Abstract
To understand the process by which antibiotic resistance genes are acquired by human pathogens, we functionally characterized the resistance reservoir in the microbial flora of healthy individuals. Most of the resistance genes we identified using culture independent sampling have not been previously identified and are evolutionarily distant from known resistance genes. By contrast, nearly half of the resistance genes we identified in cultured aerobic gut isolates (a small subset of the gut microbiome) are identical to resistance genes harbored by major pathogens. The immense diversity of resistance genes in the human microbiome could contribute to future emergence of antibiotic resistance in human pathogens.
Multiple antibiotic resistance in human pathogens has increased over the past decades and challenged our ability to treat bacterial infections (1, 2). For example, methicillin-resistant Staphylococcus aureus (MRSA) caused 18,964 mortalities in the USA in 2006 (3). The comparison with 14,627 AIDS-related mortalities that occurred in the same year (4) highlights the public health significance of just one multiresistant bacterial pathogen in an industrialized nation. Whole genome sequencing of bacteria has revealed that many of the resistance genes harbored by these strains have not evolved within the sequenced strain, but were acquired by lateral gene transfer events (5). Antibiotic resistance determinants encoded on mobilizable elements move between diverse bacteria to disseminate resistance genes into a variety of interacting microbial communities (6, 7). Consequently, there is an increasing interest in elucidating reservoirs of mobile antibiotic resistance genes that may be accessible to clinically relevant pathogens (8-10).
The human microbiome significantly impacts human health and plays beneficial roles in dietary processing and prevention of pathogen intrusion (11-15). The widespread use of antibiotics in human medicine and agriculture has likely induced substantial responsive changes in this community. Phylogenetic analysis of human and mice gut microbiomes reveal significant transient increases in the Proteobacteria at the expense of Firmicutes and Bacteroidetes during antibiotic treatment (16, 17). While the phylogenetic distribution of the human gut microbiome was found to largely re-establish after ciprofloxacin treatment (18), it has likely been enriched in resistance to this antibiotic. Indeed, persistent clarithromycin resistance in commensal enterococci has been observed years after cessation of antibiotic therapy (19), and resistance towards amoxicillin has been observed in cultured oral bacterial isolates from children not exposed to antibiotic therapy (20). Previous work has also shown that the abundance of ermB and tetQ, encoding resistance to erythromycin and tetracycline, respectively, has increased in frequency in cultured human Bacteriodes isolates during the past three decades, and that ermB has been exchanged between distantly related cultured commensal gut bacteria (21). These findings suggest that the human microflora could constitute a significant reservoir of antibiotic resistance genes accessible to pathogens (22, 23). We used functional selections coupled with metagenomic analysis to characterize the antibiotic resistance genes in oral and gut microflora of healthy individuals.
We isolated DNA directly from saliva and fecal samples from two unrelated healthy humans who had not been treated with antibiotics for at least 1 year (24). 1-3 kilobase fragments of the purified metagenomic DNA were cloned into an expression vector, and transformed into an Escherichia coli host strain, resulting in libraries of a total size of 9.3×109 base pairs. Antibiotic resistant clones from each library were selected by plating on Luria Broth agar containing one of 13 antibiotics belonging to the classes: amino-acid derivatives, aminoglycosides, amphenicols, beta-lactams, and tetracyclines, at concentrations where the wild-type host strain is susceptible (Table S1)(24). Inserts conferring resistance to all 13 antibiotics tested were sequenced and annotated, enabling the discovery of 95 unique inserts containing functional antibiotic resistance genes (Table S3)(24).
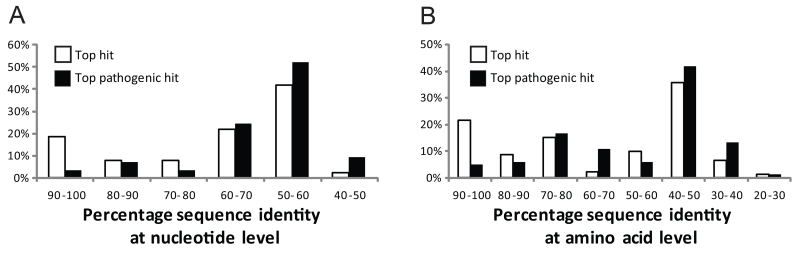
On average, the sequence similarity of the resistance genes we obtained and the closest related gene in GenBank is 69.5 % at the nucleotide level (Figure 1A) and 65.3 % at the amino acid level (Figure 1B)(24). A minority (19 %) had high homology (> 90 % amino acid identity) to previously known genes, including identical matches to the tet(Q)-3 gene previously identified in cultured Bacteriodes isolates from the human colon (21), and the CTX-M-15 enzyme, which over the past decade has become one of the most prevalent extended-spectrum beta-lactamases (Table 1)(25). Most of the closely related homologs are derived from commensals including non pathogenic species such as Bifidobacterium longum (27), as well as commensals with the capacity to become opportunistic pathogens such as Bacteroides fragilis and Bacteroides uniformis (Table S3)(21). Interestingly, we also identified genes which encode proteins that are 100 % identical to hypothetical proteins of unverified function in GenBank, e.g. BACUNI_02013 from Bacteroides uniformis ATCC 8492, which we show encodes resistance to broad-spectrum beta-lactams such as amoxicillin and carbenicillin, and the third-generation oxyimino-cephalosporin cefdinir (Table S3)(26). This highlights the utility of a functional selection approach to improve annotation of genomic and metagenomic sequencing data from the human microbiome project (31).
Figure 1.
Distributions of (A) nucleotide identities and (B) amino acid identities for 93 resistance genes identified from DNA extracted directly from saliva and fecal samples to the most similar resistance gene from any organism (white bars) as well as the most similar resistance gene harbored by a pathogenic isolate (black bars) in GenBank (Table S3)(24). Two resistance genes with no significant similarity to sequences in GenBank are not shown.
Table 1.
Unique beta-lactamase genes identified from gut and oral microbiomes from healthy humans. Gene ID refers to unique identifier in Table S3 and S4, and enzyme names use established nomenclature (24, 32). Percentage amino acid identity to the closest related gene in GenBank is calculated using the global alignment program clustalW (24, 33).
| Beta- Lactamase Family |
Enzyme name |
Gene ID | Genbank ID |
Source | % amino acid identity to NCBI |
|---|---|---|---|---|---|
| AmpC | AmpC- EcolK12 |
AX_iG2_08 | GQ343010 | Aerobic gut isolate | 100.0 |
| AmpC-EC6 | PE_iG1_02 | GQ343155 | Aerobic gut isolate | 100.0 | |
| AmpC-EC31 | PE_iG2_05 | GQ343162 | Aerobic gut isolate | 100.0 | |
| AmpC-HG1 | AX_iG2_21 | GQ343018 | Aerobic gut isolate | 99.7 | |
| AmpC-HG2 | CA_iG2_12 | GQ343059 | Aerobic gut isolate | 99.5 | |
| AmpC-HG3 | CF_iG2_01 | GQ343071 | Aerobic gut isolate | 90.0 | |
| AmpC-HG4 | CF_iG2_06 | GQ343073 | Aerobic gut isolate | 97.4 | |
| AmpC-HG5 | CA_iG1_06 | GQ343055 | Aerobic gut isolate | 99.2 | |
| TEM | TEM-1b | AX_iG1_01 | GQ343004 | Aerobic gut isolate | 100.0 |
| TEM-168 | PE_iG2_13 | GQ343167 | Aerobic gut isolate | 99.7 | |
| TEM-169 | PI_iG2_05 | GQ343173 | Aerobic gut isolate | 99.1 | |
| CTX-M | CTX-M-15 | AX_iG1_04 | GQ343005 | Aerobic gut isolate and metagenomic gut sample |
100.0 |
| CblA | CblA-1 | AX_iG2_02 | GQ343019 | Aerobic gut isolate and metagenomic gut sample |
100.0 |
| CblA-2 | AX_mG2_03 | GQ342999 | Metagenomic gut sample | 99.7 | |
| CblA-3 | PE_mG2_02 | GQ343154 | Metagenomic gut sample | 99.0 | |
| CfxA | CfxA6 | AX_mG2_01 | GQ342996 | Metagenomic gut sample | 87.2 |
| HGA | HGA-1 | CA_mG1_02 | GQ343038 | Metagenomic gut sample | 61.4 |
| HGB | HGB-1 | AX_mG2_05 | GQ343000 | Metagenomic gut sample | 58.5 |
| HOA | HOA-1 | AX_mO1_01 | GQ343035 | Metagenomic saliva sample | 49.5 |
| HGC | HGC-1 | CA_mG1_01 | GQ343037 | Metagenomic gut sample | 48.1 |
| HGC-2 | CA_mG1_04 | GQ343039 | Metagenomic gut sample | 51.0 | |
| HGD | HGD-1 | CA_mG2_04 | GQ343044 | Metagenomic gut sample | 52.9 |
| HGE | HGE-1 | AX_mG2_11 | GQ343003 | Metagenomic gut sample | 37.1 |
| HGF | HGF-1 | AX_mG2_09 | GQ343002 | Metagenomic gut sample | 43.3 |
| HGG | HGG-1 | PE_mG1_01 | GQ343153 | Metagenomic gut sample | 38.8 |
| HGH | HGH-1 | PI_mG1_01 | GQ343170 | Metagenomic gut sample | 34.5 |
| HGI | HGI-1 | CA_mG2_07 | GQ343045 | Metagenomic gut sample | 42.6 |
Most of the antibiotic resistance genes harbored by the human microflora were distantly related (60.7 % at the nucleotide level and 54.9 % at the amino acid level) to antibiotic resistance genes so far detected in pathogenic isolates (Figure 1 and Table S3)(24). In total we identified 78 unique inserts with genes with low homology (<90 % amino acid identity) to proteins in GenBank, encoding resistance to the 13 antibiotics profiled (Table S1 and S3). This may imply that the resistance genes of the human microbiome are inaccessible or infrequently exchanged with human pathogens; however, all the resistance genes we identified in this study were functional in E. coli, which suggests that if a barrier to gene transfer exists between the constituents of the human microbiome and pathogens, it must stem from processes other than functional compatibility.
Phylogenetic analysis of the inserts using PhyloPythia (28) indicates that they predominantly originate from the phyla Bacteroidetes and Firmicutes, which dominate the gut flora (14). However, the majority of the genes we discovered have low sequence identity to resistance genes previously identified in pathogens from these phyla (e.g. Staphylococcus aureus and Streptococcus pneumoniae), as well as from the numerous pathogens that are readily culturable facultative anaerobic bacteria from the phylum Proteobacteria. While commensal Proteobacteria constitute less than 1 % of the human gut microflora (14), they increase in abundance during antibiotic treatment at the expense of the normally abundant Bacteroidetes and Firmicutes (16, 17). As a consequence of their normal low abundance in healthy individuals, they are not well represented in unbiased metagenomic libraries.
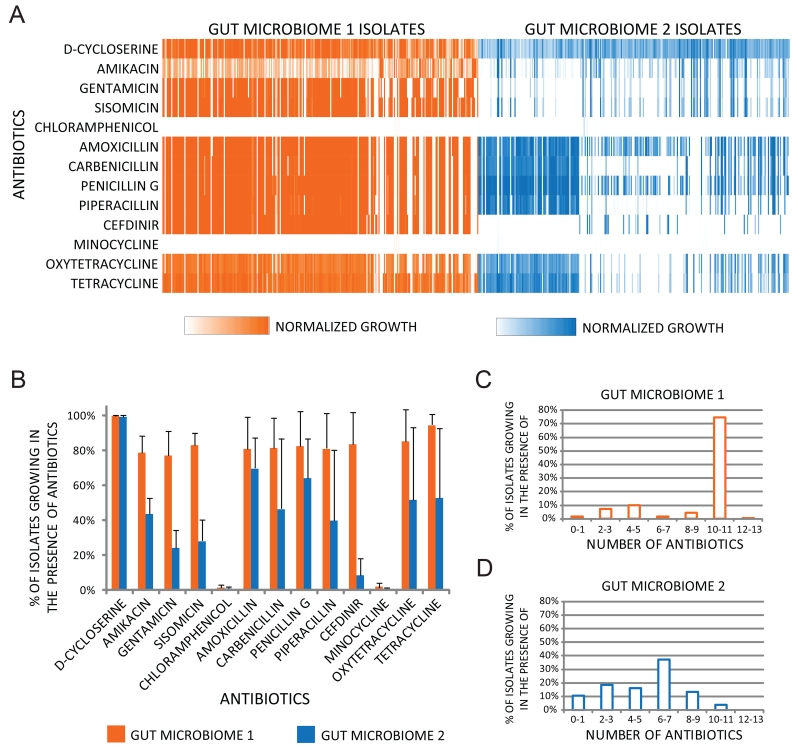
We isolated 572 bacterial strains on rich media under aerobic conditions from fecal samples from two healthy individuals (24). Phylogenetic profiling revealed that they belonged primarily to Proteobacteria, with a few Firmicutes and Actinobacteria (Figure S1). The isolates from individuals 1 and 2 were on average resistant to 9 and 5 out of 13 antibiotics, respectively (Figure 2C and D). Chloramphenicol and minocycline were the only antibiotics tested that were able to prevent the growth of more than 99 % of the isolates (Figure 2A, 2B, S2, S3).
Figure 2.
Antibiotic resistance profiles of cultured aerobic gut microbiome isolates. (A) Heat map displaying resistance profiles of 572 aerobic bacterial isolates obtained across 3 different sampling times from two human gut microbiomes. 7436 growth measurements of aerobic cultured microbiome isolates after 24 hrs at 37 °C in Luria Broth containing one of 13 antibiotics at concentrations between 20 and 100 mg/mL that prevent the growth of wild type E.coli (Table S1) are displayed as linear color-scaled bars. White denotes no growth, and color intensity is proportional to growth in the presence of antibiotic, scaled to growth in the absence of antibiotic per individual isolate. (B) Percentage of aerobic gut isolates resistant to each of 13 antibiotics. Each data point represents the mean number of isolates resistant to each antibiotic, and error bars represent the standard deviation of this mean value from each of the three sampling times. Histogram depicting the distribution of the number of different antibiotics that aerobic (C) gut microbiome 1 and (D) gut microbiome 2 isolates are resistant to.
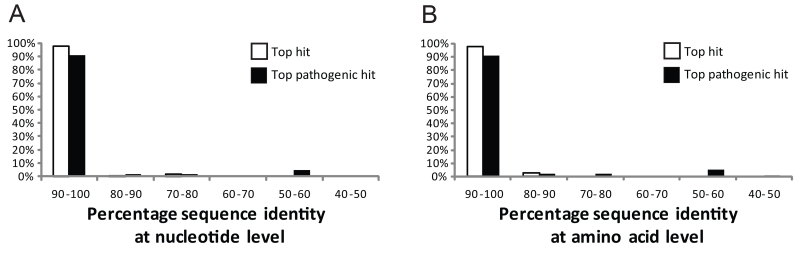
Functional selections identified 115 unique inserts encoding transferable antibiotic resistance genes from the cultured aerobic gut microbiome isolates (Figure 3 and Table S4)(24). We found that 95 % of these genes are over 90 % identical at the nucleotide level to resistance genes in pathogenic isolates, and almost half of these genes were 100% identical (Figure 3A), indicating an evolutionarily close relationship to the resistance genes harbored by clinical pathogens.
Figure 3.
Distributions of (A) nucleotide identities and (B) amino acid identities for 114 resistance genes identified from cultured aerobic gut isolates to the most similar resistance gene from any organism (white bars) as well as the most similar resistance gene harbored by a pathogenic isolate (black bars) in GenBank (Table S4)(24). One resistance gene with no significant similarity to sequences in GenBank is not shown.
The group of resistance genes identical to those in pathogens belong to one class of tetracycline efflux pumps (TetA), two classes of aminoglycoside modifying enzymes (AAC(3)-II and AAC(6)-Ib), and three classes of beta-lactam inactivating enzymes (TEM, AmpC, and CTX-M) (Figure 4). We identified a TEM-1 gene variant (Figure 4 and Table 1) in cultured isolates from one gut microbiome on every sampling time (24) that has recently been reported in pathogenic strains of Salmonella enterica, Escherichia coli, Klebsiella pneumonia, Haemophilus parainfluenzae, Serratia marcescens, Pseudomonas aeruginosa, and Neisseria meningitidis, isolated around the globe (Table S2). Nearly 80 % of the depositions of this TEM-1 variant to Genbank have occurred between 2007 and 2008, which seems to indicate a relationship between the emergence of this resistance gene variant in the clinic and its occurrence in healthy humans.
Figure 4.
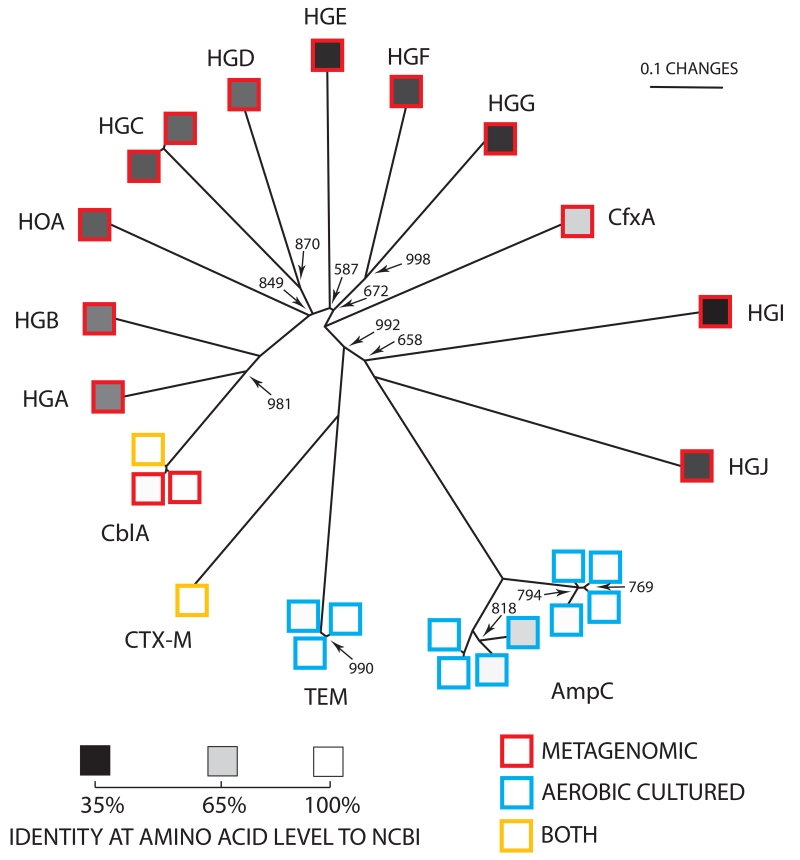
The phylogenetic relationship of unique beta-lactamases derived from gut and oral microbiomes from healthy humans is displayed as an unrooted neighbor-joining tree (24). Except for nodes indicated, bootstrap values = 1000. Scale bar is in fixed amino acid substitutions per sequence position. Border colors of squares denote sequences derived from cultured aerobic gut isolates (blue), metagenomic DNA (red) or both (yellow). Internal shading of each square represents percentage amino acid identity to the most similar sequence in GenBank, with a linear gradient between 100% identity (white) and 35% identity (black). Sequence groups are labeled according to standard nomenclature (Table 1)(24, 32).
The AmpC and CTX-M family of enzymes are extended-spectrum beta-lactamases which hydrolyze a wider variety of later generation beta-lactams (Table 1)(29). We identified the CTX-M-15 beta-lactamase (Figure 4 and Table 1) in libraries from cultured gut microbiome isolates across multiple sampling days, as well as in our metagenomic libraries from the same microbiome.
Global sequence alignments of each of the 27 unique beta-lactamase sequences from our study identified 15 distinct sequence groups (Figure 4). Of these groups, 5 were previously characterized (CblA, CfxA, CTX-M, TEM, and AmpC) (30), while 10 constitute entirely new beta-lactamase sequence families as they are between only 35 % and 61 % identical at the amino acid level to any gene products in GenBank (Figure 4). Interestingly, the known beta-lactamase genes we identified were from the cultured microbiome isolates, but the 10 novel gene families were identified solely by our culture independent characterization (Figure 4). In general we found that the metagenomically derived resistance genes in our study were more distantly related to previously identified genes than those derived from aerobic gut isolates (Figure 1, 3, S5, S6).
Of our 210 unique microbiome derived inserts encoding antibiotic resistance, we found a subset of 29 that also contained genes similar (> 96 % nucleotide sequence identity) to previously characterized transposases (Table S6). Of these, 14 transposases were identical to those previously identified on resistance and conjugative plasmids from Bacteroides fragilis, and clinical pathogenic isolates of Klebsiella pneumoniae, Escherichia coli and Salmonella enterica (Table S6). Interestingly, the proportion of identified transposases derived from the culturable aerobic isolates (90%) was significantly higher than those derived from metagenomic sampling (Pearson’s chi-squared test, p<0.0005).
Nearly half of the resistance genes identified in the cultured human gut isolates were identical at the nucleotide level to resistance genes from human pathogenic isolates. While this identity provides no information regarding the direction or mechanism of transfer, we can offer some speculation regarding the implications of our findings. First, the human microbiome may constitute a mobilizable reservoir of antibiotic resistance genes which are accessed by a pathogenic bacterium to acquire antibiotic resistance, although direct experimental proof of in vivo transfer of antibiotic resistance genes within the human microbiome remains to be shown. Second, in spite of selecting samples from untreated healthy humans, the aerobic cultured isolates may be dormant pathogens inhabiting the human microbiome. Third, by contrast with the cultured isolates, the resistance genes discovered by the culture independent approach were distantly related to resistance genes from even closely related pathogenic isolates, which may reflect an unappreciated barrier to lateral gene transfer in vivo between the dominant commensals in healthy humans and disease causing isolates. Fourth, this work exposes previous significant under-sampling of antibiotic resistance genes in the human microbiome.
We found many microbial DNA fragments encoding resistance genes that have never before been described, and our analysis suggests that we have just begun to scratch the surface of the immense diversity of antibiotic resistance machinery in the human microbiome. Over half of our inserts derived from both metagenomic libraries as well as libraries from cultured gut aerobes were sequenced only once in our experiment (Figure S4), and we estimate complete sequencing of these libraries would yield hundreds more resistance genes (24). Interestingly, when we compared the resistance genes derived from the microbiomes of the two different individuals, we find that over 65 % of the resistance genes derived from cultured aerobes are highly similar (>90 % nucleotide sequence identity) between the two individuals, whereas less than 10 % of the metagenomically derived resistance genes were highly similar between the individuals (Table S7)(24).
Many commensal bacterial species, which were once considered relatively harmless residents of the human microbiome, have recently emerged as multidrug resistant disease causing organisms (7). In the absence of in-depth characterization of the resistance reservoir of the human microbiome, the process by which antibiotic resistance emerges in human pathogens will remain unclear.
Supplementary Material
Acknowledgments
We acknowledge Julian Davies, John Aach and Michael Strong for helpful discussions regarding this manuscript, the expert assistance of George Jacoby and Karen Bush for beta-lactamase enzyme family nomenclature, Laurens Kraal and Graham Rockwell for sequence manipulations and similarity computations, and Sebastian Caliri, Aubrey Elison, and Tyler Elison for microbial culturing and DNA processing. We acknowledge NHGRI CEGS, the Personal Genome Project, The Bill and Melinda Gates Foundation, Harvard Biophysics Program, The Hartmann Foundation, and Det Kongelige Danske Videnskabernes Selskab for funding. Genbank accession numbers GQ342978-GQ343187 are listed in tables 1, S3, S4, and S7.
Footnotes
References and Notes
- 1.Walsh C. Nature. 2000 Aug;406:775. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 2.Alekshun MN, Levy SB. Cell. 2007 Mar;128:1037. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 3.CDC . Centers for Disease Control and Prevention - Active Bacterial Core Surveillance Report, Emerging Infections Program Network, Methicillin-Resistant Staphylococcus aureus, 2006. 2006. [Google Scholar]
- 4.CDC Centers for Disease Control and Prevention HIV/AIDS Surveillance Report, 2006. 2008;18 [Google Scholar]
- 5.Ochman H, Lawrence JG, Groisman EA. Nature. 2000 May;405:299. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 6.Davies J. Science. 1994 Apr;264:375. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 7.Marshall BM, Ochieng DJ, Levy SB. Microbe. 2009;4:231. [Google Scholar]
- 8.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Science. 2006 Jan;311:374. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 9.Dantas G, Sommer MOA, Oluwasegun RD, Church GM. Science. Apr 4;3202008:100. 2008. [Google Scholar]
- 10.Riesenfeld CS, Goodman RM, Handelsman J. Environmental Microbiology. 2004 Sep;6:981. doi: 10.1111/j.1462-2920.2004.00664.x. [DOI] [PubMed] [Google Scholar]
- 11.Gill SR, et al. Science. 2006 Jun 2;312:1355. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, et al. Nature. 2009 Jan 22;457:480. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia W, Li H, Zhao L, Nicholson JK. Nat Rev Drug Discov. 2008 Feb;7:123. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- 14.Eckburg PB, et al. Science. 2005 Jun 10;308:1635. [Google Scholar]
- 15.Ley RE, Peterson DA, Gordon JI. Cell. 2006 Feb 24;124837 doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 16.Young VB, Schmidt TM. J Clin Microbiol. 2004 Mar;42:1203. doi: 10.1128/JCM.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antonopoulos DA, et al. Infect Immun. 2009 Jun;77:2367. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dethlefsen L, Huse S, Sogin ML, Relman DA. PLoS Biology. 2008 Nov 01;6:e280. doi: 10.1371/journal.pbio.0060280. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sjolund M, Wreiber K, Andersson DI, Blaser MJ, Engstrand L. Annals of Internal Medicine. 2003 Sep;139483 doi: 10.7326/0003-4819-139-6-200309160-00011. [DOI] [PubMed] [Google Scholar]
- 20.Ready D, et al. Antimicrob Agents Chemother. 2004 Aug;48:2883. doi: 10.1128/AAC.48.8.2883-2887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Appl Environ Microbiol. 2001 Feb;67:561. doi: 10.1128/AEM.67.2.561-568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salyers AA, Gupta A, Wang Y. Trends Microbiol. 2004 Sep;12:412. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Dethlefsen L, McFall-Ngai M, Relman DA. Nature. 2007 Oct 18;449:811. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Materials and methods are available as supporting material on Science Online
- 25.Canton R, Coque TM. Curr Opin Microbiol. 2006 Oct;9:466. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Jacoby GA, Bush K. J Clin Microbiol. 2005 Dec;43:6220. doi: 10.1128/JCM.43.12.6220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Florez AB, Ammor MS, Alvarez-Martin P, Margolles A, Mayo B. Appl Environ Microbiol. 2006 Nov;72:7377. doi: 10.1128/AEM.00486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McHardy AC, Martin HG, Tsirigos A, Hugenholtz P, Rigoutsos I. Nat Methods. 2007 Jan;4:63. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- 29.Jacoby GA, Munoz-Price LS. N Engl J Med. 2005 Jan 27;352:380. doi: 10.1056/NEJMra041359. [DOI] [PubMed] [Google Scholar]
- 30.Amino Acid Sequences for TEM, SHV and OXA Extended-Spectrum and Inhibitor Resistant ß- Lactamases
- 31.Turnbaugh PJ, et al. Nature. 2007 Oct 18;449:804. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacoby GA. Antimicrob Agents Chemother. 2006 Apr;50:1123. doi: 10.1128/AAC.50.4.1123-1129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chenna R, et al. Nucleic Acids Res. 2003 Jul 1;31:3497. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.