Summary
Itaconic acid is an important biomass‐derived chemical building block but has also recently been identified as a metabolite produced in mammals, which has antimicrobial activity. The biosynthetic pathway of itaconic acid has been elucidated in the ascomycetous fungus A spergillus terreus and in human macrophages. In both organisms itaconic acid is generated by decarboxylation of the tricarboxylic acid (TCA) cycle intermediate cis‐aconitate. Here, we show that the basidiomycetous fungus U stilago maydis uses an alternative pathway and produces itaconic acid via trans‐aconitate, the thermodynamically favoured isomer of cis‐aconitate. We have identified a gene cluster that contains all genes involved in itaconic acid formation. Trans‐aconitate is generated from cis‐aconitate by a cytosolic aconitate‐Δ‐isomerase (Adi1) that belongs to the PrpF family of proteins involved in bacterial propionate degradation. Decarboxylation of trans‐aconitate is catalyzed by a novel enzyme, trans‐aconitate decarboxylase (Tad1). Tad1 displays significant sequence similarity with bacterial 3‐carboxy‐cis,cis‐muconate lactonizing enzymes (CMLE). This suggests that U . maydis has evolved an alternative biosynthetic pathway for itaconate production using the toxic intermediate trans‐aconitate. Overexpression of a pathway‐specific transcription factor (Ria1) or a mitochondrial tricarboxylic acid transporter (Mtt1) resulted in a twofold increase in itaconate yield. Therefore, our findings offer new strategies for biotechnological production of this valuable biomass‐derived chemical.
Introduction
The unsaturated dicarboxylic acid itaconate (Fig. 1A) is commercially used in the production of pharmaceuticals, adhesives and as a copolymer for synthetic resins. Microbial itaconate production was first described for the ascomycetous fungus Aspergillus itaconicus (Kinoshita, 1932) and the related species Aspergillus terreus is still used for industrial production of itaconate via fermentation (Willke and Vorlop, 2001; Okabe et al., 2009; Klement and Büchs, 2013). Itaconic acid is also an interesting starting material for biofuel production because it can easily be converted into 3‐methyltetrahydrofuran (3‐MTHF), a fuel with excellent physical and chemical combustion properties (Geilen et al., 2010). Therefore, itaconic acid is regarded as one of the ‘top value added chemicals from biomass’ that can be subsequently converted to a number of high‐value chemicals or materials in the industry (Werpy and Petersen, 2004).
Figure 1.
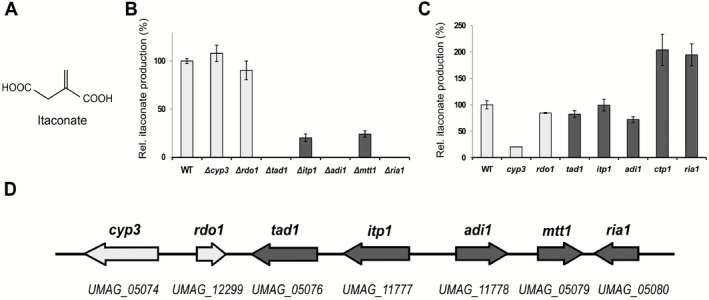
Gene cluster involved in itaconate biosynthesis.
A. Chemical structure of itaconic acid.
B. Deletion analysis of genes involved in itaconate formation in U . maydis. Itaconate levels in the supernatant were determined by HPLC after 96 h of cultivation.
C. Relative itaconate production upon overexpression of individual cluster genes in U . maydis after 68 h in screening medium. Error bars indicate standard deviation of the mean (n = 2).
D. Schematic overview of the itaconate biosynthesis gene cluster of U . maydis. Dark colour indicates genes directly involved in itaconate production and transport.
Quite recently, itaconate production by mammalian macrophages during inflammation has also been observed (Sugimoto et al., 2012; Michelucci et al., 2013; Cordes et al., 2015). There, it serves as an immune‐supportive metabolite that blocks the bacterial glyoxylate shunt by inhibition of isocitrate lyase (McFadden and Purohit, 1977). Numerous human pathogenic bacteria have evolved itaconate degradation and detoxification pathways as counter‐response, emphasizing the medical relevance for itaconate and its biochemistry (Sasikaran et al., 2014). For fungi, itaconate production is likely beneficial in an ecological context through several effects. In general, production of organic acids enables the liberation of micronutrients, such as phosphate and metals through chelating properties of the acids and a decrease of pH (Whitelaw, 1999; Landeweert et al., 2001). This pH decrease, combined with a high tolerance to low pH of the producing organism, provides a competitive advantage in carbon‐rich environments (Cray et al., 2013), complemented by the specific action of itaconate as inhibitor of isocitrate lyase (McFadden and Purohit, 1977).
A biosynthetic route for microbial itaconate formation via enzymatic decarboxylation of the tricarboxylic acid (TCA) cycle intermediate cis‐aconitate has been proposed already in 1957 (Bentley and Thiessen, 1957a, 1957b). The responsible genes, however, were identified only recently. The A. terreus cadA gene encodes a cis‐aconitate decarboxylase, which converts cis‐aconitate into itaconate (Dwiarti et al., 2002; Kanamasa et al., 2008). The cadA gene is part of a gene cluster that also comprises a mitochondrial TCA transporter, a membrane permease and a transcription factor proposed to regulate pathway‐specific expression of cluster genes (Li et al., 2011). The biochemical elucidation of itaconate production, including its genetic background, has led to a surge of metabolic engineering efforts in recent years with itaconate production established in several heterologous hosts (Kanamasa et al., 2008; Li et al., 2012; Blumhoff et al., 2013; Klement and Büchs, 2013; Blazeck et al., 2014; Huang et al., 2014; Okamoto et al., 2014; Chin et al., 2015; Vuoristo et al., 2015).
Itaconate production has also been observed in the phytopathogenic basidiomycete Ustilago maydis in a range of up to 45 g l−1 (Haskins et al., 1955; Guevarra and Tabuchi, 1990; Klement and Büchs, 2013; Maassen et al., 2014). Ustilago maydis serves as an important model organism due to its well‐characterized genome to study fungal virulence in plants (Kaemper et al., 2006; Brefort et al., 2009). In addition, the single‐celled (yeast‐like) morphology of this fungus in liquid media provides a major advantage in submerged fermentations by circumventing several problems of filamentous fungi, such as elevated viscosity and clogging, hindered oxygen transfer, sensitivity to hydro‐mechanical stress and laborious handling of spores (Klement et al., 2012). Therefore, we sought to investigate the itaconate biosynthesis pathway in U. maydis.
Results
Identification of an itaconate biosynthesis gene cluster in U . maydis
Genome analysis revealed that U. maydis contains no orthologue of the A. terreus cis‐aconitate decarboxylase CadA, which belongs to the PrpD family of proteins. This suggests that U. maydis uses an alternative biosynthetic route for itaconic acid production. We noticed that the U. maydis genome contains three genes (UMAG_02807, UMAG_06058 and UMAG_11778 ) coding for PrpF family proteins related to the 3‐methylitaconate‐Δ‐isomerase Mii. Mii converts methylitaconate into dimethylmaleate during nicotinate fermentation by Eubacterium barkeri and also shows activity towards itaconate (Velarde et al., 2009). Gene disruption experiments revealed that deletion of UMAG_11778 abolished itaconate production (Fig. 1B and Supporting Information Fig. S1), pointing to an important role for this gene during itaconate biosynthesis. Interestingly, several neighbouring genes are co‐regulated with UMAG_11778 during pathogenic development of U. maydis (Zheng et al., 2008). To test whether these genes also contribute to itaconate biosynthesis, we performed a systematic deletion analysis of the putative gene cluster (Fig. 1D). Itaconate production was completely abolished in mutants disrupted for UMAG_05076, UMAG_11778 or UMAG_05080, whereas deletion of UMAG_11777 and UMAG_05079 resulted in decreased itaconate production (Fig. 1B). Deletion of the two leftmost genes of the potential gene cluster (UMAG_05074 and UMAG_12299) did not affect itaconate production significantly (Fig. 1B), although they appear to be co‐regulated in planta (Zheng et al., 2008). The fully annotated sequence of the cluster is deposited in NCBI GenBank under accession number (NCBI GenBank KT852988).
Identification of the itaconate pathway‐specific regulator Ria1
Under itaconate producing conditions, transcription of all potential cluster genes with exception of UMAG_05074 and UMAG_12299 was at least 3.5‐fold induced (Supporting Information Fig. S2). UMAG_05074 encodes a P450 monooxygenase of the CYP504 family and UMAG_12299 a ring cleaving dioxygenase (Table 1). Therefore, we named the respective genes cyp3 and rdo1. UMAG_05080 is predicted to encode a basic helix–loop–helix (bHLH) transcription factor (Table 1). Deletion of UMAG_05080 resulted in significant reduction of expression (UMAG_05079 90%, UMAG_11778 85%, UMAG_11777 70%, UMAG_05076, UMAG_05074 and UMAG_12299 each 30%), whereas the overexpression of UMAG_05080 was sufficient to trigger expression of most cluster genes (Supporting Information Fig. S3). Therefore, we assume that UMAG_05080 encodes a pathway‐specific regulator of itaconate biosynthesis and renamed the corresponding gene ria1 (regulator of itaconic acid).
Table 1.
Itaconic acid biosynthesis gene cluster in U . maydis.
| Gene (locus tag) | Putative function | Size (aa) | Closest homologue with known function or structure | E‐value | Signature pattern |
|---|---|---|---|---|---|
|
cyp3
(UMAG_05074) |
Hydroxylation of itaconate | 540 | Phenylacetate hydroxylase (CYP504 family) A. nidulans (Mingot et al., 1999) | 9e‐121 | P450 Monooxygenase |
|
rdo1
(UMAG_12299) |
unknown | 180 | Macrophage colonization factor Salmonella enterica (Shi et al., 2006) | 1e‐37 | Ring‐cleaving dioxygenase |
|
tad1
(UMAG_05076) |
Decarboxylation of trans‐aconitate | 493 | 3‐Carboxy‐cis,cis‐muconate lactonizing enzyme Pseudomonas putida (Yang et al., 2004) | 3e‐101 | 3‐carboxy‐cis,cis‐muconate lactonizing enzyme (pCMLE) |
|
itp1
(UMAG_11777) |
Export of itaconate | 491 | EmrD multi drug resistance E. coli (Yin et al., 2006) | 2e‐11 | Major facilitator superfamily (MFS) |
|
adi1
(UMAG_11778) |
Isomerization of aconitate | 443 | Oxalo‐mesaconate tautomerase Pseudomonas putida (Nogales et al., 2011) | 5e‐72 | PrpF‐superfamily |
|
mtt1
(UMAG_05079) |
Mitochondrial cis‐aconitate export | 300 | Citrate transport protein S. cerevisiae (Kaplan et al., 1995) | 3e‐80 | Mitochondrial carrier protein |
|
ria1
(UMAG_05080) |
Regulator of itaconate biosynthesis genes | 362 | Upstream stimulatory factor 2 (USF2) (Gregor et al., 1990) | 0.003 | Helix–loop–helix transcriptional regulator |
In vivo characterization of itaconate biosynthesis genes
Two of the genes essential for itaconate biosynthesis (Fig. 1B) code for proteins with similarity to known enzymes (Table 1). The gene product of UMAG_05076 is highly similar to prokaryotic 3‐carboxy‐cis,cis‐muconate lactonizing enzyme (CMLE) and the product of UMAG_11778 belongs to the PrpF family of isomerases (Grimek and Escalante‐Semerena, 2004; Velarde et al., 2009; Nogales et al., 2011) (see Table 1). To test whether these two genes are directly responsible for itaconate biosynthesis, we expressed both in Saccharomyces cerevisiae. Although single expression of either gene did not trigger itaconate production, co‐expression of UMAG_05076 and UMAG_11778 resulted in itaconate production in this heterologous host (Fig. 2).
Figure 2.
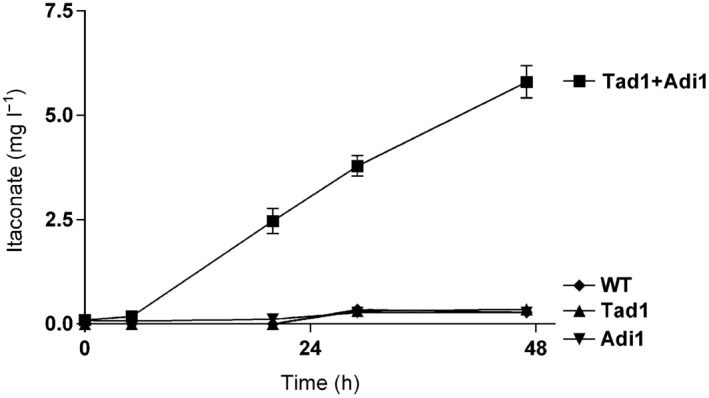
Heterologous itaconate production by S . cerevisiae expressing U . maydis' Adi1 and Tad1. Graph shows itaconate production over time by expression of the U . maydis itaconate biosynthetic enzymes Adi1 and Tad1 under control of the GAL4 promoter in the heterologous host S . cerevisiae in inducing medium (2% (w/v) galactose). Values are the arithmetic mean of two biological determinations. Error bars indicate standard deviation of the mean.
To elucidate the biosynthetic route of itaconate production in U. maydis, permeabilized cell assays were performed. Remarkably, itaconate formation was observed upon incubation of permeabilized cells with trans‐aconitate but not with cis‐aconitate, citrate or isocitrate (Fig. 3). Itaconate production from trans‐aconitate was not observed in ΔUMAG_05076 cells, indicating that the catalytic activity of UMAG_05076 is required for itaconate formation.
Figure 3.
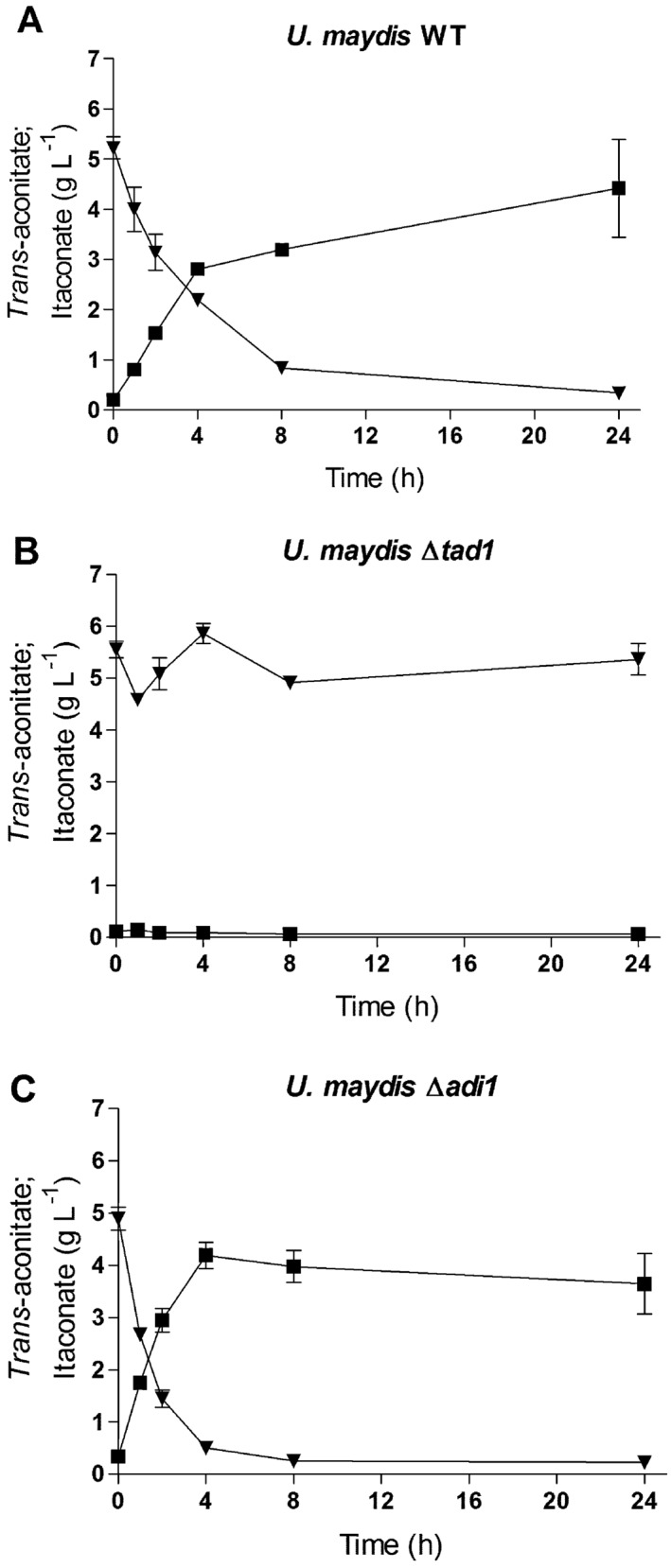
Permeabilized cell assay. Conversion of trans‐aconitate (▲) to itaconate (■) by permeabilized U . maydis cells. WT and mutant cells were incubated for indicated times after addition of 5 g l−1 of trans‐aconitate, (A) WT, (B) Δtad1 and (C) Δadi1. Error bars indicate standard deviation of the mean (n = 2).
In vitro characterization of trans‐aconitate decarboxylase (UMAG_05076) and aconitate‐Δ‐isomerase (UMAG_11778)
In order to determine the catalytic activity of UMAG_05076, we heterologously expressed it as GST fusion protein in E. coli and tested the activity of the purified protein with different substrates in vitro. UMAG_05076 was able to convert trans‐aconitate completely into itaconate within 15 min (Fig. 4A), whereas cis‐aconitate was not converted (Fig. 4B). This indicates that the gene product of UMAG_05076 acts as a decarboxylase specific for trans‐aconitate (Fig. 4C). Therefore, we termed the enzyme trans‐aconitate decarboxylase (Tad1) and renamed the corresponding gene tad1. To our knowledge, such enzymatic activity has not been described yet.
Figure 4.
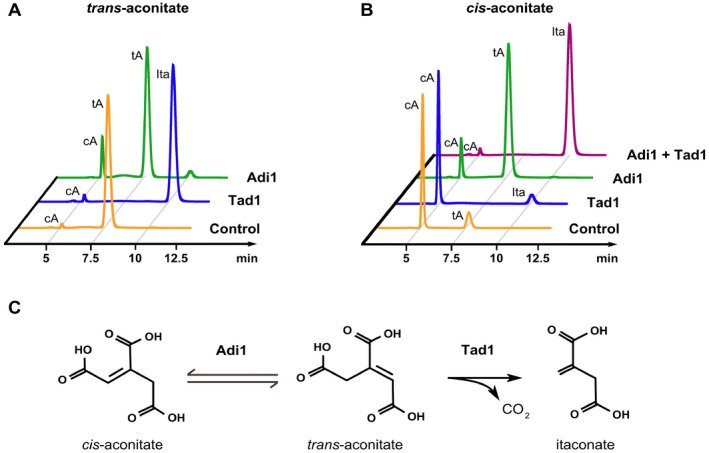
Enzymatic activities of Adi1 and Tad1.
A. Conversion of trans‐aconitate (tA) into itaconate (Ita) or cis‐aconitate (cA) by purified Tad1 or Adi1.
B. Conversion of cis‐aconitate into trans‐aconitate or itaconate by purified Tad1 and/or Adi1. Shown are HPLC chromatograms measured at 210 nm after 15 min of incubation with the indicated enzyme at room temperature.
C. Schematic description of the itaconate biosynthesis pathway in U . maydis. Adi1 catalyzes isomerization of cis‐aconitate and trans‐aconitate. Trans‐aconitate is decarboxylated by Tad1 giving rise to itaconate.
Trans‐aconitate is not a regular intermediate of cellular metabolism but acts as competitive inhibitor of the TCA cycle enzymes aconitase and fumarase (Saffran and Prado, 1949). We reasoned that during itaconate formation, trans‐aconitate might be generated from cis‐aconitate by the gene product of UMAG_11778. Indeed, recombinant GST‐UMAG_11778 fusion protein purified from E. coli was able to isomerize cis‐aconitate into trans‐aconitate in vitro (Fig. 4B). The enzyme catalyzed interconversion of cis‐aconitate and trans‐aconitate, resulting in equilibrium formation of 88% trans to 12% cis. Because we propose that cis/trans‐isomerization occurs via an allylic re‐arrangement (see below), we termed this enzyme aconitate‐Δ‐isomerase (Adi1) and renamed the respective gene adi1.
We were able to reconstitute the complete transformation of cis‐aconitate into itaconate in vitro upon incubation of cis‐aconitate with both Adi1 and Tad1 (Fig. 4B). This indicates that the coupled reaction appears to be irreversible. This may help to prevent any deleterious accumulation of the toxic intermediate trans‐aconitate during itaconate biosynthesis.
Intracellular localization of the itaconate biosynthesis proteins
To determine the intracellular localization of the enzymes involved in itaconate biosynthesis, we followed green fluorescent protein (GFP) fusion proteins by fluorescence microscopy. Both Adi1 and Tad1 were located in the cytosol but were also found in the nucleus (Fig. 5A). The gene product of UMAG_05079 is similar to mitochondrial TCA transporters (Table 1) and accumulated at distinct locations in mitochondria (Fig. 5B). UMAG_11777 is annotated as major facilitator superfamily (MFS) transporter (Table 1) and was found at the plasma membrane. Based on these data, we propose that the gene product of UMAG_05079 actively exports cis‐aconitate from mitochondria into the cytosol. There it is converted into itaconate and secreted across the plasma membrane by the major facilitator UMAG_11777 (Fig. 6). Accordingly, we termed these transport proteins Mtt1 (mitochondrial TCA transporter) and Itp1 (itaconate transport protein) respectively. Taken together, our results demonstrate that U. maydis uses an alternative metabolic pathway to generate itaconate via the unusual intermediate trans‐aconitate.
Figure 5.
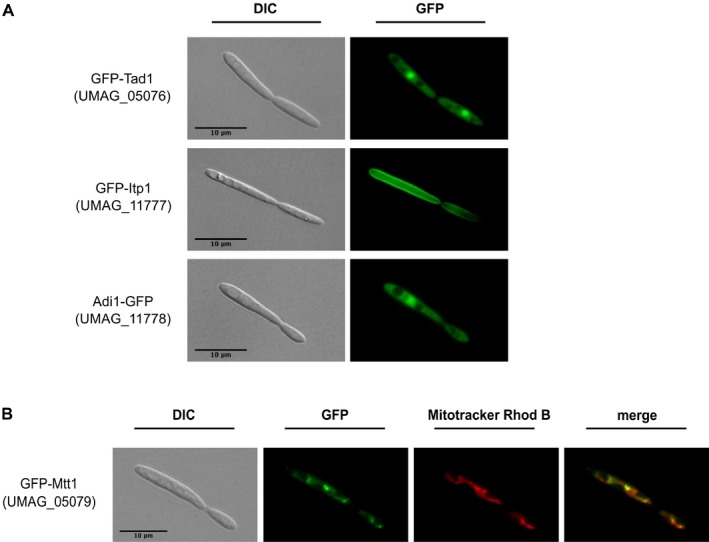
Intracellular localization of itaconate biosynthetic enzymes.
A. Indicated GFP‐fusion proteins were analyzed by fluorescence microscopy.
B. Mitochondrial localization of Mtt1 was confirmed by co‐localization with Mitotracker Rhodamine B (Mitotracker Rhod B). Scale bar = 10 μm.
Figure 6.
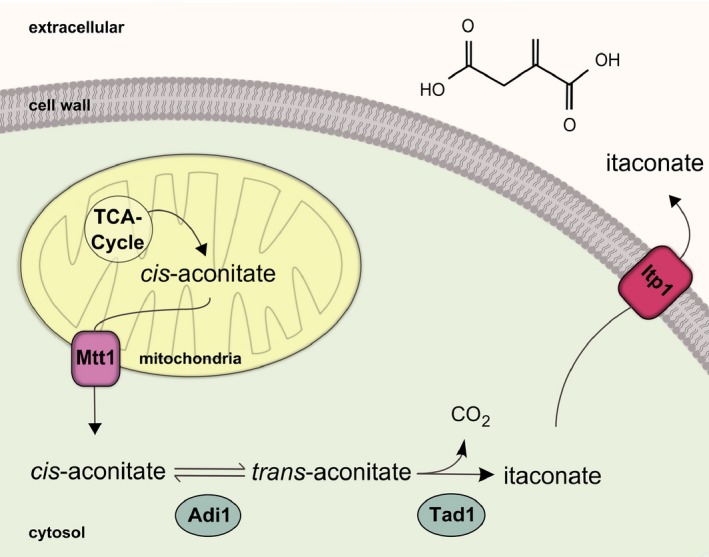
Proposed intracellular organization of the itaconate biosynthesis pathway in U . maydis. Cis‐aconitate is secreted by the mitochondrial tricarboxylate transporter Mtt1. In the cytosol cis‐aconitate is converted into itaconate via the intermediate trans‐aconitate. Secretion of itaconate into the medium is mediated by the major facilitator Itp1.
Improvement of itaconate production by overexpression of itaconate cluster genes
To test whether this newly gained knowledge of itaconate biosynthesis can be used for metabolic engineering of U. maydis, we overexpressed all cluster genes individually. More than a twofold increase in itaconate production was observed upon overexpression of either the pathway‐specific transcription factor Ria1, or the mitochondrial transporter Mtt1 (Fig. 1C). Surprisingly, overexpression of the P450 monooxygenase Cyp3 (Fig. 1C) caused a significant decrease in itaconate production to 20% of the wildtype level, most likely caused by P450‐dependent oxidation of itaconate (see Discussion).
Discussion
So far, the molecular basis of itaconate biosynthesis has been elucidated only in the ascomycetous fungus A. terreus and in human macrophages (Kanamasa et al., 2008; Li et al., 2011; Michelucci et al., 2013). In both organisms, itaconate is generated by decarboxylation of cis‐aconitate. In A. terreus, the key enzyme CadA is part of a gene cluster that also contains mitochondrial and plasma membrane transporters and a transcription factor (Li et al., 2011). Here, we could show that in the basidiomycete U. maydis itaconate is produced by decarboxylation of trans‐aconitate. The critical enzyme Tad1 is remarkably different to A. terreus CadA. While CadA is related to pro‐ and eukaryotic methylcitrate‐dehydratases of the PrpD family, which catalyze an unusual syn‐elimination of water from (2S,3S)‐2‐methylcitrate, U. maydis Tad1 is highly similar to prokaryotic CMLE. Prokaryotic CMLE catalyze γ‐lactonization of 3‐carboxy‐cis,cis‐muconate during degradation of aromatic compounds (Harwood and Parales, 1996) via an anti‐1,2‐addition‐elimination reaction similar to that of class II fumarases (Williams et al., 1992). Therefore, we propose that Tad1 catalyzes decarboxylation of trans‐aconitate by addition of a proton to the double bond leading to transient formation of a carbenium ion and subsequent anti‐elimination of the carboxyl group (Supporting Information Fig. S4).
In U. maydis, trans‐aconitate is produced by isomerization of the TCA intermediate cis‐aconitate. We identified the aconitate‐Δ‐isomerase Adi1, which catalyzes interconversion of cis‐ and trans‐aconitate. Such isomerase activity has been described previously for plants that accumulate trans‐aconitate as antifeedant (Thompson et al., 1990) and for bacteria that are able to grow on trans‐aconitate (Altekar and Raghaven Rao, 1963). Although the responsible proteins have not been identified yet, biochemical studies in Pseudomonas putida indicated that isomerization proceeds via an allylic rearrangement (Klinman and Rose, 1971). The U. maydis aconitate isomerase Adi1 is closely related to methylitaconate‐Δ‐isomerase Mii from E. barkeri (Velarde et al., 2009) and to Shewanella oneidensis PrpF (Grimek and Escalante‐Semerena, 2004). For both enzymes, allylic mechanisms have been proposed. Therefore, we assume that Adi1 uses a similar mechanism to catalyse delta‐isomerization of cis‐aconitate into trans‐aconitate.
Overexpression of the P450 monooxygenase Cyp3 led to a drastic decrease of itaconate production, whereas deletion of cyp3 resulted in a slight but not significant increase (P = 0.08). This suggests that itaconate might be a substrate for oxidation by Cyp3. We propose that Cyp3‐dependent oxidation results in the formation of 2‐hydroxyparaconate, which is a known byproduct of itaconate production in Ustilago species (Guevarra and Tabuchi, 1990). The presence of 2‐hydroxyparaconate, however, could not be confirmed yet because standards are not commercially available. The exact action and role of Cyp3 in relation to itaconate production will be studied in more detail in future experiments.
Overexpression of itaconate cluster genes mtt1 and ria1 resulted in significant improvement of itaconate yield. The effect of ria1 is most probably due to direct upregulation of all cluster genes including mtt1 (see Supporting Information Fig. S3). Together, this suggests that the putative mitochondrial cis‐aconitate transporter Mtt1 appears to be a bottleneck for itaconate production in U. maydis. There is an interesting analogy to fungal citrate production, where the ability of the mitochondrial citrate transporter to compete with the TCA cycle enzyme aconitase is critical for efficient citrate production (Karaffa and Kubicek, 2003).
In this manuscript, we describe a novel and alternative pathway for itaconate biosynthesis in U. maydis involving isomerization of cis‐aconitate into trans‐aconitate and its subsequent decarboxylation to generate itaconate. The molecular identification of Tad1 and Adi1 allows further biochemical and structural characterization of these interesting enzyme activities. In addition, our findings open up new avenues for metabolic engineering of homologous and heterologous itaconate production strains. Initial overexpression experiments already led to a twofold increase in itaconate production. Combinatorial overexpression of multiple genes will likely lead to further improvements, enabling to use U. maydis for sustainable large‐scale itaconate production.
Experimental procedures
Strains and culture conditions
Escherichia coli strain Top10 (Invitrogen) was used for all DNA manipulations. Cells were grown in dYT liquid medium (1% (w/v) yeast extract, 1.6% (w/v) tryptone, 0.5% (w/v) NaCl) at 37°C and maintained on dYT‐agar plates (dYT components + 1.3% (w/v) agar) at 4°C. Ustilago maydis strain MB215 (DSM17144) was used for all experiments. U. maydis cells were grown in YEPS medium (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) sucrose) at 30°C and maintained on potato dextrose agar (PDA, Difco) at 4°C.
Generation of deletion and overexpression strains
Deletion of single genes was performed by replacing the complete open reading frame (ORF) with a hygromycin resistance marker derived from plasmid pMF1‐H (Brachmann et al., 2004) by restriction with SfiI. Briefly, 1 kb recombination flanks were generated by polymerase chain reaction (PCR) using the primers listed in Supporting Information Table S1. Flanks, hygromycin‐cassette and vector pRS426 (Sikorski and Hieter, 1989) were assembled using homologous recombination in S. cerevisiae. Prior to transformation of U. maydis, the derived plasmid was restricted with SspI to isolate the deletion construct. Plasmid p123, which carries the strong constitutive otef promoter (Spellig et al., 1996) was used for generation of overexpression constructs by PCR and conventional cloning using primers indicated in Supporting Information Table S2. Plasmids were linearized with SspI prior to U. maydis transformation and all constructs were integrated into the ip‐locus by homologous recombination (Loubradou et al., 2001). Transformation of U. maydis was performed using standard protocols (Schulz et al., 1990). For selection of transformants, PDA plates with 200 μg ml−1 of hygromycin (Duchefa Biochemie) or 2 μg ml−1 of carboxin (Sigma‐Aldrich) were used. Correct integration of constructs was verified by Southern analysis or by PCR. For all deletion mutants itaconate production could be restored by complementation with the respective genes in trans, with the exception of deletion mutant UMAG_05080. This might be possibly related to lack of expression of this transcriptional regulator due to the ectopic insertion at the ip‐locus. For localization studies GFP‐fusion proteins generated in plasmid p123 were overexpressed in the respective deletion strains.
Itaconate production procedure
Itaconate production cultures with U. maydis strains were performed in the System Duetz® (24 well plates) with a filling volume of 1.5 ml (shaking diameter = 50 mm, agitation speed = 300 r.p.m., temperature = 30°C, and relative air humidity = 80%) (Duetz et al., 2000). The screening medium contained 50 g l−1 of glucose, 0.8 g l−1 of NH4Cl, 0.2 g l−1 of MgSO4·7H2O, 0.01 g l−1 of FeSO4·7H2O, 0.5 g l−1 of KH2PO4, 33 g l−1, 1 ml l−1 of vitamin solution, 10 ml l−1 of trace element solution and as buffer 19.5 g l−1 2‐(N‐morpholino)ethanesulfonic acid (MES) (Geiser et al., 2014). The pH of the MES stock solution was adjusted to 6.5 with NaOH. The vitamin solution contained (per litre) 0.05 g of D‐biotin, 1 g of D‐calcium panthotenate, 1 g of nicotinic acid, 25 g of myo‐inositol, 1 g of thiamine hydrochloride, 1 g of pyridoxol hydrochloride, and 0.2 g of para‐aminobenzoic acid. The trace element solution contained (per litre) 1.5 g of EDTA, 0.45 g of ZnSO4·7H2O, 0.10 g of MnCl2·4H2O, 0.03 g of CoCl2·6H2O, 0.03 g of CuSO4·5H2O, 0.04 g of Na2MoO4·2H2O, 0.45 g of CaCl2·2H2O, 0.3 g of FeSO4·7H2O, 0.10 g of H3BO3, and 0.01 g of KI.
Itaconate, citrate/isocitrate, cis‐aconitate and trans‐aconitate in the supernatants were analysed in a Beckmann Coulter System Gold High Performance Liquid Chromatography (Beckmann Coulter GmbH, Germany) with an Organic Acid Resin 300 × 8 mm column (CS‐Chromatography, Germany) and a differential refractometer LCD 201 (MELZ, Germany). As solvent, 5 mM H2SO4 with a flow rate of 0.6 ml min−1 and a temperature of 30°C was used. All samples were filtered with Rotilabo® syringe filters (CA, 0.20 μm, Ø 15 mm) and afterwards diluted 1:5 with 5 mM H2SO4. All relative components were identified via retention time and Ultraviolet (UV)/Refractive index (RI) quotient compared with corresponding standards. All values are the arithmetic mean of at least two biological replicates. Error bars indicate the deviation from the mean. Statistical analysis of significant difference was performed using Welch's test (level of significance of P ≤ 0.01).
Permeabilized cell assay with U . maydis cells
The U. maydis strains were cultivated in 50 ml of screening medium in 500 ml Erlenmeyer flasks. Cells were grown for 24 h at 30°C and 250 r.p.m. Ten millilitres of culture broth of each strain was centrifuged (2200 × g, 5 min) and washed twice with 0.9% (w/v) sterile NaCl solution. Cells were re‐suspended in screening medium without carbon source. For permeabilization, cells were frozen and thawed twice. After addition of different substrate solutions (trans‐aconitate, cis‐aconitate, citrate and isocitrate) to a final concentration of 5 g l−1, samples of 900 μl were taken at different time points during incubation at 30°C. The enzymatic reaction was stopped by adding 100 μl of concentrated H2SO4. Substrate and product concentrations in the supernatants were analysed via high‐performance liquid chromatography (HPLC) as described before.
Recombinant protein expression
Overexpression of GST fusion proteins UMAG_05076 (Tad1) and UMAG_11778 (Adi1) was performed in E. coli strain Rosetta 2 (DE3) (Novagen, Madison, WI, USA) using plasmids derived from pGEX4T‐1 (GE Healthcare, Waukesha, WI, USA). Plasmids were constructed via PCR and conventional cloning using primers listed in Supporting Information Table S3 and S4. Cells were pre‐cultivated in 200 ml of dYT medium to an OD600 of 0.6 at 37°C and protein production was induced by addition of 0.1 mM IPTG followed by overnight cultivation at 22°C. Cells were harvested by centrifugation, resuspended in lysis buffer (50 mM Tris‐HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, 1 mM PMSF) and lysed three times in a French pressure cell (max. 10.000 psi). After ultra‐centrifugation (1 h, 115 000 × g, 4°C), the supernatant was loaded on 500 μl of GSH Sepharose beads (Protino Glutathione Agarose 4B, Macherey‐Nagel) prior equilibrated with lysis buffer and incubated rolling at 4°C for 1 h. Beads were washed five times with lysis buffer (2 min, 700 × g, 4°C) and then eluted in 700 μl of elution buffer 30 min rolling at 4°C (100 mM Tris‐HCl, pH 9.0, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT and 50 mM reduced glutathione). Protein concentration was determined using the Bradford test (Bradford, 1976).
Enzyme activity assay
Ten micrograms of the corresponding purified protein was incubated with substrate (5 mM) for 15 min at room temperature in assay‐buffer (50 mM Tris‐HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT) in a final volume of 100 μl in a microtitreplate. The reaction was stopped by freezing samples at −20°C. Five microlitres reaction products were analysed by HPLC (Agilent Technologies 1200 Infinity series) at room temperature (22°C) with the following conditions. Column: organic acid resin, 250 × 8 mm (CS Chromatographie); detector: VWD 1260 (Agilent Technologies) at 210 nm; solvent: 2.5 mM sulfuric acid; flow rate: 1.0 ml min–1.
Expression of itaconic acid biosynthesis genes in S . cerevisiae
Plasmids were constructed using the pGREG‐series published by Jansen et al., 2005 and the drag&drop‐cloning method described therein (Jansen et al., 2005) using primers listed in Supporting Information Table S5. Yeast strain ESM356 was used as host (Pereira et al., 2001). Transformants were pre‐cultured at 30°C and 250 r.p.m. overnight in 10 ml of SC dropout medium complemented with 2% (w/v) galactose. Main culture contained 50 ml of SC dropout medium complemented with 2% (w/v) galactose and cells from pre‐culture were diluted to a final OD600 of 0.5. Cultures were incubated for 48 h at 30°C and 250 r.p.m.. Itaconic acid production in the supernatant was detected by HPLC as previously described.
RNA preparation from U . maydis, cDNA synthesis and reverse transcription polymerase chain reaction (qRT‐PCR)
Cells were grown under various conditions to determine gene expression. For itaconate‐producing conditions, cells were diluted from a pre‐culture to a final OD600 of 0.5 in screening medium (Geiser et al., 2014). After 12 h cells were harvested and diluted to a final OD600 of 0.8. Cells containing constructs with the arabinose‐inducible crg‐promoter were diluted from a pre‐culture to a final OD600 of 0.1 in minimal medium containing either 2% (w/v) glucose (control) or 2% (w/v) arabinose (induced) and harvested after 6 h. Cells grown in rich medium (YEPS) were diluted from a pre‐culture to a final OD600 of 0.1 and incubated for 6 h. All cultures were incubated at 30°C and 150 r.p.m. Cells were frozen in liquid nitrogen immediately after harvest.
For RNA extraction, cells were thawed and extracted using TRIzol (Chomczynski and Sacchi, 1987). After extraction, first‐strand cDNA synthesis kit (Thermo Scientific) was used to reverse‐transcribe 1 μg of total RNA to cDNA with oligo(dT) Primers. The amount of produced cDNA was determined using NanoDrop ND‐1000 (PEQLab). The qRT‐PCR analysis was performed using an iCycler (Bio‐Rad) using primers listed in Supporting Information Table S6. For the reaction Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific) was used as recommended in the protocol. Gene expression levels were calculated relative to ppi expression levels using the cycle threshold (CT) 2‐ΔCt method (Livak and Schmittgen, 2001).
Average expression and standard error of three independent experiments were calculated, and expression of the control (WT in YEPS) was set to 1. P‐values were estimated using an unpaired Student's t‐test. P‐values: *P < 0.05; **P < 0.01; ***P < 0.001.
Microscopy
For microscopy, cells were incubated shaking overnight in 10 ml of YEPS at 30°C. Cells from pre‐culture were washed three times in water and diluted to a final OD600 of 0.2 in low fluorescent medium (YNB + 2% (w/v) glucose + 0.5% (w/v) (NH4)2SO4). Cells were grown to OD600 = 0.8 at 30°C and 150 r.p.m. For mitochondria staining 1 ml of cells at OD600 of 0.8 were harvested and re‐suspended in 10 mM Hepes pH 7.4 + 2% (w/v) glucose. Mitotracker Rhodamine B (1 μM in dimethylsulphoxide) was added to a final concentration of 0.1 μM and cells were incubated for 30 min at 30°C. Afterwards cells were washed three times in 10 mM Hepes pH 7.4. Cells were placed on agarose cushions and visualized by DIC and epifluorescence microscopy (GFP: 395 nm; Rhodamine: 500 nm) using a Zeiss Axiovert 200 microscope. Images were taken using a CCD camera (Hamamatsu Orca‐ER) with an exposure time of 30–300 ms. Image acquisition was performed using improvision volocity software and processing was done using imagej.
Conflict of Interest
All authors have seen and approved the manuscript. All authors have contributed significantly to the work, and the manuscript has not been published and is not being considered for publication elsewhere. All authors declare that they have no competing financial interests.
Author Contributions
E.G. and S.K.P. contributed equally to this manuscript. N.W., L.M.B., W.B. and M.B. conceived the project. N.W., L.M.B. and M.B. designed experiments and analysed results. M.B., S.K.P. and N.W. wrote the manuscript with the help of E.G. and L.M.B. S.K.P. generated U. maydis, S. cerevisiae and E. coli strains and performed enzyme activity assays and fluorescence microscopy. E.G. performed cultivation experiments, permeabilized cell assays and analysed itaconic acid production. A.F. and S.K.P. performed expression analysis of cluster genes.
Supporting information
Fig. S1. A PrpF‐like protein is involved in itaconate formation.
Fig. S2. Expression of cluster genes depends on the pathway‐specific transcription factor Ria1.
Fig. S3. Induction of cluster genes by overexpression of Ria1.
Fig. S4. Hypothetical reaction mechanism of Tad1.
Table S1. Oligonucleotides for U. maydis gene deletion constructs.
Table S2. Oligonucleotides for U. maydis overexpression constructs.
Table S3. Oligonucleotides for recombinant protein expression in E. coli.
Table S4. Oligonucleotides for intron removal of UMAG_11778.
Table S5. Oligonucleotides for gene expression in S. cerevisiae.
Table S6. Oligonucleotides for quantitative RT‐PCR.
Acknowledgements
We thank Marco Hornung (SYNMIKRO, Marburg) for excellent technical assistance, Björn Sandrock (Philipps‐University Marburg) for contributions to strain construction and Vincent Wiebach (RWTH Aachen University) for contributions to strain characterization.
Funding Information This work was supported by SFB 987 ‘Microbial Diversity in Environmental Signal Response’ (M.B., S.K.P. and A.F.) and the Excellence Initiative of the German federal and state governments as a part of the Cluster of Excellence ‘Tailor‐Made Fuels from Biomass’ (TMFB, E.G.).
References
- Altekar, W.W. , and Raghavendra Rao, M.R. (1963) Microbiological dissimilation of tricarballylate and trans‐aconitate. J Bacteriol 85: 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, R. , and Thiessen, C.P. (1957a) Biosynthesis of itaconic acid in Aspergillus terreus I Tracer studies with C‐14‐labeled substrates. J Biol Chem 226: 673–687. [PubMed] [Google Scholar]
- Bentley, R. , and Thiessen, C.P. (1957b) Biosynthesis of itaconic acid in Aspergillus terreus. III The properties and reaction mechanism of cis‐aconitic acid decarboxylase. J Biol Chem 226: 703–720. [PubMed] [Google Scholar]
- Blazeck, J. , Miller, J. , Pan, A. , Gengler, J. , Holden, C. , Jamoussi, M. , and Alper, H.S. (2014) Metabolic engineering of Saccharomyces cerevisiae for itaconic acid production. Appl Microbiol Biotechnol 98: 8155–8164. [DOI] [PubMed] [Google Scholar]
- Blumhoff, M.L. , Steiger, M.G. , Mattanovich, D. , and Sauer, M. (2013) Targeting enzymes to the right compartment: metabolic engineering for itaconic acid production by Aspergillus niger . Metab Eng 19: 26–32. [DOI] [PubMed] [Google Scholar]
- Brachmann, A. , Konig, J. , Julius, C. , and Feldbrugge, M. (2004) A reverse genetic approach for generating gene replacement mutants in Ustilago maydis . Mol Genet Genomics 272: 216–226. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Brefort, T. , Doehlemann, G. , Mendoza‐Mendoza, A. , Reissmann, S. , Djamei, A. , and Kahmann, R. (2009) Ustilago maydis as a pathogen. Annu Rev Phytopathol 47: 423–445. [DOI] [PubMed] [Google Scholar]
- Chin, T. , Sano, M. , Takahashi, T. , Ohara, H. , and Aso, Y. (2015) Photosynthetic production of itaconic acid in Synechocystis sp PCC6803. J Biotechnol 195: 43–45. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P. , and Sacchi, N. (1987) Single‐step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal Biochem 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Cordes, T. , Michelucci, A. , and Hiller, K. (2015) Itaconic acid: the surprising role of an industrial compound as a mammalian antimicrobial metabolite. Annu Rev Nutr 35: 451–473. [DOI] [PubMed] [Google Scholar]
- Cray, J.A. , Bell, A.N.W. , Bhaganna, P. , Mswaka, A.Y. , Timson, D.J. , and Hallsworth, J.E. (2013) The biology of habitat dominance; can microbes behave as weeds? Microb Biotechnol 6: 453–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duetz, W.A. , Ruedi, L. , Hermann, R. , O'Connor, K. , Buchs, J. , and Witholt, B. (2000) Methods for intense aeration, growth, storage, and replication of bacterial strains in microtiter plates. Appl Environ Microbiol 66: 2641–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwiarti, L. , Yamane, K. , Yamatani, H. , Kahar, P. , and Okabe, M. (2002) Purification and characterization of cis‐aconitic acid decarboxylase from Aspergillus terreus TN484‐M1. J Biosci Bioeng 94: 29–33. [DOI] [PubMed] [Google Scholar]
- Geilen, F.M.A. , Engendahl, B. , Harwardt, A. , Marquardt, W. , Klankermayer, J. , and Leitner, W. (2010) Selective and flexible transformation of biomass‐derived platform chemicals by a multifunctional catalytic system. Angew Chem 49: 5510–5514. [DOI] [PubMed] [Google Scholar]
- Geiser, E. , Wiebach, V. , Wierckx, N. , and Blank, L.M. (2014) Prospecting the biodiversity of the fungal family Ustilaginaceae for the production of value‐added chemicals. BMC Fungal Biol Biotechnol 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor, P.D. , Sawadogo, M. , and Roeder, R.G. (1990) The adenovirus major late transcription factor USF is a member of the helix–loop–helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev 4: 1730–1740. [DOI] [PubMed] [Google Scholar]
- Grimek, T.L. , and Escalante‐Semerena, J.C. (2004) The acnD genes of Shewenella oneidensis and Vibrio cholerae encode a new Fe/S‐dependent 2‐methylcitrate dehydratase enzyme that requires prpF function in vivo . J Bacteriol 186: 454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevarra, E.D. , and Tabuchi, T. (1990) Accumulation of itaconic, 2‐hydroxyparaconic, itatartaric, and malic‐acids by strains of the genus Ustilago . Agric Biol Chem 54: 2353–2358. [Google Scholar]
- Harwood, C.S. , and Parales, R.E. (1996) The beta‐ketoadipate pathway and the biology of self‐identity. Annu Rev Microbiol 50: 553–590. [DOI] [PubMed] [Google Scholar]
- Haskins, R.H. , Thorn, J.A. , and Boothroyd, B. (1955) Biochemistry of the Ustilaginales. 11. Metabolic products of Ustilago zeae in submerged culture. Can J Microbiol 1: 749–756. [DOI] [PubMed] [Google Scholar]
- Huang, X. , Lu, X. , Li, Y. , Li, X. , and Li, J.J. (2014) Improving itaconic acid production through genetic engineering of an industrial Aspergillus terreus strain. Microb Cell Fact 13: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, G. , Wu, C.L. , Schade, B. , Thomas, D.Y. , and Whiteway, M. (2005) Drag&Drop cloning in yeast. Gene 344: 43–51. [DOI] [PubMed] [Google Scholar]
- Kaemper, J. , Kahmann, R. , Boelker, M. , Ma, L.J. , Brefort, T. , Saville, B.J. , et al (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis . Nature 444: 97–101. [DOI] [PubMed] [Google Scholar]
- Kanamasa, S. , Dwiarti, L. , Okabe, M. , and Park, E.Y. (2008) Cloning and functional characterization of the cis‐aconitic acid decarboxylase (CAD) gene from Aspergillus terreus . Appl Microbiol Biotechnol 80: 223–229. [DOI] [PubMed] [Google Scholar]
- Kaplan, R.S. , Mayor, J.A. , Gremse, D.A. , and Wood, D.O. (1995) High level expression and characterization of the mitochondrial citrate transport protein from the yeast Saccharomyces cerevisiae . J Biol Chem 270: 4108–4114. [DOI] [PubMed] [Google Scholar]
- Karaffa, L. , and Kubicek, C.P. (2003) Aspergillus niger citric acid accumulation: do we understand this well working black box? Appl Microbiol Biotechnol 61: 189–196. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K. (1932) Über die Produktion von Itaconsäure und Mannit durch einen neuen Schimmelpilz Aspergillus itaconicus. Acta Phytochimica 5: 271–287. [Google Scholar]
- Klement, T. , and Büchs, J. (2013) Itaconic acid – a biotechnological process in change. Bioresour Technol 135: 422–431. [DOI] [PubMed] [Google Scholar]
- Klement, T. , Milker, S. , Jäger, G. , Grande, P.M. , de Maria, P.D. , and Büchs, J. (2012) Biomass pretreatment affects Ustilago maydis in producing itaconic acid. Microb Cell Fact 11: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinman, J.P. , and Rose, I.A. (1971) Purification and kinetic properties of aconitate isomerase from Pseudomonas putida . Biochemistry 10: 2253–2259. [DOI] [PubMed] [Google Scholar]
- Landeweert, R. , Hoffland, E. , Finlay, R.D. , Kuyper, T.W. , and van Breemen, N. (2001) Linking plants to rocks: ectomycorrhizal fungi mobilize nutrients from minerals. Trends Ecol Evol 16: 248–254. [DOI] [PubMed] [Google Scholar]
- Li, A. , van Luijk, N. , ter Beek, M. , Caspers, M. , Punt, P. , and van der Werf, M. (2011) A clone‐based transcriptomics approach for the identification of genes relevant for itaconic acid production in Aspergillus . Fungal Genet Biol 48: 602–611. [DOI] [PubMed] [Google Scholar]
- Li, A. , Pfelzer, N. , Zuijderwijk, R. , and Punt, P. (2012) Enhanced itaconic acid production in Aspergillus niger using genetic modification and medium optimization. BMC Biotechnol 12: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. , and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(‐Delta Delta C) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Loubradou, G. , Brachmann, A. , Feldbrugge, M. , and Kahmann, R. (2001) A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis . Mol Microbiol 40: 719–730. [DOI] [PubMed] [Google Scholar]
- Maassen, N. , Panakova, M. , Wierckx, N. , Geiser, E. , Zimmermann, M. , Bolker, M. , et al (2014) Influence of carbon and nitrogen concentration on itaconic acid production by the smut fungus Ustilago maydis . Eng Life Sci 14: 129–134. [Google Scholar]
- McFadden, B.A. , and Purohit, S. (1977) Itaconate, an isocitrate lyase‐directed inhibitor in Pseudomonas indigofera . J Bacteriol 131: 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelucci, A. , Cordes, T. , Ghelfi, J. , Pailot, A. , Reiling, N. , Goldmann, O. , et al (2013) Immune‐responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci USA 110: 7820–7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingot, J.M. , Penalva, M.A. , and Fernandez‐Canon, J.M. (1999) Disruption of phacA, an Aspergillus nidulans gene encoding a novel cytochrome P450 monooxygenase catalyzing phenylacetate 2‐hydroxylation, results in penicillin overproduction. J Biol Chem 274: 14545–14550. [DOI] [PubMed] [Google Scholar]
- Nogales, J. , Canales, A. , Jimenez‐Barbero, J. , Serra, B. , Pingarron, J.M. , Garcia, J.L. , and Diaz, E. (2011) Unravelling the gallic acid degradation pathway in bacteria: the gal cluster from Pseudomonas putida . Mol Microbiol 79: 359–374. [DOI] [PubMed] [Google Scholar]
- Okabe, M. , Lies, D. , Kanamasa, S. , and Park, E.Y. (2009) Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus . Appl Microbiol Biotechnol 84: 597–606. [DOI] [PubMed] [Google Scholar]
- Okamoto, S. , Chin, T. , Hiratsuka, K. , Aso, Y. , Tanaka, Y. , Takahashi, T. , and Ohara, H. (2014) Production of itaconic acid using metabolically engineered Escherichia coli . J Gen Appl Microbiol 60: 191–197. [DOI] [PubMed] [Google Scholar]
- Pereira, G. , Tanaka, T.U. , Nasmyth, K. , and Schiebel, E. (2001) Modes of spindle pole body inheritance and segregation of the Bfa1p‐Bub2p checkpoint protein complex. EMBO J 20: 6359–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffran, M. , and Prado, J.L. (1949) Inhibition of aconitase by trans‐aconitate. J Biol Chem 180: 1301–1309. [PubMed] [Google Scholar]
- Sasikaran, J. , Ziemski, M. , Zadora, P.K. , Fleig, A. , and Berg, I.A. (2014) Bacterial itaconate degradation promotes pathogenicity. Nat Chem Biol 10: 371–377. [DOI] [PubMed] [Google Scholar]
- Schulz, B. , Banuett, F. , Dahl, M. , Schlesinger, R. , Schäfer, W. , Martin, T. , et al (1990) The b alleles of Ustilago maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain‐related motif. Cell 60: 295–306. [DOI] [PubMed] [Google Scholar]
- Shi, L. , Adkins, J.N. , Coleman, J.R. , Schepmoes, A.A. , Dohnkova, A. , Mottaz, H.M. , et al (2006) Proteomic analysis of Salmonella enterica serovar Typhimurium isolated from RAW 264.7 macrophages – identification of a novel protein that contributes to the replication of serovar Typhimurium inside macrophages. J Biol Chem 281: 29131–29140. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S. , and Hieter, P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellig, T. , Bottin, A. , and Kahmann, R. (1996) Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis . Mol Genet Genomics 252: 503–509. [DOI] [PubMed] [Google Scholar]
- Sugimoto, M. , Sakagami, H. , Yokote, Y. , Onuma, H. , Kaneko, M. , Mori, M. , et al (2012) Non‐targeted metabolite profiling in activated macrophage secretion. Metabolomics 8: 624–633. [Google Scholar]
- Thompson, J.F. , Schaefer, S.C. , and Madison, J.T. (1990) Determination of aconitate isomerase in plants. Anal Biochem 184: 39–47. [DOI] [PubMed] [Google Scholar]
- Velarde, M. , Macieira, S. , Hilberg, M. , Broker, G. , Tu, S.M. , Golding, B.T. , et al (2009) Crystal structure and putative mechanism of 3‐methylitaconate‐delta‐isomerase from Eubacterium barkeri . J Mol Biol 391: 609–620. [DOI] [PubMed] [Google Scholar]
- Vuoristo, K.S. , Mars, A.E. , Sangra, J.V. , Springer, J. , Eggink, G. , Sanders, J.P. , and Weusthuis, R.A. (2015) Metabolic engineering of itaconate production in Escherichia coli . Appl Microbiol Biotechnol 99: 221–228. [DOI] [PubMed] [Google Scholar]
- Werpy, T. and Petersen, G. (2004). Top value added chemicals from biomass. 2004 US‐DoE report PNNL‐16983.
- Whitelaw, M.A. (1999) Growth promotion of plants inoculated with phosphate‐solubilizing fungi In Advances in Agronomy. Sparks D.L. (ed.). Waltham, MA, USA: Academic Press, pp. 99–151. [Google Scholar]
- Williams, S.E. , Woolridge, E.M. , Ransom, S.C. , Landro, J.A. , Babbitt, P.C. , and Kozarich, J.W. (1992) 3‐carboxy‐cis,cis‐muconate lactonizing enzyme from Pseudomonas putida is homologous to the class‐II fumarase family – a new reaction in the evolution of a mechanistic motif. Biochemistry 31: 9768–9776. [DOI] [PubMed] [Google Scholar]
- Willke, T. , and Vorlop, K.D. (2001) Biotechnological production of itaconic acid. Appl Microbiol Biotechnol 56: 289–295. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Wang, Y. , Woolridge, E.M. , Arora, V. , Petsko, G.A. , Kozarich, J.W. , and Ringe, D. (2004) Crystal structure of 3‐carboxy‐cis,cis‐muconate lactonizing enzyme from Pseudomonas putida, a fumarase class II type cycloisomerase: enzyme evolution in parallel pathways. Biochemistry 43: 10424–10434. [DOI] [PubMed] [Google Scholar]
- Yin, Y. , He, X. , Szewczyk, P. , Nguyen, T. , and Chang, G. (2006) Structure of the multidrug transporter EmrD from Escherichia coli . Science 312: 741–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y. , Kief, J. , Auffarth, K. , Farfsing, J.W. , Mahlert, M. , Nieto, F. , and Basse, C.W. (2008) The Ustilago maydis Cys(2)His(2)‐type zinc finger transcription factor Mzr1 regulates fungal gene expression during the biotrophic growth stage. Mol Microbiol 68: 1450–1470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. A PrpF‐like protein is involved in itaconate formation.
Fig. S2. Expression of cluster genes depends on the pathway‐specific transcription factor Ria1.
Fig. S3. Induction of cluster genes by overexpression of Ria1.
Fig. S4. Hypothetical reaction mechanism of Tad1.
Table S1. Oligonucleotides for U. maydis gene deletion constructs.
Table S2. Oligonucleotides for U. maydis overexpression constructs.
Table S3. Oligonucleotides for recombinant protein expression in E. coli.
Table S4. Oligonucleotides for intron removal of UMAG_11778.
Table S5. Oligonucleotides for gene expression in S. cerevisiae.
Table S6. Oligonucleotides for quantitative RT‐PCR.


