Significance
Parasites of phylum Apicomplexa cause significant morbidity and mortality on a global scale. Central to the pathogenesis of these parasites is their ability to invade host cells through a junction formed by members of the apical membrane antigen (AMA) and rhoptry neck protein 2 (RON2) families localized to the parasite surface and host outer membrane, respectively. Here we structurally and functionally characterize Toxoplasma gondii AMA4 (TgAMA4), a highly divergent AMA protein. Structural analyses of TgAMA4 in the apo and RON2L1 bound forms reveal a previously underappreciated level of molecular diversity at the parasite–host-cell interface that offers important insight into stage-dependent invasion strategies and yields a more comprehensive model of apicomplexan invasion.
Keywords: Apicomplexa, Toxoplasma gondii, invasion, moving junction, X-ray crystallography
Abstract
Plasmodium falciparum and Toxoplasma gondii are widely studied parasites in phylum Apicomplexa and the etiological agents of severe human malaria and toxoplasmosis, respectively. These intracellular pathogens have evolved a sophisticated invasion strategy that relies on delivery of proteins into the host cell, where parasite-derived rhoptry neck protein 2 (RON2) family members localize to the host outer membrane and serve as ligands for apical membrane antigen (AMA) family surface proteins displayed on the parasite. Recently, we showed that T. gondii harbors a novel AMA designated as TgAMA4 that shows extreme sequence divergence from all characterized AMA family members. Here we show that sporozoite-expressed TgAMA4 clusters in a distinct phylogenetic clade with Plasmodium merozoite apical erythrocyte-binding ligand (MAEBL) proteins and forms a high-affinity, functional complex with its coevolved partner, TgRON2L1. High-resolution crystal structures of TgAMA4 in the apo and TgRON2L1-bound forms complemented with alanine scanning mutagenesis data reveal an unexpected architecture and assembly mechanism relative to previously characterized AMA–RON2 complexes. Principally, TgAMA4 lacks both a deep surface groove and a key surface loop that have been established to govern RON2 ligand binding selectivity in other AMAs. Our study reveals a previously underappreciated level of molecular diversity at the parasite–host-cell interface and offers intriguing insight into the adaptation strategies underlying sporozoite invasion. Moreover, our data offer the potential for improved design of neutralizing therapeutics targeting a broad range of AMA–RON2 pairs and apicomplexan invasive stages.
Phylum Apicomplexa comprises >5,000 parasitic protozoan species, many of which cause devastating diseases on a global scale. Two of the most prevalent species are Toxoplasma gondii and Plasmodium falciparum, the causative agents of toxoplasmosis and severe human malaria, respectively (1, 2). The obligate intracellular apicomplexan parasites lead complex and diverse lifestyles that require invasion of many different cell types. Despite this diversity of target host cells, most apicomplexans maintain a generally conserved mechanism for active invasion (3). The parasite initially glides over the surface of a host cell and then reorients to place its apical end in close contact to the host-cell membrane. After this initial attachment, a circumferential ring of adhesion (termed the moving or tight junction) is formed, through which the parasite actively propels itself while concurrently depressing the host-cell membrane to create a nascent protective vacuole (4).
Formation of the moving junction relies on a pair of highly conserved parasite proteins: rhoptry neck protein 2 (RON2) and apical membrane antigen 1 (AMA1). Initially, parasites discharge RON2 into the host cell membrane where an extracellular portion (domain 3; D3) serves as a ligand for AMA1 displayed on the parasite surface (5–8). Intriguingly, recent studies have shown that the AMA1–RON2 complex is an attractive target for therapeutic intervention (9–12). The importance of the AMA1–RON2 pairing is also reflected in the observation that many apicomplexan parasites encode functional paralogs that are generally expressed in a stage-specific manner (13–15). We recently showed that, in addition to AMA1 and RON2, T. gondii harbors three additional AMA paralogs and two additional RON2 paralogs (14, 15): TgAMA2 forms a functional invasion complex with TgRON2 (15), TgAMA3 (also annotated as SporoAMA1) selectively coordinates TgRON2L2 (14), and TgAMA4 binds TgRON2L1 (15). Despite substantial sequence divergence, structural characterization of all AMA–RON2D3 complexes solved to date [TgAMA1–TgRON2D3 (16), PfAMA1–PfRON2D3 (17), and TgAMA3–TgRON2L2D3 (14)] reveal a largely conserved architecture and binding paradigm. Intriguingly, however, sequence analysis indicates that TgAMA4 and TgRON2L1 are likely to adopt substantially divergent structures with an atypical assembly mechanism.
To investigate the functional implications of the AMA4–RON2L1 complex in T. gondii, we first established that TgAMA4 is part of a highly divergent AMA clade that includes the functionally important malaria vaccine candidate Plasmodium merozoite apical erythrocyte-binding ligand (MAEBL) (18–20) and that TgRON2L1 displays a similar divergence consistent with coevolution of receptor and ligand. We then show that TgAMA4 and TgRON2L1 form a high-affinity binary complex and probe its overall architecture and underlying mechanism of assembly using crystal structures of TgAMA4 in the apo and TgRON2L1D3 bound forms. Finally, we show proof of principle that TgAMA4 and TgRON2L1 form a functional pairing capable of supporting host-cell invasion. Collectively, our study reveals exceptional molecular diversity at the parasite–host-cell interface that we discuss in the context of the unique invasion barriers encountered by the sporozoite.
Results
TgAMA4 and TgRON2L1 Are Divergent Members of the Apicomplexan-Specific AMA and RON2 Families.
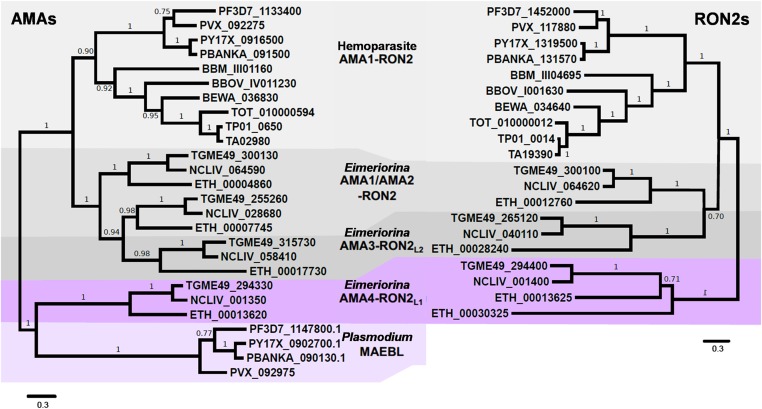
To place TgAMA4 and TgRON2L1 in the broader context of AMA and RON2 family members, we first performed a phylogenetic analysis. Custom homology searches of TgAMA1 and TgRON2 against predicted protein sets of apicomplexans from EuPathDB (eupathdb.org/eupathdb/) recovered sequences of AMA and RON2-like proteins from diverse lineages, except Cryptosporidium, consistent with the lack of a moving junction-dependent invasion mechanism in this atypical apicomplexan (21). Phylogenetic analysis of AMA-like proteins recovered two deeply branching clades. One clade is subdivided into AMA1 of apicomplexan hemoparasites (Plasmodium, Babesia, and Theileria) and of Eimeriorina (Toxoplasma, Eimeria, and Neospora), with the latter having two duplications of the AMA1 ancestor resulting in AMA1, AMA2, and AMA3 paralogs (Fig. 1, Left) (14, 15). In contrast, the second AMA clade consists of AMA4 from Toxoplasma and its orthologs in Eimeria and Neospora. Intriguingly, although the functionally important Plasmodium MAEBLs (18–20) have been described with different relationships to AMA1 (13, 22), our analysis indicates that MAEBL is most closely related to AMA4 (Fig. 1, Left).
Fig. 1.
Phylogenetic analysis reveals coevolution of a divergent set of AMA (AMA4/MAEBL) and RON2 (RON2L1) proteins. Unrooted maximum-likelihood-based phylogeny of AMA (Left) and RON2 (Right) protein families in apicomplexans is shown. Predicted protein annotations and species and strain identifiers correspond to accessions from EuPathDB (SI Appendix). AMA1 and RON2 from hemoparasites (Plasmodium, Babesia, and Theileria) are shaded in light gray; AMA1/AMA2 and RON2 from Eimeriorina (Toxoplasma, Neospora, Eimeria) are medium gray; AMA3 and RON2L2 from Eimeriorina are dark gray; AMA4 and RON2L1 from Eimeriorina are purple; and MAEBL from Plasmodium spp. is light purple.
RON2 homologs showed a broadly similar pattern of diversification across apicomplexans and were only recovered from species with AMAs (Fig. 1, Right). Notably, evolutionary analysis reveals a correlation between MAEBL and the RhopH1/Clag proteins that mimics the AMA–RON2 pairs. However, temporal and spatial expression patterns appear to be inconsistent with formation of a functional complex (23, 24), and thus the RhopH1/Clag proteins were not included in the final figure. The RON2 ancestor underwent a duplication in the Eimeriorina into RON2 and RON2L2, consistent with both AMA1 and AMA2 binding RON2 and only AMA3 binding RON2L2 (14, 15). Interestingly, the putative partner of AMA4, RON2L1, was only recovered from lineages with AMA4 and underwent an Eimeria-specific duplication (Fig. 1, Right).
A Restructured Apical Surface on TgAMA4 Suggests a Unique Binding Mechanism with its Putative Partner, TgRON2L1.
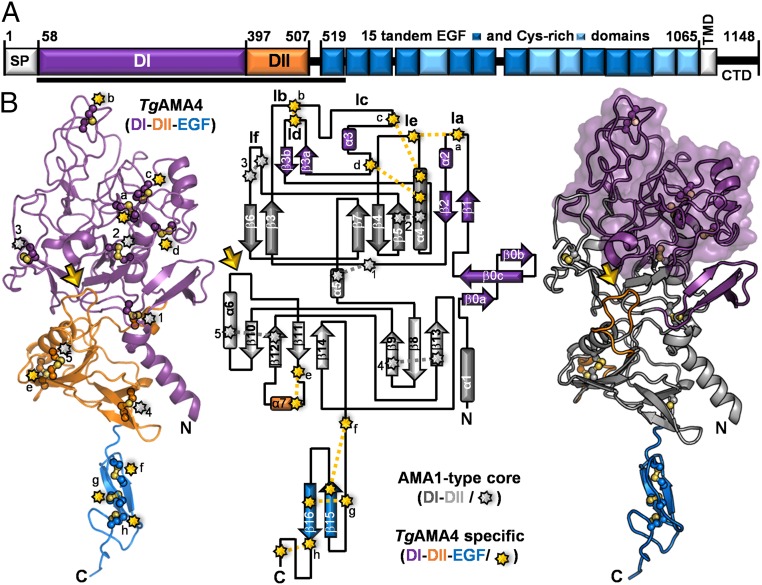
Sequence analysis indicates that TgAMA4 is a type I integral membrane protein with an ectodomain comprised of an expanded N-terminal AMA1 DI-like domain (∼50% larger), an AMA1 DII domain, and 15 tandem epidermal growth factor (EGF) and Cys-rich modules (10 noncalcium binding EGF domains and 5 Cys-rich regions; Fig. 2A). The lack of structural information for any member of the divergent AMA4/MAEBL clade, however, limited predictions to these general features that yielded little mechanistic insight. To overcome this limitation, we determined the structure of a selenomethionine derivatized form of TgAMA4 produced in insect cells that included DI, DII, and the first EGF domain (SI Appendix, Table S1). The 2.05-Å resolution structure was modeled completely with the exception of four disordered residues (Arg-252 to Thr-255) in an apical surface loop.
Fig. 2.
Structural characterization of TgAMA4 reveals an AMA1-type core with extensive additions localized to the apical surface. (A) Schematic of TgAMA4 domain architecture with numbering based on the initiation methionine in the signal sequence. CTD, cytoplasmic C-terminal domain; EGF, EGF-like domain; SP, signal peptide; TMD, transmembrane domain. Black bar indicates construct used for structural studies. (B, Left) Cartoon depiction of tertiary structure of TgAMA4 DI (purple), DII (orange), and first EGF (blue) domains. Disulfide bonds are indicated with starbursts (gray, conserved with other AMAs; yellow, unique to TgAMA4). Yellow arrow indicates the truncated DII loop. (B, Center) Topology diagram of TgAMA4 showing the conserved AMA core (dark gray, DI; light gray, DII) and TgAMA4-specific additions to the core (colored as in Left). Disulfide bonds and truncated DII loop are indicated and colored as in Left. Loops that comprise the apical surface are annotated according to previously established nomenclature (30). (B, Right) TgAMA4 tertiary structure colored as in Center with a transparent surface over the expanded apical surface loops.
Structural analysis revealed that the DI and DII cores of TgAMA4 are stabilized by five conserved disulfide bonds and, along with the EGF domain, are vertically stacked with an architecture globally reminiscent of the AMA1 and AMA3 structures (Fig. 2B) (14, 25, 26). The similarity between TgAMA4 and other AMAs, however, is restricted to the DI and DII cores (rmsd with TgAMA1 of 2.4 Å over 265 Cα, representing approximately half of the Cαs in the TgAMA4 structure). A key area of divergence is the apical surface, which is substantially expanded in TgAMA4 to include a three-stranded beta-sheet (27 residues) that packs against the side of the DI core, and large insertions into apical surface loops such as loops Ia, Ib, Ic, and Id, which are 16, 14, 46, and 25 residues longer in TgAMA4, respectively (Fig. 2B and SI Appendix, Fig. S1). Together, the DI loop insertions contain an additional eight cysteines that pin together the apical loops with four TgAMA4-specific disulfides (Ia-Ie, Ib-Id, Ic-Ie, Ic-Ie) (Fig. 2B and SI Appendix, Fig. S1). Importantly, the restructured surface in TgAMA4 lacks the deep surface cleft that coordinates RON2 partners in other structurally characterized AMAs. Furthermore, a large deletion in the TgAMA4 DII loop (TgAMA4, 14 residues; TgAMA1, 35; PfAMA1, 52), which governs access to the TgRON2D3 binding groove in TgAMA1 (27), reduces its size such that it does not contribute to the apical surface (Fig. 2B and SI Appendix, Fig. S1). These first structural insights into the AMA4/MAEBL clade suggest an unusual mechanism of complex formation consistent with the evolutionary divergence.
A Divergent Binding Interface Supports Assembly of the TgAMA4–TgRON2L1D3 Binary Complex.
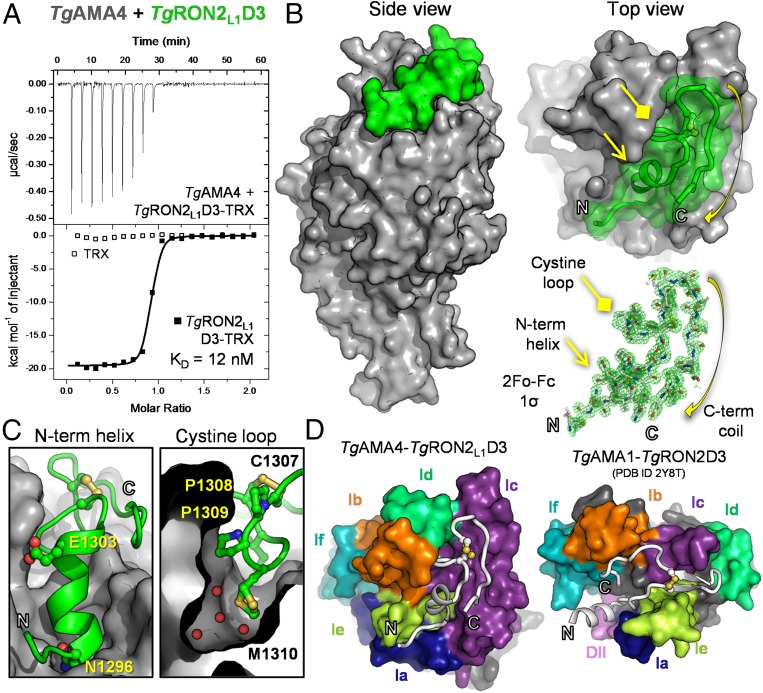
Next, we used isothermal titration calorimetry (ITC) to show that TgAMA4 forms a high-affinity (KD = 12.0 ± 2.1 nM) complex with TgRON2L1D3 with a 1:1 stoichiometry (0.87 ± 0.01) in an enthalpy driven process (ΔH = −19.6 ± 0.1 kcal/mol and −TΔS = 8.8 kcal/mol) (Fig. 3A). By comparison, the KD of TgAMA1–TgRON2D3 was previously measured to be 6 nM (27). To establish the detailed mechanism underlying complex formation, we determined the structure of the TgAMA4–TgRON2L1D3 complex to 1.53-Å resolution (SI Appendix, Table S1).
Fig. 3.
Biophysical and structural characterization of the TgAMA4–TgRON2L1D3 complex reveals a divergent binding paradigm. (A) ITC thermogram of TgRON2L1D3-TRX or TRX titrated into TgAMA4. (B) Side (Left) and apical (Upper Right) views of the TgAMA4–TgRON2L1D3 complex, with the overall surface of TgAMA4 colored gray and TgRON2L1D3 colored green. (Lower Right) Sigma-A weighted 2Fo − Fc electron density map contoured at 1.0 σ for TgRON2L1D3. (C) Zoomed in view of the N-terminal helix (Left) and cystine loop (Right; red spheres indicate water molecules) packing against the surface of TgAMA4 (gray). Residues investigated by mutagenesis are labeled and shown in ball-and-stick form. (D) Apical view of surface representations of the TgAMA4–TgRON2L1D3 and TgAMA1–TgRON2D3 (PDB ID code 2Y8T) complexes oriented with the conserved AMA DI-DII cores aligned and the TgAMA apical DI loops colored individually (navy blue, Ia; orange, Ib; purple, Ic; lime green, Id; yellow-green, Ie; teal, If) and the DII loop colored pink.
Analysis of the costructure revealed that TgRON2L1D3 (modeled from I1293 to S1324) packs against the shallow apical surface of TgAMA4 (Fig. 3 B and C), resulting in a buried surface area of 2,485 Å2, which is notably less than other AMA–RON2D3 complexes that range in buried surface area from 3,200 to 3,700 Å2 (14, 16, 17). Minimal structural rearrangement of TgAMA4 is required to accommodate TgRON2L1D3, consistent with both the lack of an extended DII loop and the lack of a deep groove, as observed for other AMAs (Fig. 3D) (14, 25). As initially predicted from our evolutionary analyses, the structural diversity of TgAMA4 and TgRON2L1D3 gives rise to a markedly divergent binary complex within the AMA–RON2 families; TgRON2L1D3 extends from an N-terminal helix, through a kinked cystine loop that forms a knob-in-hole interaction with a surface pocket in TgAMA4 (Fig. 3C), to a hairpin turn that leads into C-terminal coil. Based on buried surface area and specific interactions, we identified six residues from TgRON2L1D3 to individually probe by alanine substitution (SI Appendix, Table S2) and also truncated the C terminus of the peptide by nine residues (G1316 to S1324); ELISAs showed that none of these alterations substantially affected binding (SI Appendix, Fig. S2), consistent with the lack of a “hot spot” residue such as R2041 in the PfAMA1–PfRON2D3 complex (17). In contrast, ITC analysis of a TgRON2L1D3 N1296A/P1309A double mutant (N1296, base of helix; and P1309, cysteine loop) revealed no detectable interaction with TgAMA4, whereas a C1307S/C1313S variant displayed similar affinity to native TgRON2L1D3, yet coordinated TgAMA4 through a different mechanism (SI Appendix, Fig. S3). Together with additional double mutants tested by ELISA (SI Appendix, Fig. S2), these data suggest that the N-terminal helix and kinked cystine loop, mediated by P1308-(cis)-P1309, act synergistically to ensure the appropriate conformation for binding (Fig. 3C and SI Appendix, Figs. S2 and S3). Intriguingly, the TgAMA4 pocket that accommodates the knob-like TgRON2L1D3 cystine loop is not exquisitely specific because several buried water molecules line the bottom of the pocket and two conformations are observed for TgRON2L1 M1310 that follow the proline pair (Fig. 3C). These data reveal that the TgAMA4–TgRON2L1D3 complex diverges markedly from previously characterized AMA–RON2 pairs.
The TgAMA4–TgRON2L1 Complex Is Functional to Support Host-Cell Invasion.
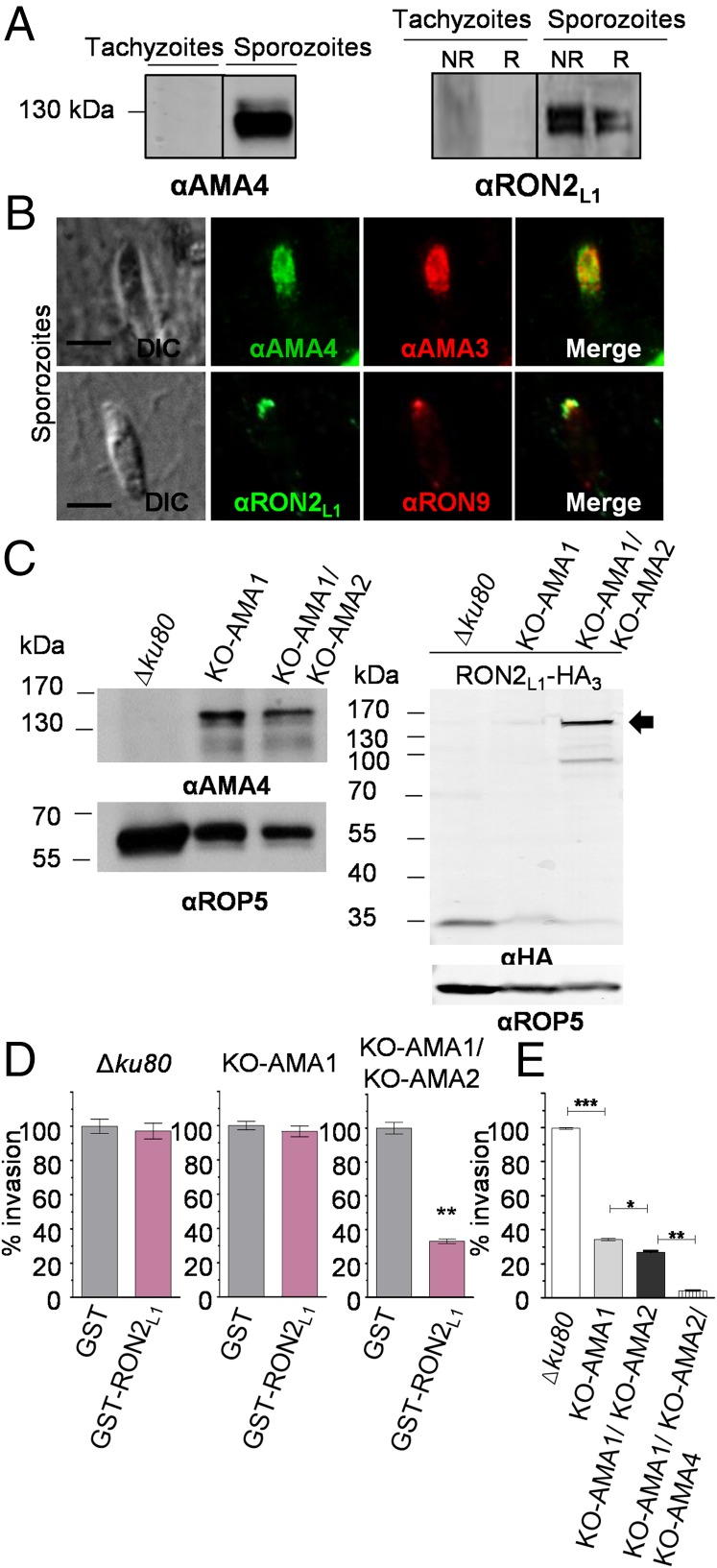
To investigate the biological role(s) of the TgAMA4–TgRON2L1 pair, we first established that both proteins are strongly up-regulated in sporozoites (Fig. 4A). Immunofluorescence on intracellular sporozoites showed an apical distribution of TgAMA4 similar to TgAMA3 (Fig. 4B, Upper) that is slightly posterior to standard tachyzoite microneme markers (SI Appendix, Fig. S4A) (14). Consistent with a microneme protein, AMA4 is redistributed on the entire surface of the sporozoite before invasion (SI Appendix, Fig. S4B), and the TgAMA4 staining disappeared in the tachyzoite stage after days of intracellular growth, confirming sporozoite specificity (SI Appendix, Fig. S4C). Immunofluorescence of TgRON2L1 on sporozoites showed colocalization with a rhoptry neck protein (RON9; Fig. 4B, Lower), consistent with TgRON2 localizing to the rhoptry necks in tachyzoites (5) and with a role early in invasion, such as formation of the moving junction (6, 8).
Fig. 4.
TgAMA4 and TgRON2L1 are up-regulated in the absence of TgAMA1 and TgAMA2 and form a functional complex for parasite invasion. (A) Western blot of 1 million tachyzoite and sporozoite stage parasites using anti-TgAMA4 (Left) and anti-TgRON2L1 (Right) antibodies. NR, nonreduced; R, reduced. (B) Immunofluorescence on sporozoites using anti-TgAMA4 and -TgRON2L1 antibodies revealed colocalization of TgAMA4 with TgAMA3 (Upper) and colocalization of TgRON2L1 with a rhoptry neck protein (Lower). DIC, differential interference contrast. (Scale bars, 5 µm.) (C, Left) Western blot of TgAMA4 in wild-type (Δku80), KO-AMA1, and KO-AMA1/KO-AMA2 strains showing up-regulation of the complex in mutant parasites. (C, Right) Western blot on TgRON2L1–HA3-tagged parasites using anti-HA antibodies. TgRON2L1 has two transcription initiation sites yielding proteins of different sizes, both containing the same C terminus, but only the larger one (166 kDa indicated by the arrow) is targeted to the secretory organelles (15). TgRON2L1 full-length protein is substantially up-regulated in the double mutant. (D) Invasion assay of Δku80, KO-AMA1 and KO-AMA1/KO-AMA2 parasites in the presence of 200 µg/mL GST or GST-TgRON2L1D3. An inhibitory effect of GST-TgRON2L1D3 is observed only in the double mutant. **P = 0.0032 (t test). (E) Invasion assay of mutant strains KO-AMA1, KO-AMA1/KO-AMA2, and KO-AMA1/KO-AMA2/KO-AMA4 relative to the parental Δku80 strain. Invasions are normalized to 100% in Δku80 strain. *P = 0.0195; **P = 0.0014; ***P = 0.0002 (t test).
To investigate the role played by the TgAMA4–TgRON2L1D3 complex in sporozoite invasion of fibroblasts, we attempted to block invasion using soluble TgRON2L1D3-GST (SI Appendix, Fig. S5). No significant reduction in invasion was observed. This finding may reflect a mechanism whereby parasites rely on the sporozoite-expressed TgAMA3 and TgRON2L2 (14) as the primary pair (much like TgAMA1–TgRON2 vs. TgAMA2–TgRON2 in tachyzoites), especially in a fibroblast-based invasion model that likely does not mimic the cells targeted by sporozoites in natural intestinal infections. To address this hypothesis, we took advantage of the versatile, engineered tachyzoite line depleted in both TgAMA1 and TgAMA2 that overexpresses TgAMA4 (15), but not TgAMA3. We first confirmed using anti-TgAMA4 antibodies that TgAMA4 is considerably up-regulated in both KO-AMA1 and KO-AMA1/KO-AMA2 parasites (Fig. 4C, Left), although comparably much lower than in sporozoites (SI Appendix, Fig. S6). TgRON2L1 is expressed at very low levels in tachyzoites and shows only minimal overexpression in the KO-AMA1 mutant (15), so we next endogenously tagged TgRON2L1 in the KO-AMA1/KO-AMA2 line (SI Appendix, Fig. S7) and compared the expression of TgRON2L1–HA in the different strains. TgRON2L1 showed higher expression in the KO-AMA1/KO-AMA2 strain relative to the KO-AMA1 and wild-type strains (Fig. 4C, Right), suggesting an important role for the TgAMA4–TgRON2L1 pair in the absence of both TgAMA1 and TgAMA2. Consistent with these observations, incubation with recombinant TgRON2L1D3–GST yielded a significant invasion inhibitory effect in the double mutant, while not affecting invasion of wild-type or single KO-AMA1 parasites (Fig. 4D). This finding suggests that the compensatory mechanisms for invasion rely on overexpression of TgAMA2 in the single KO-AMA1 mutant (15) and on the TgAMA4–TgRON2L1 complex in the KO-AMA1/KO-AMA2 double mutant. To further probe this invasion defect, we engineered a triple mutant (KO-AMA1/KO-AMA2/KO-AMA4; SI Appendix, Fig. S8) that showed significantly reduced invasion relative to the KO-AMA1/KO-AMA2 double mutant (Fig. 4E). Collectively, these results are a proof of principle that the TgAMA4–TgRON2L1 complex is functional to support parasite invasion.
Discussion
The junctional interface between the invasive stage of an apicomplexan parasite and target host cell is dependent on binary AMA–RON2 complexes; AMAs are present on the surface of the parasite, whereas RON2s are discharged from the parasite and embedded in the host-cell membrane to serve as the ligands for AMAs. An intriguing feature of T. gondii is its arsenal of stage-specific AMA and RON2 paralogs that all show a generally conserved architecture and binding mode. Sequence analysis, however, reveals a significant level of diversity in the recently identified TgAMA4 and TgRON2L1 that suggests an atypical assembly mechanism with the potential for intriguing functional consequences.
Phylogenetic analyses revealed a clear coevolution of TgAMA4 and TgRON2L1, suggesting the potential for complex formation (Fig. 1), which we ultimately confirmed and characterized using ITC (Fig. 3). We then defined a detailed molecular blueprint and mechanism of assembly of the binary complex by determining the crystal structures of TgAMA4 in the apo (Fig. 2) and TgRON2L1D3-bound (Fig. 3) forms. These structures revealed a significantly expanded and completely restructured apical surface on TgAMA4 that largely relies on a shallow seat for the TgRON2L1D3 helix and a deep pocket to anchor a knob-like projection formed by the TgRON2L1D3 cystine loop. The absence of the DII loop in TgAMA4, which is displaced in other AMAs to coordinate RON2 (14, 16, 17), reveals a binding mechanism that does not rely on conformational flexibility. This finding is an intriguing departure from all previously structurally characterized AMA–RON2 pairs, where conformational flexibility is absolutely required for complex formation. One potential consequence of dispensing with regulatory and selectivity determinants is to endow TgAMA4 with the ability to coordinate additional, as-yet-unidentified partners. The potential for an increased ligand repertoire is also consistent with the imperfect fit of the TgRON2L1D3 cystine loop into the TgAMA4 surface pocket (Fig. 3C). Notably, structure-guided sequence alignments (SI Appendix, Fig. S9) and molecular modeling (SI Appendix, Fig. S10) of the functionally important Plasmodium MAEBLs (18, 19) indicate a well-conserved DI-DII core scaffold and truncated DII loop similar to AMA4. Considerable divergence of the DI loops, however, suggests the potential for a divergent interaction surface; additional structure-based insights into the MAEBLs will likely require identification of the MAEBL binding partner.
To probe the functionality of the TgAMA4–TgRON2L1D3 complex, we next showed that both proteins are highly expressed in T. gondii sporozoites and display similar localization patterns to other AMA and RON2 proteins that form functional invasion complexes (Fig. 4) (5, 14). Despite TgAMA4 and TgRON2L1 being appropriately localized to support invasion and forming the requisite high-affinity complex, preincubation with TgRON2L1D3 did not significantly reduce sporozoite invasion in a standard fibroblast-based invasion assay (SI Appendix, Fig. S5) (14). This finding may reflect a functional complexity in TgAMA4–TgRON2L1 that is not effectively captured by the in vitro assay. Importantly, however, we were able to definitively show that the addition of TgRON2L1D3 to a tachyzoite cell line depleted in TgAMA1 and TgAMA2 and expressing higher amounts of TgAMA4 and TgRON2L1 significantly reduced invasion of fibroblasts (Fig. 4D), indicating that the peptide competes with endogenous TgAMA4–TgRON2L1 complex formation. To validate these data, we generated a TgAMA4 knockout in the KO-AMA1/KO-AMA2 background and showed that invasion was significantly reduced (Fig. 4E). Collectively, these observations indicate that TgAMA4 and TgRON2L1 are fundamentally capable of forming a functional invasion complex.
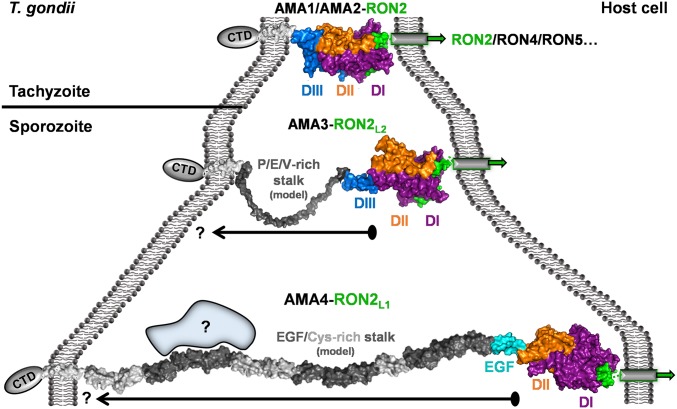
In addition to the apical surface of AMAs that coordinate the RON2 partners, the disposition of the binding domains relative to the parasite membrane are likely to have a profound effect on function. Thus, we used molecular modeling to investigate the regions that connect the functional head group to the transmembrane domain (TMD). The TgAMA1 and TgAMA2 head groups are connected to the TMD through linkers of <10 residues, resulting in close proximity to the parasite surface (Fig. 5, Top) (25). TgAMA3 presents a considerably longer, 93-residue linker rich in Pro, Glu, and Val residues, which we modeled in a semiextended, kinked conformation to be ∼175 Å from the TMD (Fig. 5, Middle). Notably, the TgAMA4 linker is the most extended of all TgAMAs by a significant margin (547 residues from the base of DII) and comprises 15 tandem EGF and Cys-rich modules (Fig. 2A). Whereas calcium-bound tandem EGF domains are often rigid rod-like structures [e.g., Protein Data Bank (PDB) ID code 1EMN], tandem noncalcium binding EGF domains such as those in TgAMA4 are likely to display increased flexibility. Thus, if the TgAMA4 EGF/Cys-rich domains are connected in a relatively linear fashion, we predict the apical surface of TgAMA4 to be ∼450 Å from the TMD (Fig. 5, Bottom). A ratcheting effect, however, could lead to a more compact organization, approximating the C-terminal tandem EGF pair of Plasmodium spp. merozoite surface protein 1 (e.g., PDB ID code 1B9W) and may be influenced by shear flow in the intestinal environment (28).
Fig. 5.
Variable stalk regions on AMA family members likely result in substantially different distances between the RON2-binding head group and the parasite cell membrane. (Top) TgAMA1 (purple/orange/blue)–TgRON2D3 (green) complex (PDB ID code 2Y8T). AMA1 TMD (generic seven-turn alpha-helix) is shown as a white surface. CTD, C-terminal domain. (Middle) TgAMA3–TgRON2L2D3 complex (PDB ID code 3ZLD) colored the same as in Top. Model of the Pro/Glu/Val rich stalk region is shown as a gray surface; the presented model represents a semiextended form, with the arrow reflecting the potential for considerable flexibility. (Bottom) TgAMA4–TgRON2L1D3 complex colored the same as in Top, with tandem EGF and Cys-rich domains connected head to tail and displayed as dark gray (EGF) and light gray (Cys-rich) surfaces. Blue shape with question mark indicates the possibility for the TgAMA4 stalk to recruit additional proteins. Arrow with question mark indicates potential for compaction.
Although it is common for apicomplexan adhesive micronemal proteins to have long linkers comprising tandem small modules or low-complexity sequences, the roles of these divergent stalk regions are poorly defined. Extended stalks may support protein– protein interactions [e.g., TgMIC2–TgM2AP (29)], enable signaling processes, or facilitate proteolytic processing. Intriguingly, the presence of an extended linker correlates with the invasive stage, because both TgAMA3 and TgAMA4 are predominately expressed on sporozoites (Fig. 4A) (14). This finding may reflect the need for a more flexible junction or the need to span expansive glycocalyx-like surface structures on host cells encountered by sporozoites during the course of natural infection, such as the intestinal epithelial cells. Accordingly, AMA4s may function initially as an extended tether, with the stalk able to compact to promote subsequent, more intimate coordination mediated by AMA3s reflecting a staged process, which would be consistent with TgRON2L1D3 not significantly reducing sporozoite invasion in a standard fibroblast-based invasion assay (SI Appendix, Fig. S5). Based on this model, some important questions are raised, including whether truncation of the TgAMA4 EGF/Cys-rich modules would impair the parasite’s ability to invade intestinal cells? Also, are the TgAMA4–TgRON2L1 complexes capable of stabilizing more expansive sporozoite junctional interfaces that could be visualized by electron microscopy?
Unraveling the complexity of the junctional interface between apicomplexan parasite and host cell is a crucial step toward establishing a comprehensive model of invasion. Key to this process is defining the detailed structural and functional contributions of the AMA–RON2 binary complexes. Here, we report, to our knowledge, the first structural dissection of the highly divergent TgAMA4–TgRON2L1 complex and show that these proteins form an overall architecture and use an assembly mechanism unique among AMA–RON2 pairs. Although further studies will be required to precisely establish the biological function of the TgAMA4–TgRON2L1 complex, it is clear that these two proteins significantly enhance the molecular diversity of the AMA–RON2 family at the parasite–host-cell interface. These data have important implications both for designing broad-based therapeutics targeting the moving junction and for understanding the mechanisms by which the sporozoite may overcome the unique barriers of the intestinal invasion environment.
Materials and Methods
Animal studies were conducted according to European Union guidelines for handling laboratory animals. Immunizations for antibody production in rabbits were conducted at the Centre de Recherches de Biochimie Macromoléculaire (CRBM) animal house (Montpellier, France) and approved by the Committee on the Ethics of Animal Experiments (Languedoc-Roussillon, Montpellier, France) (Permit D34-172-4, delivered on September 20, 2009). Immunizations for antibody production in mice were carried out at the Istituto Superiore di Sanità and authorized by the Italian Ministry of Health, according to Legislative Decree 116/92 that implemented the European Directive 86/609/EEC. Phylogenetic trees were constructed with 1,000 bootstrap replicates. TgAMA4 DIDIIEGF1 (S58 to D553) was produced in insect cells and TgRON2L1D3 constructs in Escherichia coli. Primers are listed in SI Appendix, Table S3, and all plasmids were sequenced. ITC data were processed by using a one-site model. X-ray diffraction data were collected at the Canadian Light Source. The TgAMA4–TgRON2L1D3 structure was solved by selenomet phasing. TgAMA4 and native TgAMA4–TgRON2L1D3 structures were solved by molecular replacement. Data collection and refinement statistics are presented in SI Appendix, Table S1. Atomic coordinates and structure factors have been deposited in the Protein Data Bank [PDB ID codes 4Z81 (TgAMA4DIDIIEGF1) and 4Z80 (TgAMA4DIDIIEGF1 in complex with TgRON2L1D3)]. Antibodies against TgAMA4 and TgRON2L1 were produced in rabbits and mice, respectively. For immunofluorescence assay on sporozoites, confluent human foreskin fibroblast monolayers were infected with excysted sporozoites, and fixed, washed, permeabilized, blocked, and stained with primary and secondary antibodies (SI Appendix, Table S4). ELISAs and tachyzoite invasion inhibition assay were performed as described (15).
Detailed methods are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff at the Canadian Light Source and Stanford Synchrotron Radiation Lightsource. This work was supported by Canadian Institutes for Health Research Grant MOP82915 (to M.J.B.); Agence Nationale de la Recherche Grant ANR-12-BSV3-0012-01; Laboratoire d'Excellence ParaFrap ANR-11-LABX-0024; and Fondation pour la Recherche Médicale Equipe FRM DEQ20130326508 (to M.L.). M.J.B. received salary support from the Canada Research Chair program. S.J.P. was supported by the Natural Sciences and Engineering Research Council of Canada and the Canadian Institute for Advanced Research–Integrated Microbial Biodiversity Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.D.S. is a guest editor invited by the Editorial Board.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4Z81 (TgAMA4DIDIIEGF1) and 4Z80 (TgAMA4DIDIIEGF1 in complex with TgRON2L1D3)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515898113/-/DCSupplemental.
References
- 1.Robert-Gangneux F, Dardé ML. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev. 2012;25(2):264–296. doi: 10.1128/CMR.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . World Malaria Report. World Health Organization; Geneva: 2014. [Google Scholar]
- 3.Sharma P, Chitnis CE. Key molecular events during host cell invasion by Apicomplexan pathogens. Curr Opin Microbiol. 2013;16(4):432–437. doi: 10.1016/j.mib.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Aikawa M, Miller LH, Johnson J, Rabbege J. Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J Cell Biol. 1978;77(1):72–82. doi: 10.1083/jcb.77.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besteiro S, Michelin A, Poncet J, Dubremetz JF, Lebrun M. Export of a Toxoplasma gondii rhoptry neck protein complex at the host cell membrane to form the moving junction during invasion. PLoS Pathog. 2009;5(2):e1000309. doi: 10.1371/journal.ppat.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamarque M, et al. The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS Pathog. 2011;7(2):e1001276. doi: 10.1371/journal.ppat.1001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srinivasan P, et al. Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion. Proc Natl Acad Sci USA. 2011;108(32):13275–13280. doi: 10.1073/pnas.1110303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyler JS, Boothroyd JC. The C-terminus of Toxoplasma RON2 provides the crucial link between AMA1 and the host-associated invasion complex. PLoS Pathog. 2011;7(2):e1001282. doi: 10.1371/journal.ppat.1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller LH, Ackerman HC, Su XZ, Wellems TE. Malaria biology and disease pathogenesis: Insights for new treatments. Nat Med. 2013;19(2):156–167. doi: 10.1038/nm.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srinivasan P, et al. Disrupting malaria parasite AMA1-RON2 interaction with a small molecule prevents erythrocyte invasion. Nat Commun. 2013;4:2261. doi: 10.1038/ncomms3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srinivasan P, et al. Immunization with a functional protein complex required for erythrocyte invasion protects against lethal malaria. Proc Natl Acad Sci USA. 2014;111(28):10311–10316. doi: 10.1073/pnas.1409928111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pihan E, et al. Computational and biophysical approaches to protein-protein interaction inhibition of Plasmodium falciparum AMA1/RON2 complex. J Comput Aided Mol Des. 2015;29(6):525–539. doi: 10.1007/s10822-015-9842-7. [DOI] [PubMed] [Google Scholar]
- 13.Chesne-Seck ML, et al. Structural comparison of apical membrane antigen 1 orthologues and paralogues in apicomplexan parasites. Mol Biochem Parasitol. 2005;144(1):55–67. doi: 10.1016/j.molbiopara.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Poukchanski A, et al. Toxoplasma gondii sporozoites invade host cells using two novel paralogues of RON2 and AMA1. PLoS One. 2013;8(8):e70637. doi: 10.1371/journal.pone.0070637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamarque MH, et al. Plasticity and redundancy among AMA-RON pairs ensure host cell entry of Toxoplasma parasites. Nat Commun. 2014;5:4098. doi: 10.1038/ncomms5098. [DOI] [PubMed] [Google Scholar]
- 16.Tonkin ML, et al. Host cell invasion by apicomplexan parasites: Insights from the co-structure of AMA1 with a RON2 peptide. Science. 2011;333(6041):463–467. doi: 10.1126/science.1204988. [DOI] [PubMed] [Google Scholar]
- 17.Vulliez-Le Normand B, et al. Structural and functional insights into the malaria parasite moving junction complex. PLoS Pathog. 2012;8(6):e1002755. doi: 10.1371/journal.ppat.1002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preiser P, et al. Antibodies against MAEBL ligand domains M1 and M2 inhibit sporozoite development in vitro. Infect Immun. 2004;72(6):3604–3608. doi: 10.1128/IAI.72.6.3604-3608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saenz FE, Balu B, Smith J, Mendonca SR, Adams JH. The transmembrane isoform of Plasmodium falciparum MAEBL is essential for the invasion of Anopheles salivary glands. PLoS One. 2008;3(5):e2287. doi: 10.1371/journal.pone.0002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leite JA, et al. Immunization with the MAEBL M2 domain protects against lethal Plasmodium yoelii infection. Infect Immun. 2015;83(10):3781–3792. doi: 10.1128/IAI.00262-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borowski H, Clode PL, Thompson RC. Active invasion and/or encapsulation? A reappraisal of host-cell parasitism by Cryptosporidium. Trends Parasitol. 2008;24(11):509–516. doi: 10.1016/j.pt.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Michon P, Stevens JR, Kaneko O, Adams JH. Evolutionary relationships of conserved cysteine-rich motifs in adhesive molecules of malaria parasites. Mol Biol Evol. 2002;19(7):1128–1142. doi: 10.1093/oxfordjournals.molbev.a004171. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko O, et al. Apical expression of three RhopH1/Clag proteins as components of the Plasmodium falciparum RhopH complex. Mol Biochem Parasitol. 2005;143(1):20–28. doi: 10.1016/j.molbiopara.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Nguitragool W, et al. Malaria parasite clag3 genes determine channel-mediated nutrient uptake by infected red blood cells. Cell. 2011;145(5):665–677. doi: 10.1016/j.cell.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crawford J, Tonkin ML, Grujic O, Boulanger MJ. Structural characterization of apical membrane antigen 1 (AMA1) from Toxoplasma gondii. J Biol Chem. 2010;285(20):15644–15652. doi: 10.1074/jbc.M109.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tonkin ML, Crawford J, Lebrun ML, Boulanger MJ. Babesia divergens and Neospora caninum apical membrane antigen 1 structures reveal selectivity and plasticity in apicomplexan parasite host cell invasion. Protein Sci. 2013;22(1):114–127. doi: 10.1002/pro.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker ML, Boulanger MJ. An extended surface loop on Toxoplasma gondii apical membrane antigen 1 (AMA1) governs ligand binding selectivity. PLoS One. 2015;10(5):e0126206. doi: 10.1371/journal.pone.0126206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tonkin ML, Boulanger MJ. The shear stress of host cell invasion: Exploring the role of biomolecular complexes. PLoS Pathog. 2015;11(1):e1004539. doi: 10.1371/journal.ppat.1004539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song G, Springer TA. Structures of the Toxoplasma gliding motility adhesin. Proc Natl Acad Sci USA. 2014;111(13):4862–4867. doi: 10.1073/pnas.1403059111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai T, et al. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc Natl Acad Sci USA. 2005;102(36):12736–12741. doi: 10.1073/pnas.0501808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.