Abstract
The cellular mechanisms that control protein degradation may constitute a non-oncogenic cancer cell vulnerability and, therefore, a therapeutic target. Although this proposition is supported by the clinical success of proteasome inhibitors in some malignancies, most cancers are resistant to proteasome inhibition. The ATPase valosin-containing protein (VCP; p97) is an essential regulator of protein degradation in multiple pathways and has emerged as a target for cancer therapy. We found that pharmacological depletion of VCP enzymatic activity with mechanistically different inhibitors robustly induced proteotoxic stress in solid cancer and multiple myeloma cells, including cells that were insensitive, adapted, or clinically resistant to proteasome inhibition. VCP inhibition had an impact on two key regulators of protein synthesis, eukaryotic initiation factor 2α (eIF2α) and mechanistic target of rapamycin complex 1 (mTORC1), and attenuated global protein synthesis. However, a block on protein translation that was itself cytotoxic alleviated stress signaling and reduced cell death triggered by VCP inhibition. Some of the proteotoxic effects of VCP depletion depended on the eIF2α phosphatase, protein phosphatase 1 regulatory subunit 15A (PPP1R15A)/PP1c, but not on mTORC1, although there appeared to be cross-talk between them. Thus, cancer cell death following VCP inhibition was linked to inadequate fine-tuning of protein synthesis and activity of PPP1R15A/PP1c. VCP inhibitors also perturbed intracellular amino acid levels, activated eukaryotic translation initiation factor 2α kinase 4 (EIF2AK4), and enhanced cellular dependence on amino acid supplies, consistent with a failure of amino acid homeostasis. Many of the observed effects of VCP inhibition differed from the effects triggered by proteasome inhibition or by protein misfolding. Thus, depletion of VCP enzymatic activity triggers cancer cell death in part through inadequate regulation of protein synthesis and amino acid metabolism. The data provide novel insights into the maintenance of intracellular proteostasis by VCP and may have implications for the development of anti-cancer therapies.
The intracellular degradation of proteins that are damaged, misfolded, or no longer required is essential for normal cellular function. To maintain protein homeostasis (proteostasis), cells orchestrate a delicate balance between protein degradation and protein synthesis. Cancer cells may have a heightened dependence on protein degradation pathways, as their numerous genomic mutations often effect an imbalance in protein levels or the production of defective proteins.1, 2 Moreover, cancer cells may hyperactivate pathways that control protein synthesis, placing additional strain on the cellular mechanisms that govern protein degradation.3, 4 Therefore, drugs that disrupt protein breakdown pathways have considerable potential for anticancer therapy.
The ubiquitin–proteasome system (UPS) is the major mechanism in eukaryotic cells by which cytosolic, nuclear, and endoplasmic reticulum (ER)-derived proteins are degraded.5 Cells maintain physiological protein levels and an adequate intracellular amino acid pool by balancing protein synthesis with the activity of the UPS, and that of proteasome-independent degradation pathways.6, 7 The clinical use of proteasome inhibitors in multiple myeloma (MM) and mantle cell lymphoma has demonstrated that it is in principle possible to disrupt protein degradation in the UPS with fatal consequences for cancer cells, while largely sparing healthy cells. However, proteasome inhibitors are largely ineffective in other cancers.
VCP (valosin-containing protein; also known as p97) is an abundant ATPase that is conserved across all eukaryotes and is essential for life in budding yeast and mice.8, 9, 10, 11 VCP has the ability to use the energy derived from ATP hydrolysis to unfold client proteins, or to extract them from cellular structures. This allows VCP to engage in a range of cellular processes, but its role is best understood in the context of ER-associated degradation (ERAD).12, 13, 14, 15, 16, 17, 18, 19 As a key component of ERAD, VCP mediates the extraction of misfolded proteins across the ER membrane and their delivery to the proteasome.20, 21, 22 However, VCP has also been linked to the proteasome-independent handling of protein aggregates and autophagy.23, 24, 25, 26, 27, 28 Moreover, VCP has been implicated in proteasome recovery after proteasome inhibition, which may underlie the resistance of some cancers to proteasome inhibitors.29, 30, 31 Thus, VCP is fundamental for proteostasis. This broad involvement of VCP in intracellular protein turnover, combined with observations of aberrant VCP expression in different cancers,32, 33, 34, 35, 36, 37, 38, 39, 40 suggests that VCP inhibitors may overcome some limitations of proteasome inhibitors by affecting multiple proteostatic mechanisms simultaneously. Indeed, VCP-targeting compounds activate caspases and have an impact on both ubiquitin-dependent and autophagic pathways in cancer cells in vitro and in vivo.27, 41, 42 In contrast, primary rat hepatocytes and mouse skeletal muscle cells do not undergo apoptosis on VCP depletion and non-malignant human cells appear to be less susceptible to VCP inhibition than cancer cells.27, 43, 44 Phase I clinical trials of one VCP inhibitor are currently underway in patients with advanced solid cancers and relapsed/refractory myeloma (https://clinicaltrials.gov, NCT02243917 and NCT02223598). However, the mechanisms by which pharmacological VCP depletion induces cancer cell death, and how they differ from those mediated by proteasome inhibition, remain incompletely understood.
ER stress is potentially fatal to cells and can be brought about by various insults to the ER, such as the accumulation of misfolded proteins. It is linked to a diverse range of illnesses and is thought to be a key mechanism by which proteasome inhibitors effect their toxicity in MM cells.45 Cells normally respond to ER stress by activating the unfolded protein response (UPR), which comprises a number of cellular adaptations that aim to restore protein homeostasis.46 Phosphorylation of eukaryotic initiation factor 2α (eIF2α) on a single serine is central to one arm of the UPR and rebalances proteostasis by temporarily attenuating global messenger RNA (mRNA) translation.47 However, as a prolonged decrease in protein translation would prove fatal, phosphorylation of eIF2α also simultaneously triggers a negative feedback loop that promotes protein synthesis. This feedback loop begins with the preferential translation of the transcription factor activating transcription factor 4 (ATF4), which induces CCAAT/enhancer-binding proteins homologous protein (CHOP; encoded by the DNA damage-inducible transcript 3 (DDIT3) gene), another transcription factor. Both ATF4 and CHOP promote the expression of protein phosphatase 1 regulatory subunit 15A (PPP1R15A; also known as growth arrest and DNA damage-inducible protein, GADD34), the regulatory subunit of a stress-induced phosphatase that consists of PPP1R15A and PP1c. PPP1R15A/PP1c dephosphorylates eIF2α to reinstate physiological protein synthesis following the resolution of ER stress.48, 49, 50, 51, 52, 53, 54 However, in cells with unresolved ER stress this process generates oxidative stress and contributes to cell death.51, 53, 54 Thus, the eIF2α-ATF4/CHOP–PPP1R15A/PP1c feedback loop finely tunes protein synthesis to regulate cell viability under conditions of protein folding stress. eIF2α is also central to signaling networks that integrate oxidative stress and nutrient availability with other translation regulators such as mechanistic target of rapamycin complex 1 (mTORC1).55, 56
Here we studied the role of VCP in maintaining cancer cell proteostasis by using compounds that inhibit VCP enzymatic activity by different mechanisms.27, 42 We show that pharmacological depletion of VCP kills cancer cells of diverse tissue origins through mechanisms that regulate protein synthesis and amino acid homeostasis. Moreover, we demonstrate that the effects of VCP inhibition differ markedly from the effects of proteasome inhibition. The data provide previously unrecognized biological insights into the cellular mechanisms by which VCP governs proteostasis and may have important implications for the development of anticancer therapies.
Results
VCP inhibitors kill cancer cells independently of their tissue origins and sensitivity to proteasome inhibition
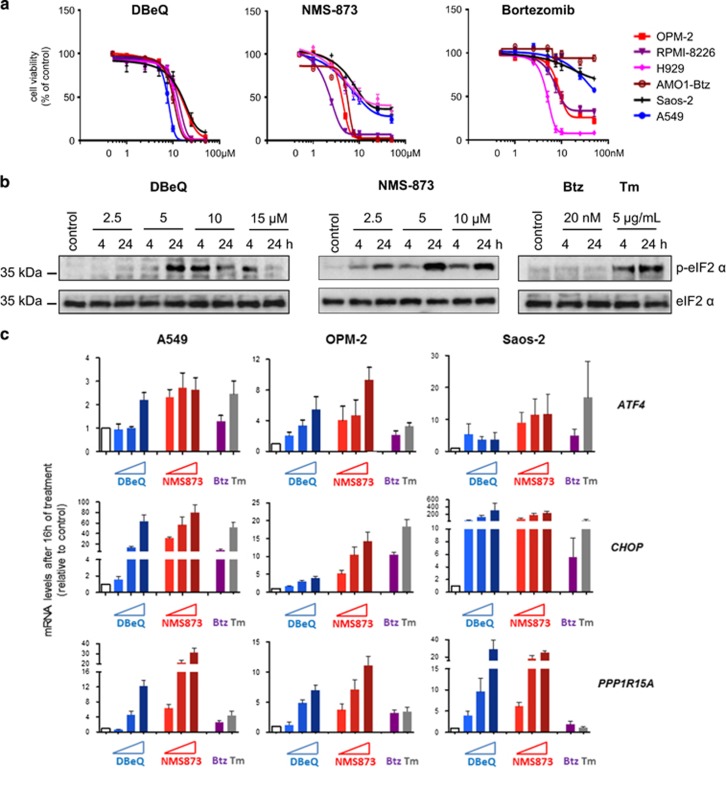
We first examined the effects of VCP inhibition by directly comparing the ability of the VCP inhibitors N2,N4-dibenzylquinazoline-2,4-diamine (DBeQ)27 and Nerviano Medical Sciences-873 (NMS-873)42 with that of the proteasome inhibitor bortezomib, to kill a range of solid cancer and MM cell lines, as well as primary MM cells. We chose these two VCP-targeting compounds based on their extensive characterization and mechanistic difference. The quinazoline DBeQ is an ATP competitive VCP inhibitor, whereas NMS-873 is a non-ATP-competitive allosteric inhibitor of VCP.1, 27, 42 Both inhibitors rapidly turn off VCP enzymatic activity and thereby avoid potential limitations of genetic targeting related to slow or incomplete depletion of the abundant VCP.1, 57 As expected, a 24-h treatment with bortezomib effectively killed the OPM-2, Roswell Park Memorial Institute (RPMI)-8226, and H929 MM cell lines at an IC50 of ~5–10 nM (Figure 1a and Supplementary Table S1). However, bortezomib in concentrations up to 50 nM had a limited or no impact on the viability of A549 lung cancer cells, Saos-2 osteosarcoma cells, and bortezomib-adapted AMO1-Btz MM cells. In contrast, DBeQ and NMS-873 killed all cell lines tested within a narrow range of IC50 levels. The moderate differences in IC50s and in the level of cell death at 24 h that we observed can probably be explained by differences in the potency and efficacy of DBeQ and NMS-873 (Figure 1a and Supplementary Table S1).1, 27, 42 We also found that DBeQ- and NMS-873-induced cell death largely amounted to caspase-dependent apoptotic death (Supplementary Figure S1). Importantly, both inhibitors also killed primary bone marrow-derived MM cells from a patient with clinically bortezomib-resistant MM at concentrations similar to those that were effective in cell lines (Supplementary Table S1). Thus, an ATP-competitive and an allosteric VCP inhibitor effectively kill cancer cell lines of different tissue origins, including bortezomib-adapted and clinically bortezomib-resistant cells. The data suggest that the effects of VCP inhibitors have different mechanisms of action from proteasome inhibitors, and that their effects are not limited to cancer cells with a distinctive secretory load, such as MM cells.
Figure 1.
VCP inhibitors kill multiple myeloma and solid cancer cells and induce eIF2α signaling. (a) Viability of cancer cell lines after treatment with DBeQ, NMS-873, or bortezomib (Btz) for 24 h relative to untreated control cells. (b) Representative immunoblots with antibodies against phosphorylated (Serine 51) and total eIF2α on whole-cell extracts from A549 cells treated with DBeQ, NMS-873, bortezomib (Btz), or tunicamycin (Tm) as indicated. Treatment with 15 μM NMS-873 for 24 h resulted in a protein yield that was too low for immunoblot analysis (not shown). (c) mRNA levels of the indicated eIF2α-regulated genes relative to control cells (white bars; solvent treated) determined by real-time quantitative PCR. The indicated lung cancer (A549), multiple myeloma (OPM-2), and osteosarcoma (Saos-2) cell lines were treated with DBeQ (5, 10, and 15 μM), NMS-873 (5, 10, and 15 μM), bortezomib (Btz, 20 nM), or tunicamycin (Tm, 5 μg/ml) for 16 h. Data shown are the mean±S.E.M. from three independent experiments
VCP inhibition activates eIF2α signaling and upregulates expression of its downstream targets
To investigate the role of eIF2α-ATF4/CHOP–PPP1R15A/PP1c signaling following VCP inhibition, we first examined the effects of DBeQ and NMS-873 on eIF2α phosphorylation. We found that both DBeQ and NMS-873 caused time- and dose-dependent phosphorylation of eIF2α (Figure 1b). Treatment with 15 μM DBeQ also resulted in a decrease in eIF2α phosphorylation at 24 h compared with 4 h, possibly reflecting its dephosphorylation by PPP1R15A (Supplementary Figure S2). In contrast, bortezomib did not have a noticeable effect on eIF2α phosphorylation. However, bortezomib did cause intracellular accumulation of ubiquitinated proteins in A549 and OPM-2 cells, confirming that it was disrupting the UPS at the concentration used (Supplementary Figure S3). By comparison, tunicamycin, which induces ER stress by blocking N-linked glycosylation of ER-resident proteins, caused eIF2α phosphorylation after both 4 and 24 h (Figure 1b).
We next examined the effects of VCP and proteasome inhibition on key targets downstream of eIF2α. Using real-time quantitative PCR, we found that DBeQ and NMS-873 upregulated CHOP, ATF4, and PPP1R15A mRNA levels in a largely dose-dependent manner (Figure 1c). Consistent with its effect on cell death and eIF2α phosphorylation, NMS-873 appeared to be slightly more potent than DBeQ. We also found that the extent to which these genes were induced was largely comparable between lung cancer, osteosarcoma, and myeloma cells, although some differences between cell lines could be noted. In contrast to the VCP inhibitors, bortezomib had only a minimal effect on PPP1R15A expression in all three cell lines tested and a minor effect on CHOP expression in Saos-2 cells (Figure 1c). By comparison, tunicamycin upregulated CHOP and ATF4 mRNA levels to a similar extent as the VCP inhibitors, but its effect on PPP1R15A expression was minimal. Both DBeQ and NMS-873 also increased CHOP and PPP1R15A protein levels, with NMS-873 again appearing more potent (Supplementary Figure S2). We then tested the ability of VCP inhibitors to induce eIF2α targets in bortezomib-adapted AMO1-Btz cells (Supplementary Figure S4). We found that NMS-873 and DBeQ strongly induced CHOP and PPP1R15A mRNA expression, and that NMS-873 also upregulated ATF4, similar to what we observed in A549, Saos-2, and OPM-2 cells. In contrast, bortezomib had no discernible effect and the effects of tunicamycin were limited.
When misfolded proteins accumulate in the ER, cells respond by creating more luminal chaperone proteins to assist in protein folding. This process is regulated by an alternative arm of the UPR from that involving eIF2α phosphorylation.46 To test whether VCP inhibitors also induced ER chaperones, we analyzed mRNA levels of binding immunoglobulin protein (BIP) and P58IPK, and found both to be largely upregulated in all three cell lines tested (Supplementary Figure S5). In line with our findings on eIF2α targets, NMS-873 had a greater effect on the transcriptional induction of BIP and P58IPK than DBeQ. Although tunicamycin also induced BIP and P58IPK, bortezomib had little effect. Finally, we tested whether VCP inhibitor-induced cell death correlated with the steady-state baseline expression of VCP, ATF4, CHOP, PPP1R15A, or BIP mRNAs in the cancer cell lines investigated and found no significant correlation (Supplementary Figure S6).
Taken together, these findings show pronounced effects of VCP inhibitors on the eIF2α-ATF4/CHOP-PPP1R15A/PP1c pathway and ER chaperones. Moreover, signaling downstream of eIF2α in response to VCP inhibitors appeared unaffected in bortezomib-adapted cells that show a blunted response to proteasome inhibition and ER stress. Our observations also demonstrate that the effects of DBeQ and NMS-873 on pathways employed by the UPR differ from those induced by bortezomib or tunicamycin, suggesting that VCP inhibition triggers a distinctive type of proteotoxic stress.
Protein synthesis and PPP1R15A/PP1c-mediated eIF2α dephosphorylation modulate proteotoxic effects of VCP inhibition
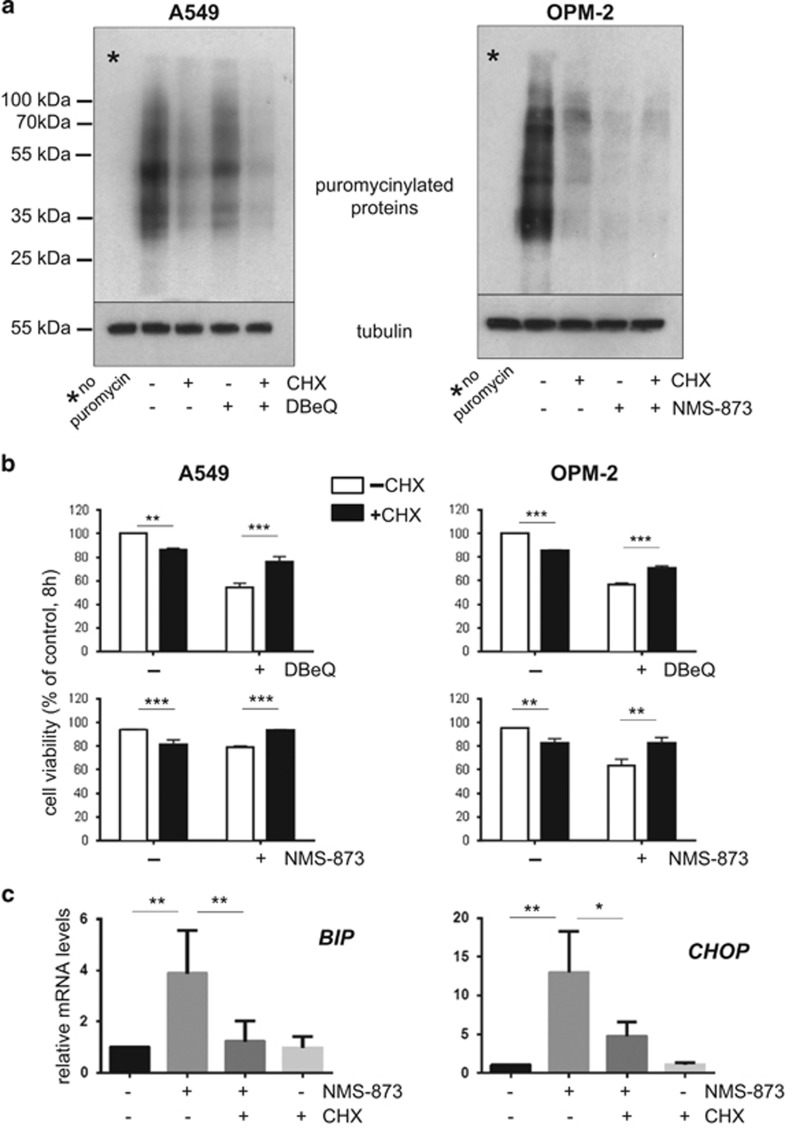
Protein synthesis can lead to cell death under conditions of ER stress.54 However, cells require controlled protein translation for normal function. This led us to investigate the role of protein synthesis following VCP inhibition, making use of the translation inhibitor cycloheximide. Immunoblotting for newly synthesized puromycinylated proteins demonstrated that a 8h treatment with cycloheximide largely abrogated protein synthesis, as expected. Our analysis also revealed that VCP inhibition triggered a considerable reduction in global protein translation (Figure 2a), a finding that is compatible with the phosphorylation of eIF2α we observed. We then determined that cycloheximide treatment resulted in cell death in a small proportion of cells (Figure 2b). However, translation inhibition with cycloheximide partially rescued DBeQ- and NMS-873-induced cell death. Moreover, cycloheximide rescue was associated with reduced induction of CHOP and BIP (Figure 2c). Thus, a block on protein synthesis that is itself cytotoxic has a cytoprotective effect on cells treated with VCP inhibitors that correlates with signs of reduced proteotoxic stress.
Figure 2.
Protein synthesis modulates cell death and proteotoxic stress after VCP inhibition. (a) VCP inhibitors attenuate protein synthesis. Protein synthesis was monitored by immunoblotting for newly synthesized puromycinylated peptides. A549 and OPM-2 cells were treated for 8 h with DBeQ (15 μM) or NMS (10 μM) in the absence or presence of the translation inhibitor cycloheximide (CHX, 1 μg/ml) before addition of puromycin for 10 min followed by the preparation of whole-cell extracts. (b) A block on protein synthesis reduces cell death following VCP inhibition. Cell viability of A549 and OPM-2 cells treated as described in (a). (c) Cycloheximide reduced VCP-inhibitor-mediated stress signaling. mRNA levels of the indicated genes following treatment of OPM-2 cells with NMS-873 (10 μM) in the absence or presence of cycloheximide (CHX, 1 μg/ml) for 8 h. Data shown in (b) and (c) are the mean±S.E.M. from three independent experiments. *P<0.05; **P<0.01; ***P<0.001
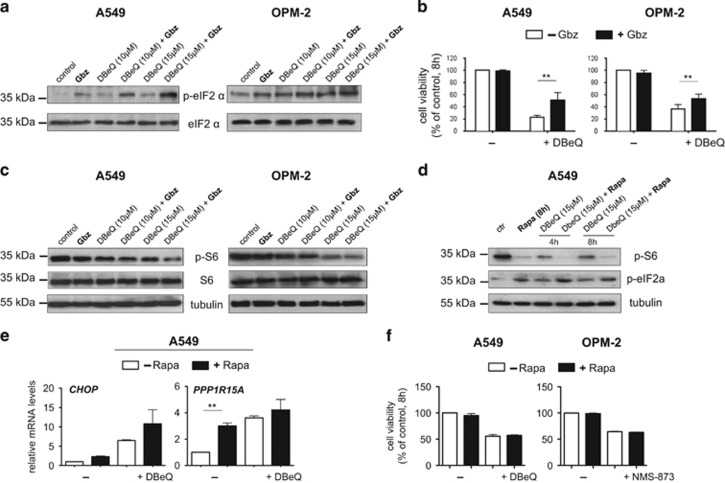
These data led us to examine whether the effects of VCP inhibition are modulated by the eIF2α phosphatase PPP1R15A/PP1c.48, 49 To this end we made use of the small molecule guanabenz, which selectively inhibits stress-induced dephosphorylation of eIF2α by PPP1R15A/PP1c.58 We first confirmed that treatment with guanabenz resulted in the expected increase in the level of eIF2α phosphorylation following VCP inhibition (Figure 3a). As expected, guanabenz largely increased expression of CHOP, ATF4, and PPP1R15A in DBeQ-treated cells but did not appear to affect expression levels of these genes in unstressed cells (Supplementary Figure S7). Next, we determined that DBeQ-induced cell death was indeed significantly attenuated in guanabenz-treated cells (Figure 3b), indicating that PPP1R15A/PP1c activity partly regulates cell death in response to VCP inhibition.
Figure 3.
Impact of PPP1R15A/PP1c and mTORC1 signaling on proteotoxicity triggered by VCP inhibition. (a) Guanabenz treatment is associated with increased levels of eIF2α phosphorylation. Representative immunoblots with antibodies against phosphorylated (Serine 51) and total eIF2α on whole-cell extracts from A549 and OPM-2 cells treated for 8 h with DBeQ as indicated in the absence or presence of guanabenz (Gbz, 2.5 μM). (b) Guanabenz reduces VCP inhibitor-mediated cell death. Cell viability after treatment with DBeQ (15 μM) in the absence or presence of guanabenz (Gbz, 2.5 μM) for 8 h. (c) Guanabenz decreases S6 phosphorylation as a readout for mTORC1 activity. Representative immunoblots with antibodies against phosphorylated (Serine 235/236) and total ribosomal protein S6 on whole-cell extracts from A549 cells after treatement with or without rapamycin (Rapa) as indicated. (d) mTORC1 inhibition has an impact on eIF2α phosphorylation. Immunoblottings demonstrating the effects of the mTORC1 inhibitor rapamycin (Rapa, 20 nM) on S6 and eIF2α phosphorylation in unstressed cells and cells treated with DBeQ. (e) The impact of mTORC1 inhibition on the indicated mRNA levels after VCP inhibition for 8 h. (f) Rapamycin does not modulate VCP inhibitor-induced cell death. Cell viability was determined after treatment with rapamycin (Rapa, 20 nM), DbeQ (15 μM), and NMS-873 (10 μM) for 8 h. Data shown in (b), (e), and (f) are the mean±S.E.M. from three independent experiments; **P<0.01
Considering its role as a regulator of protein synthesis, we then tested whether mTORC1 might be affected by PPP1R15A/PP1c. We found that DBeQ alone caused a moderate reduction in ribosomal protein S6 phosphorylation, a readout for mTORC1 activity (Figure 3c).6, 59 We also observed that co-treatment with guanabenz mildly enhanced the DBeQ-induced reduction of S6 phosphorylation. Thus, inhibition of PPP1R15A/PP1c-mediated eIF2α dephosphorylation by guanabenz decreases cell death and enhances attenuation of mTORC1 signaling following VCP inhibition.
We then tested whether inhibition of mTORC1 activity with rapamycin had an impact on eIF2α phosphorylation and the proteotoxic effects of VCP depletion. Indicating its inhibitory effect on mTORC1, rapamycin remarkably reduced phosphorylation of S6 in unstressed A459 cells and appeared to mildly enhance the reduction of S6 phosphorylation in cells treated with DBeQ (Figure 3d). Rapamycin also increased eIF2α phosphorylation both in unstressed and DBeQ-treated cells, although the effect appeared moderate (Figure 3d). Moreover, rapamycin appeared to increase mRNA levels of CHOP and PPP1R15A, in line with its effects on eIF2α phosphorylation (Figure 3e). Comparable results on the effects of rapamycin on eIF2α phosphorylation and gene expression were observed in OPM-2 cells treated with NMS-873 (not shown). Importantly, rapamycin had no effect on the viability of cells treated with either DBeQ or NMS-873 (Figure 3f). Thus, VCP inhibition has an impact on two important cellular regulators of protein synthesis, eIF2α and mTORC1, and there appears to be cross-talk between them; however, mTORC1 does not affect cell viability following VCP inhibition.
VCP inhibition disrupts intracellular amino acid homeostasis
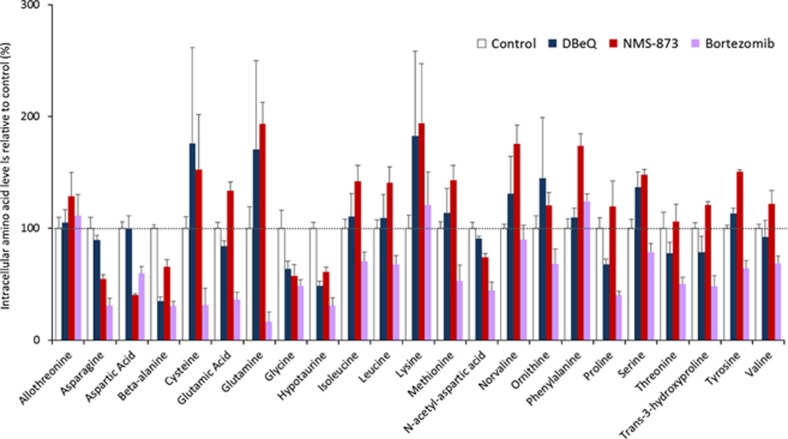
To maintain intracellular amino acid homeostasis, cells tightly co-ordinate protein synthesis with degradation.6 Proteasome inhibition has been reported to reduce intracellular amino acids levels;60, 61 however, the effects of VCP inhibitors on amino acid homeostasis have not been studied. To address this gap in knowledge, we performed unsupervised gas chromatography–mass spectrometry (GC-MS) of extracts from A549 cells treated with DBeQ or NMS-873 and compared their effects with that of bortezomib (Figure 4 and Supplementary Table S2). Bortezomib decreased intracellular levels of numerous amino acids, consistent with previous reports.6, 60, 61 However, VCP inhibitors had a different and more complex effect. Although both DBeQ and NMS-873 caused a significant decrease of β-alanine and hypotaurine levels, treatment with NMS-873 also decreased levels of asparagine, aspartic acid, and N-acetyl aspartic acid. Surprisingly, treatment with NMS-873 significantly increased the levels of four other amino acids: glutamic acid, phenylalanine, serine, and tyrosine. DBeQ appeared to increase the levels of three of these four amino acids, and those of several others, but the differences were not statistically significant compared with untreated cells (Supplementary Table S2). Although DBeQ and NMS-873 caused changes in intracellular amino acid levels that were largely comparable with each other, a larger number of statistically significant changes were observed after NMS-873 treatment, which may be explained by its higher potency. Thus, pharmacological depletion of VCP enzymatic activity elicits a complex effect on intracellular amino acid pools.
Figure 4.
Inhibitors of VCP and the proteasome have different effects on intracellular amino acid levels. Relative levels of intracellular amino acids determined by GC-MS. A549 cell extracts were prepared as described in Materials and Methods after treatment with DBeQ (10 μM), NMS-873 (10 μM), or bortezomib (20 nM) for 8 h. Data shown are the mean±S.E.M. relative to untreated (control) cells from four independent experiments
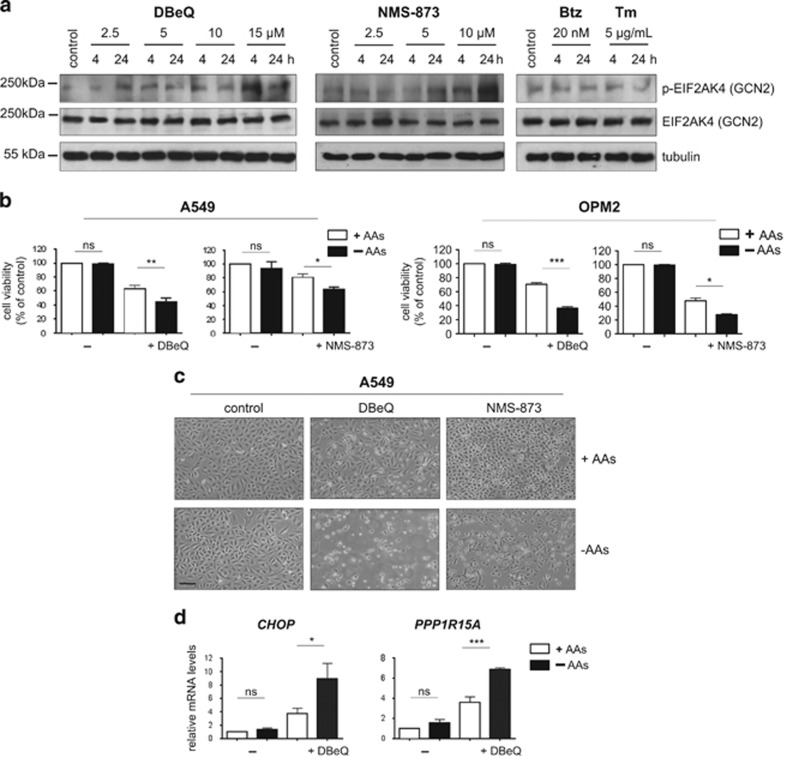
We next asked whether the effect of VCP inhibition on amino acid levels was functionally relevant to cells. To test this, we first examined activation of eukaryotic translation initiation factor 2α kinase (EIF2AK4; general control nonderepressible 2 (GCN2)), an eIF2α kinase primarily activated by amino acid deprivation.62, 63, 64 Indeed, immunoblots on extracts from A549 cells treated with DBeQ and NMS-873 showed phosphorylation of EIF2AK4 at Threonine 889, indicating its activation. In contrast, neither bortezomib nor tunicamycin appeared to trigger phosphorylation of EIF2AK4 (Figure 5a).
Figure 5.
VCP inhibitors disrupt intracellular amino acid homeostasis. (a) VCP inhibitors trigger EIF2AK4 phosphorylation. Representative immunoblots with antibodies against phosphorylated (Thr889) and total EIF2AK4 (GCN2) on whole-cell extracts from A549 cells treated with DBeQ, NMS-873, bortezomib (Btz), or tunicamycin (Tm). (b) Cell viability of A549 and OPM-2 cells growing in complete media (+AA) or media deficient in L-glutamine, L-methionine, and L-cystine (–AA). Cells were treated with DBeQ (15 μM for A549 cells and 10 μM for OPM2 cells) or NMS-873 (10 μM for A549 cells and 5 μM for OPM-2 cells) for 16 h. (c) Representative images of A549 cells treated as in (b). Images shown were taken with an ECOS core digital microscope at × 100 magnification (size bar: 25 μm). (d) mRNA levels of the indicated genes relative to untreated controls determined by real-time quantitative PCR after treatment of A549 cells as described in (b). Data shown in (b) and (d) are the mean±S.E.M. from three independent experiments. *P<0.05, **P<0.01, ***P<0.001
We then hypothesized that putting an additional strain on amino acid metabolism would enhance the toxicity of VCP inhibitors if their effects on amino acid levels were functionally relevant. Indeed, when we depleted three amino acids (L-methionine, L-cystine, and L-glutamine) from the cell culture medium, we observed increased cell death on treatment with DBeQ and NMS-873 (Figures 5b and c). Moreover, we observed that amino acid depletion enhanced the VCP inhibitor-mediated induction of CHOP and PPP1R15A (Figure 5d). However, amino acid depletion affected neither viability nor CHOP/PPP1R15A expression in unstressed cells or in cells treated with tunicamycin (Figure 5d and Supplementary Figure S8). Thus, VCP inhibitors disrupt intracellular amino acid homeostasis and enhance cellular dependence on amino acid supplies.
Discussion
Phosphorylation of the translation initiation factor eIF2α can promote cell survival under acute stress conditions by attenuating protein translation. However, eIF2α phosphorylation can also mediate death in cells with unresolved stress by promoting protein synthesis.46, 54 We found that VCP inhibitors consistently and strongly activated eIF2α signaling in cancer cells derived from different tissues, including proteasome inhibitor-adapted myeloma cells. This finding led us to investigate the role of major protein translation control pathways in VCP inhibitor-induced cell death. We found that a block on protein translation, which was toxic on its own, was cytoprotective in cells in which VCP was inhibited. How could protein synthesis have a detrimental impact on cells? The effect might be explained by a reduced cellular dependence on protein quality control mechanisms that depend on VCP, such as ERAD, when protein synthesis is reduced. Protein synthesis under conditions of VCP inhibition could also result in higher level of reactive oxygen species or loss of ATP.54 Intriguingly, protein synthesis was attenuated in response to VCP inhibition, probably as part of the proteotoxic stress response. Therefore, our data suggest that either the extent or nature of this downregulation was inadequate for optimal cytoprotection. This finding highlights the requirement for cells to precisely fine-tune protein synthesis under conditions of proteotoxic stress.
Our observations also indicate that VCP and PPP1R15A/PP1c have an impact on the mTORC1 pathway, although it remains to be established how these effects are mediated. Moreover, we found that pharmacological inhibition of mTORC1 had an impact on eIF2α and its downstream targets. These data are important, as they show that at least two key cellular signaling hubs that regulate protein synthesis, eIF2α and mTORC1, are affected by VCP inhibition, and that there is cross-talk between them, thereby adding to the growing literature on the interactions between eIF2α and mTORC1.55, 64, 65, 66, 67 However, only guanabenz, but not rapamycin, had a cytoprotective effect following VCP inhibition. This may be explained by feedback effects of mTORC1 inhibition on the PI3K/Akt pathway and mTORC2, or effects on autophagy, which is also modulated by VCP.27, 42, 55, 64 The findings may be clinically relevant, as some approaches to improve cancer therapy aim to reduce the activity of pathways that promote protein synthesis, such as mTORC1.68 Our results suggest that such a strategy may not be beneficial when combined with proteotoxic treatment approaches such as VCP inhibition.
The effects of VCP inhibitors on mTORC1, which also responds to amino acid shortage, led us to investigate their effects on intracellular amino acid homeostasis. We found that VCP inhibitors triggered both decreases and increases in individual intracellular amino acid levels. In contrast, and in line with previous reports, we found that proteasome inhibition resulted in a substantial decrease in the intracellular levels of many amino acids6, 60 The unexpected impact of VCP inhibition on amino acid levels might be related to the VCP inhibitors' complex effects on the regulation of protein synthesis and thus amino acid utilization. Given that DBeQ and NMS-873 promoted phosphorylation of EIF2AK4, a major eIF2α kinase that senses amino acid shortages,62 the complex effects on amino acid homeostasis elicited by VCP inhibitors are likely to be functionally relevant. In contrast, bortezomib did not induce EIF2AK4 phosphorylation and was less effective than the VCP inhibitors in inducing cell death, suggesting that the cells activated mechanisms that at least partly compensated for the reduction in amino acid levels triggered by proteasome inhibition.
We noted that EIF2AK4 phosphorylation occurred quite some time after the changes in mTORC1 activity and eIF2α phosphorylation. As such, the precise mechanisms and kinetics by which VCP inhibition leads to EIF2AK4 activation remain to be established. Moreover, it is still unclear whether EIF2AK4 or the ER stress sensor EIF2AK3 protein kinase R (PKR)-like endoplasmic reticulum kinase is the predominant eIF2α kinase after VCP inhibition or, indeed, whether both have a role.
We also found that the toxic effects of VCP inhibition were enhanced when extrinsic amino acid supplies were limited, whereas the same amino acid limitation had no measurable effect on unstressed cell or cells undergoing protein misfolding stress. Given that tumours may be hyperdependent on nutrient supplies and may scavenge extracellular protein to ensure sufficient amino acid availability,69 the VCP inhibitor-associated heightened dependence on amino acid supplies may be clinically relevant. Taken together, our data provide evidence that VCP inhibitors induce a functionally detrimental disruption of intracellular amino acid homeostasis.
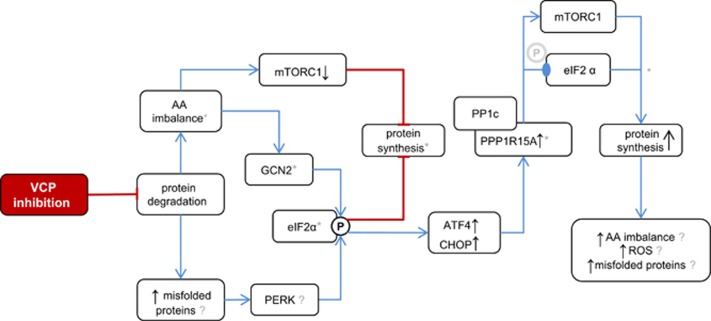
In summary (Figure 6), we demonstrate that compounds that target VCP enzymatic activity disrupt multiple aspects of intracellular protein metabolism. These findings advance our understanding of how VCP maintains proteostasis and may be relevant for the clinical development of VCP inhibitors for cancer therapy.
Figure 6.
Proposed working model of the mechanisms by which VCP inhibitors induce cancer cell death. VCP inhibition impinges on protein degradation, resulting in aberrant amino acid (AA) recycling and increased levels of misfolded proteins in the ER. This results in reduced mTORC1 activity and activates the eIF2α kinases EIF2AK4 (GCN2) and EIF2AK3 (PERK), thereby attenuating protein synthesis to alleviate proteotoxic stress. Downstream of eIF2α, the ATF4/CHOP-mediated induction of PPP1R15A governs eIF2α dephosphorylation by PPP1R15A/PP1c and increases mTORC1 activity. Together, these effects promote protein synthesis, resulting in inadequate attenuation of protein translation that aggravates cell stress and death through mechanisms that may involve amino acid imbalance, reactive oxygen species, and ER stress caused by misfolded proteins.  Phosphorylated serine 51;
Phosphorylated serine 51;  dephosphorylated serine 51; *probable qualitative or quantitative differences between VCP and proteasome inhibitors
dephosphorylated serine 51; *probable qualitative or quantitative differences between VCP and proteasome inhibitors
Materials and Methods
Cells and reagents
The human MM cell lines OPM-2, H929, and RPMI-8226 were purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). Saos-2 cells were from the European Collection of Cell Cultures (Salisbury, UK). A549 cells were a gift from Jane Mitchell (Imperial College London, London, UK). Cell culture identity was verified by short tandem repeat profiling provided by the University of Sheffield, UK (not shown). Bortezomib-adapted AMO-1 cells were a gift from Christoph Driessen (Kantonsspital St. Gallen) and have been described before.70, 71 Cell lines were regularly tested for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza, Cambridge, UK). Primary human myeloma cells were isolated from a diagnostic bone marrow aspirate performed on a patient with relapsed MM by CD138 magnetic bead selection under appropriate research ethics committee approval (REC reference 11/H0308/9) at Imperial College Healthcare NHS Trust. The cells were isolated shortly before initiation of treatment with a regimen consisting of bortezomib, cyclophosphamide, and dexamethasone, to which the patient subsequently proved to be clinically resistant. Cells were grown in RPMI (myeloma cells) or Dulbecco's modified Eagle's medium (DMEM; Saos-2, A549 cells) media (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS) and penicillin plus streptomycin. For experiments on amino acid depletion we compared DMEM and RPMI containing amino acids (Sigma-Aldrich; D1145, R8758) with DMEM and RPMI without L-methionine, L-cystine, and L-glutamine (Sigma-Aldrich; D0422, R7513), both supplemented with 10% dialyzed FBS (Sigma-Aldrich; F0392). DBeQ (Biovision, Milpitas, CA, USA), NMS-873 (Sigma-Aldrich), bortezomib (Sigma-Aldrich), tunicamycin (Sigma-Aldrich), guanabenz (Sigma-Aldrich), rapamycin (Sigma-Aldrich), cycloheximide (Sigma-Aldrich) and Z-VAD-FMK (Tocris Bioscience, Bristol, UK) were dissolved in dimethylsulfoxide and stored at −20 °C.
Cell viability
Cell viability was measured using the AlamarBlue cell viability reagent (Thermo Fisher, Waltham, MA, USA) according to the manufacturer's instructions. Briefly, cells were grown in 96-well plates, AlamarBlue was added at 1/10 of the culture volume at the end of the indicated experimental treatment times, and the fluorescence read at an emission wavelength of 590 nm following excitation at 544 nm. Apoptosis was determined by flow cytometric analysis of Annexin V-FITC and propidium iodide-stained cells (BD Biosciences, San Jose, CA, USA). All results were obtained from three independent experiments, each consisting of three technical replicates. Results are shown as mean viability relative to untreated or vehicle (solvent)-treated cells.
mRNA expression analysis by real-time PCR
Cells were collected and snap frozen using liquid nitrogen. RNA was extracted using the GeneJET RNA Purification Kit (Thermo Fisher) followed by removal of genomic DNA according to the manufacturer's instructions. cDNA synthesis was performed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher) according to the manufacturer's instructions using an Applied Biosystems 2720 Thermal Cycler (Life Technologies, Carlsbad, CA, USA). PCR reactions were performed on an Applied Biosystems StepOnePlus machine (Applied Biosystems, Foster City, CA, USA) using 10 μl SYBR Green JumpStart Taq ready Mix (Sigma-Aldrich), 0.3 μM sequence-specific primers, and 25 ng cDNA under standard conditions.
Immunoblotting
Whole-cell protein extracts were prepared on ice using a lysis buffer containing 50 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) pH 7.5, 50 mM NaF, 5 mM Na pyrophosphate, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 1% Triton, and Complete EDTA-free Protease Inhibitor Cocktail (Roche, Basel, Switzerland). Primary antibodies used were as follows: phospho-GCN2 Thr889 (Abcam, Cambridge, UK), GCN2 (Cell Signalling Technology, Danvers, MA, USA), GADD34/PPP1R15A (Santa Cruz Biotechnology, Dallas, TX, USA), phospho-eIF2α Ser51 (Cell Signalling Technology), eIF2α (Cell Signalling Technology), β-tubulin (Cell Signalling Technology), S6 ribosomal protein 5G10 (Cell Signalling Technology), and phospho-S6 ribosomal protein Ser235/236 (Cell Signalling Technology). The PageRuler Plus Prestained Protein Ladder (Thermo Fisher) was used as a molecular weight marker.
GC-MS of intracellular amino acids
Intracellular metabolites were extracted from cultured cells by cold methanol quenching. Aqueous metabolites were separated from the intracellular extract using a 2 : 1 : 3 chloroform:methanol:water extraction method. The aqueous portion of the extract was separated and lyophilized in silanized 1.5 ml glass vials before analysis. Derivatization for GC-MS was carried out by methoximation followed by trimethylsilylation according to the protocol described by Kind et al.72 Samples were analyzed on an Agilent 7890 gas chromatograph (Agilent Technologies, Santa Clara, CA, USA) connected to an Agilent 5975 MSD (Agilent Technologies) using the FiehnLib settings72 and retention time-locking to myristic acid-d27. GC-MS data were processed by deconvolution using AMDIS using the Fiehn library, followed by integration using GaVIN73 based on the quantification ion for each metabolite as taken from the Fiehn library. Data were normalized to cell number and statistical analyses were carried out in the R statistical environment.
Analysis of protein synthesis by puromycin labeling
Semi-quantitative monitoring of protein synthesis was carried out based on the previously described SUnSET method.74 Briefly, newly synthesized peptides were labeled in cultured cells by the addition of puromycin (InvivoGen, San Diego, CA, USA); 5 μg/ml for 10 min before cells were collected and whole-cell extracts were prepared for immunoblotting as described above, using anti-puromycin antibody clone 12D10 (Merck Millipore, Darmstadt, Germany) and anti-mouse IgG-HRP-linked antibody (Cell Signalling Technology).
Statistical analysis
Unless stated otherwise, data show means and S.E.M. from three independent experiments. Analysis of variance and Bonferroni posttests were used for comparisons between treatment groups. Correlation between cell viability loss after inhibitor treatment and baseline gene expression was analyzed using Pearson's correlation coefficient (r). T-test was used to establish correlation coefficient significance. Calculations were done using GraphPad PRISM Version 6.05 (GraphPad Software, La Jolla, CA, USA).
Acknowledgments
This work was funded by a Cancer Research UK Clinician Scientist Fellowship grant (C41494/A15448) to HWA and was supported by a Wellcome Trust Research Career Development Fellowship to EG and a Whitaker International Program Fellowship to TMC. The work was also supported by grants from the Imperial College London National Institute of Health Research-Biomedical Research Centre (NIHR-BRC), the Imperial College London Cancer Research UK Centre, and the Imperial College Healthcare Charity ‘Dorothy Cave Legacy Fund'. We are grateful to Christoph Driessen and Marianne Kraus for Amo-1-Btz cells; to Jane Apperley, Anne Marie Moody, and Simone Cenci for critical comments on the manuscript; and to David Ron, Heather Harding, Raymond Deshaies, Eric Lam, Valentina Caputo, Katerina Goudevenou, and Jaqueline Fok for advice and discussions.
Author contributions
HWA conceived experiments. KP, TMC, MZC, SL, FP, and GNV performed experiments. All authors analyzed data and interpreted results. HWA wrote the paper. All authors commented on the paper.
Glossary
- ATF4
activating transcription factor 4
- BIP
binding immunoglobulin protein
- CHOP
CCAAT/enhancer-binding protein homologous protein
- DBeQ
N2,N4-dibenzylquinazoline-2,4-diamine
- DMEM
Dulbecco's modified Eagle's medium
- eIF2α
eukaryotic initiation factor 2α
- EIF2AK
eukaryotic translation initiation factor 2α kinase
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- FBS
fetal bovine serum
- GADD34
growth arrest and DNA damage protein
- GC-MS
gas chromatography–mass spectrometry
- GCN2
general control nonderepressible 2
- HEPES
N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid
- MM
multiple myeloma
- mRNA
messenger RNA
- mTORC
mechanistic target of rapamycin complex
- NMS-873
Nerviano Medical Sciences-873
- PPP1R15A
protein phosphatase 1 regulatory subunit 15A
- RPMI
Roswell Park Memorial Institute
- UPR
unfolded protein response
- UPS
ubiquitin–proteasome system
- VCP
valosin-containing protein
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by M Agostini
Supplementary Material
References
- 1Deshaies RJ. Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biol 2014; 12: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Clarke HJ, Chambers JE, Liniker E, Marciniak SJ. Endoplasmic reticulum stress in malignancy. Cancer Cell 2014; 25: 563–573. [DOI] [PubMed] [Google Scholar]
- 3Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149: 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Yecies JL, Manning BD. Transcriptional control of cellular metabolism by mTOR signaling. Cancer Res 2011; 71: 2815–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67: 425–479. [DOI] [PubMed] [Google Scholar]
- 6Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature 2014; 513: 440–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 2007; 6: 472–483. [DOI] [PubMed] [Google Scholar]
- 8Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002; 418: 387–391. [DOI] [PubMed] [Google Scholar]
- 9Muller JM, Deinhardt K, Rosewell I, Warren G, Shima DT. Targeted deletion of p97 (VCP/CDC48) in mouse results in early embryonic lethality. Biochem Biophys Res Commun 2007; 354: 459–465. [DOI] [PubMed] [Google Scholar]
- 10Peters JM, Walsh MJ, Franke WW. An abundant and ubiquitous homo-oligomeric ring-shaped ATPase particle related to the putative vesicle fusion proteins Sec18p and NSF. EMBO J 1990; 9: 1757–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 2012; 14: 117–123. [DOI] [PubMed] [Google Scholar]
- 12Sasagawa Y, Higashitani A, Urano T, Ogura T, Yamanaka K. CDC-48/p97 is required for proper meiotic chromosome segregation via controlling AIR-2/Aurora B kinase localization in Caenorhabditis elegans. J Struct Biol 2012; 179: 104–111. [DOI] [PubMed] [Google Scholar]
- 13Uchiyama K, Totsukawa G, Puhka M, Kaneko Y, Jokitalo E, Dreveny I et al. p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis. Dev Cell 2006; 11: 803–816. [DOI] [PubMed] [Google Scholar]
- 14Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol Cell 2011; 41: 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell 1995; 82: 905–914. [DOI] [PubMed] [Google Scholar]
- 16Janiesch PC, Kim J, Mouysset J, Barikbin R, Lochmuller H, Cassata G et al. The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat Cell Biol 2007; 9: 379–390. [DOI] [PubMed] [Google Scholar]
- 17Cao K, Nakajima R, Meyer HH, Zheng Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 2003; 115: 355–367. [DOI] [PubMed] [Google Scholar]
- 18Verma R, Oania RS, Kolawa NJ, Deshaies RJ. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife 2013; 2: e00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Defenouillere Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L et al. Cdc48-associated complex bound to 60 S particles is required for the clearance of aberrant translation products. Proc Natl Acad Sci USA 2013; 110: 5046–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 2001; 414: 652–656. [DOI] [PubMed] [Google Scholar]
- 21Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol 2003; 162: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Ye Y, Shibata Y, Kikkert M, van Voorden S, Wiertz E, Rapoport TA. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci USA 2005; 102: 14132–14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH et al. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol 2009; 187: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP et al. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 2010; 6: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Kobayashi T, Manno A, Kakizuka A. Involvement of valosin-containing protein (VCP)/p97 in the formation and clearance of abnormal protein aggregates. Genes Cells 2007; 12: 889–901. [DOI] [PubMed] [Google Scholar]
- 26Ju JS, Miller SE, Hanson PI, Weihl CC. Impaired protein aggregate handling and clearance underlie the pathogenesis of p97/VCP-associated disease. J Biol Chem 2008; 283: 30289–30299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Chou TF, Brown SJ, Minond D, Nordin BE, Li K, Jones AC et al. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc Natl Acad Sci USA 2011; 108: 4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 2006; 126: 361–373. [DOI] [PubMed] [Google Scholar]
- 29Steffen J, Seeger M, Koch A, Kruger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol Cell 2010; 40: 147–158. [DOI] [PubMed] [Google Scholar]
- 30Radhakrishnan SK, den Besten W, Deshaies RJ. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. eLife 2014; 3: e01856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Sha Z, Goldberg AL. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr Biol 2014; 24: 1573–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Tsujimoto Y, Tomita Y, Hoshida Y, Kono T, Oka T, Yamamoto S et al. Elevated expression of valosin-containing protein (p97) is associated with poor prognosis of prostate cancer. Clin Cancer Res 2004; 10: 3007–3012. [DOI] [PubMed] [Google Scholar]
- 33Yamamoto S, Tomita Y, Uruno T, Hoshida Y, Qiu Y, Iizuka N et al. Increased expression of valosin-containing protein (p97) is correlated with disease recurrence in follicular thyroid cancer. Ann Surg Oncol 2005; 12: 925–934. [DOI] [PubMed] [Google Scholar]
- 34Yamamoto S, Tomita Y, Hoshida Y, Iizuka N, Kidogami S, Miyata H et al. Expression level of valosin-containing protein (p97) is associated with prognosis of esophageal carcinoma. Clin Cancer Res 2004; 10: 5558–5565. [DOI] [PubMed] [Google Scholar]
- 35Yamamoto S, Tomita Y, Hoshida Y, Iizuka N, Monden M, Yamamoto S et al. Expression level of valosin-containing protein (p97) is correlated with progression and prognosis of non-small-cell lung carcinoma. Ann Surg Oncol 2004; 11: 697–704. [DOI] [PubMed] [Google Scholar]
- 36Yamamoto S, Tomita Y, Hoshida Y, Nagano H, Dono K, Umeshita K et al. Increased expression of valosin-containing protein (p97) is associated with lymph node metastasis and prognosis of pancreatic ductal adenocarcinoma. Ann Surg Oncol 2004; 11: 165–172. [DOI] [PubMed] [Google Scholar]
- 37Yamamoto S, Tomita Y, Hoshida Y, Sakon M, Kameyama M, Imaoka S et al. Expression of valosin-containing protein in colorectal carcinomas as a predictor for disease recurrence and prognosis. Clin Cancer Res 2004; 10: 651–657. [DOI] [PubMed] [Google Scholar]
- 38Yamamoto S, Tomita Y, Hoshida Y, Takiguchi S, Fujiwara Y, Yasuda T et al. Expression level of valosin-containing protein is strongly associated with progression and prognosis of gastric carcinoma. J Clin Oncol 2003; 21: 2537–2544. [DOI] [PubMed] [Google Scholar]
- 39Yamamoto S, Tomita Y, Hoshida Y, Toyosawa S, Inohara H, Kishino M et al. Expression level of valosin-containing protein (VCP) as a prognostic marker for gingival squamous cell carcinoma. Ann Oncol 2004; 15: 1432–1438. [DOI] [PubMed] [Google Scholar]
- 40Yamamoto S, Tomita Y, Nakamori S, Hoshida Y, Nagano H, Dono K et al. Elevated expression of valosin-containing protein (p97) in hepatocellular carcinoma is correlated with increased incidence of tumor recurrence. J Clin Oncol 2003; 21: 447–452. [DOI] [PubMed] [Google Scholar]
- 41Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S et al. Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell 2015; 25: 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Magnaghi P, D'Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat Chem Biol 2013; 9: 548–556. [DOI] [PubMed] [Google Scholar]
- 43Acharya P, Liao M, Engel JC, Correia MA. Liver cytochrome P450 3A endoplasmic reticulum-associated degradation: a major role for the p97 AAA ATPase in cytochrome P450 3A extraction into the cytosol. J Biol Chem 2011; 286: 3815–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Piccirillo R, Goldberg AL. The p97/VCP ATPase is critical in muscle atrophy and the accelerated degradation of muscle proteins. EMBO J 2012; 31: 3334–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Aronson LI, Davies FE. DangER: protein ovERload. Targeting protein degradation to treat myeloma. Haematologica 2012; 97: 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 2011; 334: 1081–1086. [DOI] [PubMed] [Google Scholar]
- 47Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 2011; 13: 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol 2001; 153: 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Choy MS, Yusoff P, Lee IC, Newton JC, Goh CW, Page R et al. Structural and functional analysis of the GADD34:PP1 eIF2alpha phosphatase. Cell reports 2015; 11: 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2000; 2: 326–332. [DOI] [PubMed] [Google Scholar]
- 51Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000; 6: 1099–1108. [DOI] [PubMed] [Google Scholar]
- 52Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 2000; 5: 897–904. [DOI] [PubMed] [Google Scholar]
- 53Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 2004; 18: 3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 2013; 15: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55Rajesh K, Krishnamoorthy J, Kazimierczak U, Tenkerian C, Papadakis AI, Wang S et al. Phosphorylation of the translation initiation factor eIF2alpha at serine 51 determines the cell fate decisions of Akt in response to oxidative stress. Cell Death Dis 2015; 6: e1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 2003; 11: 619–633. [DOI] [PubMed] [Google Scholar]
- 57Chou TF, Deshaies RJ. Development of p97 AAA ATPase inhibitors. Autophagy 2011; 7: 1091–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58Tsaytler P, Harding HP, Ron D, Bertolotti A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 2011; 332: 91–94. [DOI] [PubMed] [Google Scholar]
- 59Meyuhas O. Physiological roles of ribosomal protein S6: one of its kind. Int Rev Cell Mol Biol 2008; 268: 1–37. [DOI] [PubMed] [Google Scholar]
- 60Suraweera A, Munch C, Hanssum A, Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell 2012; 48: 242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science 2005; 310: 1960–1963. [DOI] [PubMed] [Google Scholar]
- 62Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci 2013; 70: 3493–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63Romano PR, Garcia-Barrio MT, Zhang X, Wang Q, Taylor DR, Zhang F et al. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol Cell Biol 1998; 18: 2282–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64Wengrod J, Wang D, Weiss S, Zhong H, Osman I, Gardner LB. Phosphorylation of eIF2alpha triggered by mTORC1 inhibition and PP6C activation is required for autophagy and is aberrant in PP6C-mutated melanoma. Sci Signal 2015; 8: ra27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65Guan BJ, Krokowski D, Majumder M, Schmotzer CL, Kimball SR, Merrick WC et al. Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2alpha. J Biol Chem 2014; 289: 12593–12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66Tenkerian C, Krishnamoorthy J, Mounir Z, Kazimierczak U, Khoutorsky A, Staschke KA et al. mTORC2 balances Akt activation and eIF2alpha serine 51 phosphorylation to promote survival under stress. Mol Cancer Res 2015; 13: 1377–1388. [DOI] [PubMed] [Google Scholar]
- 67Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev 2003; 17: 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68Wander SA, Hennessy BT, Slingerland JM. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J Clin Invest 2011; 121: 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res 2015; 75: 544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70Ruckrich T, Kraus M, Gogel J, Beck A, Ovaa H, Verdoes M et al. Characterization of the ubiquitin-proteasome system in bortezomib-adapted cells. Leukemia 2009; 23: 1098–1105. [DOI] [PubMed] [Google Scholar]
- 71Auner HW, Moody AM, Ward TH, Kraus M, Milan E, May P et al. Combined inhibition of p97 and the proteasome causes lethal disruption of the secretory apparatus in multiple myeloma cells. PLoS One 2013; 8: e74415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72Kind T, Wohlgemuth G, Lee DY, Lu Y, Palazoglu M, Shahbaz S et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem 2009; 81: 10038–10048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73Behrends V, Tredwell GD, Bundy JG. A software complement to AMDIS for processing GC-MS metabolomic data. Anal Chem 2011; 415: 206–208. [DOI] [PubMed] [Google Scholar]
- 74Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 2009; 6: 275–277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.