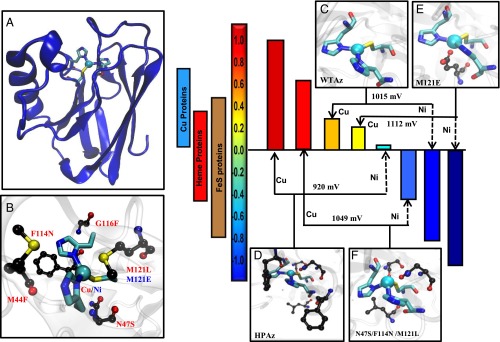
Significance
Nature spent millions of years to evolve electron transfer proteins that span a wide range of reduction potentials (E°′) under physiological conditions, from −1 V to 1 V vs. standard hydrogen electrode. Understanding the rules that govern such tuning within similar classes of metalloproteins enables scientists to rationally tune the E°′of their catalysts without changing the active site. An ultimate test of such understanding is to achieve the entire range of E°′ within a single protein, a feat that has not been achieved yet. Herein, we conclusively found that we can cover the entire 2-V range of E°′ using a single protein with five mutations and two metal ions. We have also provided explanations for structural features responsible for such high potential.
Keywords: reduction potential, secondary coordination sphere, azurin, electron transfer, cupredoxins
Abstract
The reduction potential (E°′) is a critical parameter in determining the efficiency of most biological and chemical reactions. Biology employs three classes of metalloproteins to cover the majority of the 2-V range of physiological E°′s. An ultimate test of our understanding of E°′ is to find out the minimal number of proteins and their variants that can cover this entire range and the structural features responsible for the extreme E°′. We report herein the design of the protein azurin to cover a range from +970 mV to −954 mV vs. standard hydrogen electrode (SHE) by mutating only five residues and using two metal ions. Spectroscopic methods have revealed geometric parameters important for the high E°′. The knowledge gained and the resulting water-soluble redox agents with predictable E°′s, in the same scaffold with the same surface properties, will find wide applications in chemical, biochemical, biophysical, and biotechnological fields.
Redox reactions are at the heart of a majority of biological functions and chemical transformations, from electron transfer (ET) in photosynthesis and respiration to catalytic activations of C–H and other bonds in molecules (1–6). For these redox reactions the redox potential (E°′) is the most important parameter in determining efficiency of the reactions and the accessibility of oxidation states through the catalytic cycle (5–10). Being able to controllably tune the E°′s would allow scientists and engineers to take advantage of various redox reactions and oxidation states for numerous applications from alternative energy generation to synthesis of intermediates or products for pharmaceuticals. One primary example is the overpotential issue for most fuel cell catalysts, which is a major barrier that has prevented them from being applied in industrial-scale applications (11). Being able to tune the E°′ of catalytic and electron transfer sites to lower the overpotential will have a major impact on this and other areas.
For millions of years, ET in biology has operated within the range of physiological E°′s, defined by the highest E°′ [∼ +1 V vs. standard hydrogen electrode (SHE); all potentials mentioned in this study are vs. SHE] at which water is oxidized, and the lowest E°′ (∼−1 V) at which protons are reduced to H2 (5, 6). Amazingly, nature has found a way to cover this wide range using a strikingly limited set of not only metal cofactors but also protein folds and metal center geometries, such as cupredoxins that cover 100 to 800 mV, cytochromes that cover −500 to 350 mV, and Fe–S proteins that cover −700 to 500 mV (Fig. 1) (5). Despite many years of research into these proteins and efforts to change the E°′ (12–22), it is still not well understood how the E°′s can be tuned systematically in a wide range using a limited number of metal cofactors. An ultimate test of our understanding of this process is to design redox centers in a single protein, using a minimum number of metal cofactors and mutations that can cover the entire 2-V range of physiological E°′. Furthermore, there are very few water-soluble/stable chemical redox agents within the physiological E°′ range. Even for those redox agents that can cover a wide range of E°′ in nonaqueous solutions, combining different redox agents with different scaffolds or surface properties makes it difficult to carry out systematic studies of the effect of E°′ on ET or catalytic functions, as it is difficult to deconvolute different factors in the redox process. We report herein the design of the ET protein azurin to cover a range from +970 ± 20 mV to ∼−954 ± 50 mV, by mutating only five residues and using two metal ions (Fig. 1).
Fig. 1.
By mutating only five residues and using two metal ions (Cu and Ni) in a single protein, Az, the entire range of physiological potential can be covered. (A) Overall structure of Az. (B) The metal-binding site of azurin and the residues around the site investigated in this study. (C) WTAz active site structure, Protein Data Bank (PDB) ID code 4AZU. (D) Minimized structural model of HPAz. (E) Structure of M121E-Az, PDB ID code 4QLW. (F) Minimized structural model of N47S/F114N/M121L-Az.
Results
The type 1 (T1) Cu center in wild-type azurin (WTAz) resides in a Greek key β-barrel fold (Fig. 1A) commonly found in cupredoxins, such as rusticyanin and laccase (23–26). The active site consists of a Cu(SCys)(NHis)2 core in a trigonal plane, common to all T1 Cu centers, with a thioether group from a Met residue (SMet) in the axial position (Fig. 1 B and C) (5, 18). This axial Met is found in most cupredoxins, except those with high E°′; rusticyanin and laccase contain a Leu residue at the axial position instead (5, 24). We have previously shown that the E°′ of Az can be predictably tuned from 90 to 640 mV at pH 7 by tuning hydrogen-bonding interactions to the copper ligands through Asn47Ser and Phe114Asn mutations and the hydrophobicity of the axial ligand through Met121Leu mutation (Fig. 1C) (17). To the best of our knowledge there is no report of a mononuclear cupredoxin with E°′s above or below this range.
To tune the E°′ of the T1 Cu center in azurin to approach the highest E°′ under physiological conditions, we examined the T1 Cu centers in cupredoxins that exhibit extremely high E°′, such as those in multicopper laccase (E°′ = 790 mV at pH 6). We found that these centers are highly desolvated and buried within the hydrophobic interior of the proteins (25). The area surrounding the Cu center is also typically filled with bulky, hydrophobic residues such as Phe (23, 25). Previous studies have shown that introducing such hydrophobic residues near the metal-binding sites in several metalloproteins, including azurin, has resulted in increased E°ˊ (5, 12–22, 27–31). Based on these observations, we scanned the secondary coordination sphere of the T1 Cu center in Az for mutations to Phe residue that would not interfere with the Asn47Ser/Phe114Asn/Met121Leu mutations we have already shown to increase the E°′, and we found two mutations (Met44Phe and Gly116Phe, Fig. 1B) that met the requirement.
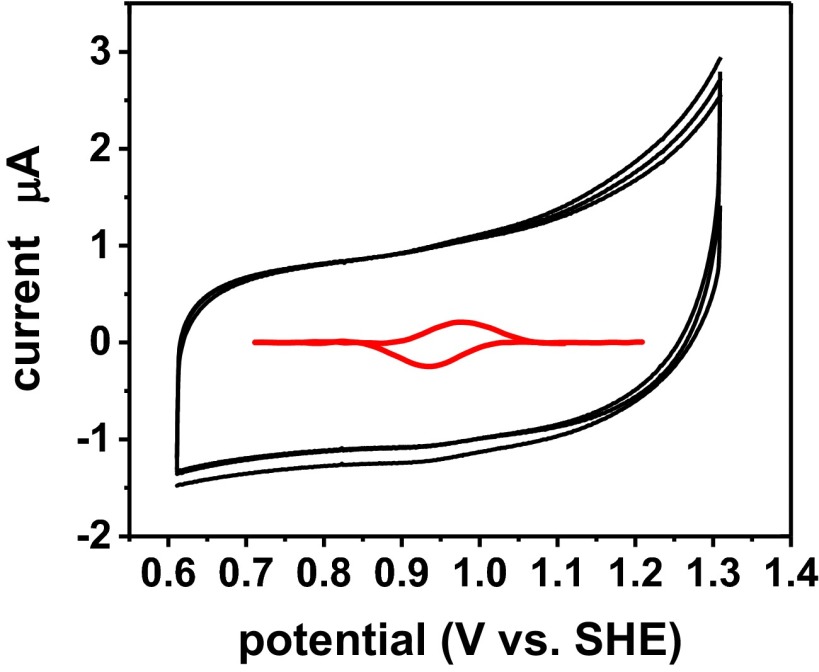
We combined the Met44Phe and Gly116Phe mutations with the Asn47Ser/Phe114Asn/Met121Leu mutations that had resulted in E°′ = 640 ± 1 mV at pH 7.0 (17). This quintuple variant of Az [Met44Phe/Asn47Ser/Phe114Asn/Gly116Phe/Met121Leu Az (Fig. 1D, called HPAz hereafter)] was constructed, expressed, and purified to homogeneity as a metal-free apo protein using a slightly modified procedure from a previous study (SI Appendix) (17). After loading Cu(I) into the apo-HPAz [Cu(I)-HPAz], we used cyclic voltammetry (CV) with a pyrolytic graphite “edge” plane electrode and observed a signal centered at 970 ± 20 mV at pH 5.0 in 50 mM ammonium acetate buffer (Fig. 2). The faradaic current from the CV is much lower than those of inorganic complexes due to much slower diffusion rates, and is similar to those of other metalloproteins reported in the literature (9, 10, 32). To ensure that the observed CV signal is from the Cu-HPAz variant, we performed several control experiments including repeating the CV experiment under the same conditions in the absence of any protein (Blank), in the presence of apo-HPAz (SI Appendix, Fig. S1A), and HPAz loaded with redox inactive Zn2+ [Zn(II)-HPAz, SI Appendix, Fig. S1B]. None of these control experiments exhibited a signal near 970 mV, which strongly suggests that the observed CV signal is from the Cu(I)/Cu(II) redox couple of Cu-HPAz, instead of any other components present in the solution.
Fig. 2.
Cyclic voltammogram showing the reversible redox couple of HPAz. The potential was obtained at scan rate of 0.1 V/s at pH 5.0 in 50 mM ammonium acetate buffer (black line). The signal with the capacitive current subtracted is shown as an inset in the middle (red line). (Em = 0.97 ± 0.02 V vs. SHE).
To provide additional support to the E°′ value determined by the CV, we probed the ability of Cu-HPAz to react with other redox agents that have different potentials (SI Appendix). Ferricyanide (E°′ = 0.424 V, pH 7.0), which is known to rapidly oxidize Cu(I)-WTAz (E°′ = 0.31 V at pH 7) (5, 33), caused only minimal reoxidation (∼2%) of 0.2 mM Cu(I)-HPAz under the same condition within the first 1 s, even when in the presence of a 10-fold molar excess of the oxidant (SI Appendix, Fig. S2). At time scales longer than 1 s, a slower process similar to previously reported ferricyanide-induced protein unfolding and subsequent reacting with Cu(I) was observed (SI Appendix, Fig. S2). The inability of ferricyanide to oxidize Cu(I)-HPAz suggests that the E°′ of the T1 Cu center in HPAz is at least higher than 0.424 V. After trying several other oxidants of varying potentials to oxidize the HPAz, we found that 1 molar equivalent of Na2IrCl6 (E°′ = 0.96 V at pH 5, SI Appendix, Fig. S3) was able to oxidize 0.2 mM Cu(I)-HPAz at pH 5 with ∼25% loss of the Na2IrCl6 absorption bands (SI Appendix, Figs. S4 and S5). These results, together with the result obtained by replacing Cu with Ni on E°′ of the same protein (vide infra), indicate that the Cu-HPAz has an extremely high E°′.
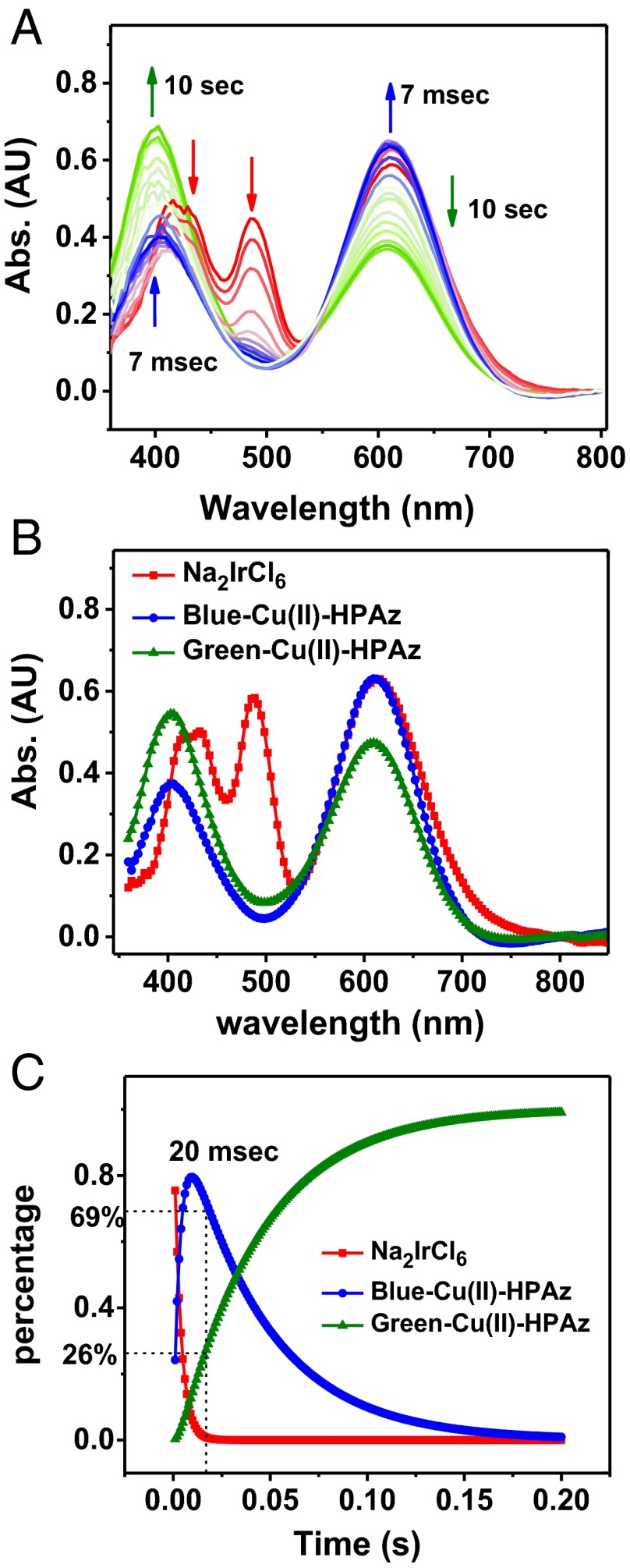
To characterize the reaction between Cu(I)-HPAz and Na2IrCl6, we used stopped-flow UV-vis absorption spectroscopy, analyzed by global spectral fit using the SpecFit program (Fig. 3). Upon mixing 0.7 mM Cu(I)-HPAz with 1 molar equivalent of Na2IrCl6 at pH 6.5 in 50 mM phosphate buffer and 0.1 mM NaCl, the absorption bands at 420, 485, and 600 nm due to Na2IrCl6 (red line) rapidly decayed, while new absorbance bands at 404 and 610 nm (blue line), typical of a T1 Cu(II) center (34–37), grew in (Fig. 3 A and B). This species [henceforth referred to as “Blue Cu(II)-HPAz”] reached a maximum at ∼7 ms and then decreased (Fig. 3A). As the Blue Cu(II)-HPAz started to decrease, a new species displaying absorption bands at 400 and 607 nm (green line, Fig. 3 A and B) concurrently increased in intensity with time (Fig. 3C). The final spectrum is identical to that observed after direct addition of CuSO4 to apo-HPAz (SI Appendix, Fig. S6), suggesting that it is the final product of the oxidation and the stable form of Cu(II)-HPAz [henceforth called “Green Cu(II)-HPAz”]. Because the decay of Na2IrCl6 signal is concomitant with formation of Blue Cu(II)-HPAz and the decay of Blue Cu(II)-HPAz results in growth of Green Cu(II)-HPAz, we used a global spectral fit to confirm the following kinetic process and obtained the following rate constants:
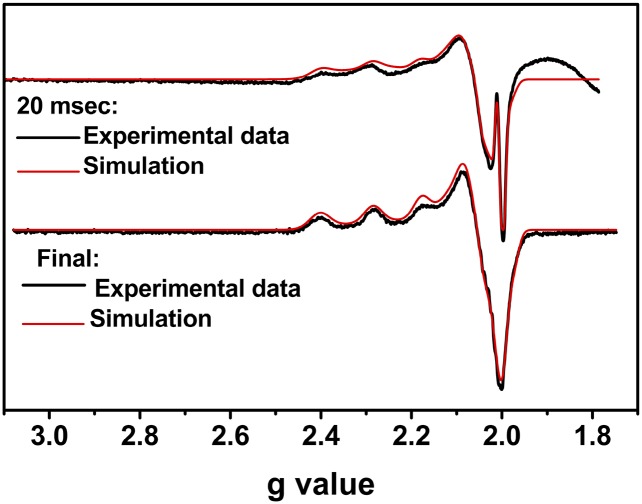
To further probe the nature of the products of the Na2IrCl6 oxidation in the aforementioned study, we used rapid freeze-quench electron paramagnetic resonance (EPR) spectroscopy under the same conditions as those used in the stopped-flow UV-vis study. The minimal time achievable by our freeze-quench apparatus was 20 ms, which is slightly slower than the time necessary to form the maximal Blue Cu(II)-HPAz intermediate observed in the stopped-flow UV-vis (7 ms). At 20 ms after addition of Na2IrCl6 to Cu(I)-HPAz, we observed an EPR spectrum (Fig. 4, Top) that can be best simulated as a mixture consisting of 60% T1 Cu (gz = 2.230 and Az = 65.19 cm−1), 39% type 2 (T2) Cu (gz = 2.309 and Az = 144.81 cm−1), and 1% radical (gaverage = 2.0021) (SI Appendix, Table S1). The percentage values for the individual species observed in EPR are consistent with those obtained from our stopped-flow UV-vis study (Fig. 3C); at 20 ms in the stopped-flow experiment, 69% Blue Cu(II)-HPAz and 26% Green Cu(II)-HPAz were formed. At longer time scales, the percentage of the T1 Cu species decreased whereas that of the T2 Cu species increased. The final EPR spectrum is identical to that of addition of Cu(II) to apo-HPAz (Fig. 4, Bottom; see SI Appendix, Fig. S7 and Table S2). Based on the similar kinetic behavior between the UV-vis and EPR studies, we assigned the Blue Cu(II)-HPAz to a T1 Cu center and the Green Cu(II)-HPAz to a T2 Cu center.
Fig. 3.
Stopped-flow UV-vis spectra of oxidation of 0.7 mM Cu(I)-HPAz by 1 eq Na2IrCl6 at pH 6.5 in 50 mM phosphate buffer and 100 mM NaCl. (A) Spectral changes in first 10 s of reaction. (B) Simulated spectra of individual species as determined by the SpecFit program. (C) Time-dependent transitions of Na2IrCl6, Blue Cu(II)-HPAz and Green Cu(II)-HPAz, as simulated by the SpecFit program.
Fig. 4.
EPR spectra of Na2IrCl6 oxidation of Cu(I)-HPAz. (Top) Spectrum shows freeze-quenched sample after 20 ms of the reaction. (Bottom) Spectrum is the final product of the reaction. The broad feature at values smaller than g = 2.0 in the 20-ms sample is due to packing of the tube during the freeze-quench process. Black line: experimental spectra; red line: simulated spectra.
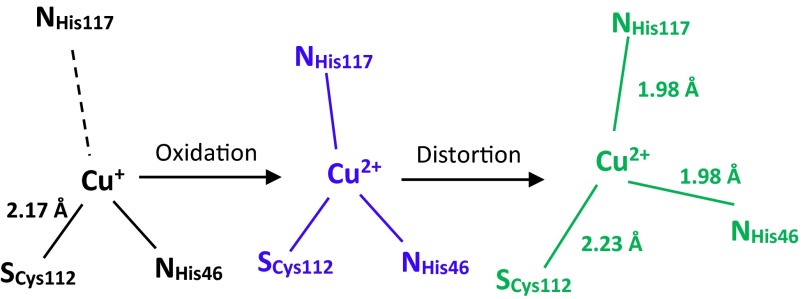
Thus far, we have been unable to crystallize the Cu(I) or Cu(II) form of the HPAz. Consequently, extended X-ray absorption fine structure (EXAFS) was used to obtain structural information. The spectra of the Cu(I)-HPAz and Cu(II)-HPAz species show a clear difference in their edges when overlaid. The reduced HPAz exhibits a strong feature at 8,983.2 eV (SI Appendix, Fig. S8D) that implicates Cu(I)-HPAz as a two-coordinate species, particularly compared with the edge obtained for other biologically relevant sites, most notably that of the Cys to Ala mutant of Bacillus subtilis Sco (38). The EXAFS spectrum of Cu(I)-HPAz at pH 6.5 (SI Appendix, Fig. S8A) was best simulated as one SCys at 2.17 Å, and one multiple scattering NHis at 1.92 Å (Table 1). The short Cu(I)–SCys and Cu–N distances are typical of 2-coordinate Cu(I) species (38). In contrast, the EXAFS spectrum of Cu(II)-HPAz at the same pH (SI Appendix, Fig. 8 A and C) required the addition of a second NHis at 1.98 Å, and the SCys–Cu bond was lengthened to 2.23 Å. This Cu(II)–SCys distance is significantly longer than that of Cu(II)–WTAz (2.17 Å) but similar to other thiolate-containing T2 Cu proteins such as nitrosocyanin (39) and Met121Hcy azurin (40). Interestingly, we observed that the EXAFS of Cu(I)-HPAz in phosphate buffer at pH 8 (SI Appendix, Fig. S8B and Table 1) showed an increase in the His contribution to 1.3. This change can be readily observed by comparing the intensity of the His peak in the Fourier-transformed spectrum of Cu(I)-HPAz at pH 8 with that of Cu(I)-HPAz at pH 6.5 (Table 1). These results suggest that at high pH there is stronger binding of His ligand to the Cu center due to possible deprotonation of the His ligand as suggested by previous studies (41).
Table 1.
EXAFS fitting parameters
| Cu–S | Cu–N(His) | Cu–S(Met) | |||||||||
| Sample/Fit | F | N | R, Å | DW, Å2 | N | R, Å2 | DW, Å | N | R, Å2 | DW, Å | E0 |
| Cu(I)-WTAz | 0.00039 | 1 | 2.19 | 0.002 | 2 | 1.97 | 0.010 | 1 | 2.68 | 0.003 | −0.231 |
| Cu(I)-HPAz (pH 6.5) | 0.00022 | 1 | 2.17 | 0.004 | 1 | 1.92 | 0.010 | −0.387 | |||
| Cu(I)-HPAz (pH 8) | 0.00023 | 1 | 2.17 | 0.003 | 1.3 | 1.93 | 0.003 | −0.591 | |||
| Cu(II)-WTAz | 0.00051 | 1 | 2.17 | 0.003 | 2 | 1.93 | 0.008 | 1 | 2.70 | 0.006 | −2.346 |
| Cu(II)-HPAz | 0.00056 | 1 | 2.23 | 0.010 | 2 | 1.98 | 0.005 | −2.905 | |||
Putting these spectroscopic results together, we propose a scheme in which Cu(I)-HPAz exists as a bidentate copper center with His117 no longer interacting with the copper, as it is the most solvent accessible copper ligand (Scheme 1). This result may also explain why the Cu-HPAz showed such a high E°′, because HPAz enforces a coordination geometry that is preferred by Cu(I). Upon reaction with Na2IrCl6, the colorless Cu(I)-HPAz was rapidly oxidized within the first 7 ms to a blue species that exhibited UV-vis (Fig. 3) and EPR (Fig. 4) spectra that are typical of a T1 Cu(II) center. To complete the T1 Cu(II) coordination sphere, the His117 rapidly bound to the Cu(II) as indicated by the combined stopped-flow, EPR, and EXAFS data. However, because this Blue Cu(II)-HPAz with E°′ of ∼1 V is so oxidizing, it became unstable under physiological conditions and changed its coordination sphere into a more stable Green Cu(II)-HPAz whose UV-vis (Fig. 2) and EPR (Fig. 4) are typical of a T2 Cu(II) center. Although the CV results are obtained at pH 5.0 whereas other characterizations are performed at pH 6.5, the similarity between intermediates observed upon reaction of Cu(I)-HPAz with Na2IrCl6 at pH 6.5 (Fig. 3) and 5.0 (SI Appendix, Fig. S4) suggests that the proposed scheme applies to pH 5.0 as well. The reason we chose pH 6.5 for the majority of characterizations was the slower kinetics and better recovery of the visible absorption signals at this pH. Interestingly, the Green Cu(II)-HPAz displays a reductive peak around 650 mV vs. SHE at pH 7.0 (SI Appendix, Fig. S9), which is consistent with the fact that the relatively lower E°′ of the Green Cu(II)-HPAz makes it more stable than the Blue Cu(II)-HPAz. To investigate the integrity of the protein, we rereduced the Green Cu(II)-HPAz and then reoxidized it. The UV-vis spectrum of the resulting protein is almost identical to that of the starting protein (SI Appendix, Fig. S10), suggesting that there is little damage to the protein from such a redox process, thus making it possible to use the protein as a redox agent.
Scheme 1.
Proposed mechanism of the oxidation of Cu(I)-HPAz with Na2IrCl6.
After tuning the T1 Cu center in Az to reach the highest possible E°′ under physiological conditions (∼1 V), we explored the next challenge which is to design azurin to reach the lowest E°′ under physiological conditions (∼−1 V). To achieve this goal, we replaced the Cu ion in azurin with other physiologically relevant metal ions. Azurin, like other cupredoxins, has been shown to be amenable to metal substitutions with minimal perturbation to either the overall structure or the metal-binding site (5, 42). Among the transition metal ions used previously, the T1 Cu in WTAz has been replaced with Ni(II) (43–46); the Ni(II)/Ni(I) redox pair has been shown to have much lower potential than the Cu(II)/Cu(I) pair within the same ligand set (46, 47). Therefore, we added Ni(II) to the apo-HPAz. Successful incorporation of Ni(II) into HPAz was first confirmed by UV-vis, which displayed similar spectral features to those of Ni(II)-WTAz (43, 46) (SI Appendix, Fig. S11). The CV of this protein indicated a potential of 50 mV from the reductive peak of the sample in pH 8.0 phosphate buffer (SI Appendix, Fig. S12), which is ∼900 mV lower than that of Cu(II)/Cu(I)-HPAz. Encouraged by the result, we added Ni(II) to an apo-azurin variant (Met121Glu-Az) whose Cu(II)/Cu(I) redox couple has a low E°′ (184 mV at pH 8.0) (13). The reductive potential of Ni(II)-M121EAz was observed to be −945 mV in pH 8.0 phosphate buffer, which is close to the lowest E°′ possible under physiological conditions. To ensure that the Ni(I) species formed under this condition, we performed cryoreduction EPR on the Ni(II)-M121E sample and observed an EPR spectrum consistent with previously reported Ni(I) species in azurin and other proteins (SI Appendix, Fig. S13) (46, 48).
Interestingly, replacing Cu ion with Ni ion in multiple variants of Az nearly always resulted in the lowering of E°′ by ∼1,000 mV, making the E°′ of the engineered proteins additive and predictable. Consistent with this trend, the E°′ of Ni-HPAz measured at 50 mV is similarly 900 mV below that of Cu-HPAz, providing additional support for the high E°′ of Cu-HPAz. By using Ni- and Cu derivatives of different Az variants we have prepared here and previously (17), we have now demonstrated the coverage of the entire range of physiological E°′ using a single protein (Fig. 1).
Conclusions
In this article, we have achieved fine-tuning of a single protein, Az, whose potentials can span the entire range of physiological E°′, from ∼+970 ± 20 mV to ∼−954 ± 50 mV, by mutating only five residues in the secondary coordination sphere of the mononuclear T1 Cu center of the protein and using two metal ions (Cu and Ni). Before this work, the highest observed E°′ for a natural mononuclear metalloprotein was that of the T1 Cu center in rusticyanin (670 mV at pH 7) (49, 50), whereas the highest observed E°′ for a multinuclear metalloprotein was that of the T1 Cu center next to the trinuclear Cu cluster in laccase (E°′ = 790 mV at pH 6) (25, 51). The positive charge from the other Cu ions in the multicopper cluster is known to play a role in raising the E°′ of the T1 Cu center in laccase over that of mononuclear cupredoxins such as rusticyanin (23). The E°′ of 970 mV represents the highest E°′ observed under physiological condition, very close to the theoretical limit of 1 V, before water itself is oxidized. We achieved this goal by tuning hydrogen-bonding networks near the Cu center (Asn47Ser and Phe114Asn), adding a hydrophobic residue in the axial position to the Cu center (Met121Leu), and desolvating the Cu center by adding Phe residues to surround it (Met44Phe and Gly116Phe).
This result is remarkable especially when considering that it has been accomplished on a mononuclear site without the aid of other positively charged metal ions nearby, as observed in the trinuclear copper center in laccase; such an E°′ exceeds the highest E°′ of a natural mononuclear cupredoxin (rusticyanin) by ∼300 mV. The spectroscopic studies indicate that the Cu(II)-HPAz displays an unusual geometric parameter from those of Az derivatives that have lower or more normal E°′. This is not unexpected for a protein with an E°′ that is so high that it comes close to the physiological E°′ limit at which H2O can be oxidized. To capture and study metal complexes with very high or low E°′, very low temperature and nonaqueous solutions were often used to stabilize the unusual states. It is significant that such an unusually high E°′ can be captured in a protein at room temperature and under physiological condition. Furthermore, by replacing the Cu ion with Ni ion in the above azurin mutants, we can tune the E°′ to as low as −954 mV at pH 8.0 and anywhere in between the two extreme E°′s, by different combinations of these mutations and the metal ions.
To cover the entire range of physiological E°′, nature must use at least three different classes of proteins and metal ions (Fig. 1). This ability to cover the entire range, which even surpasses the observed E°′ range for all ET proteins combined, using two metal ions and mutating only five residues in a single protein scaffold (Fig. 1) is a testimony to how much we now understand the structural features responsible for tuning E°′ of metalloproteins. Given the wide range of potentials attainable from a single protein possessing the same overall fold and surface properties, the azurin variants reported in this study may enable scientists and engineers to take advantage of these water-soluble redox agents for biochemical and biotechnological applications such as solar energy transfer and other alternative energy conversions. Because tuning the potentials of many inorganic, bioinorganic, and organometallic catalysts can result in catalysts with different oxidation states with dramatically different catalytic efficiency for different substrates (52), the knowledge gained from this study may also allow others to use the same principle to tune redox properties of numerous catalysts for even wider applications, such as small molecule activation and synthesis of important intermediates or products for pharmaceutical applications.
Materials and Methods
All azurin variants reported in this study are purified based on previously reported methods to obtain apo-variants with some slight modifications and were confirmed using electrospray ionization mass spectroscopy. CV experiments were performed by applying proteins on a graphite edge plane electrode as reported and measured vs. Ag/AgCl reference. The potentials were changed to SHE by adding 210 mV. EPR spectra were obtained at 30 K as a glass. The cryoreduced samples were prepared upon 3-h irradiation of samples by gamma rays at liquid nitrogen temperature. EXAFS data were collected at Beamline 7–3 at Stanford Synchrotron Radiation Lightsource. For a more detailed description of methods, readers should refer to the SI Appendix.
Supplementary Material
Acknowledgments
We thank Mr. Kevin Harnden for help with protein purification, Dr. Zhou Dai for initial EXAFS data collection, Dr. Rodney Burton for initial help in freeze-quench EPR in early time points, Mr. Bryant Kearl for collecting preliminary CV data, Mr. Khosro Khosravi for revising Fig. 1, and Mr. Shiliang Tian for discussions and comments. This material is based on work supported by the US National Science Foundation under Award CHE 14-13328 (to Y.L.) and US National Institutes of Health under Award NIH R01GM054803 (to N.J.B). K.N.C. was supported by National Science Foundation Graduate Research Fellowship DGE-0925180. We acknowledge the use of facilities at the Stanford Synchrotron Radiation Lightsource (SSRL), supported by the US Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, the National Institutes of Health, and by the National Institute of General Medical Sciences (including P41GM103393).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 248.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515897112/-/DCSupplemental.
References
- 1.Beinert H, Holm RH, Münck E. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science. 1997;277(5326):653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 2.Choi M, Davidson VL. Cupredoxins--a study of how proteins may evolve to use metals for bioenergetic processes. Metallomics. 2011;3(2):140–151. doi: 10.1039/c0mt00061b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warren JJ, Ener ME, Vlček A, Jr, Winkler JR, Gray HB. Electron hopping through proteins. Coord Chem Rev. 2012;256(21-22):2478–2487. doi: 10.1016/j.ccr.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abriata LA, et al. Alternative ground states enable pathway switching in biological electron transfer. Proc Natl Acad Sci USA. 2012;109(43):17348–17353. doi: 10.1073/pnas.1204251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, et al. Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers. Chem Rev. 2014;114(8):4366–4469. doi: 10.1021/cr400479b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosseinzadeh P, Lu Y. Design and fine-tuning redox potentials of metalloproteins involved in electron transfer in bioenergetics. Biochim Biophys Acta. August 21, 2015 doi: 10.1016/j.bbabio.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcus RA. 1993. Electron-transfer reactions in chemistry: Theory and experiment (Nobel lecture). Angew Chem 105(8):1161–1172.
- 8.Marcus RA. 1993. Electron-transfer reactions in chemistry: Theory and experiment Angew Chem Int Ed Engl 1132(1168):1111–1121.
- 9.Elliott SJ, et al. Detection and interpretation of redox potential optima in the catalytic activity of enzymes. Biochim Biophys Acta. 2002;1555(1-3):54–59. doi: 10.1016/s0005-2728(02)00254-2. [DOI] [PubMed] [Google Scholar]
- 10.Hudson JM, et al. Electron transfer and catalytic control by the iron-sulfur clusters in a respiratory enzyme, E. coli fumarate reductase. J Am Chem Soc. 2005;127(19):6977–6989. doi: 10.1021/ja043404q. [DOI] [PubMed] [Google Scholar]
- 11.Gewirth AA, Thorum MS. Electroreduction of dioxygen for fuel-cell applications: Materials and challenges. Inorg Chem. 2010;49(8):3557–3566. doi: 10.1021/ic9022486. [DOI] [PubMed] [Google Scholar]
- 12.Varadarajan R, Zewert TE, Gray HB, Boxer SG. Effects of buried ionizable amino acids on the reduction potential of recombinant myoglobin. Science. 1989;243(4887):69–72. doi: 10.1126/science.2563171. [DOI] [PubMed] [Google Scholar]
- 13.Pascher T, Karlsson BG, Nordling M, Malmström BG, Vänngård T. Reduction potentials and their pH dependence in site-directed-mutant forms of azurin from Pseudomonas aeruginosa. Eur J Biochem. 1993;212(2):289–296. doi: 10.1111/j.1432-1033.1993.tb17661.x. [DOI] [PubMed] [Google Scholar]
- 14.Hwang HJ, Berry SM, Nilges MJ, Lu Y. Axial methionine has much less influence on reduction potentials in a CuA center than in a blue copper center. J Am Chem Soc. 2005;127(20):7274–7275. doi: 10.1021/ja0501114. [DOI] [PubMed] [Google Scholar]
- 15.Garner DK, et al. Reduction potential tuning of the blue copper center in Pseudomonas aeruginosa azurin by the axial methionine as probed by unnatural amino acids. J Am Chem Soc. 2006;128(49):15608–15617. doi: 10.1021/ja062732i. [DOI] [PubMed] [Google Scholar]
- 16.Miller AF. Redox tuning over almost 1 V in a structurally conserved active site: Lessons from Fe-containing superoxide dismutase. Acc Chem Res. 2008;41(4):501–510. doi: 10.1021/ar700237u. [DOI] [PubMed] [Google Scholar]
- 17.Marshall NM, et al. Rationally tuning the reduction potential of a single cupredoxin beyond the natural range. Nature. 2009;462(7269):113–116. doi: 10.1038/nature08551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuris JA, et al. Engineering the redox potential over a wide range within a new class of FeS proteins. J Am Chem Soc. 2010;132(38):13120–13122. doi: 10.1021/ja103920k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lancaster KM, et al. Electron transfer reactivity of type zero Pseudomonas aeruginosa azurin. J Am Chem Soc. 2011;133(13):4865–4873. doi: 10.1021/ja1093919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hadt RG, et al. Spectroscopic and DFT studies of second-sphere variants of the type 1 copper site in azurin: Covalent and nonlocal electrostatic contributions to reduction potentials. J Am Chem Soc. 2012;134(40):16701–16716. doi: 10.1021/ja306438n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.New SY, Marshall NM, Hor TSA, Xue F, Lu Y. Redox tuning of two biological copper centers through non-covalent interactions: Same trend but different magnitude. Chem Commun (Camb) 2012;48(35):4217–4219. doi: 10.1039/c2cc30901g. [DOI] [PubMed] [Google Scholar]
- 22.Bhagi-Damodaran A, Petrik ID, Marshall NM, Robinson H, Lu Y. Systematic tuning of heme redox potentials and its effects on O2 reduction rates in a designed oxidase in myoglobin. J Am Chem Soc. 2014;136(34):11882–11885. doi: 10.1021/ja5054863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solomon EI, Sundaram UM, Machonkin TE. Multicopper oxidases and oxygenases. Chem Rev. 1996;96(7):2563–2606. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- 24.Gray HB, Malmström BG, Williams RJ. Copper coordination in blue proteins. J Biol Inorg Chem. 2000;5(5):551–559. doi: 10.1007/s007750000146. [DOI] [PubMed] [Google Scholar]
- 25.Davies GJ, Ducros V. 2001. Laccases. Handbook of Metalloproteins, eds Messerschmidt A, Huber R, Poulos T, Wieghardt K (Wiley, Chichester, UK), Vol 2, pp 1359–1368.
- 26.Lu Y. 2004. Electron transfer: Cupredoxins. Biocoordination Chemistry, Comprehensive Coordination Chemistry II: From Biology to Nanotechnology, eds Que JL, Tolman WB, Meyer TJ (Elsevier, Oxford), Vol 8, p 91.
- 27.Dey A, et al. Solvent tuning of electrochemical potentials in the active sites of HiPIP versus ferredoxin. Science. 2007;318(5855):1464–1468. doi: 10.1126/science.1147753. [DOI] [PubMed] [Google Scholar]
- 28.Berry SM, Baker MH, Reardon NJ. Reduction potential variations in azurin through secondary coordination sphere phenylalanine incorporations. J Inorg Biochem. 2010;104(10):1071–1078. doi: 10.1016/j.jinorgbio.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Hong G, Ivnitski DM, Johnson GR, Atanassov P, Pachter R. Design parameters for tuning the type 1 Cu multicopper oxidase redox potential: Insight from a combination of first principles and empirical molecular dynamics simulations. J Am Chem Soc. 2011;133(13):4802–4809. doi: 10.1021/ja105586q. [DOI] [PubMed] [Google Scholar]
- 30.Kennedy ML, Gibney BR. Metalloprotein and redox protein design. Curr Opin Struct Biol. 2001;11(4):485–490. doi: 10.1016/s0959-440x(00)00237-2. [DOI] [PubMed] [Google Scholar]
- 31.Lu Y, Berry SM, Pfister TD. Engineering novel metalloproteins: Design of metal-binding sites into native protein scaffolds. Chem Rev. 2001;101(10):3047–3080. doi: 10.1021/cr0000574. [DOI] [PubMed] [Google Scholar]
- 32.Dryhurst G, Kadish KM, Scheller F, Renneberg R. 1982. Proteins. Biological Electrochemistry, ed Dryhurst G (Academic, New York), Vol 1, p 398.
- 33.Hwang HJ, Lu Y. Spectroscopic evidence for interactions between hexacyanoiron(II/III) and an engineered purple CuA azurin. J Inorg Biochem. 2004;98(5):797–802. doi: 10.1016/j.jinorgbio.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Solomon EI. Spectroscopic methods in bioinorganic chemistry: Blue to green to red copper sites. Inorg Chem. 2006;45(20):8012–8025. doi: 10.1021/ic060450d. [DOI] [PubMed] [Google Scholar]
- 35.Solomon EI, Hadt RG. Recent advances in understanding blue copper proteins. Coord Chem Rev. 2011;255(7-8):774–789. [Google Scholar]
- 36.Warren JJ, Lancaster KM, Richards JH, Gray HB. Inner- and outer-sphere metal coordination in blue copper proteins. J Inorg Biochem. 2012;115:119–126. doi: 10.1016/j.jinorgbio.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solomon EI, et al. Copper active sites in biology. Chem Rev. 2014;114(7):3659–3853. doi: 10.1021/cr400327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siluvai GS, Mayfield M, Nilges MJ, Debeer George S, Blackburn NJ. Anatomy of a red copper center: Spectroscopic identification and reactivity of the copper centers of Bacillus subtilis Sco and its Cys-to-Ala variants. J Am Chem Soc. 2010;132(14):5215–5226. doi: 10.1021/ja910759v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieberman RL, Arciero DM, Hooper AB, Rosenzweig AC. Crystal structure of a novel red copper protein from Nitrosomonas europaea. Biochemistry. 2001;40(19):5674–5681. doi: 10.1021/bi0102611. [DOI] [PubMed] [Google Scholar]
- 40.Clark KM, et al. Transforming a blue copper into a red copper protein: Engineering cysteine and homocysteine into the axial position of azurin using site-directed mutagenesis and expressed protein ligation. J Am Chem Soc. 2010;132(29):10093–10101. doi: 10.1021/ja102632p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen JE, McBrayer MK, Robbins M, Suh Y. A pH dependence study on the unfolding and refolding of apoazurin: Comparison with Zn(II) azurin. Cell Biochem Biophys. 2002;36(1):19–40. doi: 10.1385/cbb:36:1:19. [DOI] [PubMed] [Google Scholar]
- 42.Hauenstein BLJ, McMillin DR. Copper proteins. In: Sigel H, editor. Metal Ions in Biological Systems. Vol 13. Marcel Dekker; New York: 1981. pp. 319–347. [Google Scholar]
- 43.Di Bilio AJ, et al. Electronic absorption spectra of M(II)(Met121X) azurins (M=Co, Ni, Cu; X=Leu, Gly, Asp, Glu): Charge-transfer energies and reduction potentials. Inorg Chim Acta. 1992;198–200(0):145–148. [Google Scholar]
- 44.Moratal JM, Romero A, Salgado J, Perales-Alarcón A, Jiménez HR. The crystal structure of nickel(II)-azurin. Eur J Biochem. 1995;228(3):653–657. doi: 10.1111/j.1432-1033.1995.tb20305.x. [DOI] [PubMed] [Google Scholar]
- 45.Czernuszewicz RS, Fraczkiewicz G, Zareba AA. A detailed resonance Raman spectrum of Nickel(II)-substituted Pseudomonas aeruginosa azurin. Inorg Chem. 2005;44(16):5745–5752. doi: 10.1021/ic050553g. [DOI] [PubMed] [Google Scholar]
- 46.Manesis AC, Shafaat HS. 2015. Electrochemical, spectroscopic, and density functional theory characterization of redox activity in nickel-substituted azurin: a model for acetyl-CoA synthase. Inorg Chem 56(16):7959–7967.
- 47.Pragathi M, Reddy KH. 2013. Synthesis, spectral characterization and DNA interactions of copper(II) and nickel(II) complexes with unsymmetrical Schiff base ligands. Indian J Chem, Sect A: Inorg, Bio-inorg, Phys, Theor Anal Chem 52A(7):845–853.
- 48.Bender G, et al. Infrared and EPR spectroscopic characterization of a Ni(I) species formed by photolysis of a catalytically competent Ni(I)-CO intermediate in the acetyl-CoA synthase reaction. Biochemistry. 2010;49(35):7516–7523. doi: 10.1021/bi1010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall JF, Kanbi LD, Harvey I, Murphy LM, Hasnain SS. Modulating the redox potential and acid stability of rusticyanin by site-directed mutagenesis of Ser86. Biochemistry. 1998;37(33):11451–11458. doi: 10.1021/bi980960m. [DOI] [PubMed] [Google Scholar]
- 50.Shoham M. Rusticyanin. In: Messerschmidt A, editor. Handbook of Metalloproteins. Vol 2. John Wiley & Sons; Chichester, UK: 2001. pp. 1235–1241. [Google Scholar]
- 51.Xu F, et al. Site-directed mutations in fungal laccase: Effect on redox potential, activity and pH profile. Biochem J. 1998;334(Pt 1):63–70. doi: 10.1042/bj3340063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Camasso NM, Sanford MS. Design, synthesis, and carbon-heteroatom coupling reactions of organometallic nickel(IV) complexes. Science. 2015;347(6227):1218–1220. doi: 10.1126/science.aaa4526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.