Abstract
Although sunlight is essential for life on earth, the ultraviolet (UV) wavelengths in its spectrum constitute a major threat to life. Various cellular responses have evolved to deal with the damage inflicted in DNA by UV, and the study of these responses in model systems has spawned the burgeoning field of DNA repair. Although we now know of many types of deleterious alterations in DNA, the approaches for studying them and the early mechanistic insights have come in large part from pioneering research on the processing of UV-induced bipyrimidine photoproducts in bacteria. It is also notable that UV was one of the first DNA damaging agents for which exposure was directly linked to cancer; the sun-sensitive syndrome, xeroderma pigmentosum, was the first example of a cancer-prone hereditary disease involving a defect in DNA repair. We provide a short history of advances in the broad field of genomic maintenance as they have emerged from research in photochemistry and photobiology.
Graphical Abstract
INTRODUCTION
Photochemical processes were surely essential for the early evolution of life on earth, and probably for the actual origin of life as well. They enabled living systems to convert solar energy into chemical energy for purposes of metabolism and growth; indeed, the sun is still the principal source of energy for the biosphere. However, the effects of absorbed UV photons in biological molecules are often more destructive than useful. Very early in evolution, organisms must have evolved in ways to afford protection from UV and for dealing with its damaging effects.
PHOTOCHEMICAL ORIGIN OF LIFE?
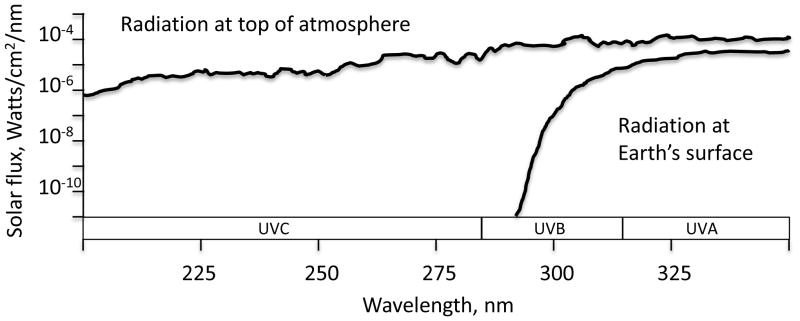
The primordial earth was continuously bombarded by a high flux of UV photons, not attenuated by an ozone layer (Figure 1), so it is likely that sunlight photochemistry played some important roles in the origin of life, while paradoxically, it was also one of the main threats to the persistence of early life forms.
Figure 1.
Wavelength distribution of sunlight impacting earth’s atmosphere and the selective attenuation of short UV wavelengths by ozone, molecular oxygen and water vapor. (Ranges: UVA, 315–400 nm; UVB 280–315 nm; UVC 100–280 nm). Adapted by Graciela Spivak from http://ozonewatch.gsfc.nasa.gov/facts/SH.html.
The popular “RNA world” hypothesis for early life is based upon two postulates: first, that RNA could fulfill all of the necessary attributes of life, including informational, structural and catalytic functions; and second, that DNA eventually entered the scene and somehow took over from RNA as the principal repository of the genetic blueprint for all living cells. A fundamental problem with this model is that the backbone instability in RNA is such that it might be difficult to maintain the lengths necessary for sufficient information storage. It is also likely that the monomers available in the primordial “stew’ for assembly of informational nucleic acids would have included both ribonucleotides and 2′-deoxyribonucleotides; so both types of monomers may have combined more or less randomly into early nucleic acids, benefitting from the DNA type for backbone stability and the RNA type for purine persistence, since purines are spontaneously lost from DNA, leaving non-instructional abasic sites. Of course, both RNA and DNA suffer spontaneous cytosine deamination, which also reduces information content, but evolving systems must have been able to survive in spite of that instability until there were mechanisms to restore the altered sites to cytosine (1).
Recent studies have implicated UV and hydrogen sulfide in key reactions in which precursors of ribonucleotides, amino acids, and lipids can all be derived from the reductive homologation of hydrogen cyanide and some of its derivatives (2). It has also been shown that ribose can be converted to 2′-deoxyribose and that a derivative of uracil can be reduced to thymine through UV photoredox chemistry (3). One of the next challenges in this “primordial soup that cooks itself” (4) would have been the polymerization of nucleotides into sets of short polymers (oligomers) with different sequences. This would have eventually yielded a large variety of oligomers; the hybridization of these to form double-stranded molecules with overlapping 3′ and 5′ ends could then have facilitated their further aggregation into much longer polynucleotides. The oligomers might have been covalently linked by UV-induced cyclobutane pyrimidine dimers (CPDs) at their abutting ends, as documented in a proof-of-principle experiment (5). Once a suitable collection of large polynucleotides became available, the “final” challenge would have been to couple them to some evolutionary processes for maintenance and replication of the most “useful” ones. Meanwhile, the accumulation of RNA species as ribozymes might have included molecules that could catalyze polynucleotide chain elongation and replication. Some simple polypeptides might also have provided important enzymatic functions. Then, all that would be needed would be for this stew to “cook” for a few billion years until something emerged that could grow and duplicate itself. Because thymine dimers can pair to some extent with two adenines in a complementary nucleic acid strand, the presence of these photoproducts in nucleic acids should not have posed a serious issue for a rudimentary level of fidelity in a relatively basic replication process. It is not our purpose to speculate further about life’s origins, but rather to move on to consider the maintenance of evolved life forms once they have emerged.
EARLY STUDIES OF INACTIVATION AND RECOVERY FROM UV EXPOSURES
Most of our early understanding of DNA damage and repair phenomena was based upon analyses of the responses of bacteria to UV irradiation. Downes and Blunt (6) reported in 1877 that bacteria were inactivated by light. Fifty years later, Gates (7) showed that the relative effectiveness of killing bacteria by different wavelengths paralleled the absorption spectrum of nucleic acid, as early evidence from action spectra that RNA and/or DNA might be the targets for lethality. A hint that “repair” of UV induced damage might occur appears in a brief report by Hollaender and Curtis (8), who accounted for the eventual growth resumption of UV-irradiated Escherichia coli by cautiously suggesting that “the possibility of recovery of the irradiated bacteria is not entirely excluded.” This observation was followed by the finding that higher survival levels of UV-irradiated fungal spores could be obtained if they were held in liquid medium for a period before plating on nutrient agar (9). Action spectra also implicated nucleic acids in UV-induced mutagenesis (10; 11). Roughly a decade later, there was a series of significant discoveries, including the phenomenon of photoreactivation of UV-irradiated bacteria and bacteriophage (12; 13), the revelation that the shapes of survival curves could be altered by varying the treatment of UV-irradiated bacteria after the irradiation (that challenged the classical “target theory” for the deleterious effects of UV photons) (14), and the enhanced survival of UV-irradiated bacteriophage if the host cells were also irradiated (an early example of what is now known as the SOS response) (15). In addition, the isolation of bacterial mutants with altered sensitivity to UV by Evelyn Witkin, Ruth Hill and Paul Howard-Flanders provided genetic evidence for recovery mechanisms (e.g., (16–20)).
In the 1950s, a number of researchers began to focus on the effects of UV on macromolecular synthesis in bacteria. Kelner (21) and Kanazir and Errera (22) reported that DNA synthesis was inhibited in UV irradiated E. coli. Iverson and Giese (23) followed DNA synthesis over a period of 8 hr after UV irradiation of E. coli, using the indole method of Dische for quantitation, and found that DNA synthesis resumed roughly in parallel with the resumption of cell division. Hanawalt and Setlow (24), using radioactive labeling, showed that monochromatic 265 nm UV inhibited DNA synthesis for a period roughly proportional to the dose and concluded that “it is evident that recovery mechanisms can eventually restore the DNA synthesis rate to nearly that of the unirradiated control except after very high doses.” Photoreactivating light had the effect of reducing the length of the lag in the recovery of DNA synthesis, and it also enhanced recovery of RNA synthesis following UV irradiation, as though inhibitors were being destroyed or blockages were being removed (25).
THE DISCOVERY THAT UV GENERATES CYCLOBUTANE THYMINE DIMERS
The landmark discovery that thymines could be dimerized by UV exposure ((26); reviewed in (27)) facilitated the next fundamental advances in our understanding of recovery from DNA damage. Studies of Hemophilus influenza transforming DNA showed that thymine dimers produced in DNA irradiated with UV in vitro caused biological damage and could be removed by photoreactivation (28; 29). Bollum and Setlow (30) found that thymine dimers interfered with synthesis by DNA polymerase in vitro. Now there was a specific chemical lesion, the CPD (31), produced by UV in DNA that needed to be processed to avoid mutagenesis and to promote the survival of UV irradiated organisms.
It was soon reported that photoreactivation requires an enzyme that can bind to thymine dimers in the dark and then split them in situ upon exposure to visible light, without breaking the phosphodiester backbone (32). The earliest form of a photoreactivating entity could have been a tripeptide of lysine-tryptophan-lysine, which has been shown to reverse dimers at low efficiency upon irradiation with visible light (33). Subsequently, the elongation of the polypeptide and the addition of a blue-light “antenna” may have led to the amazing sensitivity and specificity of current-day photoreactivating enzymes (reviewed in (34)).
In the early studies of UV irradiated bacteria (e.g., (35)), the CPD content of DNA was typically assayed by hydrolyzing DNA with a strong acid and then analyzing the hydrolysis products by chromatography. Subsequently, enzymes that specifically recognize dimers in DNA were identified (36–39). These enzymes were originally thought to be endonucleases, but later studies revealed that they were glycosylases with associated AP lyase activities (40; 41). The enzyme purified from E. coli infected with the bacteriophage T4 proved especially useful. This enzyme, called T4 endonuclease V (TEV) or T4 pyrimidine DNA glycosylase (T4 PDG) is a small, relatively stable enzyme that does not require a Mg++ cofactor. The purified enzyme specifically nicks a DNA strand at the site of a CPD in duplex DNA. This meant that a variety of assays could be developed based upon measuring the length (size) of the DNA strand before and after exposure to the enzyme, or by assessing the relative amount of DNA resistant to the enzyme. In the case of plasmids, a single nick converts superhelical molecules to open circles, which can be separated from the superhelical molecules by agarose gel electrophoresis. In the case of linear molecules, the nicked DNA can be separated from the unnicked DNA by centrifugation in alkaline sucrose gradients or by electrophoresis in alkaline gels (42–44). The sensitivity of this kind of assay depends upon the size of the DNA being analyzed; the larger the DNA, the lower the UV dose that can be used to produce detectable levels of CPDs. In some cases, a dose as low as 2 J/m2 can be studied (42; 45). By employing appropriate probes for specific genomic regions, repair in those regions (including transcriptionally active genes and inactive regions) can be studied, even in the individual DNA strands (44; 46)). The development of these techniques led to the discovery of transcription coupled repair (TCR) and its distinguishing features in comparison to global genome repair (GGR) as discussed below.
THE DISCOVERY OF NUCLEOTIDE EXCISION-REPAIR (NER)
In addition to photoreactivation, another type of repair, which did not require light, was identified in E. coli in 1963-’64. This dark recovery mode was absent in the UV sensitive E. coli BS-1, in which DNA synthesis did not recover following irradiation (47). The selective release of CPD-containing fragments from the DNA in UV resistant strains but not in UV sensitive derivatives prompted the suggestion of an excision-repair mechanism for removing these lesions (35; 48). Density labeling the replicating DNA with 5-bromouracil in UV irradiated bacteria provided evidence for the patching step in the postulated repair pathway (a non-conservative mode of synthesis in which the nascent DNA patches are much shorter than the DNA fragments containing them) (49; 50). The essential last step in nucleotide excision repair (NER) required joining the repair patch to the contiguous parental DNA strand. The needed enzyme, polynucleotide ligase, was discovered several years later (51). Subsequently, it was shown that excision deficient mutants of E. coli did not carry out repair replication (52).
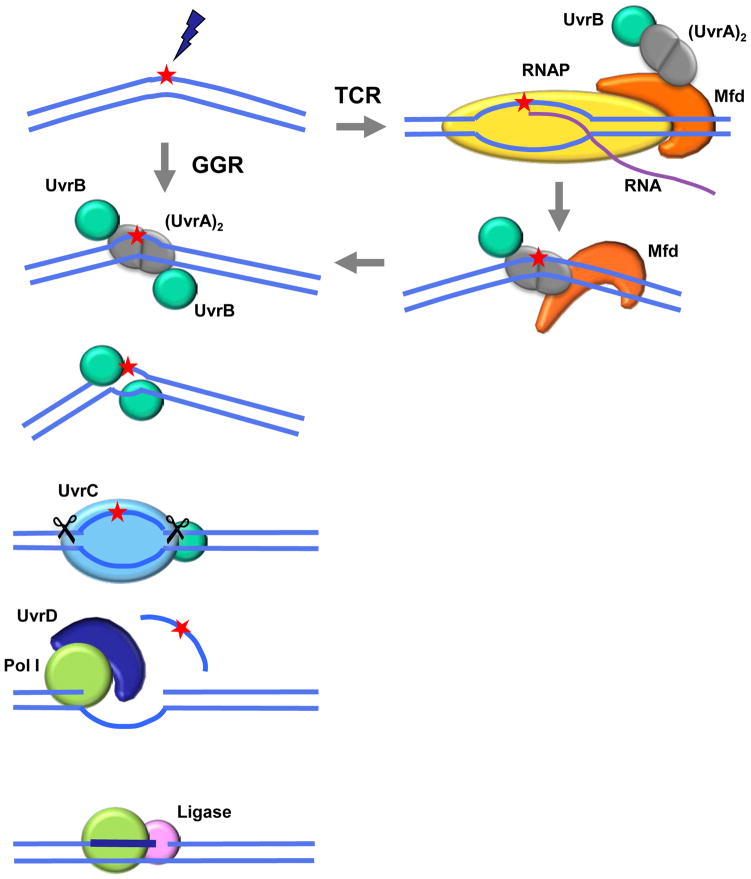
The isolation and study of mutants of E. coli K-12 led to the identification and characterization of the different biochemical steps in NER and the proteins involved (20; 53); reviewed in (54). The NER pathway in E. coli is illustrated in Figure 2, in which it is shown that only six proteins are required for global genomic repair (GGR). It is noteworthy that each step in this excision-repair pathway creates another lesion until the final ligation step has been completed. The subpathway of transcription-coupled repair (TCR) is targeted to the transcribed strands of expressed genes and is initiated by the blockage of RNA polymerase translocation at the sites of the lesions. In bacteria, all the proteins involved in GGR also participate in TCR; however, in eukaryotes, the XPC protein is needed for the initiation of GGR, but it does not participate in TCR. The number of proteins and protein complexes involved in NER in eukaryotes is much larger than that in prokaryotes and includes those involved in dealing with the chromatin structures (for reviews see (55–58).
Figure 2.
Schematic representation of NER in Escherichia coli. Damage can be recognized either by sensing instability of the duplex DNA structure (for GGR) or by the arrest of a translocating RNA polymerase upon encountering the damaged site (for TCR). In the latter case Mfd binds the arrested polymerase and recruits UvrA, which then further recruits UvrB, as the polymerase and RNA product are released. From then on the pathway proceeds in the classic manner in which the recognition proteins recruit UvrC, which catalyzes dual incisions (~ 13 nt) in the damaged strand. UvrD and DNA Pol I excise the damaged stretch and Pol I synthesizes a repair patch that is eventually ligated to the contiguous parental strand. The relative sizes of the nucleic acids and proteins depicted in this figure are not meant to reflect their actual sizes and conformations.
Several years after the identification of CPDs in DNA, another UV induced dipyrimidine lesion was discovered, the helix distorting 6-4 pyrimidine-pyrimidone photoproduct (6-4 PP) (59). It is produced in DNA with a lower quantum efficiency than that for the CPD, and it is more easily recognized by NER in bacteria and mammalian cells (60–62). Although this lesion is not recognized by T4 PDG, sensitive assays using the E. coli UvrABC incision complex or antibodies have been developed to detect it (63–65). The 6-4PP is rapidly converted to the Dewar valence isomer upon further UV irradiation and it can be sensitively assayed by high-performance liquid chromatography-electrospray ionization tandem mass spectrometry to study its repair rate, as well as that of the other bipyrimidine photoproducts (66). Yet another unique type of dipyrimidine photoproduct is generated in UV irradiated bacterial spores (67, 68).
MOST PATIENTS WITH XERODERMA PIGMENTOSUM ARE DEFICIENT IN NER
Simultaneous with the studies reporting NER in bacteria, evidence for the patching step in the NER pathway was identified in UV irradiated mammalian cells as “unscheduled DNA synthesis” (UDS) in non-S-phase human cells. This phenomenon was detected by autoradiography (69) and then validated as repair replication by carrying out DNA density labeling analyses in parallel with UDS determinations (70). James Cleaver, a post-doctoral fellow in Robert Painter’s laboratory, happened to read a feature story by David Perlman in the San Francisco Chronicle, describing a rare human hereditary disease, xeroderma pigmentosum (XP), in which the victims are sun-sensitive and suffer high levels of cancer in their sun exposed skin. Suspecting that the etiology might involve a defect in DNA repair, Cleaver studied epidermal cells from an XP patient and discovered that they were extremely sensitive to UV and deficient in repair replication (71). This was the first example of a human disease caused by a defect in DNA repair, and it stimulated studies in many laboratories to elucidate the details of the mammalian DNA repair pathways. Setlow et al. (72) confirmed Cleaver’s discovery by showing that an XP cell line cannot carry out the first nicking step in NER. Another form of XP, called the “variant”, turned out to be due to a deficiency in a translesion DNA polymerase, Pol eta, which may have evolved for the specific purpose of bypassing CPDs, since its fidelity is much higher when it copies dimerized thymines than when it replicates through adjacent thymines in undamaged DNA (73–75).
THE DISCOVERY OF POST-REPLICATION REPAIR (DAUGHTER STRAND GAP REPAIR) AND INDUCIBLE RESPONSES TO UV IRRADIATION
The isolation of recombination deficient E. coli K-12 mutants (76), which were sensitive to UV, led to the identification of another mechanism for mitigating the effects of UV. This mechanism has been referred to by several terms, including recombination repair, post-replication repair and daughter strand gap repair (77) (reviewed in (78)). Although this mechanism does not directly result in the removal of CPDs, it contributes to the survival of UV-irradiated bacteria and allows cells to tolerate persisting DNA damage. A similar mechanism has been described in the cells of other organisms, including Drosophila, mammalian cells, and yeast. Extensive analysis of the recombination deficient recA mutants of E. coli K-12 and the pleotropic RecA protein led to the characterization of a complex regulatory network originally called the LexA regulon and subsequently termed the SOS response. Although a variety of DNA damaging agents have been shown to elicit the SOS response, it was the study of UV irradiated bacteria and bacteriophage that initially revealed this network (reviewed in (79). One of the many interesting findings was the relationship of recA and lexA to NER. Originally, it was thought that daughter strand gap repair, dependent on recA, was separate from NER, which required the uvr genes. However, as the SOS hypothesis developed, it became clear that recA and lexA have important regulatory roles in NER. The RecA protein has both ATPase and co-protease activities. The co-protease activity facilitates the self-cleavage of several other proteins, including the lambda repressor and the LexA protein. The LexA protein represses the transcription of more than 40 genes in E. coli (80; 81) by binding to a sequence in the promoter region known as the LexA or SOS box (reviewed in (82)). When RecA is activated by binding to single stranded DNA generated at blocked replication forks after UV irradiation, it forms a filament that facilitates the self-cleavage of LexA, which then dissociates from the DNA allowing transcription of the regulated genes to occur. The genes repressed by LexA include uvrA and uvrB, which code for two of the subunits of the NER incision complex. The induction of these two genes facilitates GGR of CPDs but has little effect on the repair of 6-4 PPs (83).
Another protein that is part of the SOS response and subject to self-cleavage facilitated by RecA is the UmuD protein. A dimer of UmuD is activated by cleavage to UmuD′ which associates with UmuC to form an error prone polymerase, UmuD′2C (also known as polV). This polymerase is one of three polymerases in E. coli that can perform translesion synthesis (TLS); they are “error prone” and can insert nucleotides opposite lesions in DNA (reviewed in (84)). In some cases (especially opposite TT dimers) the inserted nucleotides are correct (e.g., AA), but in other cases they are not and result in mutations. In the case of polV, the active, mutagenic form of the complex (polV mut) contains RecA and ATP (UmuD′2C-RecA-ATP) (85).
INDUCIBLE RESPONSES TO UV IRRADIATION IN MAMMALIAN CELLS
Following the characterization of the SOS response in E.coli, inducible responses to DNA damage in UV irradiated mammalian cells were identified. The UV stimulated activation of the p53 tumor suppressor in human fibroblasts was shown to have an effect remarkably similar to that of the SOS response in bacteria; it was required for the efficient GGR of CPDs but not the 6-4 PPs (86; 87). It was also established that the accumulation and activation of p53 in response to DNA damage can lead to apoptosis or arrest of the mammalian cell cycle, presumably to provide time for the repair of damage before the cell divides or initiates a new round of replication (for review see (88). Skin fibroblasts derived from tumors in patients with the cancer-prone Li-Fraumeni syndrome are homozygous for mutations in the p53 gene. These fibroblasts are defective in GGR of CPDs, compared to the related heterozygous mutants and normal cells (86), and this defect can be complemented by the expression of a stably integrated tetracycline-regulated p53 cDNA (87). The p53 effect is mediated in part through the activation of p48 (DDB2), an accessory lesion-recognition factor that is upregulated in UV-irradiated human cells in a p53 dependent manner (89; 90). Upon recognition of CPDs in DNA, DDB2 recruits the essential factor XPC to initiate NER (91). Rodent fibroblasts, typically deficient in the expression of DDB2, are also deficient in repair of CPDs (92; 93). However, it is important to appreciate that the efficiency of CPD repair in cultured cells may be much less than that in the intact epidermis (94).
EFFECTS OF CHROMATIN STRUCTURE ON DNA DAMAGE AND REPAIR
Interstrand crosslinking of DNA with photoactivated psoralens contributed importantly to the early analysis of chromatin structure (95; 96). Smerdon, Tlsty and Lieberman (97) found greater amounts of repair synthesis in nuclease sensitive than in nuclease resistant regions of chromatin in UV irradiated human cells, and the rearrangement of nucleosomes in mammalian chromatin was then demonstrated during UV-induced repair replication (98; 99). It was later shown that UV-induced 6-4 PPs were largely restricted to the inter-nucleosome regions, whereas CPDs were more uniformly distributed in chromatin ((100); reviewed in (101)). The studies of the repair of UV induced DNA photoproducts in chromatin stimulated broad interest in genomic heterogeneity in repair.
GENOMIC HETEROGENEITY IN REPAIR
Zolan et al. (102) showed that CPDs in the highly-repetitive alpha DNA 179 bp sequence were almost as accessible to repair as CPDs in the bulk DNA, while psoralen monoadducts and crosslinks were repaired much less efficiently in the alpha DNA than in the overall genome. This observation supported the view that different types of lesions are recognized with different efficiencies by the proteins involved in NER, an idea suggested by the difference in the rates of repair of the two UV photoproducts, CPDs and 6-4 PPs. The difference in the rate of repair of psoralen adducts in the alpha DNA and the rest of the genome also provided the first evidence for different rates of repair in different domains of the genome. Jonathan Mansbridge discovered that in XPC mutants, deficient in GGR, there were selected domains in which CPDs were being repaired (103). Bohr et al. (44; 104) extended this finding by showing that CPDs in a transcriptionally active gene in UV irradiated Chinese hamster ovary (CHO) cells were removed from DNA more rapidly than CPDs in adjacent silent regions of the genome. The same phenomenon was confirmed in human cells (105). In addition, repair was found to be more rapid in the transcribed strands of active genes than in the non-transcribed strands or in the genome overall in mammalian cells and in bacteria (46; 106). This observation led to the identification of the two branches of NER, GGR, which depends upon recognition of DNA damage by DDB2 and XPC, and TCR, which depends upon the recognition of damage by a translocating RNA polymerase blocked at a lesion. TCR was also demonstrated in yeast (107; 108). The documentation of strand specificity of TCR in bacteria eliminated the hypothesis that the preferential repair in active genes could be fully explained by chromatin structure. However, it has become clear that there is also a component of lesion accessibility due to features of chromatin structure in actively transcribed genomic domains.
In terminally differentiated human neurons, TCR is proficient, and GGR of CPDs is generally deficient except in an expressed gene (109). Originally termed differentiation associated repair, the definition of this mode of preferential repair was then broadened when it was documented in actively growing cells that otherwise displayed poor GGR (110; 111), and it is now termed transcription domain associated repair (DAR). Within an expressed gene, DAR operates upon both strands, while TCR is superimposed upon DAR in the template strand (110).
Complementation assays in vitro (112; 113) led to the reconstitution of mammalian GGR with purified proteins (114; 115). TCR has not yet been demonstrated in eukaryotic systems in vitro, although it has been reconstituted in E. coli (116; 117).
The locations of GGR and TCR of UV induced CPDs and 6-4 PPs have recently been mapped at single-nucleotide resolution throughout the entire human genome (118).
PHOTOSENSITIVE HUMAN GENETIC DISEASES AND CANCER SUSCEPTIBILITY
As described above, NER deficient cells from victims with xeroderma pigmentosum are strikingly sensitive to UV induction of cutaneous cancers. There are a number of other photosensitive human syndromes (119; 120). Several of these are characterized by defects in the TCR pathway and the patients do not develop any cancers upon sunlight exposure. (A caveat is that these are very rare diseases with relatively few patients to study.) They include Cockayne syndrome (CS), UV sensitive syndrome (UVSS) and tricothiodystrophy (TTD). Whereas CS and TTD patients typically suffer severe developmental and neurological abnormalities, patients with UVSS only present with sunburn and freckles (for review see (121). Although cells from CS and UVSS have been shown to be equally deficient in TCR of very low levels of CPDs and 8-oxo-guanine (122), the CS cells exhibit sensitivity to reactive oxygen species, whereas UVSS cells respond like normal cells to reactive oxygen species (123; 124).
The UVA wavelengths have been shown in recent studies to generate CPDs but not 6-4PPs (66). Melanin is known to protect epidermal DNA from the UV induced generation of CPDs and other photoproducts, thereby reducing the frequency of the highly lethal cancer, melanoma. Most mutations in cells from melanoma have the signature of UV induced CPDs. Recent studies have shown that CPDs are generated in melanocytes many hours following exposure to UVA irradiation. This has been shown to be due to activation of melanin and energy transfer in the dark (125). This work provides new insights about the etiology of melanoma and raises concerns about effective protection from UV in the UVA range.
SUMMARY AND CONCLUSIONS
The study of the effects of UV on various organisms, including bacteria and humans, has been fundamental to our understanding of the biological mechanisms that ameliorate the effects of DNA damage and help to maintain genetic integrity. From the experimental point of view, UV is an extremely useful agent. Exposures can be accurately timed, and therefore the relationship between dose and lesion yield can be precisely determined. In addition, unlike some agents (e.g., ionizing radiation), UV produces a relatively restricted variety of lesions in DNA. The most frequent lesion, the CPD, is chemically stable, which has made it possible to obtain accurate estimates of the amounts present in irradiated DNA even following harsh treatments. The subsequent development of enzyme and antibody-based assays enhanced the sensitivity of detection so that the effects of very low UV doses could be studied.
The other frequent lesion produced by UV, the 6-4 photoproduct, is more easily recognized than the CPD by NER and provides an opportunity to investigate mechanisms for recognizing specific types of DNA damage with unique characteristics that differ from the canonical CPD.
Finally, from the biological perspective and relevance to human health, UV continues to be a very significant environmental threat. With the destruction of the ozone layer, a major increase in UV induced skin cancer can be anticipated.
Acknowledgments
We wish to dedicate this review to the memory of Jane Setlow and Dick Setlow, for their friendship and for their seminal contributions to UV photobiology and DNA repair. We also express our appreciation to many students and colleagues for their contributions to this field, and we apologize for the limited citations in this short article. We owe particular gratitude to Charles Allen Smith, whose ideas and generous mentoring are woven into most of the publications from our laboratory. Our research has been supported by grants from the National Institutes of Health, the American Cancer Society and the Department of Energy.
Biographies

Philip Hanawalt is the Dr. Morris Herzstein Professor of Biology at Stanford University. He graduated from Oberlin College, completed his Ph.D. at Yale and joined Stanford faculty in 1961. He co-discovered excision-repair of damaged DNA in 1964 and transcription-coupled repair was discovered in his lab several decades later. He is a member of National Academy of Sciences and a Fellow of the American Academy of Arts and Sciences, American Academy of Microbiology and American Association for Advancement of Science, as well as a Foreign Associate of EMBO. He has research awards from the American Society for Photobiology, Environmental Mutagen Society, and an AACR-Princess Takamatsu Cancer Research Lectureship.

Ann Ganesan studied genetics with Joshua Lederberg and received an MSc in Genetics from the University of Wisconsin and a PhD from Stanford University. She spent 35 years as a Senior Research Scientist in the stimulating and convivial atmosphere of Phil Hanawalt’s laboratory, where she developed a sensitive enzymatic assay for cyclobutane pyrimidine dimers in DNA, and employed this to study various aspects of DNA repair, including recombinational mechanisms and the dedicated pathway of transcription-coupled repair. She also served as an expert mentor to students and postdocs in Hanawalt’s group, and taught them how to write understandable manuscripts for publication.
Footnotes
Some of the data in this paper were presented during the 16th International Congress on Photobiology held in Cordoba, Argentina, in September (8th – 12th), 2014.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Patel BH, Percivalle C, Ritson DJ, Duffy CD, Sutherland JD. Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat Chem. 2015;7:301–307. doi: 10.1038/nchem.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ritson DJ, Sutherland JD. Conversion of biosynthetic precursors of RNA to those of DNA by photoredox chemistry. J Mol Evol. 2014;78:245–250. doi: 10.1007/s00239-014-9617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bracher PJ. Origin of life: Primordial soup that cooks itself. Nat Chem. 2015;7:273–274. doi: 10.1038/nchem.2219. [DOI] [PubMed] [Google Scholar]
- 5.Lewis RJ, Hanawalt PC. Ligation of oligonucleotides by pyrimidine dimers--a missing ‘link’ in the origin of life? Nature. 1982;298:393–396. doi: 10.1038/298393a0. [DOI] [PubMed] [Google Scholar]
- 6.Downes A, Blunt TP. Researches on the effect of light upon bacteria and other organisms. Proc Royal Soc London. 1877;26:488–500. [Google Scholar]
- 7.Gates FL. On nuclear derivatives and the lethal action of ultra-violet light. Science. 1928;6:479–480. doi: 10.1126/science.68.1768.479-a. [DOI] [PubMed] [Google Scholar]
- 8.Hollaender A, Curtis JT. Effect of sublethal doses of monochromatic ultraviolet radiation on bacteria in liquid suspensions. Exptl Biol Med. 1935;33:61–62. [PubMed] [Google Scholar]
- 9.Hollaender A, Claus WD. The bacteriocidal effect of ultraviolet radiation on Escherichis coli in liquid suspensions. J Gen Physiol. 1936;19:753–765. doi: 10.1085/jgp.19.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emmons CW, Hollaender A. The action of ultraviolet radiation on dermatophytes. II. Mutations induced in cultures of dermatophytes by exposure of spores to monochromatic ultraviolet radiation. Am J Bot. 1939;26:467–475. [Google Scholar]
- 11.Knapp E, Reuss A, Risse O, Schreiber H. Quantitative Analyse der mutationsauslösenden Wirkung monochromatischen UV-Lichtes. Naturwissenschaften. 1939;27:304. [Google Scholar]
- 12.Kelner A. Effect of visible light on the recovery of Streptomyces griseus conidia from ultra-violet irradiation injury. Proc Natl Acad Sci U S A. 1949;35:73–79. doi: 10.1073/pnas.35.2.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dulbecco R. Reactivation of ultra-violet-inactivated bacteriophage by visible light. Nature. 1949;163:949. doi: 10.1038/163949b0. [DOI] [PubMed] [Google Scholar]
- 14.Roberts RB, Aldous E. Recovery from ultraviolet irradiation in Escherichia coli. J Bacteriol. 1949;57:363–375. doi: 10.1128/jb.57.3.363-375.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weigle JJ. Induction of mutations in a bacterial virus. Proc Natl Acad Sci U S A. 1953;39:628–636. doi: 10.1073/pnas.39.7.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witkin EM. Inherited differences in sensitivity to radiation in Escherichia coli. Proc Natl Acad Sci USA. 1946;32:59–68. doi: 10.1073/pnas.32.3.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Witkin EM. Genetics of resistance to radiation in Escherichia coli. Genetics. 1947;32:221–248. doi: 10.1093/genetics/32.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill RF. A radiation-sensitive mutant of Escherichia coli. Biochim Biophys Acta. 1958;30:636–637. doi: 10.1016/0006-3002(58)90112-4. [DOI] [PubMed] [Google Scholar]
- 19.Hill RF, Simson E. A study of radiosensitive and radioresistant mutants of Escherichia coli strain B. J Gen Microbiol. 1961;24:1–14. doi: 10.1099/00221287-24-1-1. [DOI] [PubMed] [Google Scholar]
- 20.Howard-Flanders P, Boyce RP, Simson E, Theriot L. A genetic locus in E. coli K12 that controls the reactivation of UV-photoproducts associated with thymine in DNA. Proc Natl Acad Sci U S A. 1962;48:2109–2115. doi: 10.1073/pnas.48.12.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelner A. Growth, respiration, and nucleic acid synthesis in ultraviolet-irradiated and in photoreactivated Escherichia coli. J Bacteriol. 1953;65:252–262. doi: 10.1128/jb.65.3.252-262.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanazir D, Errera M. Metabolism of nucleic acids by E. coli B after ultraviolet irradiation. Biochim Biophys Acta. 1954;14:62–66. doi: 10.1016/0006-3002(54)90130-4. [DOI] [PubMed] [Google Scholar]
- 23.Iverson RM, Giese AC. Synthesis of nucleic acids in ultraviolet-treated Escherichia coli. Biochim Biophys Acta. 1957;25:62–68. doi: 10.1016/0006-3002(57)90417-1. [DOI] [PubMed] [Google Scholar]
- 24.Hanawalt P, Setlow R. Effect of monochromatic ultraviolet light on macromolecular synthesis in Escherichia coli. Biochim Biophys Acta. 1960;41:283–294. doi: 10.1016/0006-3002(60)90011-1. [DOI] [PubMed] [Google Scholar]
- 25.Hanawalt P, Buehler J. Photoreactivation of macromolecular synthesis in Escherichia coli. Biochim Biophys Acta. 1960;37:141–143. doi: 10.1016/0006-3002(60)90088-3. [DOI] [PubMed] [Google Scholar]
- 26.Beukers RT, Berends W. Isolation and identification of the irradiation product of thymine. Biochim Biophys Acta. 1960;41:550–551. doi: 10.1016/0006-3002(60)90063-9. [DOI] [PubMed] [Google Scholar]
- 27.Beukers R, Eker APM, Lohman PHM. 50 years thymine dimer. DNA Repair (Amst) 2008;7:530–543. doi: 10.1016/j.dnarep.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Setlow RB, Setlow JK. Evidence that ultraviolet-induced thymine dimers in DNA cause biological damage. Proc Natl Acad Sci U S A. 1962;48:1250–1257. doi: 10.1073/pnas.48.7.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Setlow JK, Setlow RB. Nature of the photoreactivable ultraviolet lesion in deoxyribonucleic acid. Nature. 1963;197:560–562. [Google Scholar]
- 30.Bollum FJ, Setlow RB. Ultraviolet inactivation of DNA primer activity: I. Effects of different wavelengths and doses. Biochim Biophys Acta (BBA)-Specialized Section on Nucleic Acids and Related Subjects. 1963;68:599–607. doi: 10.1016/0006-3002(63)90189-6. [DOI] [PubMed] [Google Scholar]
- 31.Mantione M-J, Pullman B. Sur le mécanisme de la photodimérisation de la thymine. Biochim Biophys Acta (BBA)-Specialized Section on Nucleic Acids and Related Subjects. 1964;91:387–398. [PubMed] [Google Scholar]
- 32.Wulff DL, Rupert CS. Disappearance of thymine photodimer in ultraviolet irradiated DNA upon treatment with a photoreactivating enzyme from baker’s yeast. Biochem Biophys Res Commun. 1962;7:237–240. doi: 10.1016/0006-291x(62)90181-x. [DOI] [PubMed] [Google Scholar]
- 33.Charlier M, Hélène C. Photosensitized splitting of pyrimidine dimers in DNA by indole derivatives and tryptophan containing peptides. Photochem Photobiol. 1975;21:31–37. doi: 10.1111/j.1751-1097.1975.tb06626.x. [DOI] [PubMed] [Google Scholar]
- 34.Sancar GB. DNA photolyases: physical properties, action mechanism, and roles in dark repair. Mutat Res/DNA Repair. 1990;236:147–160. doi: 10.1016/0921-8777(90)90002-m. [DOI] [PubMed] [Google Scholar]
- 35.Setlow RB, Carrier WL. The disappearance of thymine dimers from DNA: an error-correcting mechanism. Proc Natl Acad Sci U S A. 1964;51:226. doi: 10.1073/pnas.51.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strauss BS. Differential destruction of the transforming activity of damaged deoxyribonucleic acid by a bacterial enzyme. Proc Natl Acad Sci U S A. 1962;48:1670. doi: 10.1073/pnas.48.9.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takagi Y, Sekiguchi M, Okubo S, Nakayama H, Shimada K, Yasuda S, Nishimoto T, Yoshihara H. Nucleases specific for ultraviolet light-irradiated DNA and their possible role in dark repair. Cold Spring Harb Symp Quant Biol. 1968;33:219–227. doi: 10.1101/sqb.1968.033.01.025. [DOI] [PubMed] [Google Scholar]
- 38.Friedberg EC, King JJ. Endonucleolytic cleavage of UV-irradiated DNA controlled by the V+ gene in phage T4. Biochem Biophys Res Commun. 1969;37:646–651. doi: 10.1016/0006-291x(69)90859-6. [DOI] [PubMed] [Google Scholar]
- 39.Yasuda S, Sekiguchi M. T4 endonuclease involved in repair of DNA. Proc Natl Acad Sci U S A. 1970;67:1839–1845. doi: 10.1073/pnas.67.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haseltine WA, Gordon LK, Lindan CP, Grafstrom RH, Shaper NL, Grossman L. Cleavage of pyrimidine dimers in specific DNA sequences by a pyrimidine dimer DNA-glycosylase of M. luteus. Nature. 1980;285:634–641. doi: 10.1038/285634a0. [DOI] [PubMed] [Google Scholar]
- 41.Gordon LK, Haseltine WA. Comparison of the cleavage of pyrimidine dimers by the bacteriophage T4 and Micrococcus luteus UV-specific endonucleases. J Biol Chem. 1980;255:12047–12050. [PubMed] [Google Scholar]
- 42.Ganesan AK. A method for detecting pyrimidine dimers in the DNA of bacteria irradiated with low doses of ultraviolet light. Proc Natl Acad Sci U S A. 1973;70:2753–2756. doi: 10.1073/pnas.70.10.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkins RJ. Endonuclease sensitive sites in the DNA of irradiated bacteria: a rapid and sensitive assay. Biochim Biophys Acta. 1973;312:33–37. doi: 10.1016/0005-2787(73)90049-x. [DOI] [PubMed] [Google Scholar]
- 44.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 45.Ganesan AK. Persistence of pyrimidine dimers during post-replication repair in ultraviolet light-irradiated Escherichia coli K12. J Mol Biol. 1974;87:103–119. doi: 10.1016/0022-2836(74)90563-4. [DOI] [PubMed] [Google Scholar]
- 46.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 47.Setlow RB, Swenson PA, Carrier WL. Thymine dimers and inhibition of DNA synthesis by ultraviolet irradiation of cells. Science. 1963;142:1464–1466. doi: 10.1126/science.142.3598.1464. [DOI] [PubMed] [Google Scholar]
- 48.Boyce RP, Howard-Flanders P. Release of ultraviolet light-induced thymine dimers from DNA in E. coli K-12. Proc Natl Acad Sci USA. 1964;51:293–300. doi: 10.1073/pnas.51.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettijohn DE, Hanawalt PC. Deoxyribonucleic acid replication in bacteria following ultraviolet irradiation. Biochim Biophys Acta. 1963;72:127–129. [PubMed] [Google Scholar]
- 50.Pettijohn DE, Hanawalt PC. Evidence for repair replication of ultraviolet damaged DNA in bacteria. J Mol Biol. 1964;9:395–410. doi: 10.1016/s0022-2836(64)80216-3. [DOI] [PubMed] [Google Scholar]
- 51.Gellert M. Formation of covalent circles of lambda DNA by E. coli extracts. Proc Natl Acad Sci U S A. 1967;57:148–155. doi: 10.1073/pnas.57.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cooper PK, Hanawalt PC. Absence of repair replication following ultraviolet irradiation of an excision-deficient mutant of Escherichia coli. Photochem Photobiol. 1971;13:83–85. doi: 10.1111/j.1751-1097.1971.tb06094.x. [DOI] [PubMed] [Google Scholar]
- 53.Howard-Flanders P, Theriot L. Mutants of Escherichia coli K-12 defective in DNA repair and in genetic recombination. Genetics. 1966;53:1137–1150. doi: 10.1093/genetics/53.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 55.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 56.Ganesan A, Spivak G, Hanawalt PC. Transcription-coupled DNA repair in prokaryotes. Prog Mol Biol Transl Sci. 2012;110:25–40. doi: 10.1016/B978-0-12-387665-2.00002-X. [DOI] [PubMed] [Google Scholar]
- 57.Schärer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vermeulen W, Fousteri M. Mammalian transcription-coupled excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012625. doi: 10.1101/cshperspect.a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang SY, Varghese AJ. Cytosine-thymine addition product from DNA irradiated with ultraviolet light. Biochem Biophys Res Commun. 1967;29:543–549. doi: 10.1016/0006-291x(67)90519-0. [DOI] [PubMed] [Google Scholar]
- 60.Franklin WA, Haseltine WA. Removal of UV light-induced pyrimidine-pyrimidone(6-4) products from Escherichia coli DNA requires the uvrA, uvrB, and urvC gene products. Proc Natl Acad Sci U S A. 1984;81:3821–3824. doi: 10.1073/pnas.81.12.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitchell DL, Haipek CA, Clarkson JM. (6-4)Photoproducts are removed from the DNA of UV-irradiated mammalian cells more efficiently than cyclobutane pyrimidine dimers. Mutat Res Lett. 1985;143:109–112. doi: 10.1016/s0165-7992(85)80018-x. [DOI] [PubMed] [Google Scholar]
- 62.Koehler DR, Courcelle J, Hanawalt PC. Kinetics of pyrimidine(6-4)pyrimidone photoproduct repair in Escherichia coli. J Bacteriol. 1996;178:1347–1350. doi: 10.1128/jb.178.5.1347-1350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas DC, Morton AG, Bohr VA, Sancar A. General method for quantifying base adducts in specific mammalian genes. Proc Natl Acad Sci U S A. 1988;85:3723–3727. doi: 10.1073/pnas.85.11.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas DC, Okumoto DS, Sancar A, Bohr VA. Preferential DNA repair of (6-4) photoproducts in the dihydrofolate reductase gene of Chinese hamster ovary cells. J Biol Chem. 1989;264:18005–18010. [PubMed] [Google Scholar]
- 65.Mori T, Matsunaga T, Hirose T, Nikaido O. Establishment of a monoclonal antibody recognizing ultraviolet light-induced (6-4) photoproducts. Mutat Res. 1988;194:263–270. doi: 10.1016/0167-8817(88)90028-4. [DOI] [PubMed] [Google Scholar]
- 66.Cadet J, Mouret S, Ravanat JL, Douki T. Photoinduced damage to cellular DNA: direct and photosensitized reactions. Photochem Photobiol. 2012;88:1048–1055. doi: 10.1111/j.1751-1097.2012.01200.x. [DOI] [PubMed] [Google Scholar]
- 67.Setlow P, Li L. Photochemistry and photobiology of the spore photoproduct: a fifty year journey. Photochem Photobiol. 2015 doi: 10.1111/php.12506. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Donnellan JE, Setlow RB. Thymine photoproducts but not thymine dimers found in ultraviolet-irradiated bacterial spores. Science. 1965;149:308–310. doi: 10.1126/science.149.3681.308. [DOI] [PubMed] [Google Scholar]
- 69.Rasmussen RE, Painter RB. Evidence for repair of ultra-violet damaged deoxyribonucleic acid in cultured mammalian cells. Nature. 1964;203:1360–1362. doi: 10.1038/2031360a0. [DOI] [PubMed] [Google Scholar]
- 70.Cleaver JE, Painter RB. Evidence for repair replication of HeLa cell DNA damaged by ultraviolet light. Biochim Biophys Acta. 1968;161:552–554. doi: 10.1016/0005-2787(68)90131-7. [DOI] [PubMed] [Google Scholar]
- 71.Cleaver JE. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218:652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 72.Setlow RB, Regan JD, German J, Carrier WL. Evidence that xeroderma pigmentosum cells do not perform the first step in the repair of ultraviolet damage to their DNA. Proc Natl Acad Sci U S A. 1969;64:1035–1041. doi: 10.1073/pnas.64.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 74.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase eta. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 75.Yoon J-H, Prakash L, Prakash S. Highly error-free role of DNA polymerase η in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc Natl Acad Sci U S A. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clark AJ, Margulies AD. Isolation and characterization of recombination-deficient mutants of Escherichia coli K12. Proc Natl Acad Sci USA. 1965;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 78.Hanawalt PC, Cooper PK, Ganesan AK, Smith CA. DNA repair in bacteria and mammalian cells. Annu Rev Biochem. 1979;48:783–836. doi: 10.1146/annurev.bi.48.070179.004031. [DOI] [PubMed] [Google Scholar]
- 79.Witkin EM. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976;40:869. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kenyon CT, Walker GC. DNA damaging agents stimulate gene expression at specific loci in E. coli. Proc Natl Acad Sci U S A. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Butala M, Zgur-Bertok DD, Busby SJW. The bacterial LexA transcriptional repressor. Cell Molec Life Sci. 2009;66:82–93. doi: 10.1007/s00018-008-8378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crowley DJ, Hanawalt PC. Induction of the SOS response increases the efficiency of global nucleotide excision repair of cyclobutane pyrimidine dimers, but not 6-4 photoproducts, in UV-irradiated Escherichia coli. J Bacteriol. 1998;180:3345–3352. doi: 10.1128/jb.180.13.3345-3352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5:1–20. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang Q, Karata K, Woodgate R, Cox MM, Goodman MF. The active form of DNA polymerase V is UmuD2C-RecA-ATP. Nature. 2009;460:359–363. doi: 10.1038/nature08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ford JM, Hanawalt PC. Li-Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc Natl Acad Sci U S A. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ford JM, Hanawalt PC. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 88.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 89.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci U S A. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fitch ME, Nakajima S, Yasui A, Ford JM. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J Biol Chem. 2003;278:46906–46910. doi: 10.1074/jbc.M307254200. [DOI] [PubMed] [Google Scholar]
- 92.van Zeeland AA, Smith CA, Hanawalt PC. Sensitive determination of pyrimidine dimers in DNA of UV-irradiated mammalian cells. Introduction of T4 endonuclease V into frozen and thawed cells. Mutat Res. 1981;82:173–189. doi: 10.1016/0027-5107(81)90148-2. [DOI] [PubMed] [Google Scholar]
- 93.Vijg J, Mullaart E, van der Schans GP, Lohman PH, Knook DL. Kinetics of ultraviolet induced DNA excision repair in rat and human fibroblasts. Mutat Res. 1984;132:129–138. doi: 10.1016/0167-8817(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 94.Mullaart E, Lohman PH, Vijg J. Differences in pyrimidine dimer removal between rat skin cells in vitro and in vivo. J Invest Dermatol. 1988;90:346–349. doi: 10.1111/1523-1747.ep12456316. [DOI] [PubMed] [Google Scholar]
- 95.Hanson CV, Shen CK, Hearst JE. Cross-linking of DNA in situ as a probe for chromatin structure. Science. 1976;193:62–64. doi: 10.1126/science.935855. [DOI] [PubMed] [Google Scholar]
- 96.Cech T, Pardue ML. Cross-linking of DNA with trimethylpsoralen is a probe for chromatin structure. Cell. 1977;11:631–640. doi: 10.1016/0092-8674(77)90080-0. [DOI] [PubMed] [Google Scholar]
- 97.Smerdon MJ, Tlsty TD, Lieberman MW. Distribution of ultraviolet-induced DNA repair synthesis in nuclease sensitive and resistant regions of human chromatin. Biochemistry. 1978;17:2377–2386. doi: 10.1021/bi00605a020. [DOI] [PubMed] [Google Scholar]
- 98.Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA- repair synthesis. Proc Natl Acad Sci U S A. 1978;75:4238–4241. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zolan ME, Smith CA, Calvin NM, Hanawalt PC. Rearrangement of mammalian chromatin structure following excision repair. Nature. 1982;299:462–464. doi: 10.1038/299462a0. [DOI] [PubMed] [Google Scholar]
- 100.Gale JM, Smerdon MJ. UV induced (6-4) photoproducts are distributed differently than cyclobutane dimers in nucleosomes. Photochem Photobiol. 1990;51:411–417. doi: 10.1111/j.1751-1097.1990.tb01732.x. [DOI] [PubMed] [Google Scholar]
- 101.Smerdon MJ, Conconi A. Modulation of DNA damage and DNA repair in chromatin. Prog Nucleic Acid Res Mol Biol. 1999;62:227–255. doi: 10.1016/s0079-6603(08)60509-7. [DOI] [PubMed] [Google Scholar]
- 102.Zolan ME, Cortopassi GA, Smith CA, Hanawalt PC. Deficient repair of chemical adducts in alpha DNA of monkey cells. Cell. 1982;28:613–619. doi: 10.1016/0092-8674(82)90216-1. [DOI] [PubMed] [Google Scholar]
- 103.Mansbridge JN, Hanawalt PC. Domain-limited repair of DNA in ultraviolet irradiated fibroblasts from xeroderma pigmentosum complemetation group C. Cellular Responses to DNA Damage, UCLA Symposium on Molecular and Cellular Biology. 1983:195–207. [Google Scholar]
- 104.Bohr VA, Okumoto DS, Ho L, Hanawalt PC. Characterization of a DNA repair domain containing the dihydrofolate reductase gene in Chinese hamster ovary cells. J Biol Chem. 1986;261:16666–16672. [PubMed] [Google Scholar]
- 105.Mellon I, Bohr VA, Smith CA, Hanawalt PC. Preferential DNA repair of an active gene in human cells. Proc Natl Acad Sci U S A. 1986;83:8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 107.Sweder KS, Hanawalt PC. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc Natl Acad Sci U S A. 1992;89:10696–10700. doi: 10.1073/pnas.89.22.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smerdon MJ, Thoma F. Site-specific DNA repair at the nucleosome level in a yeast minichromosome. Cell. 1990;61:675–684. doi: 10.1016/0092-8674(90)90479-x. [DOI] [PubMed] [Google Scholar]
- 109.Nouspikel T, Hanawalt PC. Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Mol Cell Biol. 2000;20:1562–1570. doi: 10.1128/mcb.20.5.1562-1570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nouspikel TP, Hyka-Nouspikel N, Hanawalt PC. Transcription domain-associated repair in human cells. Mol Cell Biol. 2006;26:8722–8730. doi: 10.1128/MCB.01263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hsu PH, Hanawalt PC, Nouspikel T. Nucleotide excision repair phenotype of human acute myeloid leukemia cell lines at various stages of differentiation. Mutat Res. 2007;614:3–15. doi: 10.1016/j.mrfmmm.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 112.Wood RD, Robins P, Lindahl T. Complementation of the xeroderma pigmentosum DNA repair defect in cell-free extracts. Cell. 1988;53:97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]
- 113.Hansson J, Grossman L, Lindahl T, Wood RD. Complementation of the xeroderma pigmentosum DNA repair synthesis defect with Escherichia coli UvrABC proteins in a cell-free system. Nucleic Acids Res. 1990;18:35–40. doi: 10.1093/nar/18.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mu D, Park CH, Matsunaga T, Hsu DS, Reardon JT, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 115.Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protić M, Hübscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 116.Selby CP, Sancar A. Gene- and strand-specific repair in vitro: partial purification of a transcription-repair coupling factor. Proc Natl Acad Sci U S A. 1991;88:8232–8236. doi: 10.1073/pnas.88.18.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 118.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29:948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Spivak G, Hanawalt PC. Photosensitive human syndromes. Mutat Res. 2014;776:24–30. doi: 10.1016/j.mrfmmm.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oh DH, Spivak G. Hereditary photodermatoses. Adv Exp Med Biol. 2010;685:95–105. doi: 10.1007/978-1-4419-6448-9_9. [DOI] [PubMed] [Google Scholar]
- 121.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 122.Guo J, Hanawalt PC, Spivak G. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res. 2013;41:7700–7712. doi: 10.1093/nar/gkt524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair (Amst) 2006;5:13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 124.Nardo T, Oneda R, Spivak G, Vaz B, Mortier L, Thomas P, Orioli D, Laugel V, Stary A, Hanawalt PC, Sarasin A, Stefanini M. A UV-sensitive syndrome patient with a specific CSA mutation reveals separable roles for CSA in response to UV and oxidative DNA damage. Proc Natl Acad Sci U S A. 2009;106:6209–6214. doi: 10.1073/pnas.0902113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Premi S, Wallisch S, Mano CM, Weiner AB, Bacchiocchi A, Wakamatsu K, Bechara EJH, Halaban R, Douki T, Brash DE. Chemiexcitation of melanin derivatives induces DNA photoproducts long after UV exposure. Science. 2015;347:842–847. doi: 10.1126/science.1256022. [DOI] [PMC free article] [PubMed] [Google Scholar]