Summary
Uridylation occurs pervasively on mRNAs, yet its mechanism and significance remain unknown. By applying TAIL-seq, we identify TUT4 and TUT7 (TUT4/7), also known as ZCCHC11 and ZCCHC6, respectively, as mRNA uridylation enzymes. Uridylation readily occurs on deadenylated mRNAs in cells. Consistently, purified TUT4/7 selectively recognize and uridylate RNAs with short A-tails (less than ∼25 nt) in vitro. PABPC1 antagonizes uridylation of polyadenylated mRNAs, contributing to the specificity for short A-tails. In cells depleted of TUT4/7, the vast majority of mRNAs lose the oligo-U-tails, and their half-lives are extended. Suppression of mRNA decay factors leads to the accumulation of oligo-uridylated mRNAs. In line with this, microRNA induces uridylation of its targets, and TUT4/7 are required for enhanced decay of microRNA targets. Our study explains the mechanism underlying selective uridylation of deadenylated mRNAs and demonstrates a fundamental role of oligo-U-tail as a molecular mark for global mRNA decay.
Introduction
RNA tailing (nontemplated nucleotide addition to the 3′ end of RNA) is one of the most frequent types of RNA modification, with a deep evolutionary root and diverse molecular functions. In bacteria, adenylation of mRNA triggers RNA degradation whereas polyadenylation in eukaryotes increases the stability and translatability of mRNA (Dreyfus and Régnier, 2002). Tailing is catalyzed by a group of template-independent ribonucleotidyl transferases that contain DNA polymerase β-like nucleotidyl transferase domain (Aravind and Koonin, 1999). Apart from canonical poly(A) polymerases (PAPs) that generate poly(A) tail of mRNA, many noncanonical PAPs have been described from fission yeast to human (Martin and Keller, 2007; Norbury, 2013). Because some noncanonical PAPs catalyze uridylation instead of adenylation, noncanonical PAPs are also called terminal uridylyl transferases (TUTases or TUTs). Some PAPs/TUTs have more relaxed nucleotide specificity and carry out both uridylation and adenylation. Humans have seven noncanonical PAPs/TUTs with distinct substrate specificity and subcellular localization.
Uridylation of mRNA was initially noticed at the 3′ ends of miRNA-directed cleavage products in Arabidopsis and mammalian cells (Shen and Goodman, 2004). U-tails were also detected on human replication-dependent histone mRNAs that lack a poly(A) tail (Mullen and Marzluff, 2008). Histone mRNAs are uridylated and degraded at the end of S phase or upon inhibition of DNA replication (Mullen and Marzluff, 2008). TUT4 (ZCCHC11) was reported to catalyze histone mRNA uridylation (Schmidt et al., 2011; Su et al., 2013), although two other TUTs (TUT1/MTPAP/PAPD1 and TUT3/PAPD5/TRF4-2) were proposed in an earlier study (Mullen and Marzluff, 2008). Uridylation induces rapid decay of histone mRNA through both the 5′–3′ degradation by XRN1, DCP2, and LSM1 and the 3′–5′ degradation by exosome and ERI1 (3′hExo) (Hoefig et al., 2013; Mullen and Marzluff, 2008; Slevin et al., 2014).
Interestingly, uridylation occurs not only on poly(A)-lacking mRNAs but also on poly(A)+ mRNAs, as shown first with the actin (act1) mRNA in fission yeast Schizosaccharomyces pombe (Rissland et al., 2007). When six mRNAs were examined by circularized rapid amplification of cDNA ends (cRACE) technique, all of them were found to bear short U-tails (usually one or two uridines) at the end of poly(A) tails albeit at varying frequencies, indicating that mRNA uridylation may be widespread in fission yeast (Rissland and Norbury, 2009). The stability of the urg1 mRNA increased in a mutant lacking Cid1 which is one of the TUTs in fission yeast (Rissland et al., 2007; Rissland and Norbury, 2009). The uridylation frequency was enhanced in mutants defective of deadenylase and decapping enzyme (ccr4Δ and dcp1-ts). Based on these results, it was proposed that uridylation and deadenylation may act redundantly to induce decapping. A more recent study showed that Arabidopsis mRNAs are also subject to uridylation (Sement et al., 2013). Short uridyl residues (1–2 uridines) were detected on deadenylated, decapped mRNAs. The Cid1 homolog URT1 is required for uridylation. But, curiously, URT1 mutation did not have a major impact on mRNA turnover and instead inhibited trimming of mRNA from the 3′ end (Sement et al., 2013), implying that uridylation may be necessary to establish the directionality (5′–3′) rather than to control the rate of mRNA decay. Therefore, although these observations are intriguing, it was unclear if uridylation has a conserved function across species and whether animal poly(A)+ mRNAs are also uridylated. In addition, because previous studies examined a few individual mRNAs by RACE and small-scale cloning, it remained to be tested whether or not uridylation occurs globally and if the observed changes in uridylation and poly(A) length are statistically significant.
To investigate tail structures at the genomic scale, we recently developed a method called TAIL-seq that deep-sequences the 3′ most fragments of RNAs (Chang et al., 2014b). The TAIL-seq protocol begins with removal of abundant noncoding RNAs such as rRNA, tRNA, small nuclear RNA (snRNA), and small nucleolar RNA (snoRNA) by affinity-based depletion and size fractionation. To avoid any bias against unconventional tails, TAIL-seq does not use splint ligation or oligo-d(T) enrichment. The resulting RNA sample enriched with mRNA is subsequently ligated to the 3′ adaptor that contains biotin residues. Following partial fragmentation, the 3′ most fragments are purified using streptavidin beads and ligated to the 5′ adaptor. Paired-end sequencing of the cDNA library yields 51 nt from the 5′ terminus of the fragment (to identify the transcript) and 231 nt from the 3′ terminus (to examine the tail sequences).
TAIL-seq provided us with a unique opportunity to investigate poly(A) tail length and additional 3′ modifications simultaneously at the genomic scale. Surprisingly, we found that the vast majority of mRNAs are subject to uridylation in mammals. Over 85% of mRNAs are terminally uridylated at a frequency of higher than 1% in both NIH 3T3 and HeLa cells (Chang et al., 2014b). Interestingly, U-tails are found mainly on mRNAs with short A-tails (less than ∼25 nt), indicating that uridylation may occur following deadenylation. We further detected a negative correlation between uridylation frequency and mRNA half-life, suggesting a role of uridylation in general mRNA decay.
Current model for eukaryotic mRNA decay pathway is mainly based on the pioneering genetic and biochemical studies in Saccharomyces cerevisiae (Garneau et al., 2007; Houseley and Tollervey, 2009; Norbury, 2013; Parker and Song, 2004). Decay is generally initiated by deadenylation that is mediated by multiple deadenylases such as the Pan2-Pan3 complex and the Ccr4-Not complex. Subsequently, deadenylated mRNAs are subject to either of two major decay pathways. In the 5′–3′ decay pathway, the Lsm1–7 complex binds to the 3′ end of deadenylated mRNA and recruits the decapping complex (Dcp1/2) that removes 5′ cap structure. Subsequently, 5′ monophosphate-dependent exoribonuclease, Xrn1, digests mRNA processively. From the opposite orientation, a multisubunit exosome complex degrades deadenylated mRNAs from the 3′ end. This model seems to apply generally to most, if not all, eukaryotic species. However, S. cerevisiae is unusual among eukaryotes in that it does not have any known TUT homolog with uridylation activity and that mRNAs in S. cerevisiae do not carry terminal U-tails (Norbury, 2013). Thus, the current model for mRNA decay, particularly in mammals, may need to be revised to incorporate the recent findings of pervasive uridylation (Lee et al., 2014).
In this study, we aimed to identify enzyme(s) that catalyze mRNA uridylation in mammals and understand the significance of uridylation in the mRNA decay pathway. We discover TUT4 and TUT7 as uridylyl transferases for poly(A)+ mRNAs in humans and delineate in detail the action mechanism and molecular function of uridylation in the mRNA decay pathway. Based on these results, we propose a revised model for general mRNA decay in mammals.
Result
TUT4 and TUT7 Catalyze mRNA Uridylation
In order to identify enzyme(s) responsible for mRNA uridylation, we took a candidate approach by depleting seven human TUTases (Figure S1A available online). Because TUT2 (also known as GLD2 and PAPD4), TUT4 (ZCCHC11), and TUT7 (ZCCHC6) act redundantly in mono-uridylation of precursor of let-7 (pre-let-7) (Heo et al., 2012), we knocked down TUTases in two subgroups (TUT1/3/5/6 and TUT2/4/7) by transfecting siRNA mixtures into HeLa cells (Figure S1B) and carried out TAIL-seq (Figure 1A). Overall frequency of uridylation was quantified by dividing the read number of terminally uridylated mRNAs by that of total mRNAs. Because short A-tails are preferentially uridylated (Chang et al., 2014b), uridylation frequency in short A-tail range (5–25 nt) is shown in Figure 1. Interestingly, when TUT2/4/7 were depleted, terminal uridylation was significantly reduced while RNAi of TUT1/3/5/6 did not affect uridylation.
Figure 1. TUT4 and TUT7 Are Required for mRNA Uridylation in Human Cells.
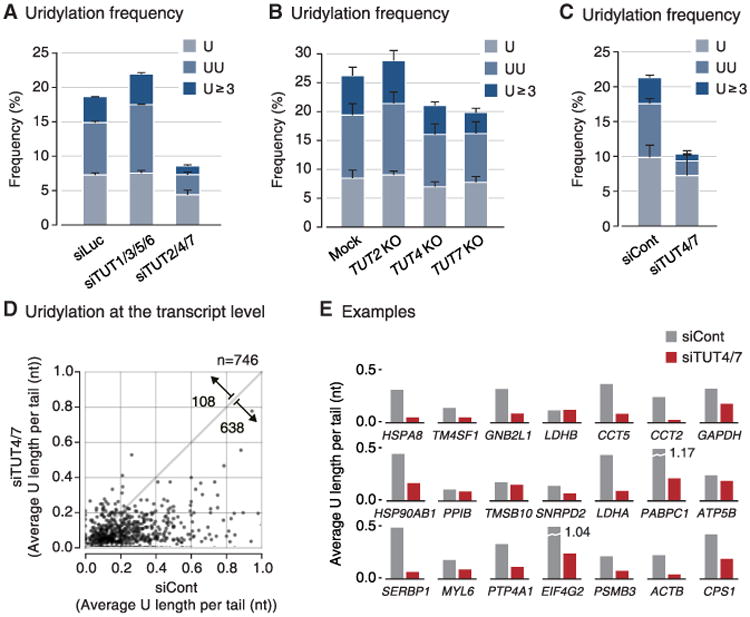
(A) Uridylation frequency measured by small-scale TAIL-seq (with Illumina MiSeq) following RNAi of the indicated genes. Frequency (y axis) is the fraction of uridylated reads among the total number of mRNA reads with short poly(A) tail (5–25 nt). Light blue refers to mono-uridylation (U), blue indicates di-uridylation (UU), and dark blue represents ≥ 3 uridines (U ≥ 3). Uridylation frequency significantly decreased in siTUT2/4/7 (p = 0.0378 for U; 0.0388 for UU; 0.0201 for U ≥ 3 by one-tailed t test). Error bar represents SEM from two biologically independent replicates (n = 2).
(B) Uridylation frequency of mRNAs with short poly(A) tails (5–25 nt) measured by small-scale TAIL-seq in knockout HeLa cell lines. Uridylation frequency was reduced modestly in TUT4 and TUT7 knockout cells (p = 0.109 for U, 0.0273 for UU, 0.142 for U ≥ 3 of TUT4 KO; p = 0.150 for U, 0.00685 for UU, 0.0713 for U ≥ 3 of TUT7 KO by one-tailed t test). Error bar represents SEM from two replicates (n = 2).
(C) Uridylation frequency of mRNAs with short poly(A) tails (5–25 nt) measured by TAIL-seq following simultaneous TUT4 and TUT7 knockdown (siTUT4/7). Uridylation was reduced when both TUT4 and TUT7 were depleted (p = 0.0941 for U, 0.00922 for UU, 0.0105 for U ≥ 3; one-tailed t test). Error bar represents SEM from three biological replicates (n = 3).
(D) Changes in uridylation of individual mRNA species upon TUT4/7 knockdown. “Average U length per tail” (y axis) is the sum of the number of all uridines on short A-tails (5–25 nt) divided by the total number of reads with short A-tails. Note that unlike “uridylation frequency,” average U length per tail weighs every uridine in oligo-U-tails. Each dot represents a transcript with ≥ 15 reads in both samples. Uridylation was significantly decreased following TUT4/7 knockdown (p = 7.69 × 10−100, one-tailed Mann-Whitney U test). The full list is shown in Table S1.
(E) Examples of gene-level uridylation changes. Twenty-one most abundant mRNAs (not including ribosomal protein mRNAs and histone mRNAs) are shown in the order of mRNA abundance.
See also Figure S1.
To narrow down on individual TUTases, we generated knockout HeLa cell lines using TALENs (transcription activatorlike effector nucleases) against the genes coding TUT2, TUT4, or TUT7 proteins (Figure S1C). We observed a modest decrease of uridylation in both TUT4 and TUT7 knockout cells, but not in TUT2 knockout cells (Figure 1B). Repeated attempts to generate double knockout of TUT4 and TUT7 by utilizing the TALEN and CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) systems have failed (Figure S1D). Although genomic deletion was effectively introduced by the nucleases, mutant clones disappeared during clonal selection processes (Figure S1D), which indicates that the combined activity of TUT4 and TUT7 is essential for cell viability.
Of note, previous studies have shown that TUT4 and TUT7 are highly similar in their domain organization and activity in pre-miRNA uridylation (Heo et al., 2012; Liu et al., 2014; Thornton et al., 2012). Thus, TUT4 and TUT7 (TUT4/7) may act redundantly in mRNA uridylation as well as in pre-miRNA uridylation. To confirm this notion, we carried out simultaneous transient RNAi against TUT4/7 by transfecting siRNAs (Figures 1C and S1E). The TUT4/7 knockdown cells looked largely normal and proliferated at a modestly reduced rate with a slight increase of apoptosis after 4 days of siRNA treatment (Figures S1F and S1G). Under this condition, uridylation of mRNA was significantly reduced when both TUT4 and TUT7 are depleted (Figure 1C). Oligo-uridylation (≥2 U) was more sensitive to TUT4/7 knockdown than mono-uridylation was (3.71-fold and 1.36-fold decrease, respectively), suggesting that a relatively high level of TUT4/7 may be required to generate oligo-U-tails on mRNA.
Gene-level analyses revealed that the majority of mRNA species (638 out of 746 genes, 85.5%) are decreased in uridylation following TUT4/7 knockdown (p = 7.69 × 10−100, one-tailed Mann-Whitney U test) (Figure 1D; Table S1). This result strongly indicates that TUT4/7 uridylate most, if not all, mRNAs. Figure 1E presents 21 most abundant mRNAs as examples, the majority of which are reduced in uridylation upon TUT4/7 knockdown. Two biological replicate experiments showed a comparable decrease of uridylation (Figure S1H).
Histone mRNAs that lack poly(A) tails are also uridylated and their uridylation is dependent modestly on TUT4/7, but not on TUT1/2/3/5/6 (data not shown). However, poly(A)− histone mRNAs were excluded from our current data analyses because we used nonsynchronous cell population for our experiments, and it is known that uridylation of histone mRNA occurs specifically at the end of S phase (Mullen and Marzluff, 2008; Schmidt et al., 2011; Su et al., 2013). It would be more appropriate to investigate histone mRNAs using synchronous cells in future studies.
TUT4/7 Selectively Oligo-Uridylate mRNAs with Short A-Tails In Vivo and In Vitro
It is intriguing that uridylation occurs preferentially on shortened A-tails in plants and animals (Chang et al., 2014b; Sement et al., 2013). Figure 2A shows the distribution of U-tails over different lengths of A-tails in HeLa cells. The frequency of uridylation on the transcripts with a short A-tails (5–25 nt) is higher than that on the rest (A-tails of >25 nt), especially when only oligo-U (≥2 U) is counted. Note that mRNAs with A-tails of shorter than 5 nt were excluded from this analysis as it is sometimes difficult to distinguish them from genomic A-rich sequences in 3′ UTR. When TUT4/7 were depleted, uridylation on short A-tails was selectively reduced (especially for oligo-U), indicating that TUT4/7 are responsible for the specific uridylation of short A-tails (Figure 2A).
Figure 2. Short A-Tails Are Selectively Uridylated by TUT4 and TUT7.
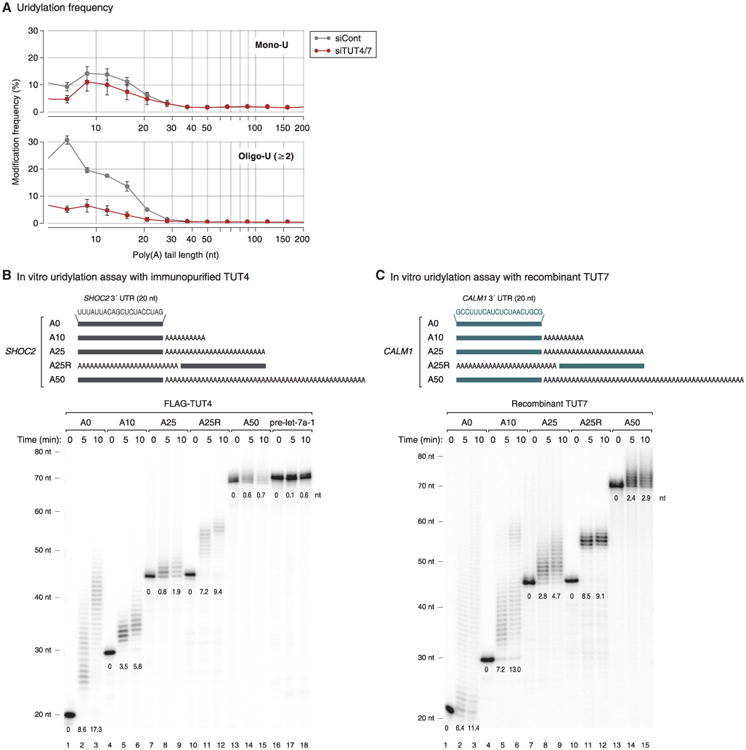
(A) Distribution of mono-uridylation (top) and oligo-uridylation (bottom) according to the length of poly(A) tails. Poly(A) tail lengths from 5 nt to 231 nt are pooled into equal-width bins in the logarithmic scale (base 2) (x axis). The left edges (inclusive) of bins are 5, 7, 9, 12, 15, 21, 28, 38, 50, 67, 89, 119, 159, and 212 nt. Uridylation frequency (y axis) indicates the percentage of uridylated reads within each poly(A) tail size range. Error bar represents SEM (n = 3).
(B) Top: illustration of chemically synthesized RNA substrates. Grey bars represent the last 20 nt of SHOC2 3′ UTR and “A” indicates an adenosine. Bottom: in vitro uridylation assay using immunopurified FLAG-TUT4. RNA (0.45 nM) was used in each reaction. The products were resolved on 6% polyacrylamide sequencing gel containing 7 M urea. The average length of uridylation is shown below each band. See Extended Experimental Procedures for quantification method.
(C) Top: illustration of chemically synthesized RNA substrates. Green bars represent the last 20 nt of CALM1 3′ UTR and “A” indicates an adenosine. Bottom: in vitro uridylation assay using recombinant TUT7 C-terminal fragment (951–1,495 aa) purified from E. coli. RNA (0.45 nM) and 14 nM of recombinant TUT7 were used in each reaction. Extension products were resolved on 6% polyacrylamide sequencing gel containing 7 M urea. The average length of uridylation was quantified as in (B).
See also Figure S2.
To understand the mechanism underlying such strong association with A-tail length, we performed in vitro uridylation assays using immunopurified full-length TUTases (Figures 2B and S2A–S2C). Substrate RNAs were chemically synthesized to contain heterogenous sequences (the last 20 nt from the SHOC2 3′ UTR) linked to A-tails of various lengths (0, 10, 25, and 50 nt) at the 3′ end (Figure 2B). We also used a “swapped” control (A25R) that has a 25 nt A segment at the 5′ side of the SHOC2 3′ UTR such that the RNA is identical to SHOC2-A25 (A25) in the overall length and base composition, but lacks an A-tail at the 3′ end (Figure 2B).
Interestingly, RNAs with no tail (A0) or a short A-tail (A10) were oligo-uridylated efficiently by TUT4 under the condition where pre-let-7a-1 is mono-uridylated weakly (Figure 2B). A25 and A50 were less efficiently uridylated than A0 and A10 were. The A25R RNA was a much better substrate than the A25 was, indicating that it is the 3′ A-tail length (not the overall RNA length) that is measured by TUT4 (Figure 2B). Comparable results were obtained with full-length TUT7 protein (Figures S2A and S2B), again demonstrating that these two related enzymes are functional paralogs. The U-tail length in Figures S2A and S2B was overall shorter than those in Figure 2B because the amount of immuno-precipitated TUT7 was smaller than that of TUT4 in Figure 2B (data not shown).
We also prepared recombinant TUT7 protein (951–1,495 aa) from Escherichia coli and used the fragment for in vitro uridylation assay (Figure S2C). Apart from the SHOC2 RNAs (Figure S2D), we synthesized and tested another series of RNAs based on the CALM1 3′ UTR sequences (Figure 2C). The purified protein fragment was fully capable of carrying out uridylation in an A-tail length-dependent manner with both RNAs (Figures 2C and S2D, see below). Thus, the C-terminal half of TUT7 is sufficient to recognize and uridylate single-stranded RNAs with a short A-tails (less than ∼25 nt), in a 3′ UTR sequence-independent manner. These results suggest that TUT4/7 possess an intrinsic ability to measure the 3′ terminal A length and avoid uridylation of long A-tails.
PABP Suppresses Uridylation of Poly(A)+ mRNA
As poly(A)+ mRNAs are associated with poly(A) binding protein (PABP) in cells, we asked if PABP has an influence on mRNA uridylation. It was previously shown that PABP preferentially interacts with poly(A) or A-rich sequences (Eliseeva et al., 2013). The binding affinity increases as the A stretch gets longer (Eliseeva et al., 2013; Khanam et al., 2006; Kühn and Pieler, 1996; Sachs et al., 1987). Full-length PABP occupies an ∼25 nt A-tail as determined by nuclease digestion assay (Baer and Kornberg, 1983; Eliseeva et al., 2013). In order to test an effect of PABP on uridylation, we carried out in vitro uridylation assays in the presence of recombinant PABPC1 (Figure 3). When PABPC1 was added to RNA, uridylation of RNAs with long poly(A) tail (A25 and A50) was suppressed even at a low concentration of PABPC1 (10 nM) while those with no or short A-tail (A0, A10, and A25R) remained largely unaffected (Figure 3). This result suggests that PABPC1 binds preferentially to long poly(A) tails and protects them from TUT4/7 and thereby enhances the selectivity of uridylation according to poly(A) tail length.
Figure 3. PABP Inhibits Uridylation of Polyadenylated mRNA.
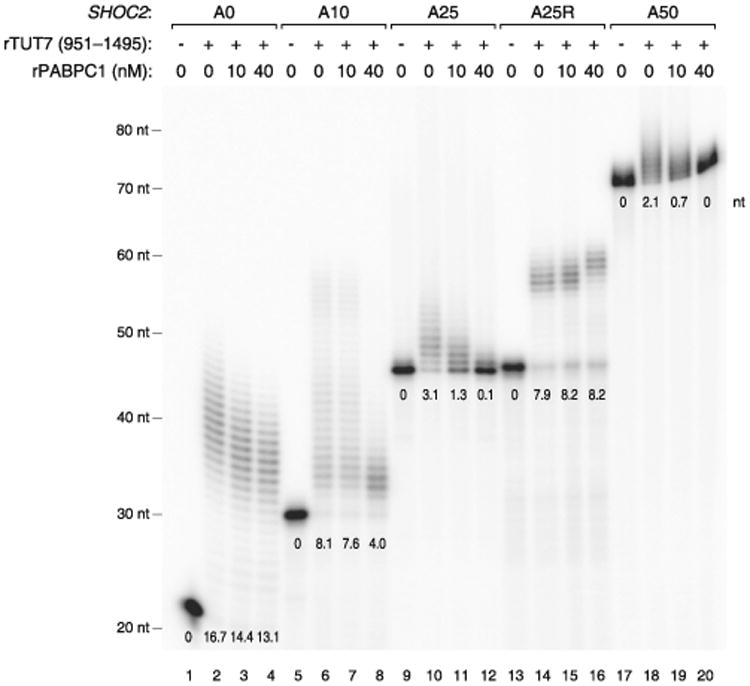
In vitro uridylation assay by using recombinant TUT7 (951–1,495 aa) with a varying concentration of recombinant PABPC1 (0, 10, or 40 nM). 0.45 nM of RNA and 160 nM of recombinant TUT7 (rTUT7) were used in the reaction. Extension products were resolved on 6% polyacrylamide sequencing gel containing 7 M urea. The average length of uridylation was quantified as described in Extended Experimental Procedures and shown below each band. See also Figure S3.
Taken together, our results suggest that the strict dependence on the A-tail length observed in vivo may be determined by the combination of two factors: (1) the intrinsic ability of TUT4/7 to measure poly(A) stretch (Figure 2), and (2) the protective activity of PABP (Figure 3).
As deadenylation is thought to occur mainly in the cytoplasm, we examined the localization of TUT4/7 by western blotting. The TUT4 and TUT7 proteins are mainly localized in the cytoplasm (Figure S3). Thus, TUT4/7 may function mainly in the metabolism of cytoplasmic, deadenylated mRNAs.
Uridylation Facilitates Global mRNA Decay
To understand the functional consequences of uridylation, we measured mRNA half-life in HeLa cells with or without TUT4/7 knockdown (Figure 4A). mRNA levels were determined by RNA-seq at 0, 1, 2, and 4 hr after actinomycin D treatment that blocks transcription. To avoid any bias from tail length variation, we omitted the oligo-dT enrichment step and instead used Ribo-Zero to remove abundant rRNAs prior to cDNA library construction. We could measure turnover rates of 1,829 mRNAs. In TUT4/7-depleted cells, the majority of mRNAs (1,426 out of 1,829 [78.0%]) showed increase stability (Figure 4A, left panel; Table S2). Half-lives were increased by ∼30% on average, and median half-life was extended from 9.45 hr to 11.2 hr (Figure 4A, right panel).
Figure 4. Uridylation Promotes mRNA Degradation.
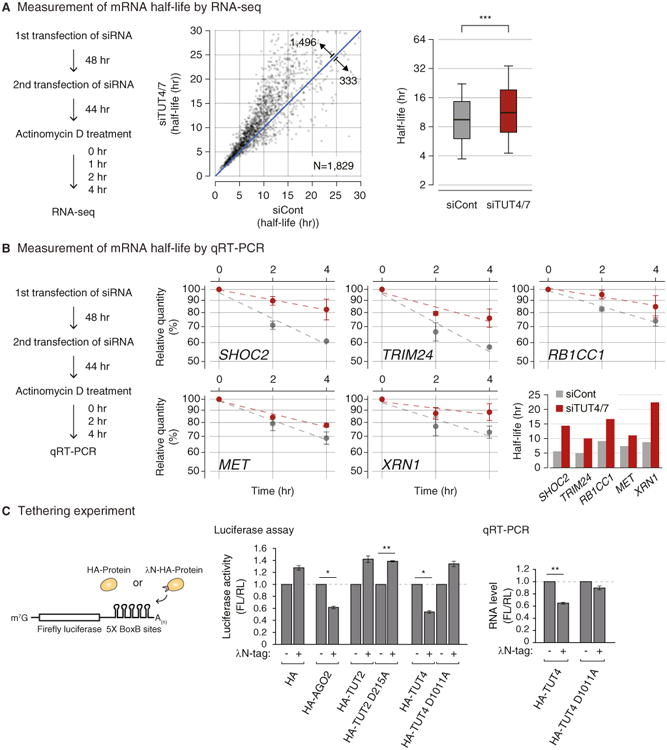
(A) Transcriptome-wide change of mRNA half-life determined by RNA-seq. Left: experimental scheme. HeLa cells were transfected twice and harvested at 0, 1, 2, and 4 hr following actinomycin D treatment. Center: changes of average mRNA half-life upon TUT4/7 knockdown from two biological replicates. The range of display is limited to between 0 and 30 hr for the better visual recognition (232 out of 1,829 mRNAs are outside of the view). The full list is available in Table S2. Right: distribution of mRNA half-lives in control or TUT4/7 knockdown cells. A box represents the first and third quartiles and an internal bar indicates median. Whiskers span between the ninth and the 91st percentiles. Half-lives of mRNAs are significantly extended by TUT4/7 knockdown (***p = 4.06 × 10−155, one-tailed paired Mann-Whitney U test). See Extended Experimental Procedures for the detailed description of procedure.
(B) Measurement of mRNA half-life by qRT-PCR. Left: the experimental scheme. Right: following 0, 2, and 4 hr of actinomycin D treatment, relative abundance (y axis) of five selected genes were measured. For normalization, GAPDH mRNA was used because it was highly stable (half-life > 24 hr, data not shown) and did not change noticeably by TUT4/7 depletion. Error bar represents SEM (n = 3). Half-lives are calculated by linear fitting of the log-transformed exponential decay function.
(C) Left: schematic representation of reporter assay system with the λN tethering. Center: reporter (firefly) luciferase activity was measured and normalized to Renilla luciferase activity (n=3). Right: reporter mRNA levels were determined by qRT-PCR (n = 4). Error bars represent SEM. Luciferase activity or RNA level were significantly reduced when AGO2 or TUT4 were tethered (*p < 0.01, **p < 0.001; two-tailed t test).
See also Figure S4.
Of note, although TUT4/7 contribute to let-7 biogenesis, double knockdown of TUT4/7 (without simultaneous knockdown of TUT2) did not substantially affect the let-7 level (Heo et al., 2012). In fact, our transcriptome analyses show that mRNAs are globally upregulated, indicating that the changes in mRNA half-life observed in this study cannot be attributed to specific regulation of let-7 biogenesis.
For validation of the impact of TUT4/7 depletion on mRNA stability, five mRNAs (SHOC2, TRIM24, RB1CC1, MET, and XRN1) were measured by quantitative RT-PCR after actinomycin D treatment (Figure 4B). None of these mRNAs contains a let-7 binding site with seed match in their 3′ UTR, yet all of them showed increased stability when TUT4/7 were depleted. Therefore, our results demonstrate that TUT4/7 play an important role in bulk mRNA degradation in a let-7 independent manner.
Next, to examine the effect of overexpressed TUTase on mRNA expression, we carried out tethering experiments in HeLa cells (Figure 4C, left panel). A related experiment was reported recently in Xenopus oocytes: when Xenopus TUT7 homolog was tethered to the 3′ UTR of luciferase reporter mRNA, luciferase activity was reduced without significant changes in mRNA, implicating translational repression (Lapointe and Wickens, 2013). However, because mRNA decay activity is generally suppressed in oocytes (Barckmann and Simonelig, 2013), it was unclear if the observation from frog oocytes can be generalized. For tethering experiments, we generated constructs that express proteins tagged with the λN peptide that interacts with its specific binding sites (BoxB sites) in the 3′ UTR of luciferase mRNA (Figures 4C and S4). Expression of λN protein modestly increased luciferase expression nonspecifically for an unknown reason (Figure 4C, middle panel). Nevertheless, tethering of AGO2 repressed luciferase reporter expression (Figure 4C), as previously shown (Pillai et al., 2004), indicating that this is a valid system to test the effect of RNA silencing factors. Neither the negative control TUT2 nor its mutant repressed luciferase reporter expression. But when wild-type TUT4 was tethered to the reporter mRNA, luciferase activity was decreased to ∼60% while such reduction was not observed with the catalytically dead point mutant (D1011A) of TUT4 (Figure 4C, middle panel), indicating that TUT4 suppressed gene expression via uridylation. Quantitative RT-PCR further showed that tethering of TUT4 induced a reduction of mRNA (Figure 4C, right panel). Thus, our results collectively indicate that TUT4/7 function as suppressors of gene expression through mRNA destabilization.
Uridylation Is Involved in miRNA-Induced Gene Silencing
Our model predicts that if a gene-specific inducer of deadenylation is introduced into cells, uridylation of the given transcript will take place, which in turn will facilitate RNA decay. To test our model, we examined the effect of miRNA as an example, which is well established to induce specific deadenylation of its complementary targets (Ameres and Zamore, 2013; Djuranovic et al., 2011; Huntzinger and Izaurralde, 2011; Krol et al., 2010).
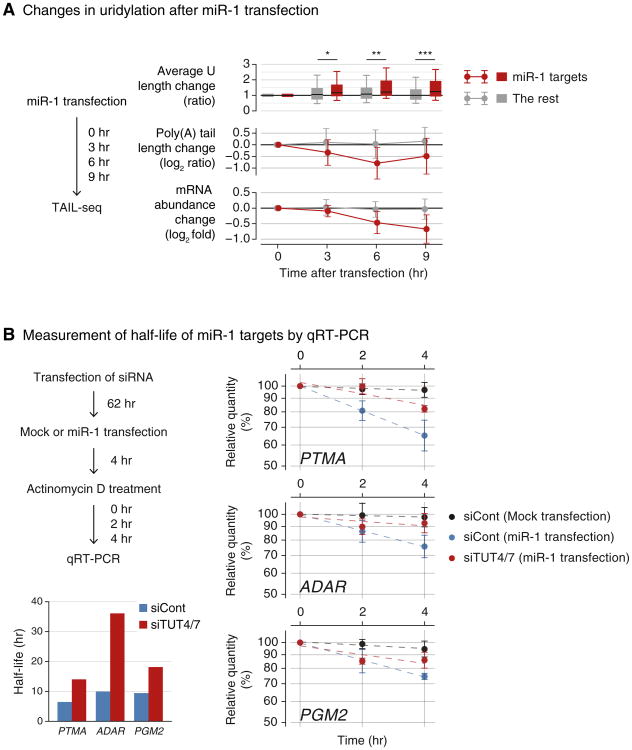
We first analyzed the TAIL-seq data from our previous experiment where miR-1 mimic was transfected into HeLa cells (Chang et al., 2014b). As expected, miR-1 targets undergo deadenylation and subsequent downregulation following miR-1 mimic transfection (Figure 5A, middle and lower panels, respectively). Importantly, we detected a specific increase of uridylation on miR-1 targets whereas the rest of genes stayed largely unaffected (Figure 5A, upper panel). This result is consistent with our model that deadenylation leads to uridylation.
Figure 5. Uridylation Facilitates miRNA-Mediated mRNA Decay.
(A) Changes in uridylation after miR-1 transfection. Left: experimental scheme. miR-1 was transfected into HeLa cells and the cells were harvested after the indicated time for TAIL-seq. Targets are the transcripts with ≥ 1 miR-1 3′ UTR site and down-regulated by ≥30% on 12 hr posttransfection of miR-1 (Guo et al., 2010). Right top: average Ulength change relative to 0 hr is shown in each time point. Average U length per tail is the number of uridines on short A-tails (5–25 nt) divided by the total number of reads with short A-tails. Box represents the interval between the first and third quartiles, and the internal bar indicates the median. Whiskers span between the ninth and 91st percentiles. Average U length of miR-1 target is significantly extended after miR-1 transfection (*p = 0.0152, **p = 0.00318, ***p = 5.79 × 10−4; one-tailed Mann-Whitney U test). Right middle: poly(A) tail length change relative to 0 hr. The length change is represented by log2 odds ratio between long tails (>25 nt) and short tails (≤25 nt) in one among 3, 6, or 9 hr and 0 hr. A negative value (<0) indicates increase of the fraction of short tails compared to 0 hr. Error bars indicate SD among mRNAs. The portion of short poly(A) tails expanded more for miR-1 targets than the others (p = 1.80 × 10−6 for 3 hr, p = 8.47 × 10−13 for 6 hr, p = 1.48 × 10−11 for 9 hr; one-tailed Mann-Whitney U test). Right bottom: mRNA abundance (poly(A)+ tag counts) change relative to 0 hr. Error bars indicate SD among mRNAs. Expression levels of miR-1 targets were decreased more than the rest transcripts (p = 2.09 × 10−4 for 3 hr, p = 2.65 × 10−14 for 6 hr, p = 5.46 × 10−18 for 9 hr; one-tailed Mann-Whitney U test).
(B) Measurement of half-life of miR-1 targets by qRT-PCR. Left: the experimental scheme. Following siRNA transfection for 62 hr, HeLa cells were transfected with miR-1 or mock transfected. After 4 hr, actinomycin D was treated and cells were harvested at 0, 1, 2, and 4 hr. Right: relative abundance (y axis) of miR-1 target mRNAs were measured. For the normalization, highly stable GAPDH mRNA was used because it did not change significantly by siTUT4/7 or miR-1 transfection. Error bar represents SEM (n = 3). Half-lives are determined by linear fitting of the log-transformed exponential decay function.
We next measured turnover rates of miR-1 targets with or without TUT4/7 knockdown. The mRNAs tested (PTMA, ADAR, and PGM2) are normally stable (half-lives >24 hr) in cells that do not contain miR-1 (Figure 5B, black line). When miR-1 was introduced, their half-lives were shortened to 6.5, 10.0, and 9.4 hr, respectively (Figure 5B, blue line). Upon TUT4/7 depletion, the miR-1 target mRNAs were stabilized (with extended half-lives of 14.0, 36.1, and 18.1 hr, respectively) (Figure 5B, red line). Therefore, TUT4/7 are necessary for the facilitated decay of miRNA targets. We propose that other factors that cause deadenylation may also induce uridylation and decay, as shown here with an example of miR-1.
mRNA Decay Factors Remove Uridylated mRNAs
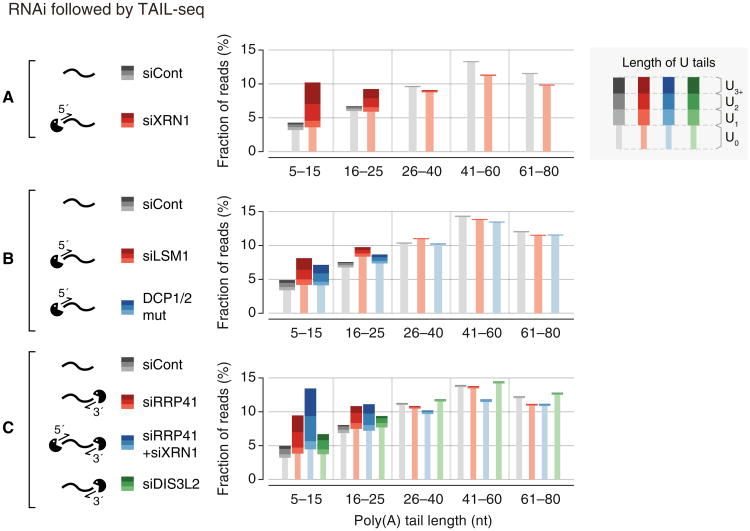
To understand downstream events of uridylation, we disrupted 5′–3′ or 3′–5′ exonucleolytic decay factors and examined the mRNA terminome (Figure S5A). The popsicle-shaped bars in Figure 6 display the relative quantity of reads with a U-tail (thick stem) or without a U-tail (thin stem). As U-tail frequencies vary depending on poly(A) tail length, different A-tail ranges are shown separately along the horizontal axis. For more information, the overall uridylation frequency and poly(A) length distribution are presented in Figures S5B and S5C, respectively.
Figure 6. The 5′ and 3′ mRNA Decay Factors Degrade Uridylated mRNAs.
(A–C) Changes of poly(A) tail and uridylation upon knockdown of decay factor(s) detected by small-scale TAIL-seq (with Illumina MiSeq). Fraction of mRNA reads out of the total poly(A)+ mRNA reads is shown in each poly(A) tail size range. Narrow bars represent reads without U-tails (U0) and wider bars indicate uridylated reads (U1–U3+). The “DCP1/2 mut” sample derived from cells coexpressed of dominant-negative mutants of DCP1 and DCP2 (DCP1a-GSSG and DCP2-E148Q, respectively).
See also Figure S5.
In order to inhibit 5′–3′ decay, we initially depleted a major 5′–3′ exoribonuclease XRN1. Interestingly, interference of XRN1 resulted in a strong accumulation of uridylated mRNAs with short A-tails (≤25 nt) (Figure 6A). Additionally, when we depleted LSM1 (a component of the LSM1-7 complex that is known to facilitate decapping) or overexpressed dominant-negative mutants of the decapping complex (DCP1 and DCP2) (Chang et al., 2014a), we detected an increase of uridylation among short A-tailed mRNAs (≤25 nt) (Figure 6B). Short A-tailed mRNAs increased in abundance (particularly, in the 5–15 nt range) when the 5′–3′ decay was suppressed. Note that the level of uridylated mRNA was upregulated substantially (U1–U3+), accounting for the overall increase of mRNA reads in this range, while mRNAs without a U-tail did not change significantly (U0). This result is consistent with a model that deadenylated, uridylated mRNAs are normally degraded rapidly by the 5′–3′ decay factors while poly(A)+ mRNAs without U-tails are relatively stable. The LSM1–7 complex is known to preferentially bind to RNAs with 3′ terminal uridyl residues (Chowdhury et al., 2007; Sharif and Conti, 2013; Song and Kiledjian, 2007; Zhou et al., 2014) and facilitate decapping through PATL1 (Pat1p in yeast) (Marnef and Standart, 2010; Wilusz and Wilusz, 2013). Thus, a short U-tail may first be recognized by the LSM1–7 complex which in turn facilitates decapping (by the DCP1/2 complex) and subsequent 5′–3′ degradation (by XRN1).
We also investigated the contribution of the 3′–5′ decay pathway by depleting 3′ exonucleolytic factors. When we knocked-down RRP41, a core subunit of human exosome, we detected a substantial accumulation of uridylated mRNAs with short A-tails (Figure 6C). Combinatorial knockdown of RRP41 and XRN1 resulted in a more pronounced increase of uridylation (Figure 6C). Therefore, both decay pathways (5′–3′ and 3′–5′) may act at the downstream of uridylation. We also tested a 3′–5′ exonuclease DIS3L2 which is related to DIS3 and DIS3L. While DIS3 and DIS3L function as components of exosome, DIS3L2 is known to work independently from exosome (Lubas et al., 2013; Malecki et al., 2013). It was recently shown that DIS3L2 preferentially binds to long U-tails of pre-let-7 and is involved in turnover of pre-let-7 and some mRNAs in yeast and human (Chang et al., 2013; Faehnle et al., 2014; Lubas et al., 2013; Malecki et al., 2013; Ustianenko et al., 2013). Our TAIL-seq experiment shows that DIS3L2 depletion results in a modest accumulation of uridylated reads (Figure 6C). Thus, although we cannot rule out the possibility of indirect effects, our results suggest that multiple decay pathways may participate in the removal of uridylated mRNAs. Due to the technical limitation of knockdown experiment, it is currently unclear which pathway plays a dominant role.
Interestingly, mRNAs with an oligo-U-tail (U2 and U3+) responded more sensitively to the suppression of decay factors than those with a mono-U-tail (U1), suggesting that oligo-uridylated mRNAs are more rapidly degraded than mono-uridylated mRNAs (Figures 6A–6C). Taken together, we propose that oligo-uridylated mRNAs are subject to degradation by multiple factors, and an oligo-U-tail may serve as a decay mark for nonfunctional, deadenylated mRNAs.
Discussion
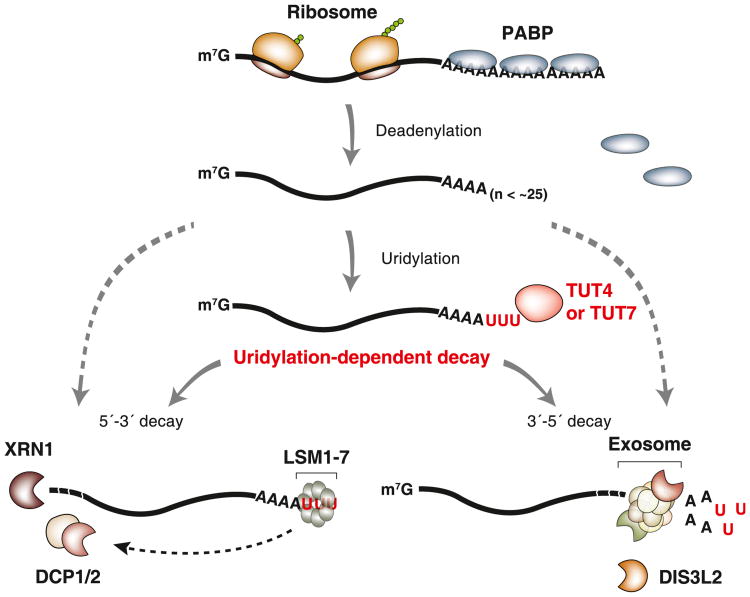
In conclusion, this study reveals an integral and general role of oligo-uridylation in mammalian mRNA decay (model shown in Figure 7). Upon deadenylation, mRNAs (with A-tails shorter than ∼25 nt) lose PABP and instead gain a U-tail by the redundant action of TUT4 and TUT7. The oligo-U-tail triggers decay by serving as a mark that is recognized by downstream decay factors. Thus, TUT4/7 function as the “writers” of the decay mark. It will be interesting in the future to identify the “readers” of the oligo-U-tail and to ask if this modification can be reversed by “erasers.” The LSM1-7 complex and DIS3L2 are likely candidates that recognize the oligo-U marks, but further investigations will be necessary to understand which factor(s) recognize the oligo-U-tails mainly, whether there is any additional factor(s) that binds to the oligo-U-tails, and what is the molecular basis of the specific recognition (Lee et al., 2014).
Figure 7. Model for Uridylation-Dependent mRNA Decay in Humans.
mRNA decay is generally initiated by deadenylation. PABP proteins are dissociated from mRNA as poly(A) tail becomes shorter (less than ∼25 nt). TUT4 and TUT7 act redundantly to uridylate mRNAs with a short A-tail. The U-tail is in turn recognized by the downstream decay factors (uridylation-dependent mRNA decay pathway). The LSM1–7 complex binds to the U-tail and facilitates decapping by the DCP1/2 complex. Decapped mRNAs are degraded by the 5′–3′ exonuclease XRN1. Alternatively, the U-tail is recognized by exosome or DIS3L2 that degrade mRNA exonucleolytically from the 3′ end. It is currently unclear if and what fraction of deadenylated mRNAs are degraded through uridylation-independent alternative pathways (indicated with gray dashed lines).
It is intriguing that TUT4/7 are capable of measuring poly(A) length (Figure 2). Poly(A) tail is unlikely to form a certain structure through base-pairing, so we do not yet understand how RNA with a poly(A) tail is discriminated by TUT4/7. It would be interesting to carry out structural studies on TUT4/7 and RNA with an A-tail of various length. Furthermore, we found that PABPC1 preferentially protects long poly(A) tails from uridylation (Figure 3). This specific inhibitory effect may come from the length-dependent binding of PABPC1 (Kühn and Pieler, 1996; Sachs et al., 1987). Thus, the combined action of TUT4/7 and PABP may selectively mark nonfunctional mRNAs while translationally active polyadenylated mRNAs are refractory to uridylation. Consequently, TUT4/7-mediated uridylation may provide the molecular basis for the tight control of mRNA stability according to poly(A) tail length.
We observed that oligo-uridylated mRNAs (with ≥2 uridines) are more sensitive than mono-uridylated mRNAs to the knockdown of TUT4/7 and decay factors. Moreover, oligo-U-tails are found in a narrow range of short A-tail length while mono-U-tails are more loosely distributed and found in polyadenylated mRNAs as well to some extent (Chang et al., 2014b). Thus, mono-uridylation appears to be less specific than oligo-uridylation and may be catalyzed in part by a TUT(s) other than TUT4/7. Furthermore, mono-U-tails may be too short to recruit decay factors effectively. Oligo-uridylated mRNAs are detected more frequently after depletion of decay factors, indicating that they are less stable than mono-uridylated mRNAs in control cells. Therefore, oligo-U-tails are likely to have a stronger effect in decay than mono-U-tails do. In fission yeast and plants, it is currently unclear if there is such a distinction between oligo-U-tails and mono-U-tails because only a small number of reads from cloning has been analyzed thus far.
Our transcriptome-wide analyses allowed us to propose a general model for the decay of poly(A)+ mRNAs. In addition, given that poly(A)− histone mRNA was also proposed to be uridylated by TUT4 (Schmidt et al., 2011; Su et al., 2013), it is possible that both poly(A)+ mRNAs and poly(A)− mRNAs are degraded by the same general principle involving uridylation although there may be some differences in details such as the choice of downstream decay factors. In fact, we detected uridylation on histone mRNAs and on trimmed decay intermediates lacking poly(A) tail and these U-tails were also dependent on TUT4/7 (data not shown).
In addition, we found that miR-1 transfection results in an increased uridylation and facilitated decay of its targets (Figure 5). These results suggest that uridylation contributes to miRNA-mediated gene silencing by removing the body of dead-enylated mRNAs. Uridylation may be involved in other decay and surveillance pathways in mammals, playing a general role. It is noted that we cannot currently assess if and to what extent uridylation-independent alternative pathway(s) contribute to bulk mRNA decay.
Tailing of mRNA is found in many eukaryotes, with some notable differences among the species. In filamentous fungus Aspergillus nidulans, mRNAs carry 3′ tails mixed with cytidine and uridine (Morozov et al., 2010). In a double deletion mutant of noncanonical PAPs, CutA and CutB, this “CUCU” modification was abrogated, and transcripts were stabilized, indicating that a CUCU tail also serves as a decay mark despite the difference in base composition (Morozov et al., 2010, 2012). In plants, although uridylation occurs similarly to mammals, mRNA half-life did not change in the urt1 mutants, and the reason underlying the difference is currently unclear (Sement et al., 2013). Another variation among the species is that uridylation occurs selectively on deadenylated mRNAs in mammals and plants whereas uridylation appears to be independent of poly(A) tail length in S. pombe and A. nidulans (Morozov et al., 2010; Rissland and Norbury, 2009). Deadenylation may not be a prerequisite for uridylation in fungi as they possess shorter poly(A) tail (20–30 nt in median) than mammals (60–100 nt in median) and plants (50–60 nt in median) (Chang et al., 2014b; Morozov et al., 2010; Subtelny et al., 2014). Thus, further investigations are clearly necessary to delineate the commonalities and differences of uridylation in diverse systems.
Tailing-mediated decay is deeply conserved and found even in prokaryotes where mRNAs typically end with stem loop structure and are degraded in an adenylation-dependent manner (Belasco, 2010; Houseley et al., 2006). An oligo-A-tail serves as a single-stranded toehold for 3′ exonucleases that are otherwise hindered by the terminal stem loop. A related phenomenon was observed in budding yeast where noncanonical PAPs, Trf4 and Trf5, adenylate defective nuclear RNAs and facilitate their degradation by exosome (Houseley et al., 2006; Norbury, 2013). Our current work shows that mammalian cytoplasmic mRNAs use uridylation, instead of adenylation, to promote mRNA decay. Together with previous findings (Morozov et al., 2010; Mullen and Marzluff, 2008; Rissland and Norbury, 2009), our study establishes a fundamental and conserved role for tailing in the mRNA decay pathways.
Experimental Procedures
Construction of TAIL-Seq Library
TAIL-seq was carried out as described previously (Chang et al., 2014b). Briefly, 25–50 μg of total RNA was extracted using TRIzol (Invitrogen), purified with RNeasy MinElute column (QIAGEN), and rRNA-depleted by using Ribo-Zero kit (Epicentre). The RNAs were ligated to the biotinylated 3′ adaptor and partially digested by RNase T1 (Ambion). The fragmented RNAs were precipitated with streptavidin beads, phosphorylated at the 5′ end, and gel purified (500–1,000 nt). The purified RNAs were ligated to the 5′ adaptor, reverse-transcribed, and amplified by PCR. The cDNA libraries were mixed with PhiX control library v3 (Illumina) and spike-in mixture and then sequenced by paired-end run (51 × 251 cycles) on Illumina MiSeq (small-scale TAIL-seq) or HiSeq 2500. Resulting data were processed as previously described (Chang et al., 2014b). See also Extended Experimental Procedures.
In Vitro Uridylation Assay
For immunoprecipitation of FLAG-TUTases, HEK293T cells grown on 10 cm dishes were collected 48 hr after transfection with FLAG-TUTase expression plasmids (full-length human TUT4 [1–1,640 aa] and human TUT7 [1–1,495 aa]). The cells were incubated in ice-cold Buffer D (200 mM KCl, 10 mM Tris-HCl [pH 8.0], 0.2 mM EDTA) for 20 min followed by sonication on ice and centrifugation twice for 15 min at 4° C. The supernatant was incubated with 5 μl of anti-FLAG antibody-conjugated agarose beads (anti-FLAG M2 affinity gel, Sigma) with constant rotation for 1 hr at 4° C. The beads were washed six times with Buffer D. Uridylation reaction was done in a total volume of 30 μl in 3.2 mM MgCl2, 1 mM DTT, 0.67 U/μl RNase inhibitor (Promega, N2515), 0.25 mM UTP, 0.45 nM of 5′ end-labeled RNA, and 15 μl of immunopurified proteins on beads or 3× Flag-peptide (Sigma) eluted proteins in Buffer D. When uridylation assay was done with recombinant TUT7 (951–1,495 aa), 14 nM of protein was used. The reaction mixture was incubated at 37° C for up to 10 min. For uridylation assay in the presence of PABPC1, 10–40 nM of recombinant human PABPC1 (Origene, TP307354) was preincubated with RNA for 10 min and then uridylation was carried out by adding 160 nM of recombinant TUT7 (951–1,495 aa). Buffer D with final 300 mM KCl was used when uridylation assay was carried out in the presence of PABPC1. The RNA was purified from the reaction mixture by phenol extraction and run on 6% polyacrylamide sequencing gel with 7 M urea (20 × 40 cm, 0.4 mm thick) at constant 1,500 V for 2 hr. The gel was exposed to phosphor imaging plate (Fujifilm) and read by Typhoon FLA 7000 (GE Healthcare). The signal intensity profile was quantified using MultiGauge v3.0 (Fujifilm). In Figure S2D, 12.5% polyacrylamide gel was used. The SHOC2 3′ UTR and CALM1 3′ UTR were selected as RNA substrates as they do not contain homopolymeric adenosines at the 3′ end. RNAs were synthesized by ST Pharm.
The list of RNA oligos is shown in Table S3.
Supplementary Material
Acknowledgments
We are grateful to members of our laboratory for discussion and technical help, especially Eunji Kim for help with plasmid cloning. We thank Dr. E. Izaurralde for insightful suggestions and the gifts of DCP1/2 mutant plasmids and Dr. G. Dreyfuss for the gift of anti-PABPC1 antibody. This work was supported by IBS-R008-D1 of Institute for Basic Science from the Ministry of Science, ICT and Future Planning of Korea (J.L., M.H., H.C., S.C.K., and V.N.K.) and the BK21 Research Fellowships from the Ministry of Education of Korea (J.L.).
Footnotes
Accession Numbers: The NCBI Gene Expression Omnibus (GEO) accession number for the sequenced reads reported in this paper is GSE59628.
Supplemental Information: Supplemental Information includes Extended Experimental Procedures, five figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2014.10.055.
Author Contributions: J.L., M.H., H.C., and V.N.K. designed experiments. J.L., M.H., and S.C.K. performed biochemical and cell biological experiments. H.C. carried out computational analyses. D.K.S. and D.J.P. prepared recombinant proteins. J.L., M.H., H.C., and V.N.K. wrote the manuscript.
References
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. DNA polymerase beta-like nucleotidyl-transferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer BW, Kornberg RD. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol. 1983;96:717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barckmann B, Simonelig M. Control of maternal mRNA stability in germ cells and early embryos. Biochim Biophys Acta. 2013;1829:714–724. doi: 10.1016/j.bbagrm.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Belasco JG. All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat Rev Mol Cell Biol. 2010;11:467–478. doi: 10.1038/nrm2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CT, Bercovich N, Loh B, Jonas S, Izaurralde E. The activation of the decapping enzyme DCP2 by DCP1 occurs on the EDC4 scaffold and involves a conserved loop in DCP1. Nucleic Acids Res. 2014a;42:5217–5233. doi: 10.1093/nar/gku129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, Kim VN. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell. 2014b;53:1044–1052. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus M, Régnier P. The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;111:611–613. doi: 10.1016/s0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- Eliseeva IA, Lyabin DN, Ovchinnikov LP. Poly(A)-binding proteins: structure, domain organization, and activity regulation. Biochemistry Mosc. 2013;78:1377–1391. doi: 10.1134/S0006297913130014. [DOI] [PubMed] [Google Scholar]
- Faehnle CR, Walleshauser J, Joshua-Tor L. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature. 2014;514:252–256. doi: 10.1038/nature13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, Kremmer E, Ansel KM, Heissmeyer V. Eri1 degrades the stem-loop of oligouridylated histone mRNAs toinduce replication-dependent decay. Nat Struct Mol Biol. 2013;20:73–81. doi: 10.1038/nsmb.2450. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Khanam T, Muddashetty RS, Kahvejian A, Sonenberg N, Brosius J. Poly(A)-binding protein binds to A-rich sequences via RNA-binding domains 1+2 and 3+4. RNA Biol. 2006;3:170–177. doi: 10.4161/rna.3.4.4075. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kühn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- Lapointe CP, Wickens M. The nucleic acid-binding domain and translational repression activity of a Xenopus terminal uridylyl transferase. J Biol Chem. 2013;288:20723–20733. doi: 10.1074/jbc.M113.455451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Kim B, Kim VN. Emerging roles of RNA modification: m(6) A and U-tail. Cell. 2014;158:980–987. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Liu X, Zheng Q, Vrettos N, Maragkakis M, Alexiou P, Gregory BD, Mourelatos Z. A MicroRNA precursor surveillance system in quality control of MicroRNA synthesis. Mol Cell. 2014;55:868–879. doi: 10.1016/j.molcel.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. Exonuclease hDIS3L2 specifies an exosome-independent 3′-5′ degradation pathway of human cytoplasmic mRNA. EMBO J. 2013;32:1855–1868. doi: 10.1038/emboj.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32:1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, Standart N. Pat1 proteins: a life in translation, translation repression and mRNA decay. Biochem Soc Trans. 2010;38:1602–1607. doi: 10.1042/BST0381602. [DOI] [PubMed] [Google Scholar]
- Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov IY, Jones MG, Razak AA, Rigden DJ, Caddick MX. CUCU modification of mRNA promotes decapping and transcript degradation in Aspergillus nidulans. Mol Cell Biol. 2010;30:460–469. doi: 10.1128/MCB.00997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov IY, Jones MG, Gould PD, Crome V, Wilson JB, Hall AJ, Rigden DJ, Caddick MX. mRNA 3′ tagging is induced by nonsense-mediated decay and promotes ribosome dissociation. Mol Cell Biol. 2012;32:2585–2595. doi: 10.1128/MCB.00316-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbury CJ. Cytoplasmic RNA: a case of the tail wagging the dog. Nat Rev Mol Cell Biol. 2013;14:643–653. doi: 10.1038/nrm3645. [DOI] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol. 2009;16:616–623. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MJ, West S, Norbury CJ. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA. 2011;17:39–44. doi: 10.1261/rna.2252511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sement FM, Ferrier E, Zuber H, Merret R, Alioua M, Deragon JM, Bousquet-Antonelli C, Lange H, Gagliardi D. Uridylation prevents 3′ trimming of oligoadenylated mRNAs. Nucleic Acids Res. 2013;41:7115–7127. doi: 10.1093/nar/gkt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif H, Conti E. Architecture of the Lsm1-7-Pat1 complex: a conserved assembly in eukaryotic mRNA turnover. Cell Reports. 2013;5:283–291. doi: 10.1016/j.celrep.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Slevin MK, Meaux S, Welch JD, Bigler R, Miliani de Marval PL, Su W, Rhoads RE, Prins JF, Marzluff WF. Deep sequencing shows multiple oligouridylations are required for 3′ to 5′ degradation of histone mRNAs on polyribosomes. Mol Cell. 2014;53:1020–1030. doi: 10.1016/j.molcel.2014.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, Kiledjian M. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Slepenkov SV, Slevin MK, Lyons SM, Ziemniak M, Kowalska J, Darzynkiewicz E, Jemielity J, Marzluff WF, Rhoads RE. mRNAs containing the histone 3′ stem-loop are degraded primarily by de-capping mediated by oligouridylation of the 3′ end. RNA. 2013;19:1–16. doi: 10.1261/rna.034470.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013;19:1632–1638. doi: 10.1261/rna.040055.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J. Lsm proteins and Hfq: Life at the 3′ end. RNA Biol. 2013;10:592–601. doi: 10.4161/rna.23695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhou Y, Hang J, Wan R, Lu G, Yan C, Shi Y. Crystal structure and biochemical analysis of the heptameric Lsm1-7 complex. Cell Res. 2014;24:497–500. doi: 10.1038/cr.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.