Abstract
Epithelial regeneration is critical for barrier maintenance and organ function after intestinal injury. The intestinal stem cell (ISC) niche provides Wnt, Notch, and epidermal growth factor (EGF) signals supporting Lgr5+ crypt base columnar ISCs for normal epithelial maintenance1,2. However, little is known about the regulation of the ISC compartment after tissue damage. Utilizing ex vivo organoid cultures, we provide evidence that innate lymphoid cells (ILCs), potent producers of Interleukin-22 (IL-22) after intestinal injury3,4, increased the growth of murine small intestine (SI) organoids in an IL-22-dependent fashion. Recombinant IL-22 directly targeted ISCs, augmenting the growth of both murine and human intestinal organoids, increasing proliferation, and promoting ISC expansion. IL-22 induced Stat3 phosphorylation in Lgr5+ ISCs, and Stat3 was critical for both organoid formation and IL-22-mediated regeneration. Treatment with IL-22 in vivo after murine allogeneic bone marrow transplantation (BMT) enhanced recovery of ISCs, increased epithelial regeneration, and reduced intestinal pathology and mortality from graft vs. host disease (GVHD). Atoh1-deficient organoid culture demonstrated that IL-22 induced epithelial regeneration independent of the Paneth cell niche. Our findings reveal a fundamental mechanism by which the immune system is able to support intestinal epithelium, activating ISCs to promote regeneration.
The epithelial layer in the gastrointestinal (GI) tract represents a fundamental line of defense against potential enteric pathogens. Paneth cells contribute to this defense by producing antimicrobial molecules and by providing an epithelial niche for Lgr5+ ISCs that maintain the epithelium2. ISCs are critical for damage-induced intestinal regeneration5, but the mechanisms regulating ISC function and inducing epithelial regeneration after tissue damage remain poorly understood. Furthermore, although epithelial barrier function is a core component of intestinal immunity, little is known about the role of the immune system in regulating the ISC compartment. Group 3 ILCs (ILC3s) are critical for maintaining GI epithelial integrity and barrier function in multiple experimental models of intestinal injury3. Tissue-resident ILC3s are potent producers of IL-22 after damage, and expression of IL-22 is associated with reduced injury in colitis as well as several non-intestinal tissue damage models3,4,6-9. However, while the IL-22 receptor (IL-22R) is present in many epithelial tissues, the specific cellular targets and mechanisms of IL-22 inducing tissue recovery are largely unknown. Utilizing an organoid model of ex vivo epithelial regeneration10, we examined if ILCs and IL-22 could regulate the ISC compartment.
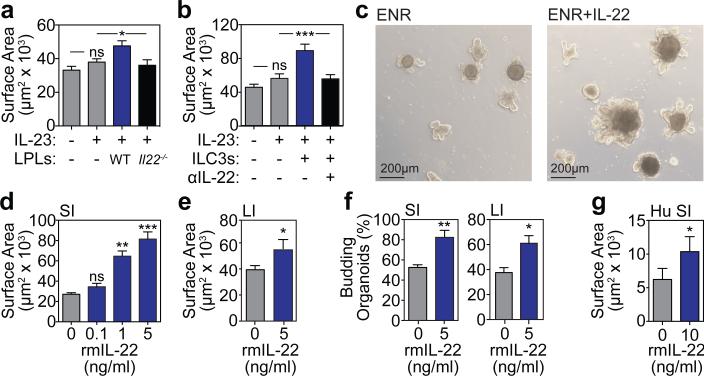
We first sorted murine SI lamina propria lymphocytes (LPLs), which include both innate and adaptive lymphoid cells capable of producing IL-224, and cultured them with freshly isolated murine SI crypts in standard organoid media containing EGF, Noggin, and R-spondin-1 (ENR). An IL-23-based cytokine cocktail was included for IL-22 induction. Two-dimensional perimeter tracing (Extended data Fig. 1a) indicated that co-culture with wild-type (WT) LPLs significantly increased organoid size (Fig. 1a). In contrast, LPLs isolated from IL-22-deficient (Il22−/−) mice failed to augment organoid size (Fig. 1a). To specifically evaluate the role of ILC3s in organoid growth, SI lamina propria CD45+CD3−RORγt+ ILC3s were isolated from RORγt-GFP reporter mice and cultured with SI crypts. ILC3s significantly increased SI organoid size, and this was inhibited by an anti-IL-22 neutralizing antibody (Fig. 1b).
Figure 1. IL-22 increases growth of intestinal organoids.
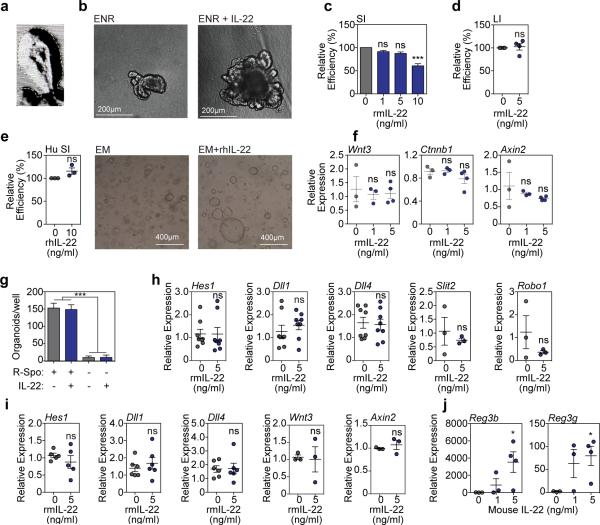
a, Size of SI organoids cultured in ENR with IL-23-containing cytokine cocktail +/− LPLs; n=62 (control), n=72 (IL-23), n=29 (WT LPLs), n=34 (Il22−/− LPLs) organoids/group; 1 of 2 experiments. b, Size of SI organoids cultured +/− ILC3s and anti-IL-22 neutralizing antibody; n=47 (control), n=55 (IL-23); n=43 (ILC3s); n=38 (anti-IL-22) organoids/group; 1 of 2 experiments. c, SI organoids, 7 days +/− rmIL-22 (5ng/ml). d-e, Size of organoids cultured +/− rmIL-22 for 7 days; n=114 (control), n=50 (0.1ng/ml), n=47 (1ng/ml), n=44 (5ng/ml) SI organoids/group (d); n=115 (control), n=61 (IL-22) LI organoids/group (e). f, New crypt formation (budding) of SI (day 4) and LI (day 7) organoids; n=6 mice/group. g, Size of human SI organoids cultured +/− rhIL-22 (10ng/ml) in standard expansion medium; n=38 (control), n=67 (IL-22) organoids/group. Bar graphs: mean+SEM; comparisons performed with t-tests (2 groups) or ANOVA (multiple groups); ns: non-significant; *p<0.05, **p<0.01, ***p<0.001. Data combined from at least three independent experiments unless otherwise stated.
Given that IL-22 was essential for ILC-mediated augmentation of organoid size, we focused on studies with recombinant murine (rm)IL-22. SI crypts cultured with rmIL-22 yielded substantially larger organoids in a concentration-dependent fashion (Fig. 1c-d and Extended Data Fig.1b). While high concentrations of IL-22 reduced the efficiency of organoid generation from SI crypts, culture with 1-5 ng/ml rmIL-22 increased organoid size without affecting organoid formation (Extended Data Fig. 1c). IL-22 also increased large intestine (LI) organoid size without affecting efficiency (Fig. 1e and Extended Data Fig.1d), and culture with IL-22 augmented crypt budding in both SI and LI organoids (Fig. 1f). Furthermore, recombinant human (rh)IL-22 significantly increased the size of human intestinal organoids generated from primary duodenal tissue (Fig. 1g and Extended Data Fig. 1e).
Wnt/β-catenin signaling is essential for ISC maintenance and organoid function ex vivo10. However, we found no evidence of enhanced production of molecules in the Wnt/β-catenin pathway within SI organoids cultured with IL-22, including no difference in expression of Wnt3, β-catenin, or the downstream target Axin2 (Extended Data Fig. 1f). Consistent with this, IL-22 could not replace R-spondin-1, an agonist of Wnt/β-catenin signaling, as its removal eliminated SI organoid growth even in the presence of IL-22 (Extended Data Fig. 1g). Additionally, we found no IL-22-induced activation of gene expression in the Notch pathway, which is also critical for ISC maintenance, or for Slit2 or Robo1, although they can regulate ISC recovery from damage induced by chemotherapy and radiation11 (Extended Data Fig. 1h). Consistently, there was also no increase in Wnt or Notch pathway gene expression in LI organoids (Extended Data Fig. 1i). However, culture with IL-22 increased SI organoid mRNA for Reg3β and Reg3γ (Extended Data Fig. 1j), innate antimicrobial molecules whose expression is dependent on Stat3 signaling12.
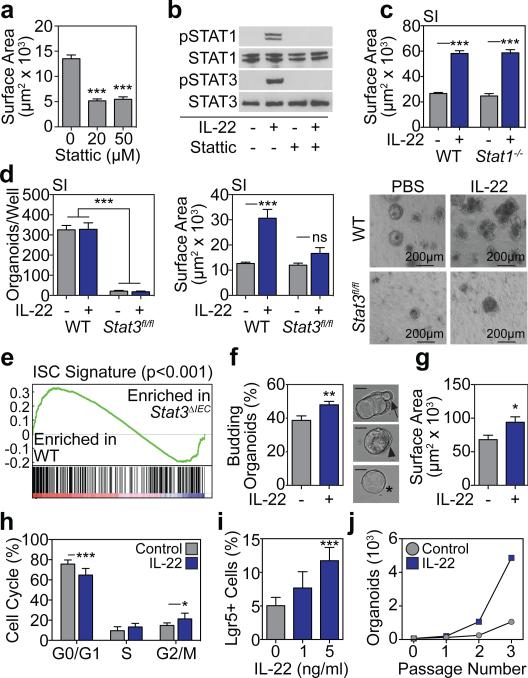
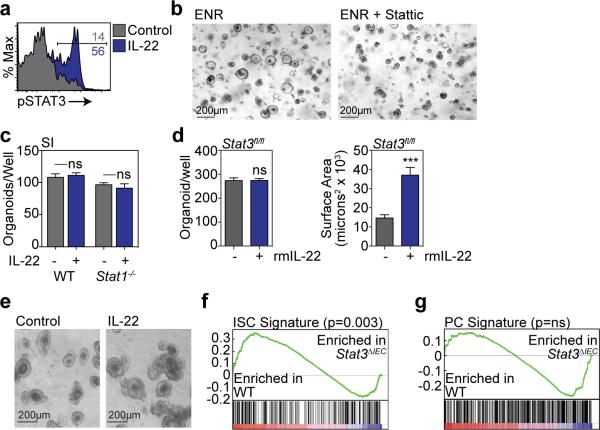
Little is known about JAK/Stat signaling within ISCs, although it has been reported that Stat3 may be important for ISC maintenance13. We evaluated Stat3 signaling in SI organoids and found that IL-22 increased phosphorylation of Stat3 Y705 (Extended Data Fig. 2a). Furthermore, treatment with the Stat3 inhibitor Stattic significantly impaired growth of SI organoids (Fig 2a and Extended Data Fig. 2b). However, IL-22 can also promote epithelial Stat1 signaling14, which Stattic has inhibitory activity against15. Indeed, organoid Stat3 and Stat1 were both phosphorylated in response to IL-22 and inhibited by Stattic (Fig. 2b). To determine their relative importance for IL-22-induced epithelial regeneration, we assessed the growth of organoids with genetic deletion of either Stat1 or Stat3. Despite the induction of phospho-(p)Stat1 by IL-22, SI crypts from Stat1−/− mice demonstrated intact organoid growth and response to IL-22 (Fig. 2c and Extended Data Fig. 2c). As Stat3−/− mice are not viable, we next cultured SI crypt cells from Stat3fl/fl mice with adenoviral-Cre (adeno-Cre) to delete Stat3. Crypt cells from WT mice demonstrated intact organoid growth and IL-22 response despite in vitro infection with adeno-Cre (Fig. 2d). Additionally, uninfected Stat3fl/fl crypt cells demonstrated normal organoid growth and response to IL-22 (Extended Data Fig. 2d-e). However, crypt cells from Stat3fl/fl mice failed to generate organoids after infection with adeno-Cre, and IL-22 failed to recover organoid growth or augment organoid size (Fig. 2d).
Figure 2. IL-22 activates organoid STAT3 signaling and augments ISC regeneration.
a, SI organoid size, 4 days +/− Stattic; n=174 (control), n=134 (20μm), n=102 (50μm) organoids/group. b Crypt pStat westerns after 30 minutes rmIL22 (5ng/ml) +/− Stattic; 1 of 3 experiments. c, Size of day 7 WT and Stat1−/− SI organoids +/−rmIL-22 (5ng/ml); n=821 (WT), n=503 (WT+IL-22), n=432 (Stat1−/−), n=269 (Stat1−/−+IL-22) organoids/group. d, Day 5 Stat3fl/fl SI organoids cultured with adeno-Cre +/− rmIL-22 (5ng/ml); numbers/well, n=6 wells/group; size, n=253 (WT), n=49 (WT+IL-22), n=38 (Stat3fl/fl), n=38 (Stat3fl/fl+IL-22) organoids/group; images representative of three experiments. e, GSEA of ISC signature genes in WT vs. Stat3fl/fl;Villin-Cre (Stat3ΔIEC) mice with DSS colitis; one analysis, nominal p-value shown. f-g, Organoids from sorted SI Lgr5-GFP+ ISCs cultured +/− rmIL-22 (1ng/ml). f, Organoid budding, percentage of total organoids/well (day 4, n=11 wells/group). Representative images of early budding indicate: *early organoid without budding; ◀polarization prior to budding; ←budding at site of polarization. Scale bar: 50μm. g, Organoid area (day 13), n=54 organoids/group. h, Cell cycle FACS of SI organoid cells cultured +/− rmIL-22 (5ng/ml); n=7 mice/group. i, FACS analysis of Lgr5-GFPhigh ISCs in organoids cultured +/− rmIL-22; n=6 mice/group. j, Organoid expansion with serial passaging +/−rmIL-22 (1ng/ml); one of two experiments. Bar graphs: mean+SEM; comparisons performed with t-tests (2 groups) or ANOVA (multiple groups); ns: non-significant; *p<0.05, **p<0.01, ***p<0.001. Data combined from at least two independent experiments unless otherwise stated. For western blot source data, see Supplementary Figure 1.
Lgr5+ ISCs can generate all cell types of mature intestinal epithelium ex vivo and in vivo1,10. To evaluate if Stat3 was important for ISCs during tissue damage in vivo, we performed a gene set enrichment analysis (GSEA), assessing expression of a published Lgr5+ ISC gene signature (GEO dataset GSE33948)16 in another dataset of WT (Stat3fl/fl) vs. epithelial Stat3-deficient (Stat3fl/fl;Villin-Cre) mice with DSS colitis (GSE15955)12. Expression of the Lgr5+ ISC gene signature was significantly reduced in Stat3-deficient mice with colitis (Fig. 2e). This was validated with a second independently established Lgr5+ ISC gene signature (GEO dataset GSE23672)16, while no significant changes were seen in a negative control Paneth cell gene signature (GEO dataset GSE39915)17 (Extended Data Fig. 2f-g). Given the induction of pStat3 by IL-22 and the importance of Stat3 for ISC gene signature maintenance, we examined the effect of IL-22 during regeneration using purified ISCs. We isolated Lgr5-GFP SI ISCs by fluorescence-activated cell sorting (FACS) and cultured single ISCs under standard conditions10 +/− IL-22. IL-22 significantly increased budding of early organoids after just four days (Fig. 2f). Furthermore, as with crypt-derived organoids, 1 ng/ml rmIL-22 augmented the size of organoids generated from purified ISCs without affecting the efficiency of organoid formation (Fig. 2g and Extended Data Fig. 3a-b).
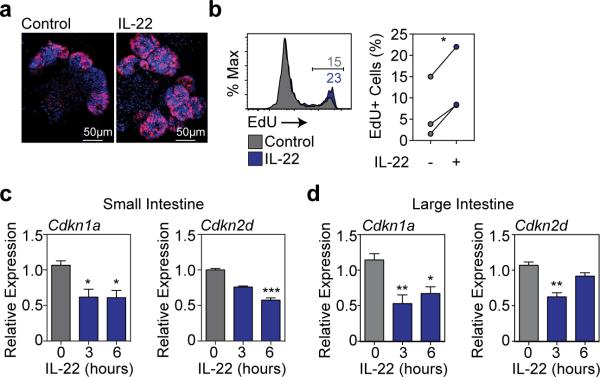
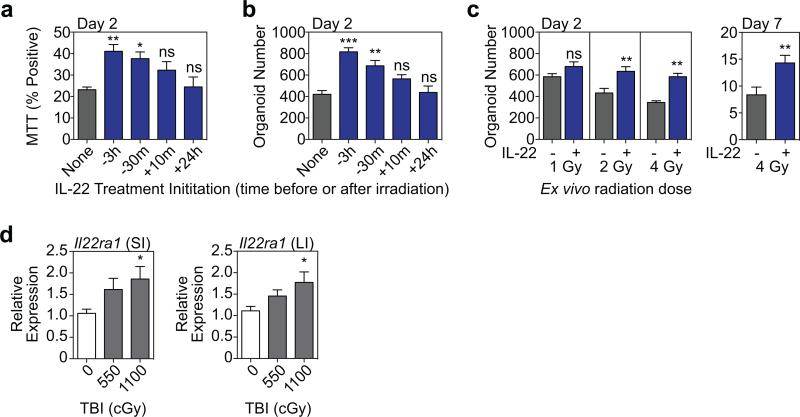
Consistent with increased size, IL-22 enhanced EdU incorporation in SI organoids, demonstrating increased proliferation (Extended Data Fig. 4a-b). Hoechst staining revealed an IL-22-dependent increase in G2/M populations after two days in culture (Fig. 2h), and IL-22 treatment rapidly reduced key cell cycle checkpoint molecules Cdkn1a and Cdkn2d in both SI and LI organoids (Extended Data Fig. 4c-d). Furthermore, IL-22 expanded Lgr5-GFPhigh ISCs in SI organoids (Fig. 2i) and increased expansion of SI organoids over multiple passages in culture (Fig. 2j). Next we evaluated the ISC compartment after radiation injury. Pre-treatment with rmIL-22 increased the percentage of dissociated SI crypt cells that were viable in culture following ex vivo irradiation, as measured by MTT reduction (Extended Data Fig. 5a). Consistent with this, IL-22 treatment increased the number of organoids that could grow from single cells two days after irradiation (Extended Data Fig. 5b). This was more evident with increasing doses of irradiation, and protection was present even seven days after irradiation (Extended Data Fig. 5c). Accordingly, irradiation was found to increase expression of IL-22R mRNA within intestinal crypts (Extended Data Fig. 5d).
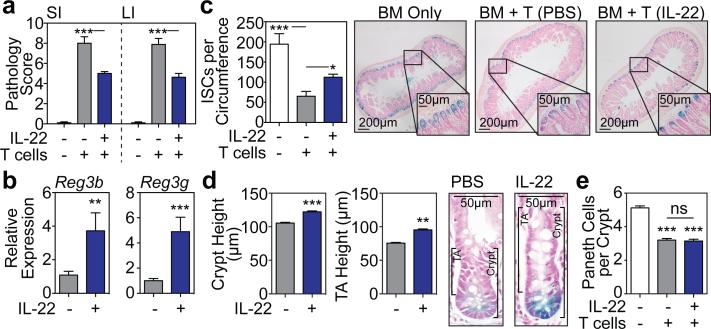
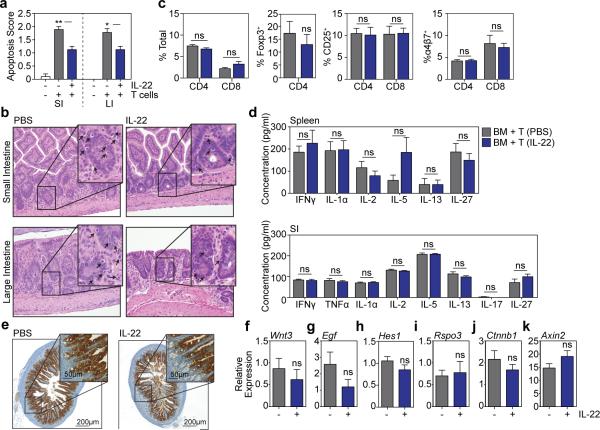
We next evaluated the effect of IL-22 in vivo after tissue damage utilizing a clinically relevant murine GVHD model. T cell-depleted (TCD) LP marrow was transplanted +/− purified LP T cells into lethally irradiated C57BL/6 (B6) recipients (H-2b→H-2b). Mice receiving allogeneic T cells for GVHD induction were treated with 4μg rmIL-22 or PBS daily via intraperitoneal (IP) injection starting seven days post-BMT. Treatment with IL-22 reduced histopathologic evidence of GVHD in SI and LI (Fig. 3a), including a reduction in apoptosis within crypt epithelium (Extended Data Fig. 6a-b). GVHD pathology was reduced despite an intact alloimmune response, as evidenced by similar T cell subset distribution, activation markers, and gut homing molecules, as well as similar systemic and GI expression of inflammatory cytokines (Extended Data Fig. 6c-d). However, IL-22 treatment did increase SI expression of Reg3β and Reg3γ mRNA (Fig. 3b). Consistent with the findings of Eriguchi et al.18, we found that Reg3β was primarily expressed by enterocytes in allogeneic BMT recipients, including after treatment with IL-22 (Extended Data Fig. 6e).
Figure 3. IL-22 reduces intestinal pathology and increases ISC recovery after in vivo tissue damage.
LP→B6 BMT; recipients treated daily with PBS or 4μg rmIL-22 IP starting day 7 post-BMT. a, Intestinal GVHD histopathology score three weeks post-BMT; n=10 (TCD BM Only), n=9 (BM+T+PBS), n=8 (BM+T+IL-22) mice/group; Kruskal-Wallis analysis. b, qPCR of Reg3b and Reg3g in SI tissue three weeks post-BMT; n=9 (PBS) and n=10 (IL-22) mice/group; Mann-Whitney analysis. c-d, B6 Lgr5-LacZ recipients. c, SI ISC frequency three weeks post-BMT; Kruskal-Wallis analysis of n=8 (TCD BM only), n=20 (BM+T+PBS), or n=20 (BM+T+IL-22) independent sections (4 sections/recipient from 2-5 mice/group); one of two experiments. d, Crypt and TA heights three weeks post-BMT, representative images on right; t-test analyses of n=285 (PBS) vs. n=324 (IL-22) crypts and n=168 (PBS) vs. n=224 (IL-22) TA compartments (1 section/mouse, >10 mice/group). e, SI lysozyme+ Paneth cell frequency; Kruskal-Wallis analysis of n=73 (TCD BM only), n=89 (BM+T+PBS), and n=88 (BM+T+IL-22) crypts (5-8 mice/group). Bar graphs: mean+SEM; ns: non-significant; *p<0.05, **p<0.01, ***p<0.001. Data combined from at least two independent experiments unless otherwise stated.
GVHD is associated with a loss of both ISCs19,20 and niche-forming Paneth cells21,22, and T cell-replete BMT led to a significant loss of Lgr5-LacZ+ SI ISCs three weeks post-transplant (Fig. 3c). However, IL-22 treatment increased the recovery of Lgr5+ ISCs (Fig. 3c). This was associated with increased regeneration as evidenced by increased crypt height, including the transit-amplifying (TA) compartment (Fig. 3d). Paneth cells support Lgr5+ ISCs through delivery of Wnt, Notch ligand, and EGF signals2. Additionally, IL-22 is thought to regulate Paneth cell production of innate antimicrobial molecules. We thus hypothesized that IL-22 could support ISC recovery post-BMT by improving the function of the stem cell niche. Consistent with prior clinical and experimental studies21-23, minor antigen-mismatched BMT led to a reduction in Paneth cells three weeks post-transplant (Fig. 3e). However, IL-22 administration did not increase recovery of Paneth cells (Fig. 3e), mRNA expression of Wnt3 or EGF (Extended Data Fig. 6f-g), or expression of Notch ligand target Hes1 (Extended Data Fig. 6h). Furthermore, although stroma can support ISC Wnt signaling in vivo independent of Paneth cells24, we found no change in expression of stromal R-spondin-3 after IL-22 treatment post-BMT and no change in expression of Wnt pathway genes regardless of the upstream source. (Extended Data Fig. 6i-k).
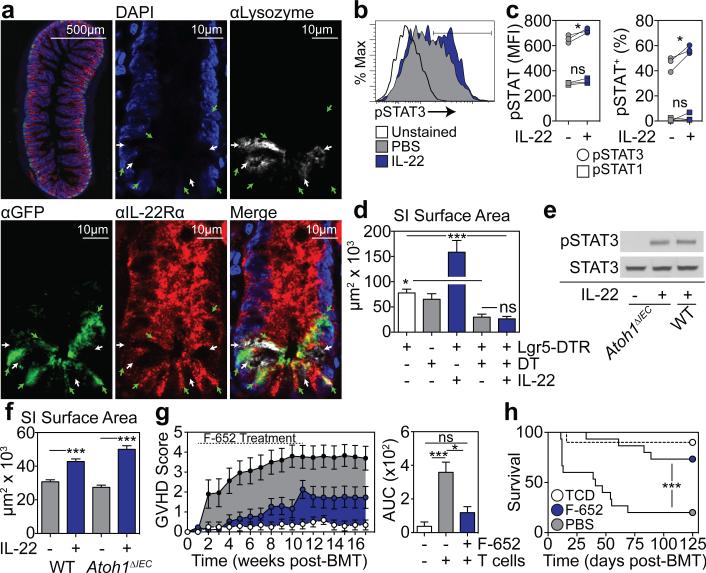
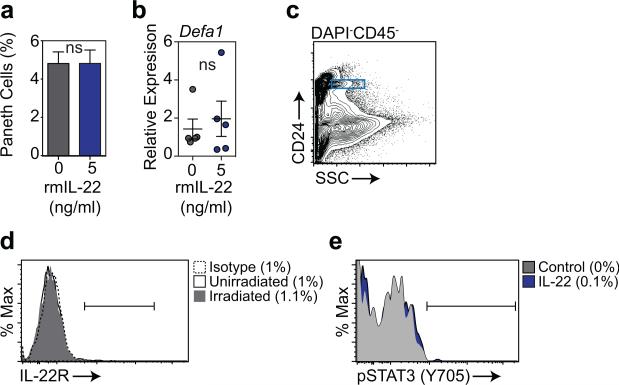
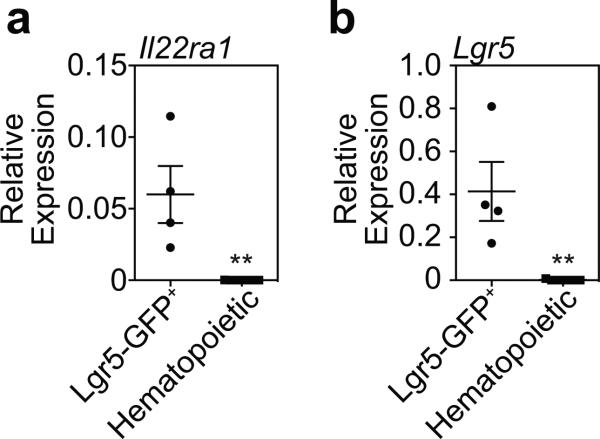
Given that IL-22 treatment appeared to improve ISC numbers without improving niche function in BMT recipients, we sought to evaluate how IL-22 was targeting the ISC compartment. Consistent with the in vivo findings, IL-22 had no effect on Paneth cell frequency or α-defensin-1 expression within SI organoids cultured ex vivo (Extended Data Fig. 7a-b). Immunofluorescent staining for IL-22Rα1 and the Paneth cell marker lysozyme using SI sections from Lgr5-GFP reporter mice indicated substantial IL-22R staining within crypts and enterocytes at the villous base, but not on lysozyme+ Paneth cells (Fig. 4a). Using flow cytometry, there was also little evidence for expression of IL-22R on Paneth cells at baseline or after radiation injury and no evidence of pStat3 in Paneth cells in response to IL-22 (Extended Data Fig. 7c-e). In contrast, IL-22R was identified in the TA progenitor compartment and on Lgr5-GFP+ ISCs (Fig. 4a). IL-22R expression in Lgr5+ cells was confirmed by qPCR after sorting for Lgr5-GFP+ cells (Extended Data Fig. 8a-b). Furthermore, SI crypt cells from Lgr5-GFP reporter mice demonstrated increased Stat3 Y705 phosphorylation within GFP+ cells after incubation with IL-22, indicating functional IL-22R signaling in ISCs (Fig. 4b-c). Stat3 phosphorylation was a specific response, as there was no effect of IL-22 on pStat1 in Lgr5+ cells (Fig. 4c).
Figure 4. IL-22 directly promotes ISC-dependent epithelial regeneration.
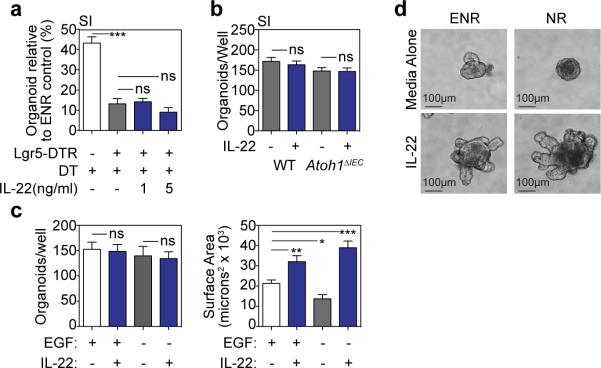
a, Immunofluorescent staining of IL-22Rα1, GFP, and lysozyme in SI sections from Lgr5-GFP mice; green arrows, Lgr5-GFP+ ISCs; white arrows; lysozyme+ Paneth cells (PCs). b-c, Phosflow analysis of Lgr5-GFP+ SI crypt cells after 30 minutes +/− rmIL-22 (20ng/ml). b, pStat3 histogram; representative of four experiments. c, pStat median fluorescence intensity (MFI) and percent+; n=3 mice/group; representative of two experiments. d, Size of WT and Lgr5-DTR day 5 SI organoids cultured with DT (1ng/μl) to deplete Lgr5+ cells +/− rmIL-22 (5ng/ml); one of three experiments; n=65 (WT), n=25 (WT+DT), n=28 (DTR+IL-22), n=18 (DTR+DT), n=40 (DTR+DT+IL-22) organoids/group. e-f, Paneth cell-deficient Atoh1ΔIEC SI organoids cultured in Wnt3-supplemented ENR +/− rmIL-22 (5ng/ml). e, Stat3 westerns after 30 minutes rmIL-22; one of four experiments. f, Day 7 organoid size; n=466 (WT), n=531 (WT+IL-22), n=197 (Atoh1ΔIEC), n=491 (Atoh1ΔIEC+IL-22) organoids/group. g-h, LP→B6 BMT +/− F-652 (100μg/kg subcutaneous, every-other-day starting day 7 post-BMT, 10-week course); n=10 (TCD BM only), n=15 (BM+T+PBS), n=15 (BM+T+IL-22). g, Clinical signs of GVHD and area under the curve (AUC) analysis of GVHD scoring. h, Survival. Bar graphs: mean+SEM; comparisons performed with t-tests (2 groups), ANOVA (multiple groups), or Log-Rank analysis (h); ns: non-significant; *p<0.05, ***p< 0.001. Data combined from at least two independent experiments unless otherwise stated. For western blot source data, see Supplementary Figure 1.
IL-22R expression and Stat3 phosphorylation suggested that IL-22 might promote regeneration via direct targeting of ISCs. To further investigate the role of ISCs and Paneth cells in IL-22-mediated regeneration, we assessed organoid growth when either cell population was depleted. Treatment of Lgr5-DTR mice with diphtheria toxin (DT) leads to a rapid deletion of Lgr5+ cells25. Deletion of Lgr5+ cells ex vivo by culturing Lgr5-DTR SI organoids with DT impaired epithelial regeneration as evidenced by a reduction in organoid size and efficiency (Fig. 4d and Extended Data Fig. 9a). Although IL-22 increased the size of Lgr5-DTR organoids cultured without DT, IL-22 failed to increase the size or maintain the numbers of Lgr5-DTR organoids cultured with DT (Fig. 4d and Extended Data Fig. 9a), indicating that Lgr5+ cells are essential for IL-22-mediated epithelial regeneration.
We next investigated the functional importance of Paneth cells for IL-22-mediated regeneration by culturing organoids with IL-22 after inducible Paneth cell depletion. Paneth cells were deleted in vivo following tamoxifen treatment of Atoh1fl/fl;Villin-CreERT2 (Atoh1ΔIEC) mice26. IL-22 led to robust Stat3 phosphorylation within Atoh1ΔIEC SI organoids, confirming that Paneth cells were not essential for IL-22-mediated intracellular signaling, and Atoh1ΔIEC organoids demonstrated an intact growth response to IL-22 (Fig. 4e-f and Extended Data Fig. 9b). Additionally, we found that IL-22 could augment the size of organoids cultured without EGF (Extended Data Fig. 9c-d). These findings indicated that the Paneth cell niche was not required for IL-22-mediated epithelial regeneration, and IL-22 could promote the growth of organoids cultured without addition of the niche-derived growth factor EGF.
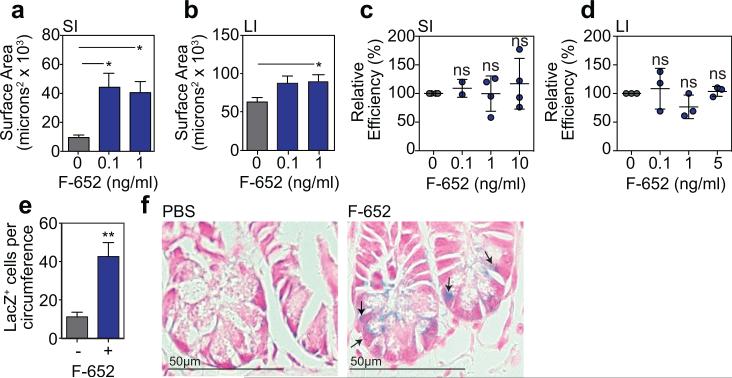
Recent reports have suggested that T cell-derived IL-22 may contribute to GVHD, as might peritransplant administration of IL-22 to MHC-mismatched BMT recipients27,28. However, IL-22-producing ILCs are eliminated in GVHD19, ILC deficiency is associated with increased clinical GVHD29, and GI damage may be central to the pathogenesis of systemic GVHD30. We thus hypothesized that stimulating regeneration with IL-22 after the initiation of GVHD-related tissue damage may be therapeutically beneficial. Given the improved pharmacologic stability of Fc-fusion molecules, we evaluated the potential of F-652, a recombinant human (rh)IL-22-dimer/Fc-fusion protein, for treatment of systemic GVHD. First, we found that F-652 had activity in murine epithelial regeneration, augmenting the growth of both SI and LI organoids without evidence of toxicity (Extended Data Fig. 10a-d). Additionally, treatment of Lgr5-LacZ reporter mice with F-652 significantly protected SI Lgr5+ crypt cells from radiation injury in vivo (Extended Data Fig. 10e-f). We next investigated an early intervention model for GVHD, treating allogeneic BMT recipients (LP→B6) with F-652 starting one week after transplant. Mice treated with F-652 demonstrated reduced systemic signs of GVHD and GVHD-related mortality compared to PBS-treated controls (Fig. 4g-h).
In summary, we found that IL-22 links immunity to epithelial regeneration by acting directly on ISCs. Purified ILCs enhanced organoid growth in an IL-22-dependent fashion, and IL-22 augmented ISC-mediated epithelial regeneration, promoting cell cycle progression, epithelial proliferation, and regeneration of the ISC pool. IL-22 induced Stat3 phosphorylation in Lgr5+ ISCs, and while IL-22 may not be its sole regulator in ISCs, Stat3 was essential for organoid growth and IL-22-dependent epithelial regeneration. Paneth cells, in contrast, were not required for IL-22-driven regeneration. Given the activation of IL-22 production and the upregulation of crypt IL-22R expression after tissue damage, these findings indicate that IL-22 contributes to damage-induced regulation of the ISC compartment. We conclude that in addition to the stromal and epithelial components of the ISC niche that are essential for normal epithelial maintenance, IL-22 provides evidence for an immunologic contribution to the ISC niche that is activated to restore the epithelium after tissue injury. By acting directly on epithelial stem cells, the immune system is thus able to regulate intestinal regeneration and support the fundamental defense system provided by the structural integrity of the epithelial barrier.
Methods
Mice
C57BL/6 (B6, H-2b) and LP (H-2b) mice were obtained from Jackson Laboratory (Bar Harbor, USA). B6 Lgr5-LacZ and B6 lgr5-gfp-ires-CreERT2 (Lgr5-GFP) mice were kindly provided by H. Clevers1,10. Mouse maintenance and procedures were done in accordance with the institutional protocol guideline of the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Animal Care and Use Committee. Mice were housed in micro-isolator cages, five per cage, in MSKCC pathogen-free facilities and received standard chow and autoclaved sterile drinking water. To adjust for differences in weight and intestinal flora among other factors, identical mice were purchased from Jackson and then randomly distributed over different cages and groups by a non-biased technician who had no insight or information about the purpose or details of the experiment. The investigations assessing clinical outcome parameters were performed by non-biased technicians with no particular knowledge or information regarding the hypotheses of the experiments and no knowledge of the specifics of the individual groups.
Crypt isolation and cell dissociation
Isolation of intestinal crypts and the dissociation of cells for flow cytometry analysis were largely preformed as previously described10. Briefly, after euthanizing the mice with CO2 and harvesting small and large intestines, the organs were opened longitudinally and washed with PBS. To dissociate the crypts, small intestine was incubated at 4°C in EDTA (10 mM) for 15 minutes and then in EDTA (5mM) for an additional 15 minutes. Large intestine was incubated in Collagenase Type 4 (Worthington) for 30 minutes at 37°C to isolate the crypts. To isolate single cells from small and large intestine crypts, the pellet was further incubated in 1x TrypLE express (Gibco, Life technologies) supplemented with 0.8KU/ml DNase1 (Roche).
Organoid culture
For mouse organoids, depending on the experiments, 200-400 crypts per well were suspended in Matrigel composed of 25% Advanced DMEM/F12 medium (Gibco) and 75% growth factor-reduced Matrigel (Corning). After the Matrigel polymerized, complete EGF/Noggin/R-spondin-1 (ENR)-medium containing advanced DMEM/F12 (Sigma), 2mM Glutamax (Invitrogen), 10 mM HEPES (Sigma), 100 U/ml penicillin/100 μg/ml streptomycin (Sigma), 1 mM N-acetyl cysteine (Sigma), B27 supplement (Invitrogen), N2 supplement (Invitrogen), 50 ng/ml mEGF (Peprotech), 100 ng/ml mNoggin (Peprotech) and 10% human R-spondin-1 conditioned medium from R-spondin-1-transfected HEK 293T cells31 was added to small intestine crypt cultures10. For experiments evaluating organoid budding, the concentration of R-spondin was lowered to 1.25-5%. For mouse large intestine, crypts were cultured in “WENR” medium containing 50% of Wnt3a-conditioned medium in addition to the aforementioned proteins and 1% BSA (Sigma) and supplemented with SB202190 (10uM, Sigma), ALK5 inhibitor A83-01(500nM, Tocris Bioscience) and Nicotinamide (10mM, Sigma). Media was replaced every 2-3 days. Along with medium changes, treatment wells received different concentrations of recombinant murine (rm)IL-22 (Genscript). We also tested the effects of F-652, a recombinant human IL-22 dimer/Fc fusion molecule provided by Generon Corporation (Shanghai) Ltd, China. In some experiments, organoids from crypts were cultured in the presence of Stattic (Tocris Bioscience). For passaging of organoids, after 5-7 days of culture, organoids were passaged by mechanically disrupting with a seropipet and cold media to depolymerize the Matrigel and generate organoid fragments. After washing away the old Matrigel by spinning down at 600 rpm, organoid fragments were replated in liquid Matrigel.
Intestinal stem cells (ISCs) were isolated from Lgr5-GFP mice using a modified crypt isolation protocol with 20 minutes of 30 mM EDTA32,33 followed by several strainer steps and a 5 minunte incubation with TrypLE and 0.8 KU/ml DNase1 under minute-to-minute vortexing to make a single cells suspension. The Lgr5-GFPhigh cells were isolated by fluorescence-activated cell sorting (FACS). 5000 ISCs were plated in 30uL Matrigel and cultured in WENR media containing Rho-kinase/ROCK inhibitor Y-27632 (10uM, Tocris Bioscience) and Jagged1 (1 μM, Anaspec). Starting from day 4 ISC were cultured without Wnt.
For lymphocyte co-culture experiments, intestinal lymphoid cells were isolated from the small intestine lamina propria. Washed small intestine fragments were incubated in EDTA/IEL solution [1xPBS with 5% FBS, 10mM Hepes Buffer, 1% Pen/strep (Corning), 1% L-glutamine (Gibco), 1mM EDTA and 1 mM DTT] in a 37°C shaker for 15 minutes. The samples were strained (100uM) and put in a Collagenase solution [RPMI 1640, 5%FCS, 10mM Hepes, 1% PS, 1% Glutamine and 1 mg/ml Collagenase D (Roche) and 1u/ml DNAse 1 (Roche)] and incubated twice for 10 minutes in a 37 °C shaker. Afterwards, the samples were centrifuged at 1500 rpm for 5 minutes and washed with RPMI solution without enzymes. After several washes, the cell suspension was transferred into a 40% Percoll solution (Percoll in PBS), which is overlaid on an 80% Percoll solution. After spinning the interface containing the lamina propria mononuclear cells was aspirated and washed in medium. Then the cell suspension was stained with extracellular markers and Topro3 for viability. Topro3−CD45+CD11b−CD11c−CD90+ lamina propria lymphocytes (LPLs) from B6 WT and IL-22−/− mice and Topro3−CD45+CD3−RORγt+ group 3 innate lymphoid cells (ILCs)34 from Rorc(γt)+ GFP mice (Jackson) were sorted for co-cultures with SI crypts. (For antibodies used, see Supplementary Table 1.) To activate and maintain LPLs and ILCs in culture, rmIL-2 (1000u/ml), rmIL-15 (10 ng/ml), rmIL-7 (50 ng/ml), and rmIL-23 (50 ng/ml) were added to the ENR medium in co-culture experiments. We have also performed co-cultures with addition of only rmIL-23 (50 ng/ml) to ENR media. LPLs and SI crypts were cultured in Matrigel with a 7:1 LPL:crypt ratio; ILCs and crypts were cultured in Matrigel with a 25:1 ILC:crypt ratio. Co-cultures were compared to crypts cultured in ENR + cytokines without LPLs/ILCs present. A neutralizing monoclonal antibody against IL-22 (8E11, Genentech)35 was used to abrogate IL-22-specific effects of ILCs.
For specific experiments, organoids were cultured from fresh crypts obtained from specific genetically modified mice, such as the Stat1−/− mice (129S6/SvEv-Stat1tm1Rds, Taconic) and Stat3fl/fl mice (Jackson). Organoids from Stat3fl/fl mice that had been grown for 7 days were dissociated as single cells and incubated with Adenoviral-Cre (University of Iowa) to cause the deletion of Stat3 from floxed organoid cells. Frozen passaged organoids from Lgr5CreER;Lgr5DTR (Lgr5GFP-DTR)25 mice were used to culture organoids in which Lgr5+ stem cells could be depleted with daily administration of Diphtheria Toxin (DT, 1 ng/ul).
For Paneth cell-deficient organoid cultures, frozen crypts from Atoh1fl/fl;Villin-CreERT2 mice36 depleted of Paneth cells were used to culture organoids. As previously described36, Atoh1fl/fl;Villin-CreERT2 mice (and littermate controls) were given an intraperitoneal (IP) injection of tamoxifen (1 mg/mouse, Sigma, dissolved in corn oil) for 5 consecutive days to achieve deletion of Atoh1 from intestinal epithelium. Animals were sacrificed on day seven after the first injection and intestinal crypts were isolated and frozen in 10% DMSO and 90% FBS.
To investigate the effect of IL-22 on human small intestine, we generated human duodenal organoids from banked frozen organoids (>passage 7) that had been previously generated from biopsies obtained during duodenoscopy of three independent healthy human donors. All human donors had been investigated for celiac disease, but turned out to have normal pathology. All provided written informed consent to participate in this study according to a protocol reviewed and approved by the review board of the UMC Utrecht, the Netherlands (protocol STEM study, METC 10-402/K). Human organoids were cultured in 10ul Matrigel drops in expansion medium containing WENR with 10nM SB202190, 500nM A83-01 and 10 mM Nicotinamide. For IL-22 stimulation experiments, rhIL-22 10 ng/ml (Genscript) was added daily. For the purpose of size measurements at D6, organoids were passaged as single cells.
Where applicable, organoid cultures were performed using conditioned media containing R-spondin-1 and Wnt3a produced by stably transfected cell lines. R-spondin-1-transfected HEK293T cells31 were kindly provided by Calvin Kuo, Department of Medicine, Stanford University. WNT3a-transfected HEK293T cells were kindly provided by Hans Clevers, Hubrecht Institute (patent WO2010090513A2). Cell lines were tested for mycoplasma and confirmed to be negative.
Organoid measurement
For size evaluation, the surface area of organoid horizontal cross sections was measured. If all organoids in a well could not be measured, multiple random non-overlapping pictures were acquired from each well using a Zeiss Axio Observer Z1 inverted microscope and then analyzed using MetaMorph or ImageJ software. Organoid perimeters for area measurements have been defined both manually and by automated determination using the Analyze Particle function of ImageJ software, with investigator verification of the automated determinations, as automated measurements allowed for unbiased analyses of increased numbers of organoids. For automated size measurements, the threshold for organoid identification was set based on monochrome images. The sizes of the largest and smallest organoids in the reference well were measured manually, and their area sizes were used as the reference values for setting the minimal and maximal particle sizes. Organoids touching the edge of the images were excluded from the counting. After five-to-seven days in culture, total organoid numbers per well were counted by light microscopy to evaluate growth efficiency. All organoid numbers were counted manually in this fashion except for the organoid counts presented in Extended Data 5b, which were counted using automated ImageJ analysis, as these organoids were too numerous to count manually. To compare organoid efficiency in different conditions, combining experiments with different organoid numbers, the percentage of organoids relative to the number of organoids in ENR-control (rmIL-22 0ng/ml) was calculated. The efficiency from sorted ISCs was presented as the percentage of cells forming organoids per number of seeded cells.
Bone marrow transplantation (BMT)
BMT procedures were performed as previously described37. A minor histocompatibility antigen-mismatched BMT model (LP→B6, H-2b→H-2b) was utilized. Female B6 WT mice were typically used as recipients for transplantation at an age of 8 to 10 weeks. Recipient mice received 1100 cGy of split-dosed lethal irradiation (550 cGy x2) 3-4 hours apart to reduce gastrointestinal toxicity. To obtain LP bone marrow cells from euthanized donor mice, the femurs and tibias were harvested aseptically and the bone marrow canals washed out with sterile media. Bone marrow cells were depleted of T cells by incubation with anti-Thy 1.2 and low-TOX-M rabbit complement (Cedarlane Laboratories). The T cell-depleted bone marrow (TCD BM) was analyzed for purity by quantification of the remaining T cell contamination using flow cytometry. T cell contamination usually was about 0.2% of all leukocytes after a single round of complement depletion. LP donor T cells were prepared by harvesting splenocytes aseptically from euthanized donor mice. T cells were purified using positive selection with CD5 magnetic Microbeads with the MACS system (Miltenyi Biotec). T cell purity was determined by flow cytometry and was routinely approximately 90%. Recipients typically received 5×106 TCD BM with or without 4 × 106 T cells per mouse via tail vein injection.
Mice were monitored daily for survival and weekly for graft vs. host disease (GVHD) scores with an established clinical GVHD scoring system (including weight, posture, activity, fur ruffling, and skin integrity) as previously described38. A clinical GVHD index with a maximum possible score of ten was then generated. Mice with a score of five or greater were considered moribund and euthanized by CO2 asphyxia.
In vivo cytokine administration
Recombinant murine IL-22 was purchased from GenScript and reconstituted as described by the manufacturer to a concentration of 40μg/ml in phosphate-buffered saline (PBS). Mice mere treated daily via IP injection with either 100μl PBS or with 100μl PBS containing 4μg rmIL-22. IL-22 administration was started on day seven post-BMT. This schedule was based on the results of rmIL-22 pharmacokinetics tested in untransplanted mice. For in vivo F-652 administration, starting from day seven post-BMT, mice were injected subcutaneously every other day for ten consecutive weeks with PBS or 100μg/kg F-652.
Histopathology analysis of GVHD target organs
Mice were euthanized for organ analysis 21 days after BMT using CO2 asphyxiation. For histopathologic analysis of GVHD, the small and large intestines were formalin-preserved, paraffin-embedded, sectioned, and stained with hematoxylin and eosin (H&E). An expert in the field of GVHD pathology assessed the sections for markers of GVHD histopathology. As described previously38, a semi-quantitative score consisting of 19 different parameters associated with GVHD was calculated.
LacZ staining
For evaluation of stem cell numbers, small intestines from Lgr5-LacZ recipient mice that were transplanted with LP bone marrow (and T cells where applicable) were harvested. β-galactosidase (LacZ) staining was preformed as previously described by Barker et al1. Washed 2.5 cm sized small intestine fragments were incubated with an ice-cold fixative, consisting of 1% formaldehyde, 0.2% NP40 and 0.2% gluteraldehyde. After removing the fixative, organs were stained for the presence of LacZ according to manufacturer's protocol (LacZ staining kit, Invivogen). Then the organs were formalin-preserved, paraffin-embedded, sectioned, and counterstained with Nuclear Fast Red (Vector Labs).
Immunohistochemistry (IHC) staining
IHC detection of Reg3β was performed at the Molecular Cytology Core Facility of Memorial Sloan Kettering Cancer Center using a Discovery XT processor (Ventana Medical Systems). Formalin-fixed tissue sections were deparaffinized with EZPrep buffer (Ventana Medical Systems), antigen retrieval was performed with CC1 buffer (Ventana Medical Systems) and sections were blocked for 30 minutes with Background Buster solution (Innovex). Slides were incubated with anti-Reg3β antibodies (R&D Systems; cat# MAB5110; 1ug/ml) or isotype (5ug/ml) for six hours, followed by a 60 minute incubation with biotinylated goat anti-rat IgG (Vector labs, cat# PK-4004) at 1:200 dilution. The detection was performed with DAB detection kit (Ventana Medical Systems) according to the manufacturer's instructions. Slides were counterstained with hematoxylin (Ventana Medical Systems) and coverslipped with Permount (Fisher Scientific). Refer to Supplementary Table 1 for full description of antibodies used.
Immunofluorescent (IF) staining and microscopic imaging
IF staining was performed at the Molecular Cytology Core Facility of Memorial Sloan Kettering Cancer Center using a Discovery XT processor (Ventana Medical Systems). Formalin-fixed tissue sections were deparaffinized with EZPrep buffer (Ventana Medical Systems), antigen retrieval was performed with CC1 buffer (Ventana Medical Systems). Sections were blocked for 30 minutes with Background Buster solution (Innovex) followed by avidin/biotin blocking for 12 minutes. IL-22R antibodies (R&D Systems cat# MAB42, 0.1ug/ml) were applied and sections were incubated for five hours followed by 60 minutes incubation with biotinylated goat anti-rat IgG (Vector labs, cat# PK-4004) at 1:200 dilution. The detection was performed with Streptavidin-HRP D (part of DABMap kit, Ventana Medical Systems), followed by incubation with Tyramide Alexa Fluor 488 (Invitrogen, cat# T20932) prepared according to manufacturer instruction with predetermined dilutions. Next, Lysozyme antibodies (DAKO; cat# A099; 2ug/ml) were applied and sections were incubated for six hours followed by incubation with biotinylated goat anti-rabbit IgG (Vector labs, cat#PK6101) for 60 minutes. The detection was performed with Streptavidin-HRP D (part of DABMap kit, Ventana Medical Systems), followed by incubation with Tyramide Alexa Fluor 594 (Invitrogen, cat# T20935) prepared according to manufacturer instruction with predetermined dilutions. Finally, GFP antibodies were applied and sections were incubated for five hours followed by incubation with biotinylated goat anti-chicken IgG (Vector labs, cat# BA-9010) for 60 minutes. The detection was performed with Streptavidin-HRP D (part of DABMap kit, Ventana Medical Systems), followed by incubation with Tyramide Alexa Fluor 647 (Invitrogen, cat# T20936) prepared according to manufacturer instruction with predetermined dilutions. Slides were counterstained with DAPI (Sigma Aldrich, cat# D9542, 5 ug/ml) for 10 minutes and coverslipped with Mowiol. For immunofluorescent and other microscopic imaging, including LacZ and IHC slides, contrast and white balance were set based on control slides for each experiment, and the same settings were used for all slides to maximize sharpness and contrast. Refer to Supplementary Table 1 for full description of antibodies used.
Cytokine multiplex assay
Spleen and small intestine were harvested from euthanized BMT recipients, then organs were homogenized and spun down. The supernatant was stored at −20°C until used for cytokine analysis. The cytokine multiplex assays were performed on thawed samples with the mouse Th1/Th2/Th17/Th22 13plex (FlowCytomix Multiplex kit, eBioscience) and performed according to the manufacturer's protocol.
Flow cytometry
For in vivo experiments, lymphoid organs were harvested from euthanized mice and processed into single cell suspension. Cells were stained with the appropriate mixture of antibodies. For intracellular analysis, an eBioscience Fixation/Permeabilization kit was used per the manufacturer's protocol. After thorough washing, the cells were stained with intracellular and extracellular antibodies simultaneously. Fluorochrome-labeled antibodies were purchased from BD Pharmingen (CD4, CD8, CD24, CD25, CD45, α4β7, and P-Stat3 Y705, P-Stat1 Y701), eBioscience (Foxp3), R&D (IL-22R), and Invitrogen (GFP). DAPI and Fixable Live/Dead Cell Stain Kits (Invitrogen) were used for viability staining. Paneth cells were identified based on bright CD24 staining and side scatter granularity as described by Sato et al2.
For flow cytometry of small intestine organoid cells, organoids were dissociated using TrypLE (37°C). After vigorously pipetting through a p200 pipette causing mechanical disruption, the crypt suspension was washed with 10ml of DMEM/F12 medium containing 10% FBS and 0.8 KU/ml DNase1 and passaged through a cell strainer. Where applicable, the cells were directly stained or first fixed (PFA4%) and permeabilized (methanol) depending on the extracellular or intracellular location of the target protein. All stainings with live cells were performed in PBS without Mg2+ and Ca2+ with 0.5% BSA. For 5-ethynyl-2’-deoxyuridine (EdU) incorporation experiments there was a 1 hour pre-incubation of EdU in the ENR medium of the intact organoid cultures before dissociating the cells with TrypLE. Cells were stained using Click-it kits for imaging and flow cytometry (Life Technologies). For cell cycle analysis, single cell suspensions obtained from dissociated organoids were fixed and stained with Hoechst 33342 (Life Technologies), then assessed with flow cytometry for DNA content and ploidy.
For intracellular phospho-Stat staining of organoids, organoids were mechanically disrupted into crypt fragments, stimulated for 20 minutes with 20 ng/ml IL-22 at 37°C, and then fixed with PFA4% (10 minutes at 37°C). To assess Stat activation in Lgr5+ cells, after freshly isolating crypts from Lgr5GFP mice, single cell suspensions including Y-27632 (10uM) were stimulated with IL-22. After obtaining a single cell suspension of stimulated and fixed cells, the samples were filtered (40uM) and permeabilized with ice-cold (−20°C) methanol. Fixed and permeabilized cells were rehydrated with PBS and thoroughly washed with PBS before staining, then stained with anti-phospho-Stat3 and anti-phospho-Stat1, plus anti-GFP or cell surface markers, for 30 minutes at 4°C.
All flow cytometry was performed with an LSRII cytometer (BD Biosciences) using FACSDiva (BD Biosciences), and the data were analyzed with FlowJo software (Treestar). Refer to Supplementary Table 1 for full description of antibodies used.
Immunoblotting analysis
Western blot analysis was carried out on total protein extracts. Free-floating crypts isolated from small intestine were treated in DMEM supplemented with Y-27632 (10 ng/ml, Tocris) and IL-22 (5ng/ml, 30 minutes). Vehicle (PBS) was added to control (CTRL) wells. Crypts were then lysed in RIPA buffer containing a cocktail of protease and phosphatase inhibitors (Sigma). After sonication, protein amount was determined using the bicinchoninic acid assay Kit (Pierce). Loading 30 μg/lane of lysate, proteins were separated using electrophoresis in a 10% polyacrylamide gel and transferred to nitrocellulose. Membranes were blocked for one hour at room temperature with 1% Blot-Qualified BSA (Promega, W384A) and 1% non-fat milk (LabScientific, M0841) and then incubated overnight at 4°c with the following primary antibodies: rabbit anti-phospho-Stat1 (7649P), rabbit anti-phospho-Stat3 (9131S), rabbit anti-Stat1 (9172P), and rabbit anti-Stat3 (4904P), all from Cell Signaling. This was followed by incubation with the secondary antibody anti-rabbit HRP (7074P2) and visualization with the Pierce™ ECL Western Blotting Substrate (Thermo Scientific, 32106).
MTT assay
Cell viability in organoids was assessed with a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) test, based on the identification of metabolically active cells. The organoids were incubated with MTT (0.9 mg/ml final concentration, Sigma) for 2 hours at 37°C. Matrigel and cells containing intracellular reduction end product formazan were solubilized with acidic isopropanol (isopropanol with HCl) and the reduction end formazan production was evaluated by spectrophotometry using the Infinite M1000 pro plate reader (Tecan).
RT qPCR
For quantitative (q)PCR, segments of small intestine or isolated crypts were harvested from euthanized mice and stored at −80°C. Alternatively, RNA was isolated from organoids after in vitro culture. Extracted RNA was also stored at −80°C. RT-PCR was performed with a QuantiTect Reverse Transcription Kit (QIAGEN) or a High-Capacity RNA-to-cDNA Kit (Applied Biosystems). qPCR was performed on a Step-One Plus or QuantStudio® 7 Flex System (Applied Biosystems) using TaqMan Universal PCR Master Mix (Applied Biosystems). Specific primers were obtained from Applied Biosystems: β-actin: Mm01205647_g1; Hprt: Mm00446968_m1; Reg3β: Mm00440616_g1; Reg3γ: Mm00441127_m1; WNT3: Mm00437336_m1 and EGF: Mm00438696_m1; Rspo3: Mm00661105_m1; Axin2: Mm00443610_m1; β-catenin: Mm00483039_m1. DEFA1: Mm02524428_g1, IL-22Ra1: Mm01192943_m1. Other primers were obtained from PrimerBank: Gapdh (ID 6679937a1), Cdnk1a/ P21 (ID 6671726a1); Cdnk2d/P19 (ID 31981844a1); WNT3a (ID 7106447a1); Axin2 (ID 31982733a1); Hes1 (ID 6680205a1) Dll4 (ID 9506547a1) Dll1 (ID 6681197a1), for which cDNAs were amplified with SYBR master mix (Applied Biosystems) in QuantStudio® 7 Flex System (Applied Biosystems). Relative amounts of mRNA were calculated by the comparative ΔC(t) method with β-Actin, Hprt or Gapdh as house keeping genes.
For Il22Ra1 qPCR on Lgr5+ cells, dissociated crypt cells from Lgr5-GFP mice were stained and isolated using the following monoclonal antibodies/parameters: EpCAM-1 (G8.8; BD Bioscience); CD45 (30F11; Life Technologies); CD31 (390; BioLegend), Ter119 (Ter119; BioLegend); GFP expression; dead cells were excluded using 7AAD. Cells were acquired on a BD ARIAIII and FACS-sorted. Cells were sorted directly into RA-1/TCEP (Macherey-Nagel) lysis buffer and stored at −80°C until further analysis. RNA of hematopoietic cells (composite of DC, ILC and B-cells) was used as negative control. RNA was extracted using the NucleoSpin RNA XS kit (Machery Nagel) and cDNA was prepared with Ovation® Pico and PicoSL WTA Systems V2 (NuGen). For quantitative PCR, a Neviti Thermal Cycler (Applied Biosystems) and DyNAmo Flash SYBR Green qPCR kit (Finnzymes) were used, with the addition of MgCl2 to a final concentration of 4 mM. All reactions were done in duplicate and normalized to Gapdh. Relative expression was calculated by the cycling threshold (CT) method as 2-ΔCT. The primer sequences were as follows: Il22ra1 forward TCGGCTTGCTCTGTTATC; il22ra1 reverse CCACTGAGGTCCAAGACA.
Gene set enrichment analysis (GSEA)
To explore the association of ISC gene signatures (GSE33948 and GSE23672)16 with Stat3-regulated genes, we performed GSEA in a Stat3ΔIEC murine DSS colitis dataset (GSE15955)12, comparing Stat3fl/fl;Villin-Cre− (WT) vs. Stat3fl/fl;Villin-Cre+ (Stat3ΔIEC) mice with DSS colitis (GSEA2-2.2.0; http://www.broadinstitute.org/gsea)39,40. A Paneth cell signature gene set was used as a negative control (DLL1+CD24hi, GSE39915)17. Nominal P-values are shown.
Statistics and software
To detect an effect size of > 50% difference in means, with an assumed coefficient of variation of 30%, common in biological systems, we attempted to have at least five samples per group, particularly for in vivo studies. All experiments were repeated at least once. No mice were excluded from experiments. Experiments that were technical failures, such as experiments in vitro where cultures did not grow or experiments in vivo where transplanted control mice (BM + T cells) did not develop GVHD, were not included for analysis. Occasional individual mice that died post-transplant prior to analysis could not be included for tissue evaluation.
All bars and error bars represent the means + standard error of the mean (SEM) for the various groups. Statistics are based on “n” biologic replicates. All tests performed are two sided. For the comparisons of two groups, a t-test or nonparametric test was performed. Adjustments for multiple comparisons were made. In most cases, non-parametric testing was performed if normal distribution could not be assumed. RT qPCRs and ordinal outcome variables were tested non-parametrically. All analyses of statistical significance were calculated and displayed vs. the reference control group unless otherwise stated.
There is large biologic variation in organoid size. Statistical analyses of organoid sizes were thus based on all evaluable organoids (at least 25 organoids/group for all experiments). Statistical analyses of organoid numbers/efficiency were based on individual wells. To take into account intra-individual and intra-experimental variation as well, all in vitro experiments were performed at least twice with multiple wells per condition and sample material coming from at least two different mice. Statistical analyses of stem cell numbers (Lgr5-LacZ mice) in vivo were performed on multiple independent sections from multiple mice. Statistics were calculated and display graphs were generated using Graphpad Prism.
Extended Data
Extended Data Figure 1.
a, Microscopic tracing of organoid to measure surface area. b, Brightfield images of SI organoids from B6 mice, after seven days of culture +/− IL-22 (5ng/ml). c-e, Organoid efficiency (%) relative to control (0 ng/ml) for (c) B6 SI organoids (statistics on data combined from n=19 wells/group from 19 individual mice), (d) B6 LI organoids (n=4 mice/group) cultured +/− rmIL-22 for 7 days, and (e) human SI organoids cultured +/− rhIL-22 for six days (n=3 donors/group). f, RT qPCR of relative mRNA expression of Wnt3, Ctnnb1, and Axin2 genes of the Wnt/β-catenin axis in SI organoids cultured +/− rmIL-22; n=3 (0-1ng/ml) and n=4 (5ng/ml) mice/group. g, Numbers of SI organoids/well +/− rmIL-22 (5ng/ml) in the presence or absence of R-spondin-1 (n=6 wells/group). h, RT qPCR-determined relative mRNA expression of notch pathway genes (Hes1, Dll1, Dll4; n=8 mice/group) as well as of Slit2 and its receptor Robo1 (n=3 mice/group) in day 7 SI organoids cultured +/− rmIL-22. i, Relative expression of Wnt3 and Axin2 (n=3 mice/group), Hes1 (n=5 mice/group), and Dll1 and Dll4 (n=6 mice/group) genes in LI organoids. j, RT qPCR for the relative mRNA expression of Reg3b and Reg3g innate antimicrobials in SI organoids cultured with rmIL-22; n=3 (0-1ng/ml) and n=4 (5ng/ml) mice/group. Organoid efficiency/number comparisons were performed with t-tests (two groups) or ANOVA (multiple groups). RT qPCRs statistics were performed with non-parametric Mann-Whitney (two groups) or Kruskal-Wallis (multiple groups) tests. Graphs demonstrate mean +/− SEM; ns: non-significant; *p<0.5, ***p< 0.001. Data combined from at least two independent experiments unless otherwise stated.
Extended Data Figure 2.
a Intracellular staining of pStat3 (Y705) in organoid cells cultured under standard EGF, Noggin, and R-spondin-1 (ENR) conditions followed by a 20 minute pulse of 20 ng/ml IL-22, evaluated by flow cytometry; data representative of two independent experiments. b, Brightfield images of SI organoids 4 days after crypt culture +/− Stattic; data representative of three experiments. c, SI organoids/well from WT and Stat1−/− mice +/− rmIL-22; n=6 wells/group; ANOVA. d-e, Day 5 organoids from Stat3fl/fl SI crypt cells cultured +/− rmIL-22 (5ng/ml) in the absence of adeno-Cre infection; (d) numbers/well (n=6 wells/group) and size (n=35 control and n=42 IL-22-treated organoids/group), t-test; (e) brightfield images representative of three experiments. f-g, GSEAs of the expression of (f) a second independent ISC signature gene set (GSE36497) and (g) a negative control DLL1+CD24hi Paneth cell gene set (GSE39915) in Stat3fl/fl;Villin-Cre− (WT) vs Stat3fl/fl;Villin-Cre+ (Stat3ΔIEC) mice with DSS colitis, using GEO database array data (GSE15955). Each GSEA represents one analysis; nominal p-values are shown. Bar graphs represent mean + SEM; ns: non-significant; ***p<0.001. Data combined from at least two independent experiments unless otherwise stated.
Extended Data Figure 3.
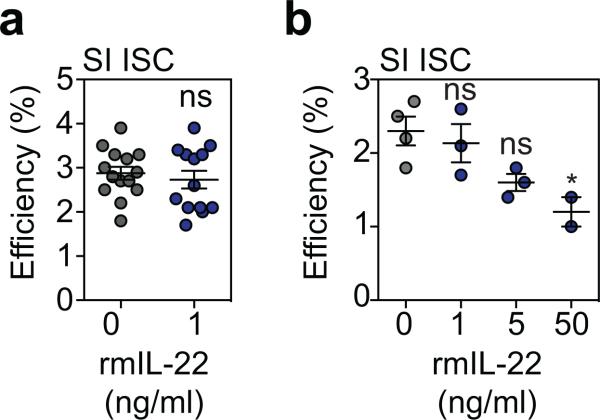
a-b Organoid efficiency as % of plated cells, in organoid cultures from sorted Lgr5+ ISCs from B6 Lgr5-GFP reporter mice using (a) a concentration of 1 ng/ml (n=14 wells/group combined from three experiments; t-test) and (b) with a concentration range (one experiment, n=3 wells/group; ANOVA). Graphs indicate mean +/− SEM; ns: non-significant; *p<0.05.
Extended Data Figure 4.
a-b, (a) Confocal images (nuclear staining, blue; and EdU staining, red; one experiment) and (b) FACS analysis of EdU incorporation (1hr) in SI organoids cultured in the presence or absence of rmIL-22 (1 ng/ml); histogram representative of two experiments, graph shows paired t-test, n=3 mice/group combined from two experiments. c-d, Cdkn1a (p21) and Cdkn2d (p19) mRNA expression (RT qPCR) in organoids cultured from (c) SI and (d) LI crypts for 24 hours with 0, 3, or 6 hours exposure to IL-22 prior to harvesting; Kruskal-Wallis analysis, n=6 mice/group combined from two independent experiments. All bar graphs represent mean + SEM; ns: non-significant; *p<0.05, **p< 0.01, ***p<.001.
Extended Data Figure 5.
a-c, Dissociated single cells from WT B6 crypts were exposed to escalating doses of irradiation ex vivo. a-b, Crypt cells were plated three hours prior to irradiation, and cultures were treated with rmIL-22 (5ng/ml) added to the culture at 3 hours before, 30 minutes before, 10 minutes after, or 24 hours after 4 Gy irradiation. Two days after irradiation, organoids were evaluated for (a) MTT viability testing (% positive, n=6 wells/group) and (b) the number of organoids generated (n=6 wells/group). c, The effect of IL-22 after irradiation was evaluated by measuring organoid number two days and seven days after irradiation (Day 2: n=9 wells/group for 1-2 Gy and n=6 wells/group for 4 Gy; Day 7: 4 Gy, n=20 wells/group). Culture +/− IL-22 was initiated three hours prior to irradiation. d, Small and large intestine crypt Il22ra1 expression determined by qPCR; RNA isolated from fresh crypts of B6 mice harvested one day (20-26 hours) after total body irradiation; n=12 control and n=11 irradiated mice/group. Comparisons performed with t-tests (two groups) or ANOVA (multiple groups). Bar graphs represent mean + SEM; ns: non-significant; *p<0.05, **p<0.01, ***p<0.001. Data combined from at least two independent experiments.
Extended Data Figure 6.
B6 recipient mice were transplanted with only TCD bone marrow from LP donors or with bone marrow and T cells from LP donors to induce GVHD (H-2b→H-2b). Mice receiving T cells were treated daily with PBS or 4μg rmIL-22 by IP injection starting day 7 post-BMT. a, Pathologic scoring of apoptosis in intestinal tissues three weeks after BMT. Data from two experiments combined: TCD BM only mice (n=10), PBS-treated mice (n=9), and IL-22-treated mice (n=8); Kruskal-Wallis analysis. b, Representative haematoxylin and eosin staining of SI and LI. Arrows indicate apoptotic cells within the intestinal epithelium. c, Splenocytes from recipients were analyzed with flow cytometry three weeks after BMT, indicating frequencies of T cell subsets, expression of activation marker CD25, and expression of gut homing molecule α4β7 integrin; n=9 PBS-treated and n=10 IL-22-treated mice/group; t-test analysis. d, Expression of inflammatory cytokines in spleen (n=9 PBS-treated and n=10 IL-22-treated mice/group) and small intestine (n=10 mice/group) was analyzed in recipient tissues three weeks post-BMT; t-test analyses, multiple comparisons corrected for with Holm-Sidak correction. e, Reg3β immunohistochemistry staining in SI samples of recipient mice three weeks post-BMT, data representative of three experiments. f-k, RT qPCR of relative mRNA expression in SI tissue samples of PBS-treated vs. IL-22-treated mice 3 weeks post-BMT for: f, Wnt3; g, Egf; h, Hes1 (from purified crypts); i, Rspo3; j, Ctnnb1 (from purified crypts); k, Axin2 (from purified crypts); n=10 mice/group for purified crypt samples; n=8 (PBS-treated) and n=9 (IL-22-treated) mice/group for whole SI tissue samples; Mann-Whitney U test. Bar graphs represent mean + SEM; ns: non-significant; *p<0.05, **p<0.01. Data combined from two independent experiments unless stated otherwise.
Extended Data Figure 7.
a, Percentage of Paneth cells in organoids cultured +/− 5 ng/ml rmIL-22 for 7 days, as evaluated by flow cytometry after dissociation into single cells; n=7 independent cultures/group (one mouse per culture); t-test. b, RT qPCR analysis of the relative mRNA expression of Paneth cell gene Defa1 in SI organoids cultured +/− 5 ng/ml rmIL-22 for 7 days; n=5 independent cultures/group (1-2 pooled mice/culture); Mann-Whitney U test. c-e, Paneth cell IL-22R expression and Stat3 phosphorylation assessed by flow cytometry. Shown are (c) gating of Paneth cells based on side scatter and CD24 expression, (d) Paneth cell IL-22R expression at baseline and five days after 1200 cGy total body irradiation (one of two experiments), and (e) Stat3 phosphorylation in Paneth cells as determined by phosflow of dissociated crypt cells after a 20 minute pulse with rmIL-22 (20 ng/ml, 37°C; one of one of two experiments). Graphs demonstrate mean +/− SEM; ns: non-significant. Data combined from at least four independent experiments unless otherwise stated.
Extended Data Figure 8.
a, Relative mRNA expression of Il22ra1 in sorted Lgr5-GFP+ cells (n=4 biological replicates), with various sorted hematopoietic populations serving as negative controls, including intestinal dendritic cells (n=4), intestinal ILC3s (n=2), and splenic B cells (n=1). b, Lgr5 mRNA relative to Gapdh expression in sorted Lgr5-GFP+ cells and hematopoietic samples described above to confirm Lgr5 expression in sorted Lgr5-GFP+ cells. Graphs indicate mean +/− SEM; Mann-Whitney U tests; **p< 0.01.
Extended Data Figure 9.
a, Efficiency of WT and Lgr5-DTR SI organoid formation after culture with DT (1ng/μl) to deplete Lgr5+ cells; one of three experiments; n=6 (WT), n=5 (Lgr5-DTR) n=6 (1ng/ml IL-22), n=6 (5ng/ml IL-22) wells/group. b, Numbers of WT and Atoh1ΔIEC day 7 SI organoids cultured +/− rmIL-22 (5ng/ml); n=6 wells/group. c-d, Omission of EGF from the standard ENR medium.c, The effect of IL-22 on organoid numbers and size in the absence of EGF; n=6 wells/group for numbers; n=45 (ENR), n=37 (ENR+IL-22), n=42 (NR), n=54 (NR+IL-22) organoids/group for size; data combined from three experiments. d, Brightfield images of WT SI organoid cultures in the presence or absence of EGF (50 ng/ml), representative of three experiments. Bar graphs represent mean + SEM. Comparisons were performed with t-tests (2 groups) or ANOVA (multiple groups); ns: non-significant; *p<0.05, **p<0.01, ***p<0.001. Data combined from three independent experiments unless otherwise stated.
Extended Data Figure 10.
a-b, Area of SI (a) and LI (b) WT B6 organoids cultured +/− rhIL-22 dimer/Fc molecule F-652; SI: n=37 (0 ng/ml), n=60 (0.1 ng/ml), and n=41 (1 ng/ml) organoids/group combined from three experiments; LI: n=137 (0 ng/ml), n=83 (0.1 ng/ml), and n=132 (1 ng/ml) organoids/group combined from two experiments; ANOVA. c-d, Organoid efficiency relative to control in cultures of (c) B6 SI organoids (n=4 wells/group combined from two experiments) and (d) B6 LI organoids (n=3 wells/group; one of two experiments) treated with different concentrations of recombinant human F-652; ANOVA. e-f, B6 Lgr5-LacZ mice were treated with PBS or F-652 (100 μg/kg), administered subcutaneously on the day of total body irradiation (10-12 Gy) and again two days later; one of three experiments. e, Lgr5-LacZ+ crypt cells per SI circumference were evaluated at Day 3.5 post-irradiation (10 Gy); statistics based on n=11 independent sections (PBS-treated) vs. n=14 independent sections (F-652-treated) from irradiated mice; independent sections were derived from 3 mice per group; first dose of PBS or F-652 was administered four hours prior to irradiation; Mann-Whitney. f, Representative crypt base images 3.5 days after irradiation (10Gy). Arrows indicate Lgr5-LacZ+ crypt cells. Graphs demonstrate mean +/− SEM; ns: non-significant; *p<0.05, **p<0.01.
Supplementary Material
Acknowledgements
We gratefully acknowledge the technical assistance of the MSKCC Research Animal Resource Center and Molecular Cytology Core Facility. We also thank Hans Clevers, Henner Farin, Sabine Middendorp, Caroline Wiegerinck, Johan van Es, Marc van de Wetering, Nobuo Sasaki, Joe Sun, and Ming Li for their advice and critical evaluation of our work. This research was supported by National Institutes of Health award numbers K08-HL115355 (A.M.H.), R01-HL125571 (A.M.H.), R01-HL069929 (M.R.M.vdB.), R01-AI100288 (M.R.M.vdB.), R01-AI080455 (M.R.M.vdB.), R01-AI101406 (M.R.M.vdB.), P01-CA023766/Project 4 (Richard J. O'Reilly/M.R.M.vdB.), K99-CA176376 (J.A.D.), and P30-CA008748 (MSKCC Core Grant). Support was also received from the U.S National Institute of Allergy and Infectious Diseases (NIAID Contract HHSN272200900059C), the European Union (award GC220918, C. Blackburn), The Experimental Therapeutics Center of MSKCC funded by Mr. William H. Goodwin and Mrs. Alice Goodwin, The Lymphoma Foundation, Alex's Lemonade Stand, The Geoffrey Beene Cancer Research Center at MSKCC, The Susan and Peter Solomon Divisional Genomics Program, MSKCC Cycle for Survival, and The Lucille Castori Center for Microbes, Inflammation & Cancer. T.C. was supported by Innovational Research Incentives Scheme Vidi grant #91710377 from the Netherlands Organization for Scientific Research (Zon-MW), and M.R. was supported by the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme FP7/2007-2013 under REA grant agreement no. 289720. A.M.M. was supported by the Bio Medical Exchange Program of the Deutscher Akademischer Austauschdienst. C.A.L. was supported by Dutch Cancer Society clinical fellowship grant #2013-5883 and by a mobility grant from the University Medical Center Utrecht. J.A.D. was supported by a C.J. Martin fellowship from the Australian National Health and Medical Research Council, a Scholar Award from the American Society of Hematology, and the Mechtild Harf Research Grant from the DKMS Foundation for Giving Life. A.M.H. was supported by a Scholar Award from the American Society of Hematology, a New Investigator Award from the American Society for Blood and Marrow Transplantation, and the Amy Strelzer Manasevit Research Program. A provisional patent application has been filed on the use of IL-22 and F-652 as ISC growth factors (US 61/901,151) with A.M.H., C.A.L., and M.R.M.vdB. listed as inventors.
Footnotes
Author Contributions
C.A.L. and M.C. designed and performed organoid experiments. A.M.M and M.H.O. performed and analyzed in vivo experiments. J.A.D, R.R.J., and E.V. provided input and helped with various assays. L.F.Y., O.M.S., and G.L. performed and monitored bone marrow transplants and maintained the mouse colonies. J.A.I. assisted with organoid quantification. YF analyzed crypt sizes and confocal microscopy. S.T. assisted with ILC co-culture experiments. G.H., M.L.M., and R.K. assisted with ISC isolation and in vivo ISC quantification experiments and provided reagents and expertise. K.P.O. and L.D. assisted with adenoviral-Cre experiments and optimizing various assays. Y.L. and N.F.S. assisted with Paneth cell deficiency experiments. M.M. and E.E.N. performed the GSEA analyses and assisted with reagents and resources. M.R.H. performed PCR analyses on purified stem cells and immune cells under the guidance of T.C., and C.L. analyzed intestinal histopathology. M.R.M.vdB. and A.M.H. supervised the research. All authors contributed to experimental design, interpretation, and manuscript editing.
The authors have no competing financial interests.
References
- 1.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 2.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2010;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med. 2015;21:698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dudakov JA, Hanash AM, van den Brink MR. Interleukin-22: immunobiology and pathology. Annu Rev Immunol. 2015;33:747–785. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metcalfe C, Kljavin NM, Ybarra R, de Sauvage FJ. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–159. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 7.Zenewicz LA, et al. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity. 2007;27:647–659. doi: 10.1016/j.immuni.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aujla SJ, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudakov JA, et al. Interleukin-22 drives endogenous thymic regeneration in mice. Science. 2012;336:91–95. doi: 10.1126/science.1218004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459 doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 11.Zhou WJ, Geng ZH, Spence JR, Geng JG. Induction of intestinal stem cells by R-spondin 1 and Slit2 augments chemoradioprotection. Nature. 2013;501:107–111. doi: 10.1038/nature12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickert G, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthews JR, Sansom OJ, Clarke AR. Absolute requirement for STAT3 function in small-intestine crypt stem cell survival. Cell Death Differ. 2011;18:1934–1943. doi: 10.1038/cdd.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez PP, et al. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13:1235–1242. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 16.Munoz J, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. Embo J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Es JH, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eriguchi Y, et al. Reciprocal expression of enteric antimicrobial proteins in intestinal graft-versus-host disease. Biol Blood Marrow Transplant. 2013;19:1525–1529. doi: 10.1016/j.bbmt.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 19.Hanash AM, et al. Interleukin-22 Protects Intestinal Stem Cells from Immune- Mediated Tissue Damage and Regulates Sensitivity to Graft versus Host Disease. Immunity. 2012;37:339–350. doi: 10.1016/j.immuni.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takashima S, et al. The Wnt agonist R-spondin1 regulates systemic graft-versus-host disease by protecting intestinal stem cells. J Exp Med. 2011;208:285–294. doi: 10.1084/jem.20101559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenq RR, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012;209:903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eriguchi Y, et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of alpha-defensins. Blood. 2012;120:223–231. doi: 10.1182/blood-2011-12-401166. [DOI] [PubMed] [Google Scholar]
- 23.Levine JE, et al. Low Paneth cell numbers at onset of gastrointestinal graft-versus-host disease identify patients at high risk for nonrelapse mortality. Blood. 2013;122:1505–1509. doi: 10.1182/blood-2013-02-485813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabiri Z, et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development. 2014;141:2206–2215. doi: 10.1242/dev.104976. [DOI] [PubMed] [Google Scholar]
- 25.Tian H, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durand A, et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc Natl Acad Sci U S A. 2012;109:8965–8970. doi: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Couturier M, et al. IL-22 deficiency in donor T cells attenuates murine acute graft- versus-host disease mortality while sparing the graft-versus-leukemia effect. Leukemia. 2013;27:1527–1537. doi: 10.1038/leu.2013.39. [DOI] [PubMed] [Google Scholar]
- 28.Zhao K, et al. Interleukin-22 Aggravates Murine Acute Graft-Versus-Host Disease by Expanding Effector T Cell and Reducing Regulatory T Cell. J Interferon Cytokine Res. 2014;34:707–715. doi: 10.1089/jir.2013.0099. [DOI] [PubMed] [Google Scholar]
- 29.Munneke JM, et al. Activated innate lymphoid cells are associated with a reduced susceptibility to graft-versus-host disease. Blood. 2014;124:812–821. doi: 10.1182/blood-2013-11-536888. [DOI] [PubMed] [Google Scholar]
- 30.Hill GR, Ferrara JL. The primacy of the gastrointestinal tract as a target organ of acute graft-versus-host disease: rationale for the use of cytokine shields in allogeneic bone marrow transplantation. Blood. 2000;95:2754–2759. [PubMed] [Google Scholar]
- 31.Ootani A, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, et al. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology. 2013;145:383–395. doi: 10.1053/j.gastro.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magness ST, et al. A multicenter study to standardize reporting and analyses of fluorescence-activated cell-sorted murine intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2013;305:G542–551. doi: 10.1152/ajpgi.00481.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spits H, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 35.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 36.Shroyer NF, et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 37.Alpdogan Ö, et al. IL-7 enhances peripheral T cell reconstitution after allogeneic hematopoietic stem cell transplantation. Journal of Clinical Investigation. 2003;112:1095–1107. doi: 10.1172/JCI17865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.KR Cooke LK, Martin TR, Brewer J, Delmonte J, Jr, Crawford JM, Ferrara JL. An experimental model of idiopathic pneumonia syndrome after bone marrow transplantation: I. The roles of minor H antigens and endotoxin. 1996 [PubMed] [Google Scholar]
- 39.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.