Redox reactions are the heart of function in biological systems. Using a deceptively simple toolbox, consisting largely of first row transition metals and the 20 canonical amino acids, nature has evolved proteins that contain cofactors with reduction potentials (E°′) between +1 V and −1 V. Chemists have long known that ligands to transition metals markedly alter E°′ of the metal, often in a systematic way. A set of such trends is much less clear in metalloproteins. In PNAS, Hosseinzadeh et al. (1) used azurin, a blue copper protein from Pseudomonas aeruginosa, as a framework that can be rationally modified to shift E°′ of the embedded metal site between +0.97 V and −0.95 V [all potentials are with respect to the normal hydrogen electrode (NHE)]. Tuning E°′ over such a large range was made possible using five (or fewer) nature-inspired mutations and substitution with CuII or NiII.
Copper sites carry out many redox roles in biological systems. Functions of type 1 sites include single electron transfer (ET) reactions (e.g., plastocyanin) and substrate oxidation coupled to dioxygen reduction (e.g., laccases) (2). The unique properties of type 1 sites have been recognized since the 1950s, and those properties have been manipulated in a great many ways (3). In addition to its striking blue color, type 1 Cu centers display E°′ values that are generally higher than for CuII/I in water (E°′ = 0.16 V), ranging from 0.18 V (stellacyanin) to almost 0.8 V for fungal laccase (4), and ≥1 V estimated for ceruloplasmin (5). This large range of E°′ is even more remarkable given the structural similarities of the proteins and the ligand sets (two His, one Cys, and a more weakly bound Met; Fig. 1). Mutations to Cu-ligating residues have major effects on spectroscopic and redox properties (6, 7), but we now know that exquisite tuning of E°′ is made possible by differences in the surrounding (outer-sphere) amino acids, mainly in terms of hydrophobicity and polarity (8). For example, introduction of several Phe groups around azurin-Cu, without changing the native Cu-ligands, can increase E°′ by almost 100 mV (9).
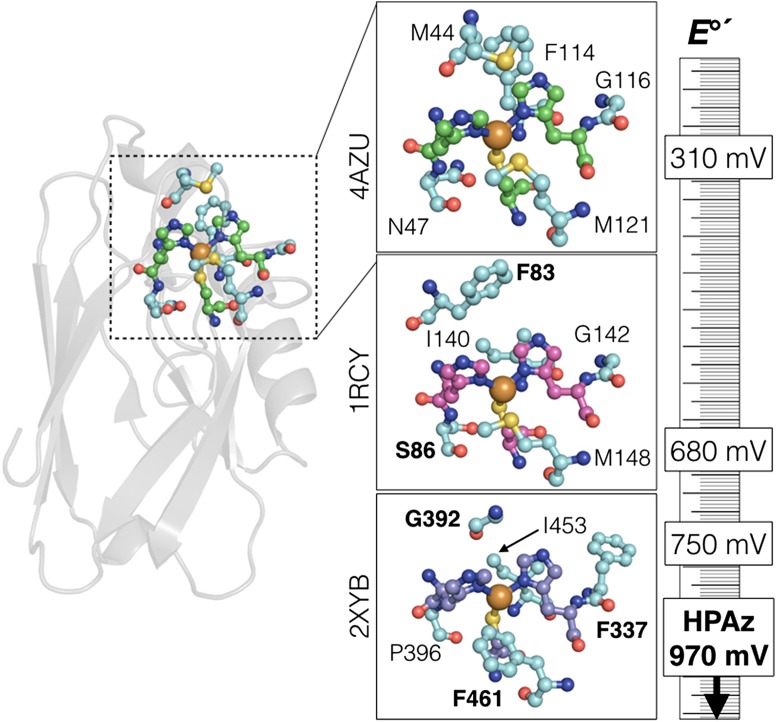
Fig. 1.
(Left) Tertiary structure of P. aeruginosa azurin, showing the location of the copper site. (Insets) Active sites of azurin (Top), Thiobacillus ferrooxidans rusticyanin (Middle), and Pycnoporus cinnabarinus laccase (Bottom). The invariant Cu-coordinating two His and one Cys are shown in green, magenta, and purple, respectively. The five residues mutated in HPAz, and those residues in the corresponding high-potential proteins, are shown in cyan. Bold lettering in the bottom two panels indicates residues that inspire the design of HPAz. The ruler shows the relative reduction potentials of each site. The Protein Data Bank codes are given to the left of each panel.
Lu and coworkers (10) previously demonstrated that E°′ (CuII/I) in azurin could be tuned between ∼0 mV and 640 mV with mutations to only three residues. The relative contributions of each mutation also were delineated in that work. Importantly, mutation to the weakly coordinating Met121 (Met121Leu and Met121Gln) induced ±50-mV changes to E°′, respectively. Those changes are modest, but other mutations to the outer-sphere residue Asn47 and Phe114 contribute additive changes to E°′, enabling tuning over a 600-mV range. This feat was impressive, but a modified protein with E°′ approaching 1 V was still out of reach.
In PNAS, Hosseinzadeh et al. (1) extend the above work to produce a modified azurin with E°′ = 0.97 V, which is the highest potential ever reported for azurin-Cu, and among the highest reported for any type 1 Cu site. A site with such a high E°′ value was produced by the addition of only two nature-inspired (Fig. 1) outer-sphere mutations (Met44Phe + Gly116Phe) to Asn47Ser/Phe114Asn/Met121Leu azurin (10). These researchers call this new protein high-potential azurin (HPAz). The mutations strongly affect the reduction potential of the embedded Cu site and have relatively small effects on other characteristic type I spectroscopic signatures (i.e., the protein’s vivid blue color and EPR properties). The ability to tune the reduction potential of embedded sites without completely changing the electronic properties of that site is a great advance.
In their original work (10), Lu and coworkers (Fig. 1) mutated azurin Met121 to a hydrophobic residue, which was inspired by P. cinnabarinus laccase. They also mutated to Asn47 to Ser, which is observed in T. ferrooxidans rusticyanin. The residue in that site forms a hydrogen bonds with the Cu-ligating Cys-sulfur, which consequently affects E°′. A unique feature of azurin is that bulky Phe114 promotes an interaction of the Cu with an oxygen from a backbone amide. Replacement of Phe114 with smaller Asn alters this interaction and further increases E°′(CuII/I). In the present work (1), the Met44Phe was made in a further analogy to rusticyanin and laccase (Fig. 1). Note that Met44 in azurin aligns with Gly392 in P. cinnabarinus laccase, but the next residue is Phe393. Finally, HPAz is completed with the Gly116Phe mutation, similar to the situation for P. cinnabarinus laccase. It is likely that the spectroscopic features of the Cu site are preserved in the quintuple mutant because all mutations derive from structurally related type 1 Cu proteins.
Reactions of HPAz-CuI with the small molecule oxidant hexachloroiridate (Na2IrCl6) are described (1). These reactions are important not only because they help confirm the high E°′ of HPAz but because the kinetics parameters also can shed light on the ET properties of the Cu site. Type 1 copper sites have rapid ET self-exchange (AzCuII + AzCuI → AzCuI + AzCuII) rate constants (11), consistent with a small amount of nuclear reorganization [λ from the semiclassical ET expression (12)]. ET reactions between proteins and small molecule redox reagents can be analyzed using the Marcus cross-relation (13) (Eq. 1). A cross-rate constant (k12) of 175 s−1 for oxidation of 0.7 mM HPAz-CuI by 1 eq of IrCl62− is reported (1). Using the equilibrium constant from the relative reduction potentials (K12 = 0.05), the known self-exchange rate constant (k11) for IrCl63−/2−, and f ∼ 1 (13), we calculate that the HPAz self-exchange rate constant (k22) is ∼3,800 s−1. This value is close to the value for WT azurin (11). In addition to maintaining spectroscopic features of the type 1 site, the outer-sphere mutations introduced by Hosseinzadeh et al. (1) do not dramatically alter the ET kinetics parameters for the metal site.
| [1] |
Little is known about the redox properties of Ni-substituted azurin mutants. The properties of the WT protein were recently investigated in detail, including work on the electronic properties of the NiI and NiII forms and electrochemical ET parameters (14). Under identical conditions, Ni-azurin undergoes ET reactions
The work of Hosseinzadeh et al. and previous work from the Lu laboratory demonstrate that we can now rationally tune reduction potentials in proteins using primarily outer-sphere mutations.
about 25-fold slower than Cu-azurin. Much more work is needed to evaluate the reactivity (or stability) of these highly reducing sites. Importantly, Hosseinzadeh et al.’s work (1) opens new doors for investigating the properties of metal-substituted azurins. For example, could these facets of outer-sphere E°′ tuning be used to create more reactive cobalt-substituted (15) or iron-substituted (16) azurin? Changing the inner-sphere (Met121Glu) allows transient access to FeIII azurin (17), but more work regarding E°′ tuning, and concomitant changes to reactivity, is needed.
Finally, the type 1 (blue) oxidized (CuII) form of HPAz is only transiently stable, changing to a secondary product (green form) over the course of seconds. The origin of this instability appears to be structural, as suggested by EPR and X-ray data. It is surprising that the high-potential site does not oxidize a nearby (9 Å) Tyr residue (Tyr72); the E°′ for Tyr at pH 6.5 is within 30 mV of CuII/I-HPAz. Instead, the CuII site appears to move away from Cys112, relaxing to a form with a lower E°′. Nature has evolved a great many ways to cope with strongly oxidizing embedded sites (18, 19). This new HPAz protein offers a unique platform to investigate, in a detailed way, the factors that stabilize and destabilize embedded metal sites with extreme (±1 V) reduction potentials.
Our collective view of how nature finely adjusts redox reactivity at a limited number of protein sites, using a limited number of transition metals, is rapidly evolving. The work of Hosseinzadeh et al. (1) and previous work from the Lu laboratory (10) demonstrate that we can now rationally tune reduction potentials in proteins using primarily outer-sphere mutations. Such outer-sphere changes can induce profound effects in E°′ while preserving most of the natural spectroscopic and ET parameters of the embedded site, which is a remarkable achievement. This work opens new doors to studying the behavior of strongly reducing or oxidizing metal sites in protein frameworks. Furthermore, we can now, in a more comprehensive manner, start envisioning modified proteins that can be designed to carry out transformations relevant to energy and the environment.
Acknowledgments
We are supported by Simon Fraser University and by Grant RGPIN-05559 of the National Sciences and Engineering Research Council of Canada (to J.J.W.).
Footnotes
The authors declare no conflict of interest.
See companion article on page 262.
References
- 1.Hosseinzadeh P, et al. Design of a single protein that spans the entire 2-Volt range of physiological redox potentials. Proc Natl Acad Sci USA. 2015;113:262–267. doi: 10.1073/pnas.1515897112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solomon EI, et al. Copper active sites in biology. Chem Rev. 2014;114(7):3659–3853. doi: 10.1021/cr400327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warren JJ, Lancaster KM, Richards JH, Gray HB. Inner- and outer-sphere metal coordination in blue copper proteins. J Inorg Biochem. 2012;115:119–126. doi: 10.1016/j.jinorgbio.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li K, Xu F, Eriksson K-EL. Comparison of fungal laccases and redox mediators in oxidation of a nonphenolic lignin model compound. Appl Environ Microbiol. 1999;65(6):2654–2660. doi: 10.1128/aem.65.6.2654-2660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machonkin TE, Zhang HH, Hedman B, Hodgson KO, Solomon EI. Spectroscopic and magnetic studies of human ceruloplasmin: Identification of a redox-inactive reduced Type 1 copper site. Biochemistry. 1998;37(26):9570–9578. doi: 10.1021/bi980434v. [DOI] [PubMed] [Google Scholar]
- 6.Garner DK, et al. Reduction potential tuning of the blue copper center in Pseudomonas aeruginosa azurin by the axial methionine as probed by unnatural amino acids. J Am Chem Soc. 2006;128(49):15608–15617. doi: 10.1021/ja062732i. [DOI] [PubMed] [Google Scholar]
- 7.Lancaster KM, DeBeer George S, Yokoyama K, Richards JH, Gray HB. Type-zero copper proteins. Nat Chem. 2009;1(9):711–715. doi: 10.1038/nchem.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lancaster KM. Biological outer-sphere coordination. Struc Bond. 2012;142:119–153. [Google Scholar]
- 9.Berry SM, Baker MH, Reardon NJ. Reduction potential variations in azurin through secondary coordination sphere phenylalanine incorporations. J Inorg Biochem. 2010;104(10):1071–1078. doi: 10.1016/j.jinorgbio.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Marshall NM, et al. Rationally tuning the reduction potential of a single cupredoxin beyond the natural range. Nature. 2009;462(7269):113–116. doi: 10.1038/nature08551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyritsis P, Dennison C, Ingledew WJ, McFarlane W, Sykes AG. Determination of the self-exchange rate constant for rusticyanin from Thiobacillus ferrooxidans and a comparison with values for other type 1 blue copper proteins. Inorg Chem. 1995;34(21):5370–5374. [Google Scholar]
- 12.Meyer TJ, Taube H. Electron transfer reactions. In: Wilkinson G, editor. Comprehensive Coordination Chemistry. Pergamon; New York: 1987. pp. 331–384. [Google Scholar]
- 13.Marcus RA, Sutin N. Electron transfers in chemistry and biology. Biochim Biophys Acta. 1985;811(3):265–322. [Google Scholar]
- 14.Manesis AC, Shafaat HS. Electrochemical, Spectroscopic, and Density Functional Theory Characterization of Redox Activity in Nickel-Substituted Azurin: A Model for Acetyl-CoA Synthase. Inorg Chem. 2015;54(16):7959–7967. doi: 10.1021/acs.inorgchem.5b01103. [DOI] [PubMed] [Google Scholar]
- 15.McMillin DR, Rosenberg RC, Gray HB. Preparation and spectroscopic studies of cobalt(II) derivatives of blue copper proteins. Proc Natl Acad Sci USA. 1974;71(12):4760–4762. doi: 10.1073/pnas.71.12.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLaughlin MP, et al. Azurin as a protein scaffold for a low-coordinate nonheme iron site with a small-molecule binding pocket. J Am Chem Soc. 2012;134(48):19746–19757. doi: 10.1021/ja308346b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, et al. Redesigning the blue copper azurin into a redox-active mononuclear nonheme iron protein: Preparation and study of Fe(II)-M121E azurin. J Am Chem Soc. 2014;136(35):12337–12344. doi: 10.1021/ja505410u. [DOI] [PubMed] [Google Scholar]
- 18.Gray HB, Winkler JR. Hole hopping through tyrosine/tryptophan chains protects proteins from oxidative damage. Proc Natl Acad Sci USA. 2015;112(35):10920–10925. doi: 10.1073/pnas.1512704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klinman JP. How do enzymes activate oxygen without inactivating themselves? Acc Chem Res. 2007;40(5):325–333. doi: 10.1021/ar6000507. [DOI] [PubMed] [Google Scholar]