Summary
We evaluated the mechanism by which neutralizing human monoclonal antibodies inhibit chikungunya virus (CHIKV) infection. Potently neutralizing antibodies (NAbs) blocked infection at multiple steps of the virus life cycle, including entry and release. Cryo-electron microscopy structures of Fab fragments of two human NAbs and chikungunya virus-like particles showed a binding footprint that spanned independent domains on neighboring E2 subunits within one viral spike, suggesting a mechanism for inhibiting low pH-dependent membrane fusion. Detailed epitope mapping identified residue E2-W64 as a critical interaction residue. An escape mutation (E2-W64G) at this residue rendered CHIKV attenuated in mice. Consistent with this data, CHIKV-E2-W64G failed to emerge in vivo under the selection pressure of one of the NAbs, IM-CKV063. As our study suggests that antibodies engaging the residue E2-W64 can potently inhibit CHIKV at multiple stages of infection, antibody-based therapies or immunogens that target this region might have protective value.
INTRODUCTION
Chikungunya virus (CHIKV) is an enveloped positive stranded RNA virus and belongs to the Alphavirus genus of the Togaviridae family. The viral structural proteins, capsid (C) and three envelope (E) glycoproteins (E1, E2 and E3), are produced from the subgenomic RNA as a polyprotein that is subsequently proteolytically processed. Alphavirus virions have T = 4 quasi-icosahedral symmetry with 240 copies of the E1-E2 heterodimer, assembled into 80 spikes on the viral surface, and 240 copies of C form an icosahedral nucleocapsid core enclosing viral genomic RNA within the lipid membrane (Cheng et al., 1995). E2 is comprised of three domains (Voss et al., 2010): domain A is located in the center of the spike surface and possesses the putative receptor binding site (Smith et al., 1995); domains B and C are located at the distal and membrane proximal end of E2, respectively. E1 is a type II membrane fusion protein and sits at the base of the trimeric spike with E2 positioned on top of E1. The ectodomain of E1 consists of three domains (Lescar et al., 2001). Domain I links distal domain II and membrane proximal domain III. A fusion loop is located at the distal end of E1 domain II, and is protected by domain B of E2 (Lescar et al., 2001; Voss et al., 2010).
Alphaviruses enter cells via receptor-mediated endocytosis (Bernard et al., 2010) and are trafficked to acidified endosomes where the low pH environment triggers conformational rearrangements in the envelope glycoproteins. E2 domain B dissociates from the tip of E1 domain II, which exposes the fusion loop (Li et al., 2010). E1 then forms a homotrimer, further exposing the fusion loops of each monomer at the end of the trimeric complex for insertion into host membrane (Gibbons et al., 2004). Compared to the well-resolved entry steps of the alphavirus life cycle, assembly and budding are less clear. The capsid and envelope glycoproteins are required for virus particle assembly and release from the surface of infected cells (Forsell et al., 2000; Garoff et al., 2004; Soonsawad et al., 2010). However, it is unclear how formation of two icosahedral layers (capsid and envelope glycoproteins) is coordinated and what viral and/or cellular factors promote virus budding.
CHIKV is transmitted to humans by Aedes species mosquitoes and causes an acute febrile illness often accompanied by severe arthralgia, with relapses for weeks to months (Couderc and Lecuit, 2015). In the past decade, CHIKV has spread from endemic areas of Africa and Asia to new parts of the world. CHIKV is now the most common alphavirus infecting humans – with millions of individuals infected during the 2000s, including several incursions into Europe (Schuffenecker et al., 2006; Watson, 2007). In the winter of 2013, autochthonous cases in the Americas were documented (Fischer et al., 2014), with the virus rapidly spreading throughout the Caribbean islands (Lanciotti and Valadere, 2014). The virus has now been disseminated to more than 40 countries in the Americas (Diaz-Quinonez et al., 2015) and likely will pose a continued threat to global human health and economy.
There are currently no approved vaccines or treatments for CHIKV infection. Several studies have demonstrated an essential role of antibodies in the control of CHIKV infection (Kam et al., 2012a; Kam et al., 2012b; Kam et al., 2012c; Lum et al., 2013). We, and others, have isolated CHIKV neutralizing antibodies (NAbs) and demonstrated their ability to protect mice and non-human primates against CHIKV infection (Fong et al., 2014; Fric et al., 2013; Goh et al., 2013; Hawman et al., 2013; Pal et al., 2013; Selvarajah et al., 2013; Smith et al., 2015). In most studies, monoclonal antibodies (mAbs) have been evaluated for their ability to block virus entry in vitro. In our current study, we demonstrate a dual block by CHIKV NAbs on both virus entry and release. Single particle CryoEM analysis demonstrated bridging of two neighboring E2 molecules by Fab fragments and revealed a common contact residue (E2-W64), which was confirmed by neutralization escape mutant selection. Overall, our functional and structural analyses explain the potent neutralizing activity and strong protection profile in vivo of single NAb against CHIKV.
RESULTS
Two potent neutralizing anti-CHIKV antibodies inhibit plasma membrane fusion
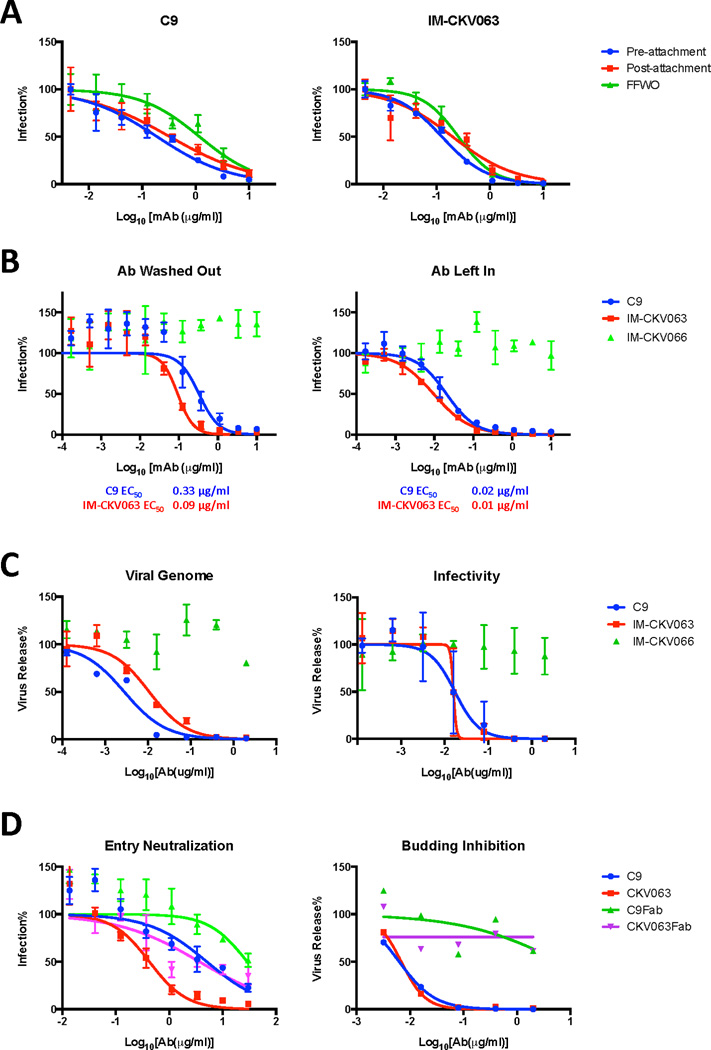
We previously reported two neutralizing monoclonal antibodies (NAbs), C9 and the ultra-potent IM-CKV063, from individuals who were infected and recovered from CHIKV during the 2006 outbreak in La Réunion, France (Fong et al., 2014) and a 2007 outbreak in northern Italy (Selvarajah et al., 2013). Antibodies can block enveloped virus entry at one or more stages including attachment, entry, and/or membrane fusion (Zinkernagel et al., 2001). To understand how C9 and IM-CKV063 inhibit CHIKV infection in cells, we performed pre- and post-attachment neutralization assays. CHIKV envelope (env) glycoprotein pseudotyped HIV luciferase reporter viruses were incubated with serially diluted C9 or IM-CKV063 before or after addition to Vero cells at 4°C. Infection was initiated by incubation at 37°C and was quantified by luciferase activity in cell lysates at 48 hours post infection. C9 and IM-CKV063 efficiently inhibited infection when incubated with virus before or after attachment to the cell surface (Figure 1A), suggesting that both C9 and IM-CKV063 can block CHIKV entry after virus engages receptor at the cell surface.
Figure 1. Mechanism of potent neutralization by CHIKV mAb C9 and IM-CKV063.
(A) Pre-and post-attachment neutralization and FFWO neutralization assays of C9 and IM-CKV063 (See also Figure S2). CHIKV Env pseudotyped HIV reporter viruses were incubated with serially diluted mAbs before binding to Vero cells (Pre-attachment), or were bound to Vero cells before incubation with serially diluted mAbs (Post-attachment). After washing, cells were incubated for 48 hours before lysis for luciferase activity detection. FFWO: Vero cells were sequentially incubated with CHIKV Env pseudotyped HIV reporter viruses and serially diluted mAbs at 4°C. Viral membrane fusion was induced by incubation with low pH buffer for 2 minutes. After pH normalization, cells were cultured for 48 hours in the presence of NH4Cl to prevent infection. Luciferase RLUs for no antibody controls in pre-attachment, post-attachment and FFWO were 38948, 27628 and 34249 respectively and were set to 100%. (B) Neutralization of replication-competent Gaussia luciferase reporter virus (CHIKV-Gluc) by C9 and IM-CKV063 (See also Figure S3 for additional mAbs). CHIKV-Gluc was incubated with serially diluted mAbs for one hour before infecting RD cells (MOI 0.01) for one hour. Left: Virus-mAbs inoculum was washed out. Right: Virus-mAb inoculum was kept in the well. Cells were cultured for 24 hours before GLuc detection in the culture supernatant. The non-neutralizing human mAb IM-CKV066 served as a control. No antibody control was set to 100%. (C) RD cells were infected with CHIKV-GLuc for three hours. After extensive washing, serially diluted mAbs in medium containing NH4Cl were added to prevent further rounds of infection. Virus release at 24 hours post infection was measured by qRT-PCR analysis of viral genomic RNA in culture supernatant (left) or by infectivity assay of the supernatant (right). Virus release (vRNA or infectivity) was normalized to the basal infection level that was measured by GLuc activity in the culture supernatant. No antibody control was set to 100%. (See also Figures S4 and S5) (D) Entry neutralization (left) and budding inhibition (right) of CHIKV-GLuc by mAb and Fab of C9 and IM-CKV063. As described in B and C, C9 and IM-CKV063 mAbs and their Fab fragments were compared for their abilities to neutralize CHIKV entry and inhibit virus release quantified by qRT-PCR of vRNA. Data shown are representative of three experiments performed in triplicate with error bars representing standard deviation.
Alphavirus mediated membrane fusion is a low-pH dependent process that occurs in endosomes after internalization. To mimic virus-endosome membrane fusion at the plasma membrane, we set up an acid-bypass viral fusion from without (FFWO) assay. Membrane fusion was induced at the plasma membrane by treating Vero cells pre-bound with CHIKV env pseudotyped HIV reporter viruses with low pH buffer for 2 minutes. Subsequently, cells were cultured in medium containing 20 mM NH4Cl to prevent further rounds of infection via the endosomal pathway. The optimal pH to induce membrane fusion for CHIKV env was first determined as pH 5.5 (Figure S1) consistent with previous reports (Pal et al., 2013). We next tested if C9 and IM-CKV063 blocked CHIKV-mediated membrane fusion by incubating CHIKV env pseudotyped HIV reporter viruses pre-bound to the cell surface, with serially diluted C9 and IM-CKV063 at 4°C before membrane fusion induction in the FFWO assay. Both C9 and IM-CKV063 inhibit CHIKV env mediated membrane fusion potently (Figure 1A). Taken together, these results suggest C9 and IM-CKV063 neutralize CHIKV entry at the low-pH dependent membrane fusion step.
Neutralizing antibodies block CHIKV release
To study neutralization by C9 and IM-CKV063 with replication-competent CHIKV, we generated a secretory Gaussia luciferase (GLuc) expressing reporter virus (CHIKV-GLuc). We observed similar pre-attachment, post-attachment and FFWO neutralizing results for C9 and IM-CKV063 using CHIKV-GLuc as using CHIKV env pseudotyped HIV reporter virus (Figure S2A–C). Consistent with this data, C9 and IM-CKV063 failed to block CHKV binding to the cell surface (Figure S2D). We next compared the neutralizing efficiency of C9 and IM-CKV063 when the mAbs were left in the culture (to allow for effects on multiple stages in the viral life cycle) or washed out (to allow mAb inhibition at a defined step). C9 and IM-CKV063 neutralized CHIKV-GLuc replication more efficiently when left in the culture medium rather than when washed out immediately after virus entry (Figure 1B). This pattern was recapitulated with other human and mouse NAbs tested (Figure S3), with ~10 fold lower EC50 values when the antibody was left in. These results suggested that NAbs might inhibit additional step(s) beyond entry in virus replication cycle. We next tested if CHIKV release was inhibited by adding NAbs to the culture after infecting cells with CHIKV-GLuc for three hours at 37°C and extensively washing away residual virus. Viral replication and release was allowed to continue for 20 hours in presence of 20 mM NH4Cl to prevent multiple rounds of infection. Virus release was quantified by measuring viral RNA levels in the supernatant by qRT-PCR or by determining levels of infectious virus in the supernatant by inoculating fresh cells after a 3,000-fold dilution (to decrease the concentration of NH4Cl and antibody to non-inhibitory levels). C9 and IM-CKV063 potently inhibited CHIKV-GLuc release in a concentration dependent manner, as quantified by both vRNA and virus infectivity assays (Figure 1C). In contrast, the control mAb IM-CKV066 did not inhibit virus release or entry. Other human and mouse NAbs also inhibited CHIKV-GLuc release with varying efficiencies (Figure S4A and B). GLuc reporter gene expression positively correlates with intracellular viral RNA level (Figure S4D). Thus, GLuc released in the culture supernatant indicates basal viral infection and replication. Addition of NAbs post infection did not affect reporter gene expression (Figure S4C), suggesting mAbs do not inhibit post-entry and pre-viral release steps of viral replication. C9 and IM-CKV063 also inhibited virus release from cells infected with wild type CHIKV ECSA strain S27 and strain LR2006 OPY1 (Figure S5).
To better understand the two anti-viral mechanisms of NAbs, we compared abilities of mAbs vs Fab fragments of C9 and IM-CKV063 to block CHIKV entry and release (Figure 1D). In contrast to efficient inhibition of virus entry and release by C9 and IM-CKV063, Fab fragments of these two NAbs could only inhibit CHIKV entry but not release, suggesting bivalent binding is critical for NAbs to block CHIKV virus release but not entry.
C9 and IM-CKV063 share an epitope centered on residue E2-W64
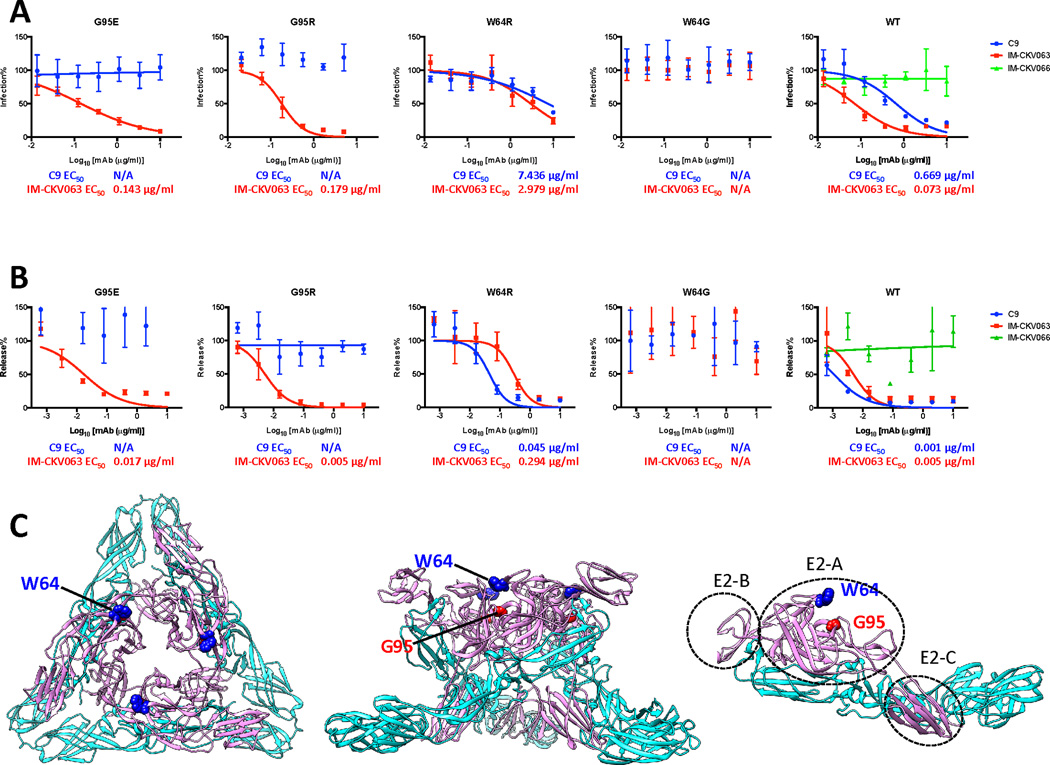
We previously used comprehensive alanine-scanning epitope mapping to identify E2-E24, G55, W64, K66, R80 and I121 as critical residues for binding by IM-CKV063 (Fong et al., 2014) and E2-A162 for C9 binding (Selvarajah et al., 2013). To identify residues that are relevant functionally for C9 and IM-CKV063 interaction with infectious virus, we generated NAb escape mutants in Vero cell culture by passaging CHIKV 37997 strain under selection of C9 or IM-CKV063. Selected escape mutant viruses were first confirmed as resistant to selecting NAb and then sequenced. E2-G95E and E2-W64G were identified as single mutations in C9 and IM-CKV063 escape mutant viruses, respectively. In addition to in vitro escape selection, we also performed in vivo escape selection. Ifnαr−/− mice were infected with 10 PFU of CHIKV 37997 strain subcutaneously and then administered C9 or IM-CKV063 at 24 hours post infection. Brain tissue was collected when mice were moribund at 2 or 3 days post infection and viral RNA was sequenced. Mutant viruses were detected in C9-treated mice with either an E2-G95E or E2-G95R substitution, whereas mutant viruses with a single E2-W64R mutation emerged in animals treated with IM-CKV063.
To establish that these single mutations conferred resistance to NAbs, they were introduced into CHIKV-GLuc reporter virus and tested for antibody inhibition of virus entry (Figure 2A) or release (Figure 2B). Consistent with sequencing results, E2-G95E and E2-G95R mutant viruses were resistant to C9 mediated entry and release inhibition. The E2-W64G mutation conferred CHIKV resistance to both IM-CKV063 mediated entry and release inhibition whereas E2-W64R was only weakly resistant to IM-CKV063. Of note, while a change at G95 rendered CHIKV resistant specifically to C9, the W64G mutation conferred resistance to both C9 and IM-CKV063. Similar results were observed when the mutations were engineered into CHIKV LR2006-OPY1 stain, with both escape viruses remaining sensitive to hCHK-152, which binds to an adjacent residue, E2-D59 (Pal et al., 2013; Sun et al., 2013) (Figure S6). These results suggest that residue E2-W64 in E2 comprises part of a common epitope shared by C9 and IM-CKV063 whereas E2-G95 is specific for C9.
Figure 2. Mapping and characterize C9 and IM-CKV063 escape mutants.
(A) CHIKV-GLuc reporter viruses carrying indicated single escape mutations are resistant to antibody-mediated neutralization of entry (See also Figures S6). Similar to Figure 1B, WT and mutants GLuc reporter viruses were incubated with serially diluted mAbs before infecting RD cells at an MOI of 0.5. Infection was presented by GLuc activity in the supernatant at 24 hpi. Virus infection was normalized to a no antibody control. Results are representative of three independent experiments in triplicate. (B) CHIKV-GLuc reporter viruses encoding indicated single escape mutations are resistant to antibody-mediated inhibition of virus release (See also Figures S6). Similar to Figure 1C, RD cells were infected with WT or mutant GLuc reporter viruses for three hours before incubated with MAbs serially diluted in medium containing NH4Cl to prevent further rounds of infection. Virus release at 24 hours post-infection was measured by an infectivity assay. No antibody treatment was set to 100%. Data shown are representative of three experiments performed in triplicate with error bars representing standard deviation. (C) Mapping of C9 and IM-CKV063 escape residues on the crystal structure of the mature CHIKV envelope glycoprotein complex (PDB code 2XFB) in top view of trimer (left), side view of trimer (middle) and E1E2 dimer (right). Pink: E2; Cyan: E1.
To define the location of the residues that potentially bound C9 and IM-CKV063, E2-W64 and E2-G95 were mapped onto the CHIKV E1-E2 atomic structure (Voss et al., 2010) (Figure 2C). Both E2-W64 and E2-G95 are solvent accessible and located in domain A and face the cavity enclosed by three E2 molecules in one spike, which consists of three E1-E2 heterodimers. E2-W64 is located at the apex of the E2 trimer, whereas E2-G95 is located below E2-W64 within the cavity. Alignment of all available 422 E2 proteins from different CHIKV strains in GenBank and 212 E2 proteins in VIPR (www.viprbrc.org) demonstrated that both E2-W64 and E2-G95 are 100% conserved.
C9 and IM-CKV063 bind similarly to Chikungunya virus-like particles
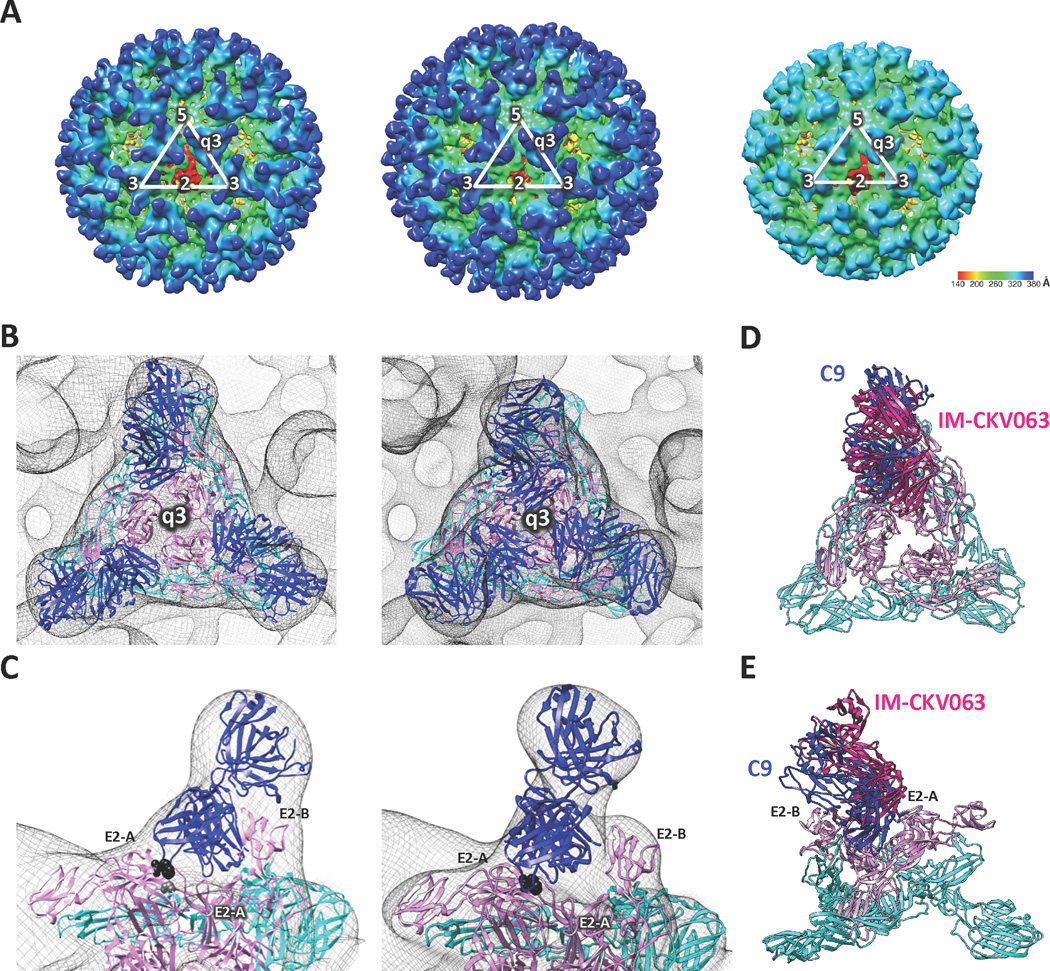
To gain further insight as to how C9 and IM-CKV063 interact and neutralize CHIKV, we determined single particle cryo-electron microscopy (cryo-EM) structures at 15.3 Å, 14.9 Å and 11.2 Å resolution for chikungunya (CHIK) virus-like particles (VLP) (Akahata et al., 2010; Sun et al., 2013), CHIK VLP in complex with C9 Fab fragment and CHIK VLP in complex with IM-CKV063 Fab fragment, respectively (Figure 3A). Three molecules of C9 Fab or IM-CKV063 Fab could be distinguished on top of each q3 or i3 CHIKV spike. We fitted a generic Fab crystal structure into the difference maps after subtracting VLP density from VLP-C9 Fab and VLP-IM-CKV063 Fab density maps, using the fitting function of UCSF Chimera. CHIK VLP pseudo-atomic resolution coordinates also were placed into the VLP-C9 Fab and VLP-IM-CKV063 Fab electron density maps (Figure 3B). Both C9 and IM-CKV063 Fab fragments bound two neighboring E2 proteins in one spike with the complementarity determining regions (CDRs) interacting with domain A of E2 and the back of the variable region appeared to lie down on an adjacent E2 (Figure 3C). The footprints of C9 and IM-CKV063 Fab fragments on the surface of the CHIK VLPs were determined with the RIVEM program (Xiao and Rossmann, 2007) (Figure 4). Both C9 and IM-CKV063 Fab spanned domain A and the β-ribbon connector of one E2 and domain A and B of a neighboring E2 within one spike. In this way, C9 and IM-CKV063 may hinder sterically the movement of the B domain that is activated by acid pH and required for membrane fusion. This structural model is consistent with our results showing that C9 and IM-CKV063 both inhibit low pH dependent plasma membrane fusion (see Figure 1A).
Figure 3. C9 and IM-CKV063 span two neighboring E2 molecules in one spike.
(A) CryoEM reconstruction of CHIKV (S27) virus like particle (VLP) (right), VLP in complex with C9 Fab fragments (left) and VLP in complex with IM-CKV063 fragments (middle). (B) Fitting of E1-E2 and Fab crystal structures into the viral spike at the q3-fold axis of the cryoEM density map of C9 Fab bound VLP (left) and IM-CKV063 Fab bound VLP (right). Blue, Fab; Pink, E2; Cyan, E1. (C) C9 Fab (left) and IM-CKV063 Fab (right) insert between domain A of one E2 (left) and domain B of a neighboring E2 (right) in one viral spike. E2-W64 was labeled in black and E2-G95 was labeled in gray. Comparison of C9 Fab (blue) and IM-CKV063 Fab (hot pink) binding to a viral spike in top view (D) and side view (E).
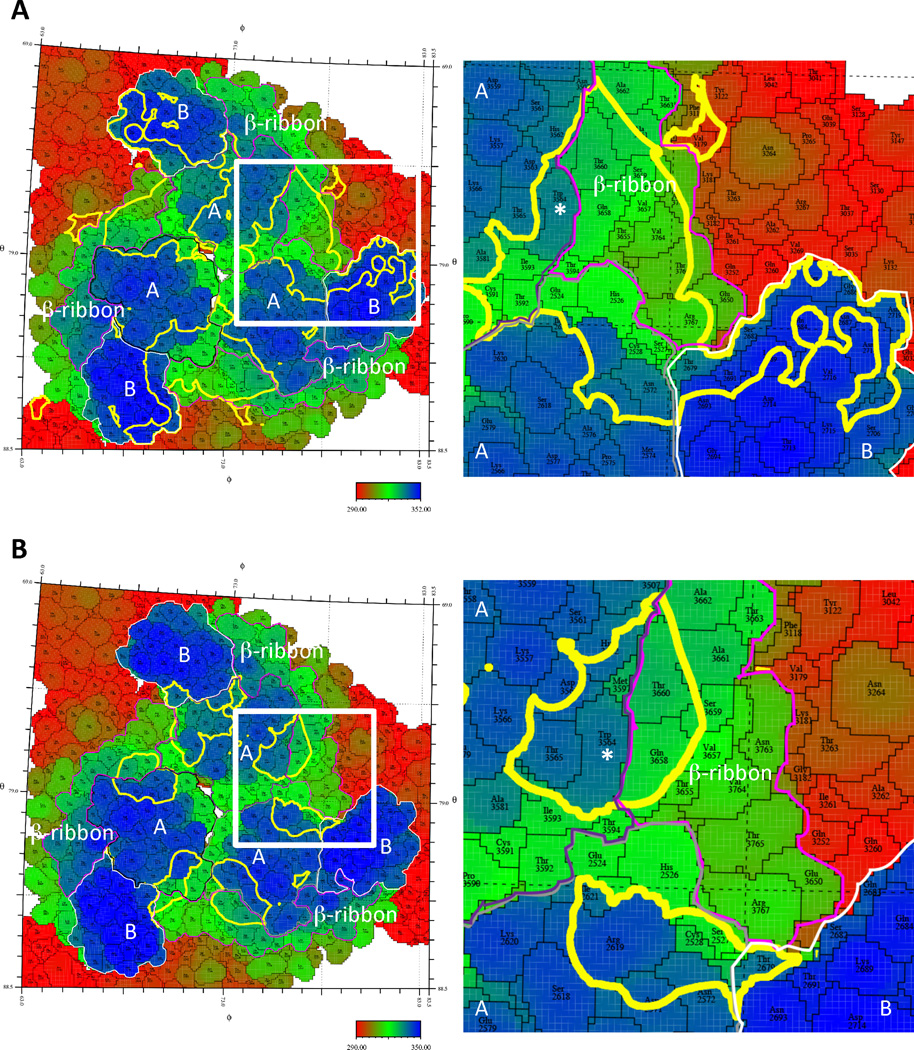
Figure 4. Footprints of C9 and IM-CKV063 span two neighboring E2.
“Roadmaps” of the footprints of C9 (A) or IM-CKV063 (B) Fabs on the VLP surface at q3 spike. To differentiate between amino acids in three different q3-fold related subunits, their identity is defined as the amino acid sequence number in E1 + 2000, 3000, and 4000, and in E2 + 2500, 3500, and 4500 individually. The surface is colored according to radial distance from the center of the VLP. Domain As of three E2 are bounded by black, grey and purple lines respectively. B domains of E2 are bounded by white lines. The β-ribbon connectors of E2 are bounded by magenta lines. The footprints of the Fabs on the VLP surface are outlined in yellow. The footprint of a single Fab is boxed in white and an enlarged image of the boxed area is shown on the right. E2-W64 is labeled with white star.
A comparison of C9 and IM-CKV063 interaction with the CHIKV spike revealed similar binding by C9 and IM-CKV063 to the crest of domain A on E2, but a deeper protrusion of C9 into the cavity of the spike (Figure 3D and E). This is consistent with C9 and IM-CKV063 sharing one contact residue (E2-W64) at the top of the cavity, whereas C9 also requires another residue (E2-G95), located at a lower position in the cavity (Figure 2C). Importantly, the location of E2-W64 was confirmed in the footprints of C9 and IM-CKV063, although E2-G95, which is situated below E2-W64, was not visible in the projection map (Figure 4). The cryo-EM determined footprints of C9 and IM-CKV063 also included residues E2-A162 and E2-I121 that were identified by alanine scanning epitope mapping for C9 and IM-CKV063, respectively (Fong et al., 2014; Selvarajah et al., 2013).
W64G mutation results in an attenuated CHIKV
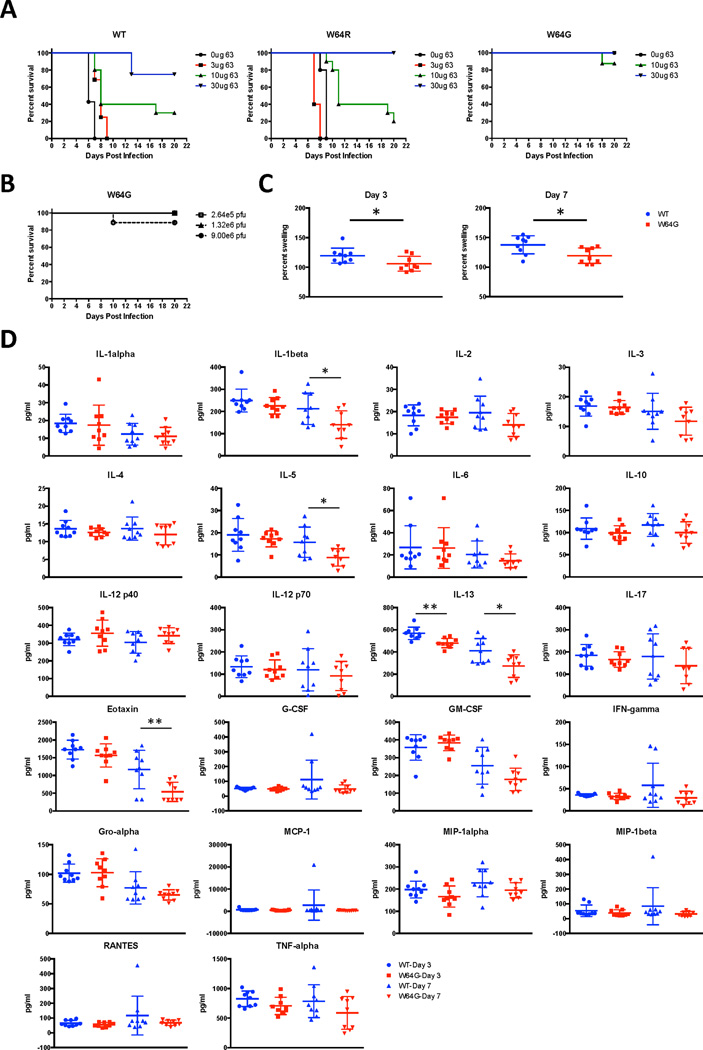
Because W64R was selected in vivo as an escape mutation yet rendered CHIKV only partially resistant to IM-CKV063 in vitro, we tested if this virus was fully resistant to IM-CKV063 in vivo in a neonatal mouse model of infection (Couderc et al., 2008). Previously, we reported that 10 µg of IM-CKV063 protected 100% of 9 day-old C57BL/6J mice against lethal CHIKV S27 (5 × 105 PFU) infection when administrated concurrently with virus (Fong et al., 2014). Here, we evaluated whether W64R and W64G mutant CHIKV 37997 were resistant to IM-CKV063 protection in neonatal mice (Figure 5A). Although administration of 5 × 105 PFU of either WT or W64R CHIKV 37997 resulted in 100% lethality in mice, the W64R mutant showed a delayed time to death (mean survival time of 6 versus 9 days, P < 0.01). IM-CKV063 provided similar protection against CHIKV 37997 WT and W64R (10 µg protected 30% of WT versus 20% W64R infected mice from lethality, P > 0.05; 30 µg protected 75% of WT versus 100% W64R infected mice, P > 0.1). Thus, and consistent with our in vitro data (Figure 2A), the W64R mutant remained sensitive to IM-CKV063 neutralization; even though it emerged as a variant under IM-CKV063 selection in Ifnαr−/− mice, it did not confer significant resistance in the WT mouse model. Somewhat unexpectedly, mice infected with 5 × 105 PFU of W64G mutant survived for at least 3 weeks (Figure 5A), and a 30-fold higher virus dose caused death in only 10% of mice (Figure 5B). These results indicated that the fully resistant W64G mutant was attenuated in neonatal mice.
Figure 5. Virulence of WT and W64 mutants in vivo.
(A) 9 day-old C57BL/6J mice were infected with 2.6 × 105 PFU of WT or the indicated escape mutant viruses via an intradermal route. Concurrently, animals were administered via an intraperitoneal injection the indicated amounts of IM-CKV063. (B) C57BL/6J mice were infected with increasing amount of CHIKV (37997) via intradermal route. The survival curves were constructed from data of at least two independent experiments with a total of between 5 and 15 mice per group. (C) Three week-old WT C57BL/6J mice were inoculated in the left rear footpad with 103 PFU of WT or W64G CHIKV. At days 3 and 7 after infection, ankle size was measured and relative swelling was calculated for each mouse. (D) At days 3 and 7 after infection, serum was collected and analyzed for the indicated cytokines and chemokines. Statistically significance was determined by unpaired t test (*, P<0.05; **, P<0.01).
We next employed a non-lethal mouse model of CHIKV-induced arthritis (Morrison et al., 2011) to compare the ability of WT and W64G mutant CHIKV to cause joint disease in 3 week-old C57BL/6J mice. 103 PFU of WT and W64G mutant CHIKV 37997 were inoculated in the left rear footpad. During the acute phase, we measured ankle swelling for each mouse at day 3 and 7 post infection and observed less swelling with the W64G mutant compared to WT virus (Figure 5C). Cytokine and chemokine levels in mouse serum at day 3 and 7 post infection also were measured. Compared to animals infected with WT virus, mice infected with CHIKV W64G mutant produced lower serum levels of several cytokines and chemokines, including IL-1β, IL-5, IL-13 and Eotaxin (Figure 5D). In a recent metaanalysis of anti-CHIKV patient signatures all of these cytokines, as well as eotaxin, were found to be elevated in acute infection (Teng et al., 2015). Thus, analogous to results in neonatal mice, the W64G mutant virus was attenuated in its ability to cause joint inflammation and swelling in juvenile 3 week-old mice.
IM-CKV063 provides strong protection against CHIKV-induced joint disease in vivo
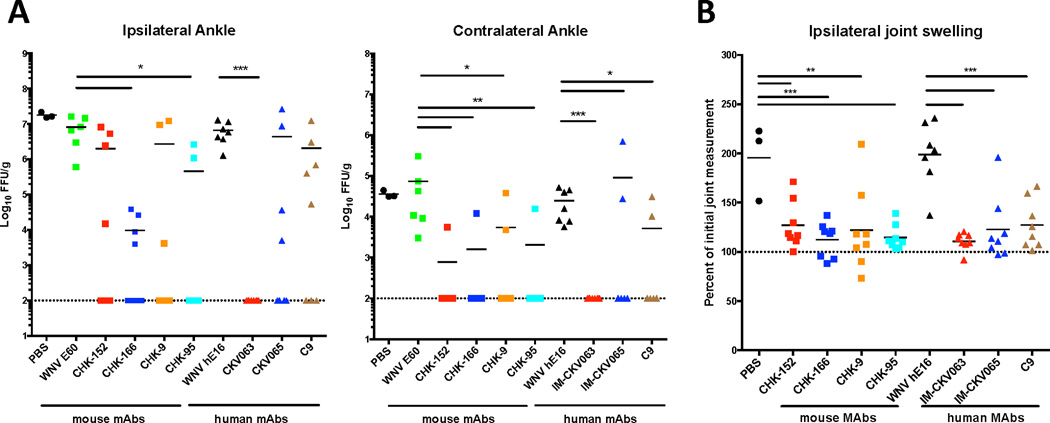
C9 and IM-CKV063 neutralized CHIKV entry and release at comparable potency, but it was more difficult for virus to escape from IM-CKV063 than C9 in vivo. To test if this correlated with their different protective activities in vivo, we compared the ability of C9 and IM-CKV063 with previously identified moderate human NAb IM-CKV065 (Fong et al., 2014) and potent mouse NAbs (Pal et al., 2013) to protect mice from arthritis caused by CHIKV-LR infection (Morrison et al., 2011). Human or mouse NAbs were administered to 4 week old-mice four hours after subcutaneous infection with 103 PFU of CHIKV in the left rear footpad. Compared to isotype control mAb, IM-CKV063 limited the CHIKV burden in the ipsilateral ankle to the limit of detection, prevented viral spread to the contralateral ankle joint (Figure 6A), and showed superior activity compared to the remainder of the panel of mouse and human NAbs. Although treatment with all tested NAbs reduced ankle joint swelling at day 3 when compared to the isotype control mAb, IM-CKV063 therapy reduced the swelling closest to the baseline (Figure 6B). These results demonstrated that IM-CKV063 provides superior protection compared to other NAbs against CHIKV-induced infection and arthritis in vivo in mice.
Figure 6. IM-CKV063 provides superior protection against CHIKV compared to other tested NAbs.
Four week-old WT mice were treated with 100 µg of indicated mAbs four hours following subcutaneous inoculation with 103 FFU CHIKV-LR in the left rear footpad. WNV E60 and WNV hE16 served as mouse and human isotype control mAbs. (A) Viral load was determined in both ipsilateral and contralateral ankles at 3 days post infection. Statistical significance was determined by a Kruskal-Wallis test with a Dunn’s multiple comparison test (*, P < 0.05; **, P < 0.01; ***, P < 0.001) (n = 6 to 7). (B) Footpad size in the ipsilateral joint was measured prior to and 3 days following infection. Statistical significance was determined by ANOVA with a Dunnett’s multiple comparison test (**, P < 0.01; ***, P < 0.001)(n = 6 to 7).
DISCUSSION
In the current study, we characterized the neutralizing mechanisms of two previously identified potently inhibitory antibodies, C9 (Selvarajah et al., 2013) and IM-CKV063 (Fong et al., 2014). A comparison of neutralization efficiency between NAbs left in culture after infection and NAbs washed out after infection led us to identify post-entry inhibition by CHIKV NAbs as a potent mechanism of inhibition. To escape from NAbs, mutant CHIKV acquired resistance to both entry and release inhibition by NAbs, suggesting the importance of these blocks to the CHIKV lifecycle. MAb-mediated inhibition of viral release has been described for influenza virus (Dowdle et al., 1974), bovine leukemia virus (Driscoll et al., 1977), herpes virus (Shariff et al., 1991), vaccinia virus (Vanderplasschen et al., 1997), rubella virus (Corboba et al., 2000), Marburg virus (MARV) (Kajihara et al., 2012) and CHIKV (Masrinoul et al., 2014). However, most of these studies utilized mAbs that do not independently neutralize virus entry. Recently, inhibition of viral release was described for two MARV glycoprotein specific mAbs, whereby these mAbs inhibited MARV release by tethering budded virus particles to the glycoproteins present on the cell surface via bivalent binding (Kajihara et al., 2012). A recent study reported a murine mAb (CK47) targeting E1-domain III that inhibited CHIKV release but not entry (Masrinoul et al., 2014), although high concentration of antibody (250 µg/ml) was required, and no in vivo studies were performed. A non-neutralizing antibody used in our study that targets E1-domain II (IM-CKV066) (Fong et al., 2014) failed to block CHIKV release. As cells used for virus release assays in our study lack Fc receptor, the inhibition of virus release by NAbs is unlikely due to the tethering of virus by IgG to cell surface. In our study, bivalent binding is critical for NAbs to block CHIKV release but not entry. It will be interesting to test if Nabs inhibit CHIKV release by tethering budded virus particles to viral glycoprotein at the cell surface or through a different mechanism. Future studies are needed to understand the mechanism by which mAbs targeting different epitopes on E1 and E2 block the same release step of the CHIKV lifecycle. In our study, all neutralizing antibodies that blocked CHIKV release also inhibited entry. The relative involvement of neutralization and viral budding inhibition, as well as other potential mechanisms of anti-viral activity, in in vivo protection will require further studies.
Single particle cryo-EM structures revealed that C9 and IM-CKV063 Fab molecules bridge adjacent individual E2 subunits within the same spike on CHIKV VLP. The fitting of a Fab fragment into the cryo-EM density map suggests that both C9 and IM-CKV063 CDRs interact with the top rim of a cavity surrounded by three E2 molecules, and tilt toward the neighboring E2 with the groove of the variable region positioned on it. This mode of binding of one Fab fragment to two neighboring E2 differs from that reported for four mouse CHIKV NAbs (CHK-152, CHK-9, m242 and m10) in which one Fab binds to one E2 without interacting with other E2 molecules in a given spike (Sun et al., 2013). Nonetheless, analogous cross-linking of neighboring E glycoproteins was reported for a human NAb against West Nile virus, CR4354 (Kaufmann et al., 2010).
By interacting with domain A and the β-ribbon connector of one E2 and domain A and B of a neighboring E2, C9 and IM-CKV063 may prevent conformational changes that are required for fusion loop exposure. This model is consistent with our results showing both C9 and IM-CKV063 blocked low-pH dependent plasma membrane fusion after virus binds to the cell. A similar mechanism, involving the constraint of CHIKV E2 domain A and domain B was proposed for a potent mouse NAb CHK-152 (Sun et al., 2013). However, for CHK-152, the CDRs interact with domain A and domain B of a single E2 subunit in a given spike, rather than adjacent subunits. The epitopes for CHK-152 (Pal et al., 2013), C9 and IM-CKV063 all were mapped to E2-domain A by escape mutant selection and confirmed by cryo-EM. Additional residues were identified by epitope mapping with alanine scanning for C9 (Selvarajah et al., 2013) and IM-CKV063 (Fong et al., 2014), and they are located in or close to the C9 and IM-CKV063 footprints on CHIKV. Small differences in the epitope mapping of the footprints between this study and Fong et al. (Fong et al., 2014) are likely due to mapping being performed using point mutations in cell-surface expressed viral glycoproteins, rather than icosahedral virus-like particles and live virus escape mutants as performed in the current study. Although no residue in domain B was found critical for CHK-152, C9 or IM-CKV063 binding based on neutralization escape or other mutagenesis studies, portions of domain B were apparent in their cryo-EM footprints. One mAb per E1-E2 trimer may be sufficient to constrain E2 and thus hinder the low-pH dependent conformational changes. This mechanism may explain the basis for their neutralizing ability, particularly of the ultrapotent IM-CKV063.
Escape mutant selection and cryoEM structure analyses suggest that C9 and IM-CKV063 share a key residue, E2-W64, as part of their epitopes. In a recent report describing another set of human NAbs against CHIKV, 5 of 18 strong NAbs and 1 of 5 moderate NAbs included W64 in their epitopes (Smith et al., 2015), suggesting E2-W64 may comprise part of dominant neutralizing epitope of CHIKV glycoprotein. E2-W64 is conserved among all the sequenced CHIKV strains available in GenBank and VIPR databases. Unexpectedly, W64 appears important for CHIKV pathogenesis, with the W64G mutation attenuating CHIKV in a neonatal mouse model of lethality and an arthritis model in juvenile mice.
Although NAbs are being explored to treat acute or chronic CHIKV infection, rapid emergence of resistant mutants could render therapeutic antibodies less useful. W64G was selected by IM-CKV063 in vitro, whereas only the less resistant mutant, W64R, was selected in vivo. The failure of CHIKV to mutate to the fully resistant W64G mutant in vivo might explain the superior protection observed from IM-CKV063 compared to other NAbs (e.g., C9 and CHK-152) to which escape mutants readily emerged (Pal et al., 2013). The relative attenuation of the fully resistant W64G mutant may contribute to the potency of IM-CKV063 to reduce inflammation induced by CHIKV infection. Although more study is warranted, it suggests that IM-CKV063 may be a good candidate for monotherapy against CHIKV infection.
In summary, our study demonstrated blocks in the CHIKV replication cycle by NAbs at both the entry and release steps. E2-W64 is a key residue targeted by multiple Nabs generated by independent groups from different subjects, and W64G mutation rendered CHIKV attenuated in vivo. The E2-W64 residue and its complete epitope may have value in structure-based vaccine design or antibody therapeutic development against CHIKV.
Experimental Procedures
Cells, viruses, antibodies and mice used in the study are described in supplemental information.
Neonatal mouse model
9 day-old mice were transferred to a biosafety level 3 facility in static disposable cages (Innovive Inc., San Diego, CA) with their mothers. Neonatal C57BL/6J mice were infected with CHIKV intradermally in the ventral thorax. Each mouse was injected via an intraperitoneal route with test mAb in 0.2 ml PBS immediately prior to CHIKV infection. Mice were then observed daily for up to 20 days. Results were analyzed using Kaplan-Meier survival curves and GraphPad Prism software (GraphPad, CA).
Arthritis mouse model
3 week-old C57BL/6J mice were inoculated with CHIKV subcutaneously in the left footpad with 103 PFU of CHIKV in PBS supplemented with 1% heat inactivated FBS. Joint swelling was monitored via left and right foot measurements (width × height) using digital calipers. Sera were collected at days 3 and 7 after infection.
NAb protection: A single 100 µg dose of human NAbs C9, IM-CKV063, CKV065, and mouse NAbs CHK-166, CHK-152, CHK-9 and CHK-95 was administered to 4 week-old C57BL/6J mice four hours after subcutaneous infection with 103 PFU of CHIKV (LR2006-OPY1) in the left rear footpad. Mouse West Nile virus (WNV) mAbs E60 and humanized hE16 were used as the isotype controls. Joint swelling was monitored via left foot measurements. Tissues were collected at day 3 after infection and virus was titered by focus forming assay (Pal et al., 2013) using chimeric human CHK-9 as the detection antibody for CHIKV.
Mutagenesis of CHIKV infectious clone
Site-directed mutagenesis was performed by standard overlapping PCR approach. Primers for site-directed mutagenesis were listed in Table S1. Molecular clones of mutant viruses confirmed by sequencing.
CryoEM reconstruction of CHIKV VLP in complex with C9 or IM-CKV063 Fab fragment
CHIKV VLPs production was described before (Fong et al., 2014). C9 and CVK063 Fab fragments were generated from purified C9 and IM-CKV063 IgG using Pierce™ Fab preparation Kit (Thermo Fisher Scientific). The purified C9 and IM-CKV063 Fab molecules were mixed with CHIKV VLP at 3:1 (Fab:E2) molar ratio for at least 30 min on ice. Samples were flash-frozen on Quantifoil copper grids (R2/1, 200 mesh) in liquid ethane with a Vitrobot Mark III. Detailed data acquiring, processing and analysis were described in supplemental information.
Accession codes
The cryo-EM density maps of CHIKV VLP, VLP in complex with C9 Fab and VLP in complex with IM-CKV063 Fab were deposited with the EM Data Bank under accession numbers EMD-6466, 6467 and 6457 respectively.
Entry neutralization assay
Pre-attachment: CHIK reporter viruses were incubated with serially diluted mAbs at 4°C for one hour before added to Vero cells for one hour binding at 4°C. After washing three times, cells were incubated in fresh medium for 48 hours at 37°C before lysis and assaying for luciferase activity. Post-attachment: CHIK reporter viruses were bound to Vero cells at 4°C for one hour. After washing three times, cells were incubated in serially diluted mAbs for one hour at 4°C. After additional washing, cells were incubated in fresh medium for 48 hours at 37°C before lysis and assaying for luciferase activity. Fusion from without (FFWO): Vero cells were incubated sequentially with CHIK reporter viruses for one hour at 4°C and serially diluted mAbs for one hour at 4°C. Viral membrane fusion was induced by incubation with buffer of pH 5.5 at 37°C for 2 minutes. After pH normalization, cells were cultured for 48 hours in the presence of 20 mM NH4Cl to prevent infection via the endosomal pathway before lysis and assaying for luciferase activity. Virus infection was normalized to a no antibody control.
Release inhibition assays
RD cells were seeded in 96 well plates at 4 × 104 cells per well. Cells were infected with CHIKV-GLuc at M.O.I 0.5 for three hours at 37°C. After washing for three times, serially diluted mAbs in medium containing 20 mM NH4Cl were added to prevent further rounds of infection. At 20 hours post infection, culture supernatants were harvested. GLuc activity in the supernatant represented the basal infection levels. Virus released in the supernatant at 24 hours post infection was measured. (a) Viral RNA in culture supernatant was extracted by the QIAamp Viral RNA Mini kit (Qiagen) following the manufacturer’s protocol. Isolated RNA was analyzed by qRT-PCR and compared to a standard curve generated from CHIKV-GLuc plasmid. Primers are listed in Table S2. (b) Viral infectivity in the supernatant was measured by infecting fresh RD cells after a 3,000-fold dilution. GLuc activities at day 2 after infection were measured. Virus release efficiency was calculated by dividing either vRNA genome copy number or virus infectivity in the supernatant by basal infection level. Virus release efficiency without antibody treatment was set to 100%.
Escape mutant selection
(a) In vitro selection: 2 × 105 PFU of CHIKV-37997 was incubated with 10 µg/ml of C9 and 1 × 106 PFU of CHIKV-37997 was incubated with 5 µg/ml of IM-CKV063 for one hour at 37°C. Virus-NAb mixtures were added to Vero cells in 96-well-plate for 24 hours of infection. At each passage, half of the supernatant was mixed with 20 µg/ml of C9 or 10 µg/ml of IM-CKV063 for one hour at 37°C. The mixtures were added to fresh Vero cells to infect 2 hours. After three passages for C9 resistant virus selection and five passages for IM-CKV063 resistant virus selection, escape mutant viruses were selected. Viral RNA was extracted from the supernatant using a QIAamp Viral RNA Mini kit (Qiagen) and reverse transcribed into cDNA with random hexamer primer using the Superscript III Reverse Transcriptase kit (Invitrogen). CHIKV structural gene was amplified by PCR and sequenced. (b) In vivo selection: Ifnαr−/− mice were infected with 10 PFU of CHIKV 37997 strain subcutaneously and received 100 ug/mouse C9 or 50 ug/mouse IM-CKV063 at 24 hours post infection. Brain tissue was collected when mice were moribund at 2 or 3 days post infection and viral RNA was extracted from the tissue using RNeasy Mini kit (Qiagen). cDNA was synthesized using the Superscript III Reverse Transcriptase kit (Invitrogen) and CHIKV structure gene was amplified by PCR and sequenced.
Mouse cytokine and chemokine assay
23 mouse cytokines and chemokines in serum were measured by the Bio-Plex Pro Mouse Cytokine 23-Plex Immunoassay (BioRad) according to the manufacturer’s instruction.
Statistical analyses
All data were analyzed using Prism software (La Jolla, CA) and statistical significance was assigned when P values were < 0.05. EC50 values were determined using non-linear regression. Cytokine and chemokine levels were analyzed using the unpaired t-test. Viral titers were analyzed using the Kruskal Wallis test with a Dunn multiple comparisons test. Joint swelling was analyzed using a one-way ANOVA adjusting for repeated measures with a Dunnett’s multiple comparisons test.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (NIH) grants R56 AI119056 (to G.S.), R01 AI089591 and R01 AI114816 (to M.S.D.) and contract HHSN272200700055C (to B.J.D.). The authors thank John Heitman, Yifan Cheng (UCSF) and and Chuan (River) Xiao (UTEP) for assistance with experimental or data analysis, S. Higgs (Kansas State University) for generous gift of molecular clones of chikungunya reporter virus, and Edgar Davidson and Manu Mabila for additional antibody analysis and preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.J., N.M.L., D-H.C., M.L., J.M.F., R.M.S. M.O.M. and G.S. performed the experiments. J.J., D.C., M.L., J.M.F., M.S.D. and G.S. designed the experiments and analyzed the data. R.H.F. and B.J.D. provided key reagents. S.B., S.K. and M.E.F. provided technique support. M.B.S. made the Fab footprint maps. J.J. and G.S. wrote the manuscript and all authors provided substantive editorial comments.
REFERENCES
- Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, Lewis MG, Higgs S, Rossmann MG, Rao S, et al. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nature medicine. 2010;16:334–338. doi: 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard E, Solignat M, Gay B, Chazal N, Higgs S, Devaux C, Briant L. Endocytosis of chikungunya virus into mammalian cells: role of clathrin and early endosomal compartments. PloS one. 2010;5:e11479. doi: 10.1371/journal.pone.0011479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng RH, Kuhn RJ, Olson NH, Rossmann MG, Choi HK, Smith TJ, Baker TS. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corboba P, Grutadauria S, Cuffini C, Zapata M. Neutralizing monoclonal antibody to the E1 glycoprotein epitope of rubella virus mediates virus arrest in VERO cells. Viral immunology. 2000;13:83–92. doi: 10.1089/vim.2000.13.83. [DOI] [PubMed] [Google Scholar]
- Couderc T, Chretien F, Schilte C, Disson O, Brigitte M, Guivel-Benhassine F, Touret Y, Barau G, Cayet N, Schuffenecker I, et al. A mouse model for Chikungunya: young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS pathogens. 2008;4:e29. doi: 10.1371/journal.ppat.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couderc T, Lecuit M. Chikungunya virus pathogenesis: From bedside to bench. Antiviral research. 2015;121:120–131. doi: 10.1016/j.antiviral.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Diaz-Quinonez JA, Ortiz-Alcantara J, Fragoso-Fonseca DE, Garces-Ayala F, Escobar-Escamilla N, Vazquez-Pichardo M, Nunez-Leon A, Torres-Rodriguez Mde L, Torres-Longoria B, Lopez-Martinez I, et al. Complete genome sequences of chikungunya virus strains isolated in Mexico: first detection of imported and autochthonous cases. Genome announcements. 2015;3 doi: 10.1128/genomeA.00300-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle WR, Downie JC, Laver WG. Inhibition of virus release by antibodies to surface antigens of influenza viruses. Journal of virology. 1974;13:269–275. doi: 10.1128/jvi.13.2.269-275.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll DM, Onuma M, Olson C. Inhibition of bovine leukemia virus release by antiviral antibodies. Archives of virology. 1977;55:139–144. doi: 10.1007/BF01314487. [DOI] [PubMed] [Google Scholar]
- Fischer M, Staples JE Arboviral Diseases Branch, N.C.f.E., and Zoonotic Infectious Diseases, C.D.C. Notes from the field: chikungunya virus spreads in the Americas -Caribbean and South America, 2013–2014. MMWR Morbidity and mortality weekly report. 2014;63:500–501. [PMC free article] [PubMed] [Google Scholar]
- Fong RH, Banik SS, Mattia K, Barnes T, Tucker D, Liss N, Lu K, Selvarajah S, Srinivasan S, Mabila M, et al. Exposure of epitope residues on the outer face of the chikungunya virus envelope trimer determines antibody neutralizing efficacy. Journal of virology. 2014;88:14364–14379. doi: 10.1128/JVI.01943-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsell K, Xing L, Kozlovska T, Cheng RH, Garoff H. Membrane proteins organize a symmetrical virus. The EMBO journal. 2000;19:5081–5091. doi: 10.1093/emboj/19.19.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fric J, Bertin-Maghit S, Wang CI, Nardin A, Warter L. Use of human monoclonal antibodies to treat Chikungunya virus infection. The Journal of infectious diseases. 2013;207:319–322. doi: 10.1093/infdis/jis674. [DOI] [PubMed] [Google Scholar]
- Garoff H, Sjoberg M, Cheng RH. Budding of alphaviruses. Virus research. 2004;106:103–116. doi: 10.1016/j.virusres.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Gibbons DL, Vaney MC, Roussel A, Vigouroux A, Reilly B, Lepault J, Kielian M, Rey FA. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427:320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- Goh LY, Hobson-Peters J, Prow NA, Gardner J, Bielefeldt-Ohmann H, Pyke AT, Suhrbier A, Hall RA. Neutralizing monoclonal antibodies to the E2 protein of chikungunya virus protects against disease in a mouse model. Clinical immunology. 2013;149:487–497. doi: 10.1016/j.clim.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Hawman DW, Stoermer KA, Montgomery SA, Pal P, Oko L, Diamond MS, Morrison TE. Chronic joint disease caused by persistent Chikungunya virus infection is controlled by the adaptive immune response. Journal of virology. 2013;87:13878–13888. doi: 10.1128/JVI.02666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajihara M, Marzi A, Nakayama E, Noda T, Kuroda M, Manzoor R, Matsuno K, Feldmann H, Yoshida R, Kawaoka Y, et al. Inhibition of Marburg virus budding by nonneutralizing antibodies to the envelope glycoprotein. Journal of virology. 2012;86:13467–13474. doi: 10.1128/JVI.01896-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam YW, Lee WW, Simarmata D, Harjanto S, Teng TS, Tolou H, Chow A, Lin RT, Leo YS, Renia L, et al. Longitudinal analysis of the human antibody response to Chikungunya virus infection: implications for serodiagnosis and vaccine development. Journal of virology. 2012a;86:13005–13015. doi: 10.1128/JVI.01780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam YW, Lum FM, Teo TH, Lee WW, Simarmata D, Harjanto S, Chua CL, Chan YF, Wee JK, Chow A, et al. Early neutralizing IgG response to Chikungunya virus in infected patients targets a dominant linear epitope on the E2 glycoprotein. EMBO molecular medicine. 2012b;4:330–343. doi: 10.1002/emmm.201200213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam YW, Simarmata D, Chow A, Her Z, Teng TS, Ong EK, Renia L, Leo YS, Ng LF. Early appearance of neutralizing immunoglobulin G3 antibodies is associated with chikungunya virus clearance and long-term clinical protection. The Journal of infectious diseases. 2012c;205:1147–1154. doi: 10.1093/infdis/jis033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG. Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18950–18955. doi: 10.1073/pnas.1011036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Valadere AM. Transcontinental movement of Asian genotype chikungunya virus. Emerging infectious diseases. 2014;20:1400–1402. doi: 10.3201/eid2008.140268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Wengler G, Rey FA. The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum FM, Teo TH, Lee WW, Kam YW, Renia L, Ng LF. An essential role of antibodies in the control of Chikungunya virus infection. Journal of immunology. 2013;190:6295–6302. doi: 10.4049/jimmunol.1300304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masrinoul P, Puiprom O, Tanaka A, Kuwahara M, Chaichana P, Ikuta K, Ramasoota P, Okabayashi T. Monoclonal antibody targeting chikungunya virus envelope 1 protein inhibits virus release. Virology. 2014;464–465:111–117. doi: 10.1016/j.virol.2014.05.038. [DOI] [PubMed] [Google Scholar]
- Morrison TE, Oko L, Montgomery SA, Whitmore AC, Lotstein AR, Gunn BM, Elmore SA, Heise MT. A mouse model of chikungunya virus-induced musculoskeletal inflammatory disease: evidence of arthritis, tenosynovitis, myositis, and persistence. The American journal of pathology. 2011;178:32–40. doi: 10.1016/j.ajpath.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal P, Dowd KA, Brien JD, Edeling MA, Gorlatov S, Johnson S, Lee I, Akahata W, Nabel GJ, Richter MK, et al. Development of a Highly Protective Combination Monoclonal Antibody Therapy against Chikungunya Virus. PLoS pathogens. 2013;9:e1003312. doi: 10.1371/journal.ppat.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, Lavenir R, Pardigon N, Reynes JM, Pettinelli F, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS medicine. 2006;3:e263. doi: 10.1371/journal.pmed.0030263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarajah S, Sexton NR, Kahle KM, Fong RH, Mattia KA, Gardner J, Lu K, Liss NM, Salvador B, Tucker DF, et al. A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus E2 protects from disease. PLoS neglected tropical diseases. 2013;7:pe2423. doi: 10.1371/journal.pntd.0002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariff DM, Davies J, Desperbasques M, Billstrom M, Geerligs HJ, Welling GW, Welling-Wester S, Buchan A, Skinner GR. Immune inhibition of virus release from human and nonhuman cells by antibody to viral and host cell determinants. Intervirology. 1991;32:28–36. doi: 10.1159/000150182. [DOI] [PubMed] [Google Scholar]
- Smith SA, Silva LA, Fox JM, Flyak AI, Kose N, Sapparapu G, Khomadiak S, Ashbrook AW, Kahle KM, Fong RH, et al. Isolation and Characterization of Broad and Ultrapotent Human Monoclonal Antibodies with Therapeutic Activity against Chikungunya Virus. Cell host & microbe. 2015;18:86–95. doi: 10.1016/j.chom.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TJ, Cheng RH, Olson NH, Peterson P, Chase E, Kuhn RJ, Baker TS. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:10648–10652. doi: 10.1073/pnas.92.23.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soonsawad P, Xing L, Milla E, Espinoza JM, Kawano M, Marko M, Hsieh C, Furukawa H, Kawasaki M, Weerachatyanukul W, et al. Structural evidence of glycoprotein assembly in cellular membrane compartments prior to Alphavirus budding. Journal of virology. 2010;84:11145–11151. doi: 10.1128/JVI.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Xiang Y, Akahata W, Holdaway H, Pal P, Zhang X, Diamond MS, Nabel GJ, Rossmann MG. Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. eLife. 2013;2:e00435. doi: 10.7554/eLife.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng TS, Kam YW, Lee B, Hapuarachchi HC, Wimal A, Ng LC, Ng LF. A Systematic Meta-analysis of Immune Signatures in Patients With Acute Chikungunya Virus Infection. The Journal of infectious diseases. 2015;211:1925–1935. doi: 10.1093/infdis/jiv049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderplasschen A, Hollinshead M, Smith GL. Antibodies against vaccinia virus do not neutralize extracellular enveloped virus but prevent virus release from infected cells and comet formation. The Journal of general virology. 1997;78(Pt 8):2041–2048. doi: 10.1099/0022-1317-78-8-2041. [DOI] [PubMed] [Google Scholar]
- Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- Watson R. Europe witnesses first local transmission of chikungunya fever in Italy. Bmj. 2007;335:532–533. doi: 10.1136/bmj.39332.708738.DB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Rossmann MG. Interpretation of electron density with stereographic roadmap projections. Journal of structural biology. 2007;158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkernagel RM, LaMarre A, Ciurea A, Hunziker L, Ochsenbein AF, McCoy KD, Fehr T, Bachmann MF, Kalinke U, Hengartner H. Neutralizing antiviral antibody responses. Advances in immunology. 2001;79:1–53. doi: 10.1016/S0065-2776(01)79001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.