Abstract
Background
Psoriasis vulgaris is an inflammatory immune-mediated disease, whose lesional skin is characterized by sharply demarcated, erythematous scaly plaques. Conversely, uninvolved psoriatic skin appears clinically similar to normal skin. However, it has been hypothesized that inflammatory cytokines, such as interleukin (IL)-17, may affect any organ or tissue having a vascular supply or blood leukocytes and, thereby, distant uninvolved skin, as well, could be exposed to increased circulating IL-17 concentrations detected in subjects affected by moderate-severe psoriasis.
Objectives
The aim of this study was to establish comparative genomic profiles between non-involved skin and normal skin, particularly, determining the immune abnormalities in distant uninvolved skin.
Methods
We performed a meta-analysis on three gene array studies in order to characterize non-lesional (NL) psoriatic skin transcriptome compared to normal gene expression profile. We investigated the immunologic features of non-involved skin, particularly linked to the IL-17 signaling pathway.
Results
Overall we detected 252 differentially expressed gene probes in uninvolved skin compared with normal skin, an upregulation of multiple immune-related genes, including IL-17- downstream genes. The increased expression of IL-17-signature genes, including DEFB4 and S100A7, was associated with an increased number of CD3+, CD8+, and DC-LAMP+ cells that we observed in NL skin vs. normal controls. Within NL skin, we detected a few T-cells that expressed the T-cell activation marker ICOS, but we did not detect ICOS expression on T-cells in normal skin.
Conclusions
Our data described the genomic profile in NL skin, characterizing the immune activation that was mainly attributed to IL-17 signaling.
Introduction
Psoriasis vulgaris is an inflammatory immune-mediated disease, characterized by sharply demarcated, erythematous scaly plaques, that results from the combination of an immune dysregulation and an altered keratinocyte differentiation1. The psoriatic phenotype reflects an altered gene expression profile and associated epigenetic changes2,3.
The pathogenic mechanism leading to the psoriatic plaque formation is driven by the activation of various antigen-presenting cells (APCs), including mature (DC-LAMP+) and inflammatory (CD11c+) DCs, followed by a wide array of T cell subsets that trigger the tissue response of fibroblasts, endothelial cells, and keratinocytes (KCs)1,4–6. Multiple pro-inflammatory and pro-proliferative products secreted by DCs (TNFα, IL-6, IL-20, IL-23, NO), T cells (IL-17, IL-22, IL21, IFN-γ), and KCs (AMPs, IL-8, IL-20, CCL20, CXCL-1,-3,-5,-9,-10,-11) are known to mediate the pathogenic circuits7–9. In this immunologic setting, mounting evidence recognizes the IL-23/T17 axis as the main pathway in psoriasis. Indeed, IL-17 was shown upregulated in both lesional psoriatic skin and bloodstream10–14. Additionally, an increased number of IL-17-producing T cells have been detected in peripheral blood and lesional skin, and their IL-17 production is mainly dependent on IL-23 stimulation, which is also overexpressed in psoriasis8,15–20.
Although non-involved skin appears clinically identical to normal skin, some considerations led us to establish comparative genomic profiles. Firstly, more than three decades ago non-lesional was reported to show some histological alterations compared to normal skin, including: a slight increase of dermal CD4+ and epidermal CD8+ cells; focal increase of CD11b+ cells; upregulation of innate immune-related genes (anti-microbial peptides); and alterations in some dendritic cell subsets21–23. More importantly, it has been hypothesized that inflammatory cytokines produced in the skin and released into the systemic circulation are linked to functional pathways associated with metabolic diseases/diabetes and cardiovascular diseases24. Indeed, any organ or tissue having a vascular supply could be exposed to circulating cytokines, thereby, the effects of pro-inflammatory cytokines such as IL-17 may impact the activation of endothelial cells, adipocytes, or blood leukocytes14,25. Similarly to other tissues, distant uninvolved skin may be also affected by circulating IL-17 with a consequent activation of IL-17 signaling and expression of IL-17 downstream pathway genes. Secondly, the increased gene expression of lesional psoriatic skin correlates with a low grade of methylation and, surprisingly, non-lesional (NL) skin also shows an overall methylation pattern similar to lesional skin26.
In this study, we sought to determine immune abnormalities in uninvolved distant skin, particularly linked to the IL-17 signaling pathway. Overall we detected 252 differentially expressed gene probes in uninvolved skin compared with normal skin, including an upregulation of both IL-17- and IL-22-downstream genes and other immune-related genes.
Methods
Patient cohort
For RT-PCR, NL skin samples were obtained from 41 psoriatic patients with moderate-to-severe psoriasis (psoriasis area severity index >12, and body surface area >10) not receiving active psoriasis treatment, while normal skin was obtained from 12 healthy volunteers.
These 41 patients, who were enrolled into an IRB-approved Phase 3, multicenter, randomized trial protocol, represent the same cohort of patients used to define the psoriasis transcriptome in a previous publication13.
Immunohistochemistry and immune cell counts were performed on patients belonging to this cohort and supplemented by additional cases of untreated moderate-to-severe psoriasis from other studies performed at The Rockefeller University.
Statistical analysis
We performed a meta-analysis on 3 previously published gene array studies13, 27,28, in order to assess the differences between normal and uninvolved psoriatic skin transcriptomes. In all studies samples from NL and normal skin were hybridized using HG_U133 plus 2 arrays. Similarly to previous meta-analysis29 comparing lesional vs. NL skin, we used the same pipeline to obtain a robust definition of uninvolved psoriatic skin transcriptome as the set of differentially expressed gens (DEGs) between NL and normal skin. Overall, meta-analysis included 167 NL psoriatic skin samples and 110 normal controls, obtained from 3 different studies previously considered by Suarez-Farinas et al.13,28,30. Succinctly, to obtain an estimate of the NL vs normal effect size, random-effect models were used which take into account the within study variation.
Additionally, Gene Set Enrichment Analysis (GSEA) was used to evaluate the enrichment of various gene sets in the NL psoriatic skin gene expression profile30.
To assess the biological meaning of uninvolved psoriatic skin transcriptome, a comparison with various psoriasis transcriptomes, in particular MAD-3 or MAD-529 psoriasis transcriptomes (defined by a meta-analysis of 3 or 5 published transcriptomes, respectively) was performed. To evaluate the impact of pathogenically relevant signaling pathways on NL psoriasis transcriptome, multiple psoriasis relevant and previously published gene sets were analyzed.
Additionally, correlations between serum levels of key-cytokines and disease severity were assessed. Serum cytokine levels were measured in clinical trials conducted on moderate-severe psoriasis patients in periods of no active treatment for psoriasis13,31.
Similarly, the correlation between disease severity and the Gene Set Variation Analysis (GSVA)-scores for a collection of gene sets was tested. As previously described32, GSVA represents a Gene Set Enrichment (GSE) method that estimates variation of any gene-set for each sample, providing evidence about the association between clinical outputs (i.e., disease severity) and the dysregulation of each gene set.
Meta Analysis
The classic application of meta-analysis is to find a single outcome using published data where only the summary statistics are typically available. With microarray experiments, however, a more fortuitous situation of having the complete set of raw data available is commonly achievable. Thus, we took advantage of this feature and modeled the differences in expression values between NL and normal skin uniformly. The general model in a meta-analysis setting is as follows. Let Yij represent the measured effect for study j (j =1, …, J) for a specific gene i. We have,
| (1) |
Where between-study variance i2 represents the variability between studies, and it is usually estimated by the DerSimonian and Laird method33. And σ2 represents the within-study variance for the ith study. Both Yij and σ2 (called as summary statistics) are already known from previous analysis/study. i is regarded as the average measure of differential expression across all datasets/studies for this gene. This is the parameter of interest and may be estimated along with its a standard error (se) as:
| (2) |
Where wij equals to the inverse of the variance of Yij.
To decided if a fixed-effect model or a random-effects model is more appropriate for the data, we used the Cochran's Q statistic to test the nule hypothesis Ho: i2 =0 (fixed effect) versus Ha: i2 >0 (random effect). Since Cochran's Q follows a χ2 distribution under the null hypothesis34, this test favored the random-effect model.. The, the overall fold changes (LFC) between NL and normal skin on the log2 scale (i.e., the parameters of interest in this analysis) and their corresponding standard errors were calculated using Equation 2. Dividing this estimate by its estimated standard error, the resultant Z-score, being assumed to follow a standard normal distribution, is used to decide statistical significance of a gene. The resulting adjusted p-values using Benjamini and Hochberg procedure, which control for false discovery rate (FDR), are used to decide the statistical significance of genes along with LFC.
Model fitting
For each study, a moderated t-test was used to analyze differences (on the log2 scale) among NL and normal samples. The summary statistics (Yij and σ2 in Equation 1) were recorded and would be the input in the meta-analysis. Then, the overall fold changes (LFC) between NL and normal skin on the log2 scale (i.e., the parameters of interest in this analysis) and their corresponding standard errors were calculated using Equation 2. The resulting adjusted p-values using Benjamini and Hochberg procedure, which control for false discovery rate (FDR), are used to decide the statistical significance of genes along with LFC.
RNA processing and reverse transcriptase–polymerase chain reaction
Skin biopsies were snap-frozen in liquid nitrogen and stored at 80°C until used. RNA was extracted using the Qiagen RNeasy Fibrous Tissue Mini Kit (QIAGEN, Valencia, CA) for either gene array or RT-PCR procedures.
To perform RT-PCR, the RNA extracted from skin samples was processed using Taqman 16-gene and 48-gene low-density array cards were used for RT-PCR analysis. The 16-gene cards were tested on NL skin biopsies and normal samples. The 48-gene cards were tested on 12 healthy/normal and 41 pairs of psoriasis skin biopsy samples, using the same gene-identifying probe set published by Suárez-Fariñas M et al.13. The resulting data were normalized to human acidic ribosomal protein expression (Gene Symbol: RPLPO).
Immunohistochemistry and immunofluorescence
Skin samples
Skin punch biopsies were obtained from normal volunteers and patients with moderate-to-severe chronic plaque psoriasis. The biopsy specimens were frozen in OTC (Sakura, Torrance, CA, U.S.A.) and stored at −80°C for immunohistochemistry and immunofluorescence.
Immunohistochemistry and Immunofluorescence
Frozen tissue sections of psoriatic NL, and normal skin were stained using standard procedures for both IHC and IF as previously described35.
Immunohistochemistry
Staining was performed with antibody targeting C/EBPδ, LCN2, DEFB4, and S100A7 (Table S1). According to the primary antibody species, either biotin-labeled horse anti-mouse antibodies (Vector Laboratories, Burlingame, CA, U.S.A.) or biotin-labeled rabbit anti-goat antibodies (Vector Laboratories, Burlingame, CA, U.S.A.) were amplified with avidin-biotin complex (Vector Laboratories) and developed using chromogen 3-amino-9-ethylcarbazole (Sigma Aldrich, St Louis, MO, U.S.A.).
Immunofluorescence
Frozen skin sections from NL psoriasis patients and controls were fixed with acetone and blocked in 10% normal chicken serum (Vector Laboratories) for 30 minutes. Primary antibodies for CD3, CD8, and ICOS (Table S3) were incubated overnight at 4°C and amplified with the appropriate secondary antibody goat anti-mouse IgG1 conjugated to Alexa Fluor 488 and chicken anti-goat Alexa Flour 594 (Invitrogen, Eugene, OR) respectively, for 30 minutes.
IF images were acquired using the appropriate filters of a Zeiss Axioplan 2 wide-field fluorescence microscope (Thornwood, NY) with a Plan Neofluar 20 × 0.7 numerical aperture lens and a Hamamatsu Orca Er-cooled charge-coupled device camera (Bridgewater, NJ), controlled by the METAVUE software (MDS Analytical Technologies, Downington, PA). Images in each figure are presented both as single-color stains (green and red) located above the merged image, so that localization of two markers on similar or different cells can be appreciated. Cells that co-express the two markers in a similar location are yellow in color. A white line denotes the dermoepidermal junction. Dermal collagen fibers gave green autofluorescence, and antibodies conjugated with a fluorochrome often gave background epidermal fluorescence.
Results
Uninvolved skin shows an increased expression of IL-17-downstream genes
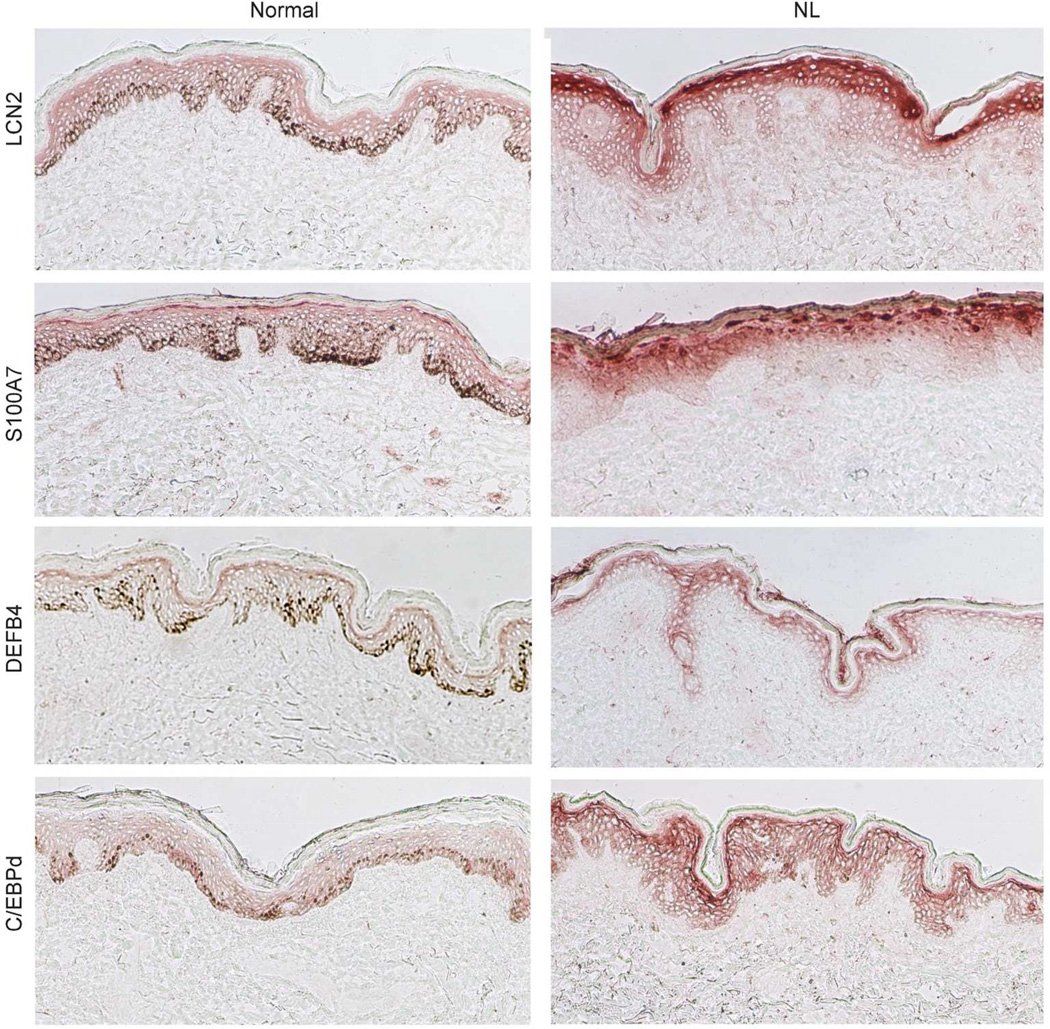
To assess possible IL-17 effects on distant uninvolved skin, we investigated the expression of proteins known to be upregulated in keratinocytes by IL-17, namely LCN2, S100A7, and DEFB4, and C/EBPδ, a key-transcription factor mediating IL-17 signaling.
We observed an enhanced staining for LCN2, S100A7, and DEFB4 in the granular and uppermost spinous layers of the epidermis of NL psoriatic skin vs. non-psoriatic controls (Fig. 1). The expression of IL-17-induced genes co-localized with an increased expression of C/EBPδ in fully differentiated keratinocytes, which are confined in the upper portion of the epidermis (Fig. 1).
Fig. 1. IL-17-signature genes are increasingly expressed by terminally differentiated keratinocytes in non-lesional skin compared to normal skin.
IL-17-signature proteins including lipocalin 2 (LCN2), S100A7, HBD2, and C/EBPδ (top) expression was predominantly localized in the spinous-granular layer of the epidermis, with a more marked staining in NL skin vs. normal skin. NL, non-lesional.
Because of this enhanced expression of IL-17-regulated proteins, we performed a comprehensive gene array analysis in order to examine the whole gene expression profile as well as selected gene expression by RT-PCR in unaffected skin.
Non-lesional skin gene profiles suggest active IL-17-, IL-22, and IFN-γ- signaling
To examine gene expression in NL psoriatic skin compared with the normal controls, we conducted a meta-analysis on three published studies13,27,28 with expression profiles from NL skin of psoriatic patients and normal skin. We followed the same methodology we used to derive a robust signature of lesional vs NL skin using these 3 studies (the MAD-3 psoriasis transcriptome)29. In total, we identified 252 gene transcripts differentially expressed in NL psoriatic skin compared to normal skin, using criteria of FCH≥1.4, and false discovery rate (FDR)≤ 0.05. Overall, 81 gene probes were up-regulated and 171 were down-regulated (Table S2 and Table S3, respectively). We found an increased expression of immune-related genes, mainly represented by AMPs (DEFB4, S100A7, S100A7A, S100A8, S100A9, and PI3), and other immune-related, KC-derived products including CXCL10, SERPINB3, SERPINB4, and RNAse7 (Table 1).
Table 1.
Upregulated immune gene transcripts and epidermal keratinocyte differentiation-related gene transcripts detected in uninvolved psoriatic skin.
| Upregulated immune gene transcripts | |||||
| Probe set ID |
Gene symbol |
Description |
NL vs Normal (Log2) |
NL vs Normal (Fold Change) |
NL vs Normal FDR |
| 207356_at | DEFB4 | defensin, beta 4 | 1.81 | 3.51 | 1.06E-13 |
| 205916_at | S100A7 | S100 calcium binding protein A7 | 1.73 | 3.32 | 7.33E-14 |
| 202917_s_at | S100A8 | S100 calcium binding protein A8 | 1.45 | 2.73 | 1.75E-12 |
| 209720_s_at | SERPINB3 | serpin peptidase inhibitor, clade B (ovalbumin), member 3 | 1.18 | 2.27 | 2.45E-09 |
| 211906_s_at | SERPINB4 | serpin peptidase inhibitor, clade B (ovalbumin), member 4 | 1.00 | 2.00 | 1.34E-06 |
| 232170_at | S100A7A | S100 calcium binding protein A7A | 0.97 | 1.96 | 3.28E-08 |
| 203535_at | S100A9 | S100 calcium binding protein A9 | 0.91 | 1.88 | 1.14E-08 |
| 203691_at | PI3 | peptidase inhibitor 3, skin-derived | 0.89 | 1.85 | 5.17E-07 |
| 204533_at | CXCL10 | chemokine (C-X-C motif) ligand 10 | 0.63 | 1.55 | 9.39E-07 |
| 233488_at | RNASE7 | ribonuclease, RNase A family, 7 | 0.51 | 1.42 | 3.82E-06 |
| Upregulated epidermal keratinocyte differentiation-related gene transcripts | |||||
|
Probe set ID |
Gene symbol |
Description |
NL vs Normal (Log2) |
NL vs Normal (Fold Change) |
NL vs Normal FDR |
| 232082_x_at | SPRR3 | small proline-rich protein 3 | 1.23 | 2.35 | 3.98E-14 |
| 208539_x_at | SPRR2D | small proline-rich protein 2D | small proline-rich protein 2B | 1.13 | 2.19 | 1.81E-11 |
| 224328_s_at | LCE3D | late cornified envelope 3D | 1.03 | 2.04 | 2.41E-12 |
| 236119_s_at | SPRR2G | small proline-rich protein 2G | 1.01 | 2.01 | 8.16E-09 |
| 224329_s_at | CNFN | cornifelin | 0.56 | 1.47 | 3.10E-09 |
The gene array analysis showed an elevated expression of IL-17 signature genes (i.e., DEFB4) and some genes, including S100A7, S100A7A, S100A8, and S100A9 that are known to be induced by both IL-17 and IL-22 (Table 1). We confirmed this increased genomic expression of AMP mRNAs by RT-PCR measurement, also detecting an upregulation of other IL-17-downstream genes playing a crucial role in psoriasis that did not appear within the microarray gene list such as IL-1β, a proinflammatory gene, and IL-19, an epidermal hyperplasia inducer (Table 2). An enhanced expression of IL-22 associated with an upregulation of AMPs was also found in NL vs. normal skin, whereas a less consistent increase in IL-17A and IL-17F mRNAs was detected in NL skin vs. normal controls (Fig. S1).
Table 2.
RT-PCR results. Immune genes showing an increased expression in uninvolved psoriatic skin compared to normal skin.
| ID | Description | NL vs Normal FCH |
NL vs Normal Raw pValue |
|---|---|---|---|
| DEFB4 | defensin beta 4 | 99.51 | 3.67E-12 |
| IL22 | interleukin 22 | 23.25 | 1.45E-05 |
| CXCL10 | chemokine (C-X-C motif) ligand 10 | 15.6 | 1.73E-06 |
| IL19 | interleukin 19 | 12.9 | 0.0012 |
| IFNA1 | interferon alpha 1 | 6,03 | 8,37E-05 |
| SERPINB3 | serpin peptidase inhibitor, clade B, member 3 | 4.91 | 1.46E-05 |
| CCL18 | chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) | 4.39 | 0.0037 |
| IL1B | interleukin 1 beta | 4.18 | 0.0016 |
| IL17F | interleukin 17F | 2.78 | 0.2007 |
| CD209 | cluster of differentiation 209 (DC-SIGN) | 2,74 | 0,056 |
| CXCL9 | chemokine (C-X-C motif) ligand 9 | 2.46 | 0.0373 |
| IL17A | interleukin 17A | 2.26 | 0.1942 |
| IFNγ | interferon gamma | 2.15 | 0.2949 |
| STAT1 | signal transducer and activator of transcription 1 | 1.81 | 0.0175 |
| CD1E | cluster of differentiation 1E | 1.78 | 0.112 |
| OASL | 2',5'-oligoadenylate synthetase-like | 1.69 | 0.0907 |
| MX1 | myxovirus resistance 1, interferon-inducible protein p78 | 1.56 | 0.1335 |
| CEBPD | CCAAT/enhancer binding protein (C/EBP), delta | 1.37 | 0.273 |
| CD69 | cluster of differentiation 69 | 1.36 | 0.4663 |
| JAK2 | janus kinase 2 | 1.33 | 0.1603 |
| CXCL11 | chemokine (C-X-C motif) ligand 11 | 1.06 | 0.9205 |
| CD207 | cluster of differentiation 207 | −1.69 | 0.711 |
| IFNB1 | Interferon beta 1 | −1.77 | 0.4957 |
| MMP9 | matrix metalloproteinase-9 | −3.27 | 0.0409 |
In addition to an IL-17 imprinting, the overall gene expression analysis, including gene array and RT-PCT data, showed an upregulation of both IL-22 and IFN-γ signaling pathway characterizing uninvolved skin. RT-PCR confirmed a higher expression of IFN-γ itself and IFN-γ-signature genes (CXCL-9, -10, -11, MX-1, STAT1, and MMP9) in NL skin compared to normal skin (Table 2). The gene array list also included some upregulated genes related to the KC differentiation process, namely SPRR2D, SPRR2G, SPRR3, LCE3D, and CNFN.
We also correlated NL psoriatic skin transcriptome with various gene sets using gene set enrichment analysis (GSEA).
Additionally, we used gene set enrichment analysis (GSEA) to identify if there was a ‘typical psoriatic lesion’ signal on the NL skin transcriptome. The differences in gene expression profiles of uninvolved psoriatic skin compared to normal skin, was strongly enriched of psoriasis genes identified in previously published lesional psoriasis transcriptomes (i.e., the DEGs presented in the original studies used in this meta-analysis and the MAD5 trancriptome) (Table 3). This analysis also detected an enrichment of: (i) genes belonging to the epidermal differentiation complex list; (ii) genes induced by IL-17 in both keratinocytes and reconstructed human epidermis (RHE); (iii) sets of genes related to various immune signaling pathways including Th22/IL-22, IL-1, and Th1; (iv) genes synergistically or additively induced by IL-17+IL-22 stimulation (Table 3). Notably, GSEA detected an enhanced Th2 signal and an enrichment of DEGs identifying the atopic dermatitis transcriptome (AD LS vs. AD NL). Additionally, a significantly higher expression of CCL18, a Th2-polarizing chemokine, was measured by RT-PCR.
Table 3.
Pathways enriched in uninvolved psoriasis skin by using GSEA
| PATHWAYS | No. of genes in pathway |
ES | NES | FDR |
|---|---|---|---|---|
| Epidermal Differentiation Complex | 33 | 0.63 | 2.07 | 4.22E-04 |
| Epidermal Differentiation Complex- Cornified Envelope | 50 | 0.59 | 2.12 | 2.85E-04 |
| Psoriasis Transcriptome Up39 | 531 | 0.43 | 2.16 | 3.56E-04 |
| Synergistic IL17 & IL22 in KC | 31 | 0.43 | 1.42 | 0.06 |
| IL17+TNF Up in KC | 33 | 0.46 | 1.55 | 0.031 |
| KC+IL1 Up | 56 | 0.43 | 1.64 | 0.016 |
| KC+IL-17 Up | 53 | 0.44 | 1.66 | 0.015 |
| KC+IL-17 Up40 | 165 | 0.39 | 1.74 | 0.009 |
| ADDITIVE IL17 & IL22 in KC9 | 26 | 0.56 | 1.78 | 0.008 |
| Psoriasis Transcriptome Up24 | 1009 | 0.34 | 1.76 | 0.008 |
| MAD5 Psoriasis Up29 | 675 | 0.35 | 1.78 | 0.008 |
| MAD3 Psoriasis Up29 | 1069 | 0.34 | 1.79 | 0.007 |
| Psoriasis Transcriptome Up13 | 597 | 0.37 | 1.90 | 0.003 |
| Psoriasis Transcriptome Up41 | 1038 | 0.34 | 1.80 | 0.007 |
| Th22 and IL-22 | 14 | 0.62 | 1.66 | 0.015 |
| Th1 | 11 | 0.58 | 1.48 | 0.045 |
| Th17 | 11 | 0.69 | 1.70 | 0.012 |
| AD LS vs. AD NL Up | 237 | 0.38 | 1.75 | 0.009 |
| Th2 | 18 | 0.61 | 1.76 | 0.009 |
ES: enrichment scores. NES: normalized enrichment scores. FDR: false discovery rate.
Increased infiltration of immune cells profiles non-inflamed skin
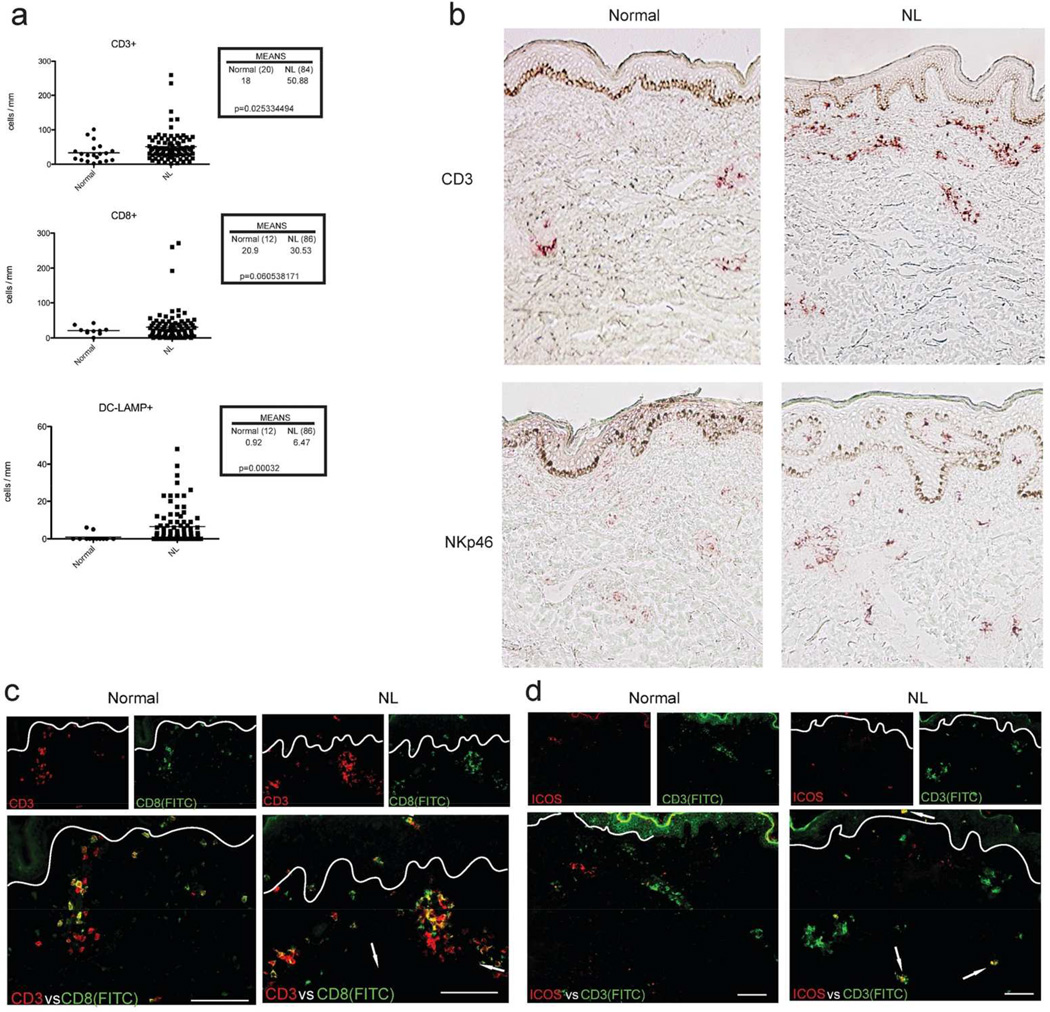
Because we detected increased expression of multiple chemokines, we examined immune cell subsets in NL skin. Immune cell markers included CD3+, CD8+, DC-LAMP+, and NKp46 for T-cell, dendritic, and NK/NKT subsets that infiltrate psoriasis skin lesions. An increased number of CD3+, CD8+, and DC-LAMP+ cells was measured in NL skin vs. normal controls, with a significant difference for CD3+ and DC-LAMP+ cell count (p=0.025 and p=0.00032, respectively) (Fig. 2a). T-cells increased in NL psoriatic skin were mainly dermal (Fig. 2b), with only a rare T-cell detected in the epidermis. Likewise, the increase in NK/NKT cells detected in NL skin was also mainly dermal. The increase in CD8+ cells in NL skin appears to be due to combined expression on T-cells (cells co-expressing CD3 and CD8) as shown in Fig. 2c and likely NK cells (CD8+ but CD3-cells). Within NL skin, we detected a few T-cells that expressed the T-cell activation marker ICOS, but we did not detect ICOS expression on T-cells in normal skin (Fig. 2d).
Fig. 2. Immune cell infiltration and activation in non-lesional skin.
Increased number of CD3+, CD8+, or DC-LAMP+ cells in NL skin vs. Normal skin (a). Enhanced dermal staining for CD3 and NKp46 markers in NL skin vs. Normal by immunohistochemistry (b). Co-expression of CD3 and CD8, showed by immunofluorescence (c), demonstrating the increased presence of CD3/CD8 + T cells in non-lesional skin mainly localized within the dermis, whereas a few cells were localized within the epidermis (white arrows). Scattered detection of T cell activation marked by ICOS expression only in NL skin (d, white arrows). NL, non-lesional; DC-LAMP, dendritic cell lysosome-associated membrane glycoprotein.
High levels of circulating cytokines correlated with the upregulation of their matching signaling pathways in non-lesional skin
In order to substantiate the hypothesis of an altered gene expression in non-lesional skin induced by cytokines released from involved areas and acting at distant skin sites, we sought to correlate disease severity with the transcritpomic analysis obtained in NL vs. normal skin, as well as serum levels of pathogenic cytokines including IL-17, IL-22, and TNF-α. As showed in Fig. S2, disease severity significantly correlated with both IL-17 (r:0.642; p=0.045) and TNF-α blood levels (r:0.435, p=0.001), while no correlation could be evaluated for other cytokines, namely IL-22 and IFN-γ, because their bloodstream concentrations were below the detection limit. As confirmatory proof about the correlation between IL-17 serum levels and disease severity, another set of previously published data was analyzed. In this proof-of-concept study, wherein 16 moderate-severe patients were treated with guselkumab31 (an anti-p19IL23 agent), IL-17 serum levels significantly correlated with disease severity (r:0.586, p=0.017) (Figure S3).
Additionally, we evaluated a potential link between disease severity and upregulated signaling pathways detected in NL skin, providing a strong correlation between individual PASI scores and various signaling pathways (Table S4) related to IL-17 (r:0.26, p=0.0199), IL-1 (r:0.31, p=0.0056), and IFN-α (r:0.28, p=0.0107).
Discussion
A major focus of transcriptomic analysis in psoriasis has been defining the disease-related transcriptome by the identification of genes that are differentially expressed between psoriasis vulgaris plaques and background NL skin or normal human skin. In some studies, comparisons have included both NL skin and skin from non-psoriatic (normal) controls. Thus, as early as 2003, gene array studies detected some differences in gene expression between normal, healthy controls and NL psoriatic skin25. Differences noted included higher expression of S100A7 (psoriasin) and STAT1 in NL skin25. In 2008, a gene array study of psoriasis28 was conducted with a focus on expression of genes that are modulated by Type 1 interferons. That study compared gene expression in psoriasis lesions to both NL skin and to normal controls. Decreased expression of several transcription factors in NL psoriatic skin vs. healthy control skin was noted, as were decreases in expression of cell adhesion and tight junction proteins. Upregulated genes were also detected, but they were not individually listed in the report. The following year, Gudjonsson et al.27 published a report with an explicit focus on comparing gene expression in NL psoriatic skin vs. healthy controls. That study identified 223 transcripts from 178 unique genes as being differentially expressed in NL skin27 using a FCH>1.4 and (unadjusted) p<0.05. Many of the dysregulated genes mapped into pathways of lipid biosynthesis, keratinocyte terminal differentiation, or innate immune pathways. Small increases in S100A7, S100A9, IL1F9, and DEFB4 (IL-17 pathway products) were detected, but the increases were less marked compared to the present report. In this study, we detected differential expression of 252 transcripts between NL psoriatic skin and healthy controls using criteria of at least a 1.4-fold change and a FDR < 0.05. Importantly, many of genes detected map into the IL-17 pathway that is strongly up-regulated in psoriasis vulgaris skin lesions and in the bloodstream of patients affected by moderate-to-severe psoriasis13,36. Compared to Gudjonsson study27, overall we detected a much stronger inflammatory signal than a metabolic signal in NL skin. It is important to note that biopsies in this previous study were taken from a cross section of mild-to-severe psoriasis patients and that patients may have been on active therapies for psoriasis at the time of the biopsy. Conversely, in our study, for immunohistochemistry, immunofluorescence, and RT-PCR, we analyzed NL skin of moderate-to-severe patients (at least 10% body surface affected with disease) who were off active systemic treatments for at least 4 weeks or topical treatments for at least 2 weeks before the biopsy was taken. Whilst meta-analysis was performed on a broader spectrum of disease severity, ranging from mild to severe psoriasis, in order to obtain a more potent signal detection. We would predict that with higher baseline severity of psoriasis and without active treatment, that T-cell activation and IL-17 synthesis in skin lesions would be higher, leading to potentially higher levels of circulating IL-17 in the blood. IL-17 effects may be observed not only on lesional skin cells but also on bloodstream cells, which showed an increased expression of pro-inflammatory mediators (i.e., CXCL5, CXCL1, IL-8) and extravasation-mediating adhesion molecules (i.e., PECAM, ITGA4, CD99) after in vitro IL-17 stimulation, and their in vivo downregulation after IL-17 blockade14. Similarly, circulating IL-17 could affect other organ, tissues, or distant uninvolved skin, and, thus, it may represent the driving cytokine for many of the gene transcription changes detected in this study. IL-17 synergistically or additively acts with another key-cytokine in psoriasis pathogenesis9, namely TNF-α, and both of them showed circulating levels correlating with disease severity. As we previously published, these two cytokines are crucial in driving the most relevant inflammatory circuits characterizing psoriasis9.
While T-cells and mature dendritic cells appear to be higher in number in NL psoriatic skin vs. healthy control skin, we found that only a subset of patients had higher expression of IL-17, IL-22, and IFN-γ compared to normal controls. Hence, we favor the hypothesis that circulating IL-17 is the inducer of IL-17 pathway gene products in NL skin, rather than requiring an endogenous T-cell activation response. Most of the immune-related genes detected in this study are AMPs, which are known to be strongly expressed in lesional psoriatic skin, and their expression is stimulated by both IL-17 and IL-22 signals. Notably, together with IL-17, we detected enhanced IL-22 expression levels in uninvolved skin compared to normal skin.
These upregulated signaling pathways significantly correlated with disease severity and blood cytokine levels. Additionally, in this study, an altered expression of some epidermal differentiation complex genes was also detected. Moreover, we noted that very few intra-epidermal T-cells are present in this study, which clearly distinguishes NL skin from active plaques. In active plaques, many CD8+ T-cells can be found in the epidermis and a subset of these cells are IL-17-producing cells (Tc17 T-cells)37. Still, T-cells appear to be somewhat more activated in NL skin based on ICOS expression in some and higher IL-17 mRNA is some samples might lead to locally produced IL-17 as an inducer of some of the changes we have noted (Fig. S1). Since IL-17 regulated genes are transcribed by the dual factors NFkB and C/EBP, it is also possible that higher levels of C/EBP β (Chiricozzi et al., 2014) and C/EBPδ in NL skin might increase sensitivity of psoriatic keratinocytes to IL-17 stimulation38. Patients with moderate-to-severe psoriasis also have highly consistent increases in circulating TNF-α13 (mean increase 2.5-fold, p = 10−50), which provides a signaling cytokine for enhanced activation of NFκB13. Many psoriasis-related genes have synergistic or additive responses to TNF-α and IL-17, so the interaction of these cytokines in NL skin could drive important sets of genes identified in this report9. Still, NL psoriatic skin is grossly normal with respect to skin structure. We do not know the threshold of cytokine levels that are needed to convert background skin into frank lesions, but in most studies, levels of key cytokine transcripts, e.g., IL-23 and IL-17A/F are elevated by 5–10-fold or more compared to NL skin31. Another factor that should be considered is that NL skin might have higher expression of regulatory pathways that prevent uncontrolled T-cell activation and expansion. In this study we detected an increased expression of Th2-related genes, which could indicate higher Th2 tone in NL skin.
Unfortunately, we do not know whether gene differences we detected here will be reversible with treatments that reverse the pathology of lesional psoriatic skin, but testing the ability of psoriasis treatments to affect pathologic gene expression in distant sites (uninvolved skin and co-morbid tissues) should be investigated in future studies. If reversible gene expression occurs in NL skin, this could be the best and most accessible “sensor” for pathologic effects of inflammatory cytokines on background tissues in psoriasis patients. Overall, the magnitude for altered gene expression in NL skin is similar to the magnitude of increased expression of inflammatory gene products in peripheral blood leukocytes of psoriasis patients, which is consistent with this possibility14.
Supplementary Material
What’s known/what’s new.
What’s already known about this topic?
Classically, uninvolved psoriatic skin has been roughly considered similar to normal skin, though a few articles showed some alterations compared to normal skin.
To our knowledge, only one article described the transcriptomic profile in non-lesional vs. normal skin, showing an altered gene expression related to lipid biosynthesis, innate immunity, and keratinocyte differentiation
What does this study add?
A complex immunologic activation is detectable in uninvolved psoriatic skin
An upregulation of key signaling pathways in psoriasis, in particular the IL-17, IL-22, and IFNγ signaling pathways, was not previously described.
Acknowledgments
JGK has consulted for/received honoraria from Janssen. KL and CB are employees of Janssen Research & Development LLC, a Johnson & Johnson pharmaceutical company. KL is stock holder of Johnson & Johnson pharmaceutical company.
AC is supported by the 2012 Leo-Pharma Research Foundation Silver Award. MSF Supported in part by grant # UL1 TR000043 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program and by the Irma T. Hirschl/Monique Weill-Caulier research award.
Footnotes
This study has been performed at the Laboratory of Investigative Dermatology, The Rockefeller University, New York City, New York, USA.
Disclosure
AC, JF-D, MS-F, ST, and IC state no conflicts of interest.
References
- 1.Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 2.Gudjonsson JE, Krueger G. A role for epigenetics in psoriasis: methylated Cytosine-Guanine sites differentiate lesional from nonlesional skin and from normal skin . J Invest Dermatol. 2012;132:506–508. doi: 10.1038/jid.2011.364. [DOI] [PubMed] [Google Scholar]
- 3.Roberson EDO, Liu Y, Ryan C, et al. A subset of methylated CpG sites differentiate psoriatic from normal skin. J Invest Dermatol. 2012;132:583–592. doi: 10.1038/jid.2011.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaba LC, Krueger JG, Lowes MA. Resident and “inflammatory” dendritic cells in human skin. J Invest Dermatol. 2009;129:302–308. doi: 10.1038/jid.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nestle FO, Turka LA, Nickoloff BJ. Characterization of dermal dendritic cells in psoriasis: autostimulation of T lymphocytes and induction of Th1 type cytokines. J Clin Invest. 1994;94:202–209. doi: 10.1172/JCI117308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson-Huang LM, McNutt NS, Krueger JG, et al. Cytokine-producing dendritic cells in the pathogenesis of inflammatory skin diseases. J Clin Immunol. 2009;29:247–256. doi: 10.1007/s10875-009-9278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowes MA, Chamian F, Abello MV, et al. Increase in TNF-alpha and inducible nitric oxide synthase-expressing dendritic cells in psoriasis and reduction with efalizumab (anti-CD11a) Proc Natl Acad Sci U S A. 2005;102:19057–19062. doi: 10.1073/pnas.0509736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowes MA, Kikuchi T, Fuentes-Duculan J, et al. Psoriasis vulgaris lesions contain discrete populations of Th1 and Th17 T cells. J Invest Dermatol. 2008;128:1207–1211. doi: 10.1038/sj.jid.5701213. [DOI] [PubMed] [Google Scholar]
- 9.Chiricozzi A, Guttman-Yassky E, Suárez-Fariñas M, et al. Integrative responses to IL-17 and TNF-α in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J Invest Dermatol. 2011;131:677–687. doi: 10.1038/jid.2010.340. [DOI] [PubMed] [Google Scholar]
- 10.Caproni M, Antiga E, Melani L, et al. Serum levels of IL-17 and IL-22 are reduced by etanercept, but not by acitretin, in patients with psoriasis: a randomized-controlled trial. J Clin Immunol. 2009;29:210–214. doi: 10.1007/s10875-008-9233-0. [DOI] [PubMed] [Google Scholar]
- 11.Arican O, Aral M, Sasmaz S, Ciragil P. Serum levels of TNF-alpha, IFN-gamma, IL-6, IL-8, IL-12, IL-17, and IL-18 in patients with active psoriasis and correlation with disease severity. Mediators Inflamm. 2005;2005:273–279. doi: 10.1155/MI.2005.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi H, Tsuji H, Hashimoto Y, et al. Serum cytokines and growth factor levels in Japanese patients with psoriasis. Clin Exp Dermatol. 2010;35:645–649. doi: 10.1111/j.1365-2230.2009.03704.x. [DOI] [PubMed] [Google Scholar]
- 13.Suárez-Fariñas M, Li K, Fuentes-Duculan J, et al. Expanding the psoriasis disease profile: interrogation of the skin and serum of patients with moderate-to-severe psoriasis. J Invest Dermatol. 2012;132:2552–2564. doi: 10.1038/jid.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang CQ, Suárez-Fariñas M, Nograles KE, et al. IL-17 induces inflammation-associated gene products in blood monocytes, and treatment with ixekizumab reduces their expression in psoriasis patient blood. J Invest Dermatol. 2014;134:2990–2993. doi: 10.1038/jid.2014.268. [DOI] [PubMed] [Google Scholar]
- 15.Martin DA, Towne JE, Kricorian G, et al. The emerging role of IL-17 in the pathogenesis of psoriasis: preclinical and clinical findings. J Invest Dermatol. 2013;133:17–26. doi: 10.1038/jid.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kagami S, Rizzo HL, Kurtz SE, et al. IL-23 and IL-17A, but not IL-12 and IL-22, are required for optimal skin host defense against Candida albicans. J Immunol. 2010;185:5453–5462. doi: 10.4049/jimmunol.1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Laurence A, Lanno Y, et al. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc Natl Acad Sci USA. 2006;103:8137–8142. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kagami S, Rizzo HL, Lee JJ, et al. Circulating Th17, Th22, and Th1 cells are increased in psoriasis. J Invest Dermatol. 2010;130:1373–1383. doi: 10.1038/jid.2009.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitch E, Harper E, Skorcheva I, et al. Pathophysiology of psoriasis: recent advances on IL-23 and Th17 cytokines. Curr Rheumatol Rep. 2007;9:461–467. doi: 10.1007/s11926-007-0075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee E, Trepicchio WL, Oestreicher JL, et al. Increased expression of interleukin 23 p19 and p40 in lesional skin of patients with psoriasis vulgaris. J Exp Med. 2004;199:125–130. doi: 10.1084/jem.20030451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker BS, Swain AD, Fry L, Valdimarsson H. Epidermal T lymphocytes and HLA-DR expression in psoriasis. Br J Dermatol. 1984;110:555–564. doi: 10.1111/j.1365-2133.1984.tb04678.x. [DOI] [PubMed] [Google Scholar]
- 22.Placek W, Haftek M, Thivolet J. Sequence of changes in psoriatic epidermis. Immunocompetent cell redistribution precedes altered expression of keratinocyte differentiation markers. Acta Derm Venereol (Stockh) 1988;68:369–377. [PubMed] [Google Scholar]
- 23.Prens EP, Benne K, van Joost T, Benner R. The autologous mixed epidermal cell–T lymphocyte reaction is elevated in psoriasis: a crucial role for epidermal HLA-DR+/CD1a) antigen-presenting cells. J Invest Dermatol. 1991;96:880–887. doi: 10.1111/1523-1747.ep12475275. [DOI] [PubMed] [Google Scholar]
- 24.Davidovici BB, Sattar N, Prinz J, et al. Psoriasis and systemic inflammatory diseases: potential mechanistic links between skin disease and co-morbid conditions. J Invest Dermatol. 2010;130:1785–1796. doi: 10.1038/jid.2010.103. [DOI] [PubMed] [Google Scholar]
- 25.Zhou X, Krueger JG, Kao MC, et al. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics. 2003;13:69–78. doi: 10.1152/physiolgenomics.00157.2002. [DOI] [PubMed] [Google Scholar]
- 26.Roberson ED, Liu Y, Ryan C, et al. A subset of methylated CpG sites differentiate psoriatic from normal skin. J Invest Dermatol. 2012;132:583–592. doi: 10.1038/jid.2011.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gudjonsson JE, Ding J, Li X, et al. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. J Invest Dermatol. 2009;129:2795–2804. doi: 10.1038/jid.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao Y, Richman L, Morehouse C, et al. Type I interferon: potential therapeutic target for psoriasis? PLoS One. 2008;3:e2737. doi: 10.1371/journal.pone.0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian S, Krueger JG, Li K, et al. Meta-analysis derived (MAD) transcriptome of psoriasis defines the “core” pathogenesis of disease. PloS One. 2012;7:e44274. doi: 10.1371/journal.pone.0044274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suárez-Fariñas M, Lowes MA, Zaba LC, et al. Evaluation of the psoriasis transcriptome across different studies by gene set enrichment analysis (GSEA) PLoS One. 2010;5:e10247. doi: 10.1371/journal.pone.0010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sofen H, Smith S, Matheson RT, et al. Guselkumab (an IL-23-specific mAb) demonstrates clinical and molecular response in patients with moderate-to-severe psoriasis. J Allergy Clin Immunol. 2014;133(4):1032–1040. doi: 10.1016/j.jaci.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 32.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 34.Choi JK, Yu U, Kim S, Yoo OJ. Combining multiple microarray studies and modeling interstudy variation. Bioinformatics. 2003;19(1):i84–i90. doi: 10.1093/bioinformatics/btg1010. [DOI] [PubMed] [Google Scholar]
- 35.Zaba LC, Fuentes-Duculan J, Steinman RM, et al. Normal human dermis contains distinct populations of CD11cBDCA-1 dendritic cells and CD163FXIIIA macrophages. J Clin Invest. 2007;117:2517–2525. doi: 10.1172/JCI32282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowes MA, Russell CB, Martin DA, et al. The IL-23/T17 pathogenic axis in psoriasis is amplified by keratinocyte responses. Trends Immunol. 2013;34:174–181. doi: 10.1016/j.it.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortega C, Fernandez AS, Carrillo JM, et al. IL-17-producing CD8+ T lymphocytes from psoriasis skin plaques are cytotoxic effector cells that secrete Th17-related cytokines. J Leukoc Biol. 2009;86:435–443. doi: 10.1189/JLB.0109046. [DOI] [PubMed] [Google Scholar]
- 38.Chiricozzi A, Nograles KE, Johnson-Huang LM, et al. IL-17 induces an expanded range of downstream genes in reconstituted human epidermis model. PLoS One. 2014;9:e90284. doi: 10.1371/journal.pone.0090284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gudjonsson JE, Ding J, Johnston A, et al. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J Invest Dermatol. 2010;130:1829–1840. doi: 10.1038/jid.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swindell WR, Johnston A, Voorhees JJ, et al. Dissecting the psoriasis transcriptome: inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC Genomics. 2013;14:527. doi: 10.1186/1471-2164-14-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jabbari A, Suárez-Fariñas M, Dewell S, et al. Transcriptional profiling of psoriasis using RNA-seq reveals previously unidentified differentially expressed genes. J Invest Dermatol. 2012;132:246–249. doi: 10.1038/jid.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.