPV1–CAPS in the absence of the ligand reveals some of the determinants of affinity. The hydroxyl group localizes in the yellow map, whereas the methoxy one is placed well inside the red region. This localization, together with the orientation of the probe waters, tells us that the hydroxyl group displaces a water to form a hydrogen bond with Glu570, whereas the carbonyl and amine with Thr550 and Tyr511, respectively, through a bridging water molecule. It is noteworthy that an analogous water-bridged interaction was found in an independent investigation based on molecular dynamics simulations (32). The aliphatic chain localizes in between red regions that account for nonpolar interactions. Two structural water molecules are found close to Glu570 (Fig. S7). Based on these water mapping calculations, modification suggestions to enhance the affinity of CAPS were generated (Fig. 7), recommending nonpolar substituents in place of the methoxy and polar in the position of the hydroxyl group that do not displace the structural water. Substitutions in the aromatic and benzylic groups that increase the van der Waals interactions with the protein supposedly will improve potency. These findings are in good agreement with results from experimentally tested compounds (Fig. S8): by substituting the methoxy at Cl, keeping the phenolic group, and adding methyl at the benzylic carbon, the activity improved from ∼2,000 (CAPS) to ∼160 nM (compound A) (26) (Fig. S8), and by masking the hydroxyl group, substituting the methoxy with Fluoro, the activity dropped to more than 10,000 nM (compound B) (26) (Fig. S8).
Fig. 6.
Water mapping of the TRPV1–CAPS complex. (A) Important water cluster with the putative orientation of individual water molecules. Note in particular the structural waters close to Glu-570. (B) Regions of space with large putative occupancy of uncharged water are represented in red, whereas the ones accommodating canonical waters are shown in yellow. (C) van der Waals region (green) is located close to nonpolar regions.
Fig. S7.
The crucial water molecules for keeping the correct geometry of the binding site of TRPV1–CAPS. The wheat surface represents the possible location of the water molecules. The two most probable water molecules are shown as ball and sticks.
Fig. 7.
(A) Modification hypothesis of CAPS. Positions, which can tolerate nonpolar/polar substituents, are shown as yellow/green spheres, respectively. (B) Polar (green) and neutral (yellow) water molecules at ligand coordinate providing clues about possible ligand modification. (C) Positions permitting polar substitutions. These modifications will be stabilized by the surrounding polar amino acids. (D) Small nonpolar substitutions such as halogens supported by hydrophobic amino acid residues. (E) van der Waals substitutions supported by hydrophobic amino acid residues.
Fig. S8.
Comparison between the modification plans and experimental results. A is ∼15 times more potent than CAPS by making favorable modifications in the proper positions, and B is ∼7 times less potent than CAPS due to nonrecommended modifications.
We analyzed the maps of TRV1–RTX to understand the reasons for the high affinity of RTX. Interestingly, the different functions of the vanillyl group are perfectly positioned in the uncharged and charged water maps. The hydroxyl and methoxy groups are localized in the yellow and red maps, respectively (Fig. 8B). Similarly, the distribution of the different groups of the other regions of RTX reveals that all polar and nonpolar groups of RTX are located in the yellow and red maps, respectively. Furthermore, modification hypotheses are suggested to improve the affinity of RTX (Fig. 9). The comparison between the details of CAPS and RTX binding reveals the determinants of the stronger affinity of RTX and highlights how slight changes in the conformation of the protein may result in different ligand binding environments.
Fig. 8.
Water mapping of RTX complex. (A) Important water clusters with the putative orientation of individual water molecules. (B) Regions of space with large putative occupancy of uncharged water are represented in red, whereas the ones accommodating canonical water are shown in yellow. (C) van der Waals region is represented in green.
Fig. 9.
(A) Modification hypothesis of RTX. Positions that can tolerate nonpolar/polar substituents are shown as yellow/green spheres, respectively. (B) Polar (green) and neutral (yellow) water molecules at ligand coordinate providing clues about possible ligand modification. (C) Positions permitting polar substitutions. These modifications will be stabilized by the surrounding polar amino acids. (D) Small nonpolar substitutions such as halogens supported by hydrophobic amino acid residues. (E) van der Waals substitutions such as alkyl groups surrounded by hydrophobic amino acid residues.
Discussion
Single particle cryoEM is becoming a routinely used technique for determining the 3D structure of large molecular complexes in their native state, such as viruses and large protein assemblies. Despite recent dramatic increase in resolution, the cryoEM density maps are still not high enough to resolve the atomic details of the interactions between protein and small organic molecules. Therefore, there is a need to develop an integrated procedure to facilitate the determination of the binding modes of ligands. The approach that we are presenting in this manuscript aims at filling this gap by describing a relatively simple procedure that allows one to overcome this limitation of the cryoEM technique. Using cavity prediction approaches and knowledge-based docking algorithms, we were able to infer a probable binding region, allowing the extraction of the ligand electron density maps and hence precisely fit of the ligand into the extracted maps.
Cavity prediction is per se a useful tool to explore the molecular cavities and identify the druggable ones. In our study, the plausible location of the vanilloid active site was known from literature and from the recent cryoEM density maps. Comparing the active site in the apo and bound states provided insights into the possible orientations of the ligands. Importantly, the three structures cannot be interchangeably used to determine the binding mode of a specific ligand. The TRPV1 apo structure has a wide and open vanilloid pocket leading to weak binding of large molecules such as phospholipid molecules. CAPS- and RTX-bound TRPV1 structures showed variable sizes of their subpockets; this flexibility highlights the relevance of induced fit in the binding of these two molecules.
The docking program that we used in this study (FRED) uses an exhaustive search algorithm to position the ligand within the active site and extract the ligand electron density maps. The docked poses overlapped nicely with the density fit poses, although the orientation of some groups was found to be slightly different. The electron density pertaining to each ligand was extracted from the total map using the docked pose. This step effectively reduced the volume of the conformational space to be explored. This knowledge-based structural fitting procedure enabled us to pinpoint unambiguously the binding modes of CAPS and RTX. These extracted densities can help to design better drugs, for instance, by enabling the use of shape-based algorithms for virtual screening.
A satisfactory understanding of the mode of action of the known TRPV1 ligands is an important first step toward a successful drug design approach. TRPV1 has a complex polymodal activation profile because it is able to sense multiple stimuli, such as noxious pain, heat, protons, ligand binding, and a number of products of cellular mechanisms. Several TRPV1 antagonist candidate drugs have failed in clinical trials because, by interfering with the detection of the aforementioned stimuli, they triggered serious side effects such as hyperthermia and impaired detection of painful heat. Thus, successful TRPV1 modulators need to interfere selectively with only a subset of these activation modalities leaving the others unperturbed.
TRPV1 desensitization by agonists such as CAPS and RTX is a potential pharmacological strategy in pain management. RTX is now in clinical trials for treatment of severe pain associated with advanced cancer, but RTX is toxic and can produce chemical burns. On the Scoville scale, RTX has a rating of 16 billion, which is about 1,000 times higher than CAPS, and ingestion of less than 10 g of RTX could be fatal. These facts motivate the need to design agonists less potent than RTX but apparently more than CAPS. By accurately determining the binding modes of CAPS and RTX and further studying the thermodynamic properties associated with them, it is possible to design more potent CAPS analogs and less toxic RTX derivatives.
Calculations of the water mapping of the active site and the thermodynamic properties at the location of the ligand atoms proved to be very useful in explaining the structure activity relationships of CAPS analogs (Fig. S9) and provided several predictions about unexplored modifications that improve ligand binding. We also propose RTX modifications. However, the extremely high potency of RTX is not well tolerated by the human body and the modification strategy involves synthesizing less potent derivatives.
Fig. S9.
Experimentally tested CAPS analogs as TRPV1 agonists. Capsaicin is shown in the upper box, illustrating the main three regions of the structure that can be modified. S1–S4 are series 1–4 of the capsaicin analogs. The three similar structural regions are described as (A) aromatic, (B) amide, ester, and (C) hydrophobic and polyring.
Our calculations indicate that polar substituents are tolerated in region B; consistently, changing the amide into thiourea derivatives or reversing the amide linkage has been shown experimentally to increase the activity (24). Several nonpolar substituents are expected to improve the activity in region C. In these series of compounds, most of the aromatic and aliphatic substituents enhanced the activity. Note, however, that some of the long chain substituents considerably decreased the activity. Region A is most sensitive to modifications; polar substituents are tolerated in the para position of the aromatic ring. Masking the hydroxyl group was experimentally found to decrease the activity of most of the synthesized compounds of these analog series. Some compounds involving allowed modifications in regions B and C but unfavorable modifications in region A; for example, replacing the hydroxyl group in the para position by aminoethyloxy were shown to be active. This kind of composite modification is not well characterized in our modification hypothesis model.
In summary, the approach we report here to extract the ligand density maps from low-resolution cryoEM density allowed us to precisely define the binding modes of CAPS and RTX. A detailed knowledge of CAPS and RTX binding modes constitutes the first step to properly understand the polymodality of TRPV1. The mutation study supported our computational predictions in this regard. Further work could involve induced fit docking (IFD) to understand the effect of ligand binding on protein structure, hybrid quantum mechanics/molecular mechanics (QM/MM) approaches to study the binding sites in detail, molecular dynamics (MD) simulations to investigate the allosteric coupling of the vanilloid site to the gate and other polymodal properties of TRPV1, and quantitative structure activity relationship (QSAR) approaches to understand the correlation between the structural features of the ligand and the different opening modalities of TRPV1.
Materials and Methods
The study was based on several steps that are described in details in the following paragraphs. In short, we extracted the ligand electron density by endowing the algorithm for the fit of the electron density map with a prior knowledge of the approximate location of the ligand obtained through computational docking. This preconditioning step greatly reduced the scope of the conformational exploration and thus made the problem computationally tractable. Next, we validated the resulting CAPS and RTX binding poses by computing the RSCC, PLP, and Chemscore scores. Then, we analyzed the predicted protein–ligand interactions in light of the phenotype of the previously studied TRPV1 mutations and predicted the behavior of novel mutants. Finally, by calculating the thermodynamic properties of each ligand chemical group and mapping the active site waters, we explained the changes in activity on ligand modification previously reported in literature and propose unexplored chemical modifications able to increase or decrease affinity of CAPS and RTX.
Database Collection and Preparation.
The single particle electron cryoEM structures of TRPV1 (9, 10) (PDB ID codes 3J5P, 3J5R, and 3J5Q) (33) and the protein electron density maps (9, 10) (accession codes: EMD-5776, EMB-5777, and EMB-5778) were downloaded. We prepared TRPV1 ligand databases, downloaded from the Binding Database (www.bindingdb.org) (34), using SPORES (35, 36), by adding missing hydrogen atoms, generating atom connectivity, correcting ionization states, adjusting hybridization and protonation state, and setting atom and bond types. The protein structures were then prepared using PrepWizard (37, 38) without applying structural minimization to avoid any geometry refinement that may incorrectly affect the resulting ligand binding mode (39).
Binding Mode Prediction.
We identified the molecular cavities of TRPV1 using the OpenEye software (www.eyesopen.com) (39) The volume and dimensions of the grid box of each protein are provided in Table S1. The molecular cavity surrounded by Tyr511, Met547, Thr550, and Leu669 was considered for docking experiments. Exploration of ligand conformational space was performed using OMEGA 2.4.6 (OpenEye) (40, 41) with the 94s variant of Merck Molecular Force Field (MMFF94s). FRED, the multiconformational rigid docking algorithm of OpenEye (42–44), was used for pose prediction of CAPS and RTX and for the docking simulation ofTRPV1 agonist databases (21, 26, 27). We kept several poses of each ligand for subsequent analysis. We gauged the consistency between the predicted binding pose and the electron density using AFITT (www.eyesopen.com) (45, 46). Before any calculation, we refined the structure of the protein by performing a rotamer search. Knowledge of the coordinates of the protein atoms was used to exclude protein density from the cryoEM density map and thus extract the ligand density. The electron density isosurface was selected to encompass points with intensities greater than 3.55 SDs from the mean value (σ level of 3.55). The blobs, which are the regions of the cryEM density maps representing the plausible location of the ligand, were pruned to 2.5 Å from the protein. To facilitate the search process for blobs, we built a box around the docked pose, and we picked the density surrounding the ligand. Then we used the automatic ligand fitting approach of AFITT. The latter is a fast and reliable approach that has been shown good performance with low-resolution densities and flexible ligands (47–49). We then analyzed the protein–ligand interaction pattern through Maestro (50) and LigPlot+ (38). The maximum distances of hydrogen–acceptor (H–A) and donor–hydrogen (D–H) were defined as 2.70 and 3.35 Å, respectively.
Table S1.
Properties of receptor grids used in docking simulations
| PDB ID code | Volume (Å3) | x (Å) | y (Å) | z (Å) | Outer contour (Å3) | Constraints |
| 3J5P | 9,744 | 18.67 | 29.00 | 18.00 | 1,900 | — |
| 3J5R | 9,639 | 17.00 | 21.00 | 27.00 | 2,228 | Tyr511 contact |
| Thr550 donor | ||||||
| 3J5R | 9,639 | 17.00 | 21.00 | 27.00 | 2,228 | Tyr511 contact |
| 3J5R | 9,639 | 17.00 | 21.00 | 27.00 | 2,228 | Thr550 donor |
| 3J5Q | 25,179 | 25.67 | 27.00 | 36.33 | 3,164 | — |
| 3J5Q | 25,179 | 25.67 | 27.00 | 36.33 | 3,164 | Tyr511 contact |
| Thr550 donor | ||||||
| 3J5Q | 25,179 | 25.67 | 27.00 | 36.33 | 3,164 | Tyr511 contact |
| 3J5Q | 25,179 | 25.67 | 27.00 | 36.33 | 3,164 | Thr550 donor |
| 3J5R | 9,639 | 17.00 | 21.00 | 27.00 | 2,228 | — |
Mapping of Active Site Waters.
We used SZMAP (51) to analyze the TRPV1 vanilloid binding site, to generate the active site’s water map, and to construct ligand modifications hypotheses. In this step, we used FIXDUPATOMNAMES to convert atoms with duplicate names to unique atom names, MKHETDICT to built PDB heterogen dictionary, and REDUCE (52) to explicitly add hydrogen atoms to the protein–ligand complex and to split the prepared protein–ligand complex into protein and ions in one file and ligand in another file. AmberFF94 (53) charges were used to assign partial charges to amino acids, and AM1BCC (40) for any other group. The stabilization and destabilization grids of the complex, apo, and ligand were calculated by SZMAP using explicit water molecules as probes. We determined the excess free energy (with respect to the bulk) of those water molecules that are displaced by the ligand. SZMAP uses a semicontinuum solvation theory, which combines a single explicit probe water with Poisson–Boltzmann continuum theory. Orientations of the probe water are sampled based on the probe interaction with the continuum solvent, protein, and ligand molecules. SZMAP uses this information to predict the solvent role in ligand binding. SZMAP is one of most accurate approaches available to calculate the position and evaluate the free energy of water molecules tightly bound to protein cavities (54). As a first step, the FRED shape-scoring function was used to determine the positions of the waters trapped in the cavity and in contact with the protein. Then, the energy of the probe at these positions was calculated by exhaustively sampling all of the possible orientations. GAMEPLAN (51) was then used to suggest modification hypotheses by defining possible replacements and modifications of the ligand that should improve the binding affinity.
Detection of Hot Spot Amino Acid Residues.
A database of 96 TRPV1 agonists (21, 26, 27) was docked into TRPV1 using FRED (42–44). To provide data-rich results, multiple poses were selected for analysis. The interactive generic evolutionary method for the molecular docking (iGEMDOCK) (28) tool was used for analysis of the results to identify the commonly interacting amino acid residues. To calculate the pharmacological interactions, we considered those residues whose interaction energy with the ligand is such as the hydrogen bonding, electrostatic, and van der Waals components are more negative than −2.5, −2.5, and −4.0 kcal/mol, respectively. All interactions that showed a z-score of 1.645 or more were considered for further analysis. We generated protein–ligand interaction tables in the form of protein–ligand interaction profiles of electrostatic (E), hydrogen-bonding (H), and van der Waals (V) interactions. The atom- and residue-based interactions were calculated to provide the individual contribution of each residue in ligand binding energy.
Xenopus Oocyte Preparation and Electrophysiology.
Oocytes from female X. laevis frogs were prepared using collagenase digestion, as describe earlier (55). Briefly, oocytes were digested using 0.2 mg/mL collagenase (Sigma) in a solution containing 82.5 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM Hepes, pH 7.4 (OR2), overnight for ∼16 h at 18 °C in a temperature-controlled incubator. Oocytes were selected and kept in OR2 solution supplemented with 1.8 mM CaCl2 and 1% penicillin/streptomycin (Mediatech) at 18 °C. Point mutants of TRPV1 in the pGEMS oocyte vector were generated using the QuikChange kit (Agilent Technologies). cRNA was generated from linearized cDNA using the mMessage mMachine kit (Ambion). cRNA (50 ng) was microinjected into each oocyte using a nanoliter injector system (World Precision Instruments). The experiments were performed 5 d after injection. Injections of lower amounts of RNA and shorter incubation periods resulted in low expression (current) levels of several mutants. Results with those low current levels were qualitatively similar to those shown in Fig. 5, but they were not included in the data summary there.
Two-electrode voltage clamp (TEVC) measurements were performed as described earlier (55) in a solution containing 97 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM 2-(N-morpholino) ethanesulfonic acid (Mes), and 5 mM Hepes, pH 7.4. Mes was added to our usual extracellular oocyte buffer to be able to set the pH to 4 with HCl in the stimulating solution. Currents were recorded with thin-wall inner filament-containing glass pipettes (World Precision Instruments) filled with 3 M KCl in 1% agarose. Currents were measured with a ramp protocol from −100 to 100 mV performed once every second from a holding potential of 0 mV, and the currents at −100, 0, and 100 mV were plotted.
Supplementary Material
Acknowledgments
We thank OpenEye Scientific Software for providing us with a free academic license. This work was supported by the National Institutes of Health via National Institute for General Medical Sciences Grant P01 GM55876 (to M.L.K.) and National Science Foundation Grants ACI-1440059 (to M.L.K. and V.C.) and R01NS055159 and R01GM093290 (to T.R.). L.D. receives funding from European Union Seventh Framework Program “Voltsens” Grant PIOF-GA-2012-329534.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517288113/-/DCSupplemental.
References
- 1.Vriens J, Nilius B, Voets T. Peripheral thermosensation in mammals. Nat Rev Neurosci. 2014;15(9):573–589. doi: 10.1038/nrn3784. [DOI] [PubMed] [Google Scholar]
- 2.Nilius B, Owsianik G. The transient receptor potential family of ion channels. Genome Biol. 2011;12(3):218. doi: 10.1186/gb-2011-12-3-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clapham DE, Julius D, Montell C, Schultz G. International Union of Pharmacology. XLIX. Nomenclature and structure-function relationships of transient receptor potential channels. Pharmacol Rev. 2005;57(4):427–450. doi: 10.1124/pr.57.4.6. [DOI] [PubMed] [Google Scholar]
- 4.Kunert-Keil C, Bisping F, Krüger J, Brinkmeier H. Tissue-specific expression of TRP channel genes in the mouse and its variation in three different mouse strains. BMC Genomics. 2006;7(1):159. doi: 10.1186/1471-2164-7-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 6.Caterina MJ, et al. The capsaicin receptor: A heat-activated ion channel in the pain pathway. Nature. 1997;389(6653):816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 7.Menigoz A, Boudes M. The expression pattern of TRPV1 in brain. J Neurosci. 2011;31(37):13025–13027. doi: 10.1523/JNEUROSCI.2589-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez-García MC, et al. Differential expression and localization of transient receptor potential vanilloid 1 in rabbit and human eyes. Histol Histopathol. 2013;28(11):1507–1516. doi: 10.14670/HH-28.1507. [DOI] [PubMed] [Google Scholar]
- 9.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504(7478):107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504(7478):113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, Liu S, Yang F, Zheng J, Wang K. Identification of a tetrameric assembly domain in the C terminus of heat-activated TRPV1 channels. J Biol Chem. 2011;286(17):15308–15316. doi: 10.1074/jbc.M111.223941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holzer P. The pharmacological challenge to tame the transient receptor potential vanilloid-1 (TRPV1) nocisensor. Br J Pharmacol. 2008;155(8):1145–1162. doi: 10.1038/bjp.2008.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nilius B. TRP channels in disease. Biochim Biophys Acta. 2007;1772(8):805–812. doi: 10.1016/j.bbadis.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Vriens J, Appendino G, Nilius B. Pharmacology of vanilloid transient receptor potential cation channels. Mol Pharmacol. 2009;75(6):1262–1279. doi: 10.1124/mol.109.055624. [DOI] [PubMed] [Google Scholar]
- 15.Nilius B, Szallasi A. Transient receptor potential channels as drug targets: From the science of basic research to the art of medicine. Pharmacol Rev. 2014;66(3):676–814. doi: 10.1124/pr.113.008268. [DOI] [PubMed] [Google Scholar]
- 16.Cui M, et al. TRPV1 receptors in the CNS play a key role in broad-spectrum analgesia of TRPV1 antagonists. J Neurosci. 2006;26(37):9385–9393. doi: 10.1523/JNEUROSCI.1246-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szallasi A, Cortright DN, Blum CA, Eid SR. The vanilloid receptor TRPV1: 10 years from channel cloning to antagonist proof-of-concept. Nat Rev Drug Discov. 2007;6(5):357–372. doi: 10.1038/nrd2280. [DOI] [PubMed] [Google Scholar]
- 18.Shaqura M, et al. New insights into mechanisms of opioid inhibitory effects on capsaicin-induced TRPV1 activity during painful diabetic neuropathy. Neuropharmacology. 2014;85:142–150. doi: 10.1016/j.neuropharm.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 19.Szallasi A, Blumberg PM. Resiniferatoxin, a phorbol-related diterpene, acts as an ultrapotent analog of capsaicin, the irritant constituent in red pepper. Neuroscience. 1989;30(2):515–520. doi: 10.1016/0306-4522(89)90269-8. [DOI] [PubMed] [Google Scholar]
- 20.Thomas KC, et al. Structure-activity relationship of capsaicin analogs and transient receptor potential vanilloid 1-mediated human lung epithelial cell toxicity. J Pharmacol Exp Ther. 2011;337(2):400–410. doi: 10.1124/jpet.110.178491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi H-K, et al. Non-vanillyl resiniferatoxin analogues as potent and metabolically stable transient receptor potential vanilloid 1 agonists. Bioorg Med Chem. 2009;17(2):690–698. doi: 10.1016/j.bmc.2008.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinberg X, Lespay-Rebolledo C, Brauchi S. A structural view of ligand-dependent activation in thermoTRP channels. Front Physiol. 2014;5:171. doi: 10.3389/fphys.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrigglesworth R, et al. Analogues of capsaicin with agonist activity as novel analgesic agents: Structure-activity studies. 4. Potent, orally active analgesics. J Med Chem. 1996;39(25):4942–4951. doi: 10.1021/jm960512h. [DOI] [PubMed] [Google Scholar]
- 24.Walpole CS, et al. Similarities and differences in the structure-activity relationships of capsaicin and resiniferatoxin analogues. J Med Chem. 1996;39(15):2939–2952. doi: 10.1021/jm960139d. [DOI] [PubMed] [Google Scholar]
- 25.Yang F, et al. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat Chem Biol. 2015;11(7):518–524. doi: 10.1038/nchembio.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho Y, et al. The SAR analysis of TRPV1 agonists with the α-methylated B-region. Bioorg Med Chem Lett. 2012;22(16):5227–5231. doi: 10.1016/j.bmcl.2012.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, et al. 3-Acyloxy-2-phenalkylpropyl amides and esters of homovanillic acid as novel vanilloid receptor agonists. Bioorg Med Chem Lett. 1999;9(20):2909–2914. doi: 10.1016/s0960-894x(99)00513-2. [DOI] [PubMed] [Google Scholar]
- 28.Hsu K-C, Chen Y-F, Lin S-R, Yang J-M. iGEMDOCK: A graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinformatics. 2011;12(Suppl 1):S33. doi: 10.1186/1471-2105-12-S1-S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter Z, et al. Functionally important amino acid residues in the transient receptor potential vanilloid 1 (TRPV1) ion channel: An overview of the current mutational data. Mol Pain. 2013;9(1):30. doi: 10.1186/1744-8069-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caterina MJ, Rosen TA, Tominaga M, Brake AJ, Julius D. A capsaicin-receptor homologue with a high threshold for noxious heat. Nature. 1999;398(6726):436–441. doi: 10.1038/18906. [DOI] [PubMed] [Google Scholar]
- 31.Jordt S-E, Julius D. Molecular basis for species-specific sensitivity to “hot” chili peppers. Cell. 2002;108(3):421–430. doi: 10.1016/s0092-8674(02)00637-2. [DOI] [PubMed] [Google Scholar]
- 32.Darré L, Domene C. Binding of capsaicin to the TRPV1 ion channel. Mol Pharm. 2015;12(12):4454–4465. doi: 10.1021/acs.molpharmaceut.5b00641. [DOI] [PubMed] [Google Scholar]
- 33.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: A web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35(Database issue) Suppl 1:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.ten Brink T, Exner TE. Influence of protonation, tautomeric, and stereoisomeric states on protein-ligand docking results. J Chem Inf Model. 2009;49(6):1535–1546. doi: 10.1021/ci800420z. [DOI] [PubMed] [Google Scholar]
- 36.ten Brink T, Exner TE. pK(a) based protonation states and microspecies for protein-ligand docking. J Comput Aided Mol Des. 2010;24(11):935–942. doi: 10.1007/s10822-010-9385-x. [DOI] [PubMed] [Google Scholar]
- 37.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, Swindells MB. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 39.Elokely KM, Doerksen RJ. Docking challenge: Protein sampling and molecular docking performance. J Chem Inf Model. 2013;53(8):1934–1945. doi: 10.1021/ci400040d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem. 2002;23(16):1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 41.Hawkins PC, Skillman AG, Warren GL, Ellingson BA, Stahl MT. Conformer generation with OMEGA: Algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J Chem Inf Model. 2010;50(4):572–584. doi: 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGann M. FRED and HYBRID docking performance on standardized datasets. J Comput Aided Mol Des. 2012;26(8):897–906. doi: 10.1007/s10822-012-9584-8. [DOI] [PubMed] [Google Scholar]
- 43.McGann M. FRED pose prediction and virtual screening accuracy. J Chem Inf Model. 2011;51(3):578–596. doi: 10.1021/ci100436p. [DOI] [PubMed] [Google Scholar]
- 44. OpenEye Scientific Software (2014) OEDOCKING version 3.0.1 (OpenEye Scientific Software, Santa Fe, NM)
- 45. OpenEye Scientific Software (2014) AFITT version 2.3.0.5 (OpenEye Scientific Software, Santa Fe, NM)
- 46.Wlodek S, Skillman AG, Nicholls A. Automated ligand placement and refinement with a combined force field and shape potential. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 7):741–749. doi: 10.1107/S0907444906016076. [DOI] [PubMed] [Google Scholar]
- 47.Fu Z, Li X, Miao Y, Merz KM., Jr Conformational analysis and parallel QM/MM X-ray refinement of protein bound anti-Alzheimer drug donepezil. J Chem Theory Comput. 2013;9(3):1686–1693. doi: 10.1021/ct300957x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peat TS, Dolezal O, Newman J, Mobley D, Deadman JJ. Interrogating HIV integrase for compounds that bind: A SAMPL challenge. J Comput Aided Mol Des. 2014;28(4):347–362. doi: 10.1007/s10822-014-9721-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amano Y, et al. Structural insights into the novel inhibition mechanism of Trypanosoma cruzi spermidine synthase. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 9):1879–1889. doi: 10.1107/S1399004715013048. [DOI] [PubMed] [Google Scholar]
- 50. Schrödinger LLC (2014) Maestro Version 9.9 (Schrödinger LLC, New York)
- 51. OpenEye Scientific Software (2014) SZMAP version 1.2.0.7 (OpenEye Scientific Software, Santa Fe, NM)
- 52.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: Using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol. 1999;285(4):1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 53.Cornell WD, et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117(19):5179–5197. [Google Scholar]
- 54.Bayden AS, Moustakas DT, Joseph-McCarthy D, Lamb ML. Evaluating free energies of binding and conservation of crystallographic waters using SZMAP. J Chem Inform Model. 2015;55(8):1552–1565. doi: 10.1021/ci500746d. [DOI] [PubMed] [Google Scholar]
- 55.Lukacs V, et al. Dual regulation of TRPV1 by phosphoinositides. J Neurosci. 2007;27(26):7070–7080. doi: 10.1523/JNEUROSCI.1866-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.