Abstract
In certain human cancers, the expression of critical oncogenes is driven from large regulatory elements, called super-enhancers, which recruit much of the cell’s transcriptional apparatus and are defined by extensive acetylation of histone H3 lysine 27 (H3K27ac). In a subset of T-cell acute lymphoblastic leukemia (T-ALL) cases, we found that heterozygous somatic mutations are acquired that introduce binding motifs for the MYB transcription factor in a precise noncoding site, which creates a super-enhancer upstream of the TAL1 oncogene. MYB binds to this new site and recruits it’s H3K27 acetylase binding partner CBP, as well as core components of a major leukemogenic transcriptional complex that contains RUNX1, GATA-3, and TAL1 itself. Additionally, most endogenous super-enhancers found in T-ALL cells are occupied by MYB and CBP, suggesting a general role for MYB in super-enhancer initiation. Thus, this study identifies a genetic mechanism responsible for the generation of oncogenic super-enhancers in malignant cells.
In cancer cells, monoallelic expression of oncogenes can occur through a variety of mechanisms including chromosomal translocation, alterations in promoter methylation, parental imprinting, and intrachromosomal deletion (1–3). A quintessential example is TAL1d, an 80-kb deletion on chromosome 1p33 that is found in 25% of cases of human T-cell acute lymphoblastic leukemia (T-ALL). The deletion results in overexpression of TAL1, an oncogene coding for a basic helix-loop-helix transcription factor, by mediating fusion of TAL1 coding sequences to the regulatory elements of the ubiquitously expressed gene “SCL-interrupting locus” (STIL) (4–6). However, we previously reported that a substantial proportion of T-ALLs, including the Jurkat T-ALL cell line, have monoallelic overexpression of TAL1 but lack either the TAL1d abnormality or a chromosomal translocation of the TAL1 locus (7, 8).
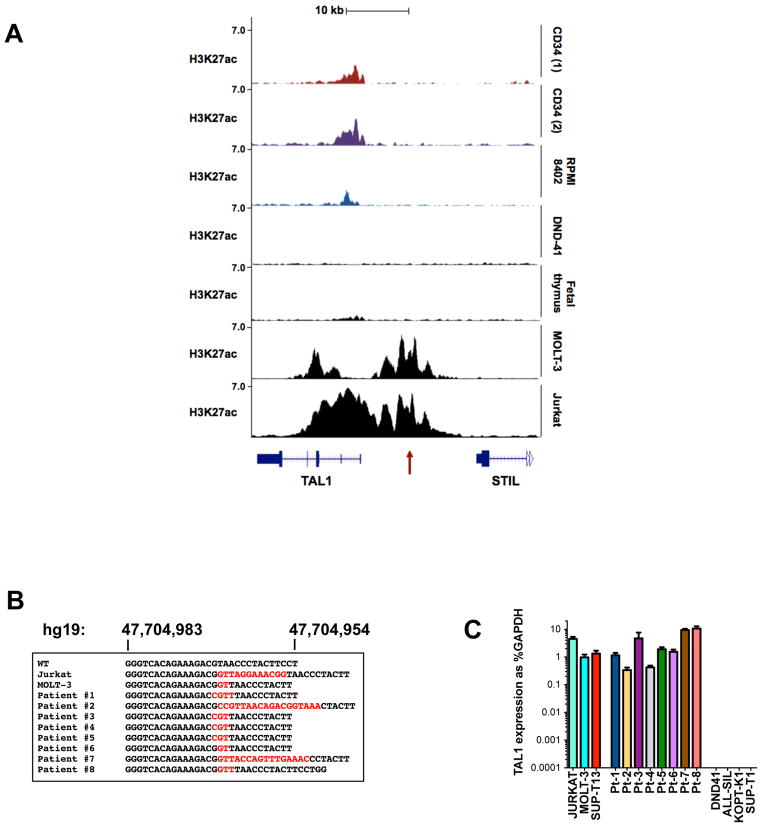
We hypothesized that cis-acting genomic lesions affecting TAL1 regulatory sequences might account for monoallelic TAL1 activation. Chromatin immunoprecipitation (ChIP) -seq analysis of Jurkat cells revealed aberrant histone H3 lysine 27 acetylation (H3K27ac), a mark of active transcription, starting upstream of the TAL1 transcriptional start site and extending across the first exons (Fig. 1A) (9, 10). Regions with such rich and broad H3K27ac marks have been termed super-enhancers (also stretch enhancers or locus control regions) and are commonly found at genes that determine cell identity in embryonic stem (ES) cells, and in tumor cells at oncogenes critical for the malignant cell state (11–17). The super-enhancer encompassing TAL1 in Jurkat cells was aberrant, in that it was not present in fetal thymocytes, normal CD34+ hematopoietic stem and progenitor cells (HSPCs) or in other T-ALL cell lines, such as TAL1d-positive RPMI-8402 cells and DND-41 T-ALL cells that lack TAL1 expression (Fig. 1A) (9). Of note, chromatin conformation capture (3C) experiments recently performed in Jurkat cells identified a looping interaction involving an enhancer site 8 kb upstream of the transcription start site (TSS), which coincides with the locations of both the aberrant super-enhancer and the positive auto-regulatory binding sites for members of the TAL1 complex in this cell line (Fig. 1A, red arrow) (9, 18).
Figure 1. Mutations at an intergenic site are associated with the TAL1 super-enhancer in T-ALL cells.
(A) Normalized ChIP-seq tracks for H3K27ac at the STIL-TAL1 locus in two human purified normal hematopoietic stem cell samples (CD34), the RPMI-8402 T-ALL cell line that over-expresses TAL1 as a result of TAL1d, DND-41 T-ALL cells that do not express TAL1, human fetal thymic tissue, and MOLT-3 and Jurkat cells that have mutations at a non-coding site 7.5 kb from the TAL1 transcriptional start site (red arrow). ChIP-seq read densities (y-axis) were normalized to reads per million reads sequenced in each sample. (B) Sequence alignments of the −7.5 kb site showing wild-type (WT) sequences in black and inserted sequences in red for Jurkat and MOLT-3 T-ALL cell lines and eight pediatric T-ALL patients. (C) TAL1 mRNA expression as determined by quantitative PCR and expressed as percentage of GAPDH. Error bars are ±SEM from two independent experiments performed in triplicate.
Sequencing of the genomic DNA region encompassing this site identified a heterozygous 12 bp insertion (GTTAGGAAACGG) that aligned precisely with the TAL1, GATA3, RUNX1, and HEB ChIP-seq peaks (Fig. 1B and 3A). Among eight additional TAL1-positive T-ALL cell lines, MOLT-3 cells also harbored an abnormal heterozygous 2 bp insertion (GT) at the same site (Fig. 1B), while none of ten TAL1-negative cell lines had a detectable genomic abnormality in this region (Table S1). Among 146 unselected pediatric primary T-ALL samples collected at diagnosis, eight cases (5.5%) had heterozygous indels 2–18 bp in length that overlapped at the same clearly defined hotspot (indels at this site are referred to here as “mutation of the TAL1 enhancer”, MuTE) (Fig. 1B). We estimate that MuTE abnormalities account for about half of the cases with unexplained monoallelic overexpression of TAL1 (7). Sequencing of DNA from remission bone marrow samples available from two mutation-positive patients showed wild-type sequences at this site, indicating that the mutations were somatically acquired in the tumor cells. All eight MuTE-positive T-ALLs markedly overexpressed TAL1 mRNA at levels comparable to those of the TAL1d-positive SUP-T13, and MuTE-positive Jurkat and MOLT-3 cells (Fig. 1C). Furthermore, MOLT-3 cells, that share the same 2 bp insertion as patient 6, also had a super-enhancer at the TAL1 locus (Fig. 1A).
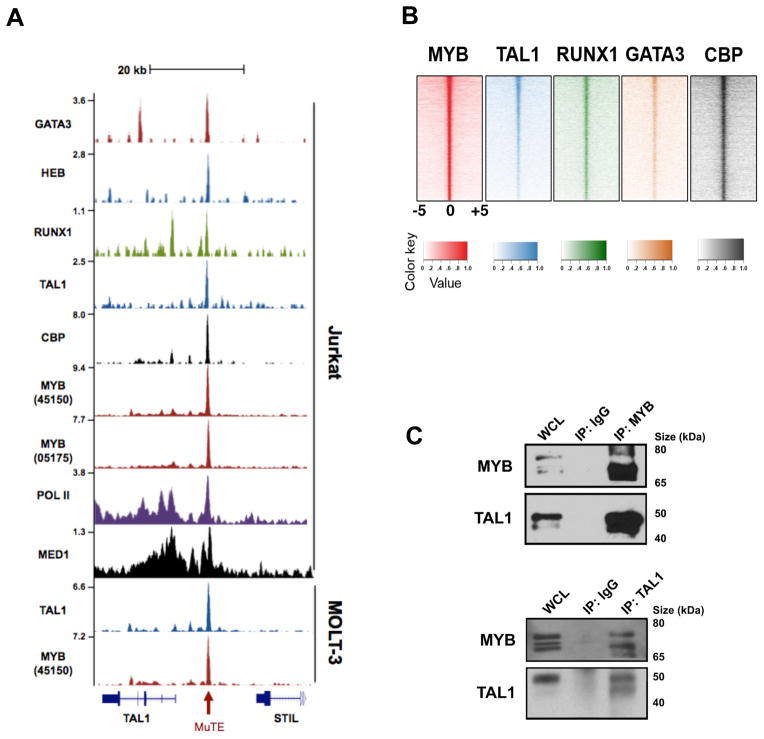
Figure 3. MYB binds the mutant TAL1 enhancer site and is a member of the TAL1 complex.
(A) ChIP-seq tracks at the STIL-TAL1 locus from Jurkat and MOLT-3 T-ALL cells for GATA3, HEB, RUNX1, TAL1, CBP, MYB (ab45150 antibody), MYB (05-175 antibody), RNA polymerase II (Pol II), and Mediator 1 (MED1). The mutation site at −7.5 kb is depicted with a red arrow. ChIP-seq read densities (y-axis) were normalized to reads per million reads sequenced in each sample. (B) Heatmaps showing genome-wide co-occupancy of MYB binding sites (+/− 1 kb) with those from TAL1, RUNX1, GATA3, and CBP sites as determined by ChIP-seq. For each region (y axis), the sequence density centered at 0 indicates overlapping bound regions. (C) Co-immunoprecipitation (Co-IP) and reciprocal Co-IP experiments performed from Jurkat lysates for MYB and TAL1. WCL, whole cell lysate; IgG, isotype control IgG antibody.
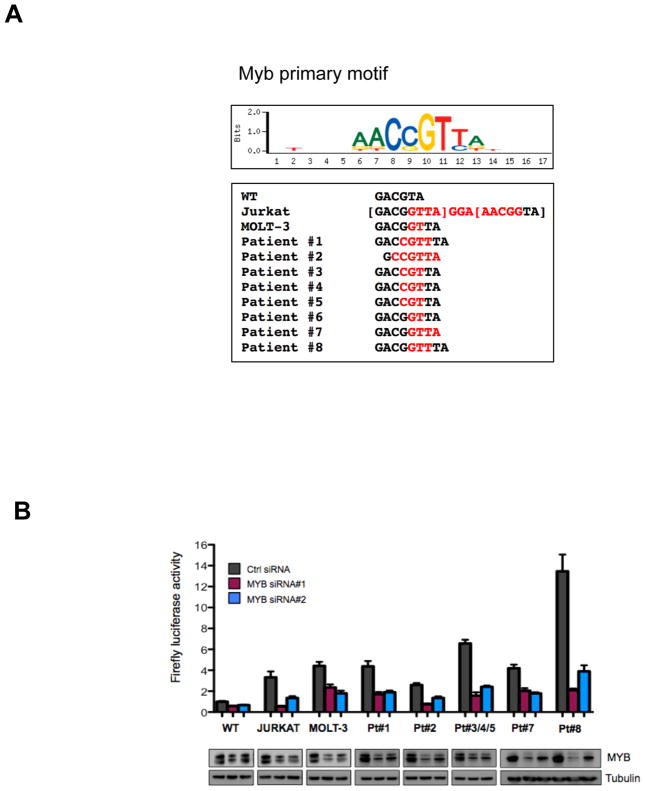
To test our hypothesis that the aberrantly formed super-enhancer drives monoallelic TAL1 expression, we analyzed the MuTE-positive samples for single nucleotide polymorphisms (SNPs) in the 3′ UTR of TAL1 gene from genomic DNA. Jurkat cells and five of the patient samples were informative, and in each case only one allele was detectable in the RNA sample, indicating monoallelic expression of TAL1 (Fig. S1). We then used the UniPROBE database to analyze whether the mutant sequences introduced new transcription factor binding sites (19). Surprisingly, all of the indels at this hotspot introduced de novo binding motifs for the MYB transcription factor, and two consecutive MYB binding motifs were generated by the 12 bp insertion in Jurkat cells (Fig. 2A; table S2).
Figure 2. Mutations of the TAL1 enhancer activate through recruitment of MYB.
(A) All TAL1 enhancer mutations introduce de novo MYB binding sites as determined by UniPROBE (19). The MYB primary binding motif is shown above the mutation-derived MYB motifs, with inserted nucleotides shown in red. (B) A 400 bp fragment of the −7.5 kb TAL1 enhancer containing either the wild-type sequence or each of the mutant alleles was cloned upstream of luciferase and a minimal promoter. Constructs were nucleofected into Jurkat cells, together with either control siRNA, or two independent siRNAs targeting MYB. Firefly luciferase activity was measured at 24 hrs, normalized to renilla luciferase to control for cell number and transfection efficiency, and expressed as a ratio relative to activity of the wild-type STIL-TAL enhancer construct. Error bars are ±SEM from two independent experiments performed in triplicate. Corresponding immunoblots for MYB and tubulin are shown below.
To ascertain if these mutations can activate gene expression, we cloned a 400 bp fragment containing either the wild-type allele or each of the TAL1-enhancer mutation alleles upstream of luciferase and tested the enhancer activity of this fragment in reporter assays. When we expressed these constructs in Jurkat cells, fragments containing each of the seven different indel mutations robustly increased reporter activity 3–14 fold more than the wild-type fragment (Fig. 2B and Fig. S2). Moreover, the activity of each of the mutant reporters was markedly reduced after MYB knockdown, indicating that the enhancer activity imparted by the mutations was indeed mediated by MYB (Fig. 2B). When we performed these experiments in HEK-293T cells, the mutant reporters had no increased activity above that of the wild-type reporter (Fig. S2), suggesting that transcription factors expressed in T-ALL, such as members of the TAL1 complex, are involved in activation of the mutant enhancer. We conclude that in T-ALL primary samples and cell lines, indel mutations that introduce MYB binding sites at a hotspot 7.5 kb upstream from the TSS of TAL1 generate a super-enhancer that drives monallelic overexpression of this oncogene.
Our recent ChIP-seq studies of the TAL1 complex in T-ALL cells identified binding of transcription factors in the core TAL1 complex (TAL1, GATA3, RUNX1, E2A and HEB) at the MYB enhancer, while knockdown of MYB generated a gene expression signature closely related to TAL1 knockdown (9, 10). We were technically unable to analyze MYB binding by ChIP-seq in our previous study, so at that time we interpreted these results to indicate that MYB is a critical downstream hub of the TAL1 complex (10). However, in the current study we used newly available MYB-specific antibodies to generate high-resolution maps of genome-wide MYB binding in Jurkat cells. Analysis of the TAL1 enhancer indel mutation site in Jurkat cells showed precise alignment of MYB binding and binding of each member of the TAL1 complex (Fig. 3A). There was also an abundance of RNA polymerase II (Pol II) and Mediator (MED1), stretching over more than 20 kb, indicating a large super-enhancer (table S3) (15). Notably, this site is also bound by MYB and TAL1 in MuTE-positive MOLT-3 cells (Fig. 3A), but not in RPMI-8402 and CCRF-CEM cells or a primary T-ALL sample, each of which overexpresses TAL1 driven by TAL1d (fig. S3). Nor were we able to detect binding of TAL1 at this site in HSPCs (fig. S3), indicating that a small insertion creating a de novo MYB binding motif at this location is required for MYB binding and subsequent binding by other members of the TAL1 complex. Accordingly, knockdown of MYB resulted in depletion of TAL1 expression in both Jurkat and MOLT-3 cells (fig. S4). Thus, MYB binding to the MuTE hotspot in a subset of TAL1-overexpressing T-ALLs results in the accumulation of an abundance of H3K27ac marks and aberrantly nucleates binding by other members of the TAL1 complex, leading to aberrant upregulation of TAL1 gene expression.
We next asked why the mutations we had identified in primary patient T-ALLs were clustered in a defined genomic location. A search for predicted transcription factor binding sites near the MuTE site identified the preferred binding sequences for RUNX1, GATA3, and ETS1, as well as E-box motifs characteristic of binding by TAL1/E-protein heterodimers (fig. S5). The absence of predicted MYB binding sites suggests that the MuTE is critical for MYB binding, and supports our hypothesis that MYB binding to its de novo motif is crucial to binding by members of the TAL1 complex at this hotspot. To explore this concept further, we extracted the raw ChIP-seq reads and determined the allelic frequency of mutant to wild-type reads of bound DNA fragments at the mutation site. Strikingly, in MOLT-3 cells, both MYB and TAL1 bound predominantly to the mutant allele, with 67 of the 68 reads, and 37 of 38 reads, revealing the mutant sequence in the bound DNA. Likewise, in Jurkat cells, 404 of 419 reads, and 12 of 14 reads, contained the mutant sequence from MYB and TAL1 ChIP, indicating that these transcription factors predominantly bind monoallelically to the mutant allele. Thus, our data indicate that indels producing MYB binding sites occur at a defined genomic location, probably because they must be in proximity to binding sites for other members of the TAL1 complex.
Given the well-established direct interaction between MYB and its potent transcriptional coactivator, CREB-binding protein (CBP) (20), we also analyzed ChIP-seq tracks for CBP in Jurkat cells, discovering that CBP was also present at the TAL1 enhancer indel mutation site (Fig. 3A). Because CBP promotes H3K27 acetylation and antagonizes Polycomb silencing (21), it seems likely that the initial event in the aberrant super-enhancer formation at this site is the recruitment of CBP by MYB, resulting in abundant H3K27 acetylation that opens the chromatin and permits binding by the other members of the TAL1 complex.
In a sense the mutations forming MYB binding sites upstream of the TAL1 gene in T-ALLs represent an “experiment of nature” that reveals the capacity of MYB binding to aberrantly nucleate a large super-enhancer that drives high levels of expression of a gene critical for the leukemic cell state. Thus, we interrogated the normal binding sites of MYB and we observed highly concordant binding of MYB with members of the TAL1 complex throughout the genome (Fig. 3B), in that 80% of TAL1 binding sites were also co-occupied by MYB. We were also able to demonstrate a strong association of TAL1 and MYB proteins biochemically in reciprocal co-immunoprecipitation experiments (Fig. 3C). Next we focused on the positive interconnected auto-regulatory loop that we had identified previously, whereby the core components of the complex, TAL1, GATA3, and RUNX1, positively regulate their own enhancers (9). We found MYB bound with TAL1, GATA3, and RUNX1 at each of their respective enhancers, including one within the MYB gene itself (fig. S6). Notably, the TAL1 complex binding sites associated with all four of these genes also contained large super-enhancer domains, and all showed CBP-MYB co-occupancy. Furthermore, siRNA knockdown of MYB was sufficient to deplete TAL1, GATA3, and RUNX1 (fig. S6). Thus, MYB is not only a core component of the TAL1 complex, but also a key factor involved in initiating the autoregulatory positive-feedback circuitry (fig. S6).
To demonstrate definitively that MuTEs are responsible for TAL1 overexpression in a subset of T-ALL patients, we employed CRISPR/Cas9 technology to disrupt the MuTE site in Jurkat cells. Initially, we had difficulty expanding single cell clones with deletion of the MuTE site, suggesting that deletion of the mutated enhancer site was diminishing TAL1 levels to a degree that impaired cell survival. Thus, we engineered Jurkat cells to express TAL1 cDNA lacking the 3′ UTR from a retroviral vector, and performed all of our CRISPR/Cas9 experiments in these cells.
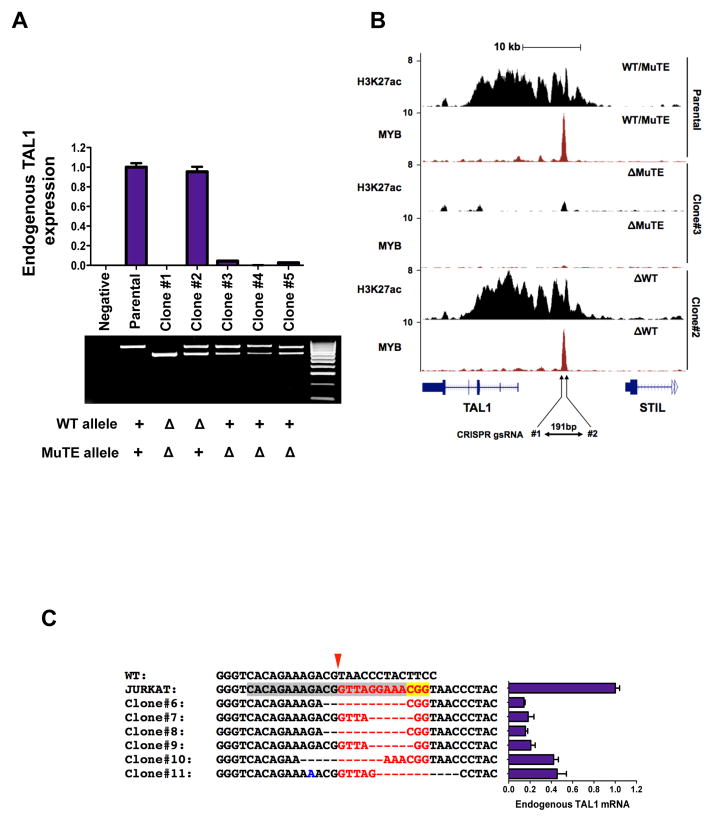
To directly target the enhancer mutation site in Jurkat cells, we first generated clones with a deletion of approximately 180 bp by targeting two guide RNAs to sites flanking either side of the enhancer mutation. Clones expanded from single cells harbored genomic deletions of 177–193 bp that involved either the wild-type allele or mutant allele, or both alleles (Fig. 4A and fig. S7). Deletion of the wild-type allele had no effect on endogenous TAL1 mRNA levels, but deletion of the mutant allele completely abrogated endogenous TAL1 expression, indicating that the enhancer mutation is responsible for TAL1 overexpression in these cells. Furthermore, ChIP-seq for H3K27ac showed complete collapse of the super-enhancer at the TAL1 locus when the deletion affected the allele with the enhancer mutation, but was not affected when the deletion involved only the wild-type allele (Fig. 4B, fig. S8 and fig. S9).
Figure 4. Targeted deletion of the TAL1 enhancer mutation collapses the TAL1 super-enhancer.
(A) Targeted deletion of 177–193 bp of the mutant but not wild-type allele in Jurkat cells abrogates expression of endogenous TAL1, as determined by qRT-PCR. Data are mean ± SD of two independent experiments performed in triplicate. Agarose gel of products from PCR amplification across the MuTE site for CRISPR/Cas9 Jurkat clones. All clones, including parental cells, express MSCV-TAL1. Hyperladder IV on right. Genotype for each clone is shown below: +, allele present; Δ, deleted allele. (B) ChIP-seq tracks for H3K27ac and MYB at the STIL-TAL1 locus from selected CRISPR/Cas9 clones. ChIP-seq read densities (y-axis) were normalized to reads per million reads sequenced in each sample. (C) Sequence alignments of Jurkat clones targeted by CRISPR/Cas9 gsRNA#3 (target sequence is highlighted in grey, PAM sequence in yellow), which targets the 12 bp insertion in Jurkat cells (red font), but not the wild-type allele. Endogenous TAL1 expression as determined by qRT-PCR for respective clones are shown. Data are mean ± SD of two independent experiments performed in triplicate.
We also targeted a single guide RNA to specific sequences that form part of the 12 bp insertion in Jurkat cells, permitting us to propagate single cell clones with a spectrum of repair-induced indel mutations directly at the insertion site (Fig. 4C). In clones with deletion of 6 bp of the 12 bp enhancer insertion, encompassing one of the two inserted MYB binding sites, endogenous TAL1 expression levels decreased by approximately 60%, while clones with more extensive deletions had endogenous TAL1 expression levels decreased by approximately 85% (Fig 4C). Thus, the MuTE is clearly responsible for TAL1 overexpression in Jurkat cells.
Importantly, our ChIP-seq results also show that MYB and CBP were bound together at 727 of the 818 (89%) super-enhancer regions that are present in Jurkat cells. When we performed shRNA knockdown for MYB, 221 of 818 (27%) super-enhancer associated genes decreased significantly in expression (9, 17), suggesting MYB has an active role in regulating their transcription. These results are consistent with the interpretation that MYB-CBP binding and the subsequent formation of abundant H3K27 acetylation marks may be broadly involved in the formation of super-enhancers in T-ALL. Thus, the role that we have shown for MYB binding in super-enhancer formation in a subset of T-ALLs with strategically placed somatic indel mutations in all likelihood provides insight into the general question of how super-enhancers are formed at the site of genes critical for the establishment of the T-ALL cell state. MYB is known to function as a master regulator of early and adult hematopoiesis, and to undergo transcriptional downregulation after lineage commitment and differentiation (22). An interesting area for future study will be to determine whether MYB acts in concert with CBP to regulate super-enhancer formation at genes critical for defining cell identity during normal hematopoietic cell differentiation (14, 16, 23, 24).
Our findings show that somatic mutation of noncoding intergenic elements can lead to binding of master transcription factors, such as MYB, which in turn aberrantly initiate super-enhancers that mediate overexpression of oncogenes. This raises the possibility that acquisition of such enhancer mutations may constitute a general mechanism of carcinogenesis employed in other types of human cancers. Mechanisms of aberrant super-enhancer formation in malignancy have broad implications not only for molecular pathogenesis, but also for clinical management. Drugs that target key components of the transcriptional machinery, such as BRD4 and CDK7 (12, 13, 17), have recently been shown to preferentially target tumor-specific super-enhancers, which provides a novel strategy to capitalize on these abnormalities for improved cancer therapy.
Supplementary Material
Acknowledgments
We thank J. Gilbert for helpful editorial comments on the manuscript, and F. Alt and F. Meng for helpful advice on the design of the CRISPR experiments. We gratefully acknowledge the children with T-ALL and their families for the samples analyzed in these studies. We would like to dedicate this paper to the memory of Michael Fayngersh. M.R.M. was supported by the Claudia Adams Barr Innovative Basic Science Research Program, the Kay Kendall Leukaemia Trust, and a Bennett Fellowship from Leukaemia and Lymphoma Research, UK. A.G. is a Research Fellow of the Gabrielle’s Angel Foundation for Cancer Research, a Clinical Investigator of the Damon Runyon Cancer Research Foundation, and is supported by grants NIH/NCI CA167124, DOD CA120215 and an award from the William Lawrence & Blanche Hughes Foundation. T.S. is supported by a grant from the National Research Foundation, Prime Minister’s Office, Singapore under its NRF Fellowship Program (Award No. NRF-NRFF2013-02). This work was funded by National Institute of Health grants 1R01CA176746-01 and 5P01CA109901-08 (A.T. Look), and 5P01CA68484 (S.E. Sallan, A. T. Look). COG cell banking and sample distribution were supported by grants CA98543, CA114766, CA98413, CA30969 and CA29139 from the National Institutes of Health. S. P. Hunger is the Ergen Family Chair in Pediatric Cancer. R.A. Y. is a founder and member of the Board of Directors of Syros Pharmaceuticals, a company developing therapies that target gene regulatory elements including super-enhancers.
References
- 1.Walker EJ, et al. Cancer Res. 2012 Feb 1;72636 [Google Scholar]
- 2.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Science. 2007 Nov 16;3181136 doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 3.Jirtle RL. Exp Cell Res. 1999 Apr 10;24818 doi: 10.1006/excr.1999.4453. [DOI] [PubMed] [Google Scholar]
- 4.Breit TM, et al. J Exp Med. 1993 Apr 1;177965 doi: 10.1084/jem.177.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown L, et al. The EMBO journal. 1990 Oct;93343 doi: 10.1002/j.1460-2075.1990.tb07535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aplan PD, et al. Science. 1990 Dec 7;2501426 [Google Scholar]
- 7.Ferrando AA, et al. Blood. 2004 Mar 1;1031909 [Google Scholar]
- 8.Leroy-Viard K, Vinit MA, Lecointe N, Mathieu-Mahul D, Romeo PH. Blood. 1994 Dec 1;843819 [PubMed] [Google Scholar]
- 9.Sanda T, et al. Cancer Cell. 2012 Aug 14;22209 [Google Scholar]
- 10.Mansour MR, et al. The Journal of experimental medicine. 2013 Jul 29;2101545 [Google Scholar]
- 11.Siersbaek R, et al. Cell reports. 2014 Jun 12;71443 [Google Scholar]
- 12.Loven J, et al. Cell. 2013 Apr 11;153320 [Google Scholar]
- 13.Filippakopoulos P, et al. Nature. 2010 Dec 23;4681067 [Google Scholar]
- 14.Parker SC, et al. Proc Natl Acad Sci U S A. 2013 Oct 29;11017921 [Google Scholar]
- 15.Hnisz D, et al. Cell. 2013 Nov 7;155934 [Google Scholar]
- 16.Whyte WA, et al. Cell. 2013 Apr 11;153307 [Google Scholar]
- 17.Kwiatkowski N, et al. Nature. 2014 Jul 31;511616 [Google Scholar]
- 18.Zhou Y, et al. Blood. 2013 Dec 19;1224199 [Google Scholar]
- 19.Newburger DE, Bulyk ML. Nucleic Acids Res. 2009 Jan;37D77 doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai P, et al. Genes Dev. 1996 Mar 1;10528 [Google Scholar]
- 21.Tie F, et al. Development. 2014 Mar;1411129 [Google Scholar]
- 22.Schulz C, et al. Science. 2012 Apr 6;33686 [Google Scholar]
- 23.Rada-Iglesias A, et al. Nature. 2011 Feb 10;470279 [Google Scholar]
- 24.Creyghton MP, et al. Proc Natl Acad Sci U S A. 2010 Dec 14;10721931 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.