Abstract
Current proteomics technologies allow substantial depth of analysis of cellular and subcellular proteomes as shown in the proteomic profiling of ovarian cancer cells. This in-depth analysis has elucidated the repertoire of proteins expressed on the cell surface and proteins released into the extracellular milieu, uncovering extensive shedding of extracellular domains of cell adhesion proteins and a highly dynamic protein secretion process. The protein sets identified provide a rich resource of potential circulating markers and targets for imaging and therapeutics for ovarian cancer.
Depth of Proteomic Analysis of Ovarian Cancer Cells
The proteome is the most functional compartment encoded for in the genome and a major source of cancer diagnostics and therapeutic targets (1, 2). Given that tumor-derived proteins are present at higher concentrations closer to their source, analysis of tumor cells, tissues, and proximal fluids to identify differentially expressed proteins is being pursued to identify diagnostic and therapeutic targets. Mass spectrometry is currently the workhorse in proteomics allowing protein identifications in cellular and subcellular compartments over a wide dynamic range of protein abundance (1, 2).
Insight into the ovarian cancer cell proteome has been gained from recent studies (3–6). In one study, the repertoire of proteins expressed in three ovarian adenocarcinoma cell lines, OVCAR3, CaOV3, and ES2, as well as in ovarian cancer cells enriched from ascites fluid has been substantially elucidated (6). To this effect, separate analyses of proteins released into culture medium and of cell surface proteins were done, using total cell lysates as a reference. The three proteomes derived from each ovarian cancer cell population were further subjected to protein fractionation before high-resolution liquid chromatography coupled with tandem mass spectrometry LC-MS/MS analysis of individual fractions. Some 6,400 proteins were identified with high confidence in this study. Similar or complementary data have been published for ovarian cell lines and ovarian ascites fluid (3, 5) with substantial concordance, thus generally confirming findings and also confirming robustness of in-depth proteomic analysis.
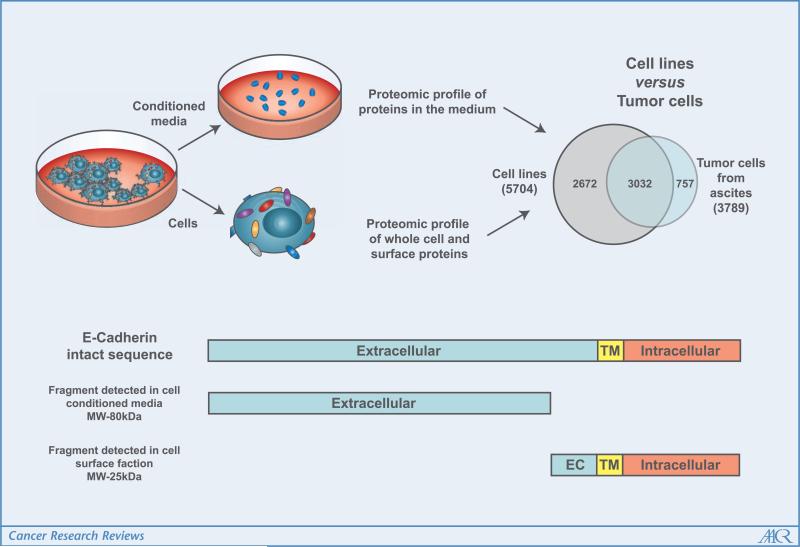
Interestingly, estimation of abundance of identified proteins based on spectral counts (7) allowed assessment of their enrichment in the subproteome compartments investigated. Substantial concordance was observed between subcellular location and predictions made from database searches and sequence analysis. Two ovarian cancer cell lines analyzed, OCVAR3 and CaOV3, were derived from human serous ovarian adenocarcinoma (8, 9), the same subtype as the ovarian cancer cells isolated from ascites fluid. An interesting finding is the close similarity of the proteomic profiles of cancer cells freshly obtained from ascites and those of ovarian cancer cell lines of the same subtype (Fig. 1).
Figure 1.
In-depth proteomic analysis of ovarian cancer cells. The overlap of protein identifications from cell lines and cancer cells obtained from patient ascites is significant, indicating good representation of ovarian cancer cells among cell lines. Analysis of proteins from the conditioned medium identified sequences spanning extracellular domains for transmembrane proteins. Sequences spanning intracellular domains were detected on the cell surface. These observations are consistent with a shedding process as indicated here for E-cadherin (CDH1).
Shedding of Surface Membrane Proteins into the Extracellular Compartment
The high mortality associated with ovarian cancer is due to tumor invasion and metastasis by the time a diagnosis is made (10, 11). Tumor metastasis has been associated with loss of cell-cell adhesion properties resulting in cell migration and tumor invasion. This process involves a wide array of signaling pathways that regulate, among others, the production of matrix-degrading proteases resulting in shedding of some key adhesion proteins (12, 13). Analysis of ovarian cancer cell–derived proteins in culture medium provided evidence for shedding of several hundred proteins annotated as membrane proteins. Related peptides identified were found to be largely derived from extracellular domains of cell surface proteins suggestive of a generalized process of cleavage and release of ectodomains. A large proportion of the shed proteins were found to be related to processes of cell adhesion and cell movement. Several integrins, cadherins, and ADAM protein family members were identified. This is illustrated for CDH1 (Fig. 1) for which peptide sequence coverage spans the extracellular domain in the culture medium, whereas peptide sequence coverage spans both intracellular and extracellular domains in the cell surface fraction. The shedding of CDH1 was recently described as a process promoted by matrix metalloproteinases (13). In contrast, some proteins were identified predominantly in the cell surface fraction, compared with the cell culture medium or to whole cell lysates, indicating that cleavage and shedding affect some proteins but not others. Included in the set of proteins resistant to cleavage are several receptors, such as integrins (ITGA6 and ITGAV) and tumor necrosis factor receptor superfamily member 10b. The interconnection of the mechanisms of loss of cell adherence by CDH1 down-regulation and/or shedding and metastasis promoted by integrins, which increases the adherence of cancer cells in the peritoneal cavity, has been recently shown (14). Additionally, CDH1 shedding and down-regulation plays an important role in the epithelial to mesenchymal transition process, which has recently been linked to ovarian cancer progression (15).
Relevance to Biomarker Discovery
Tumor cell proteins identified in culture medium or in proximal biological fluids represent a potential source of circulating markers. However, identification of tumor-derived proteins in proximal fluids is insufficient to assess whether such proteins may be detectable in circulation given the limits of sensitivity of available detection methods and uncertainty as to whether tumor-based production is sufficient to result in a measurable increment in levels in circulation (1).
Important features of secreted or shed proteins that may serve as biomarkers include a high secretion rate specifically by tumor cells and a low baseline concentration in normal plasma. A recent mathematical model has derived estimates of the balance between tumor biomarker secretion into and removal out of the intravascular compartment that take into account protein secretion rates by tumor cells (16). The model was primed based on data available for the ovarian marker CA 125 leading to estimates of the minimal tumor size required to detect elevated levels of CA125 in circulation in ovarian cancer. Applying this model to tissue inhibitor of metalloproteinase (TIMP) 1, the secretion rate of which was measured in CaOV3 and ES2 cell lines to be ~3 ng/million cells/hour (6), would lead to an estimate that a tumor ~2 cm in diameter would result to a detectable 50% increase in the level of TIMP1. Assumptions for the calculation are (a) a half-life in blood of 48 hours, which is significantly below that of CA-125 (151 hours); (b) 50% of the secreted protein reaching the blood. Delineation of a full repertoire of proteins secreted by ovarian cancer cells with measures of their secretion rates would allow prioritization of candidates based on the above variables.
Other proteins identified in the extracellular compartment of ovarian cancer cells whose corresponding genes are expressed at relatively high levels in ovarian cancer tissue relative to controls included WFDC2 (HE4), MUC16 (mucin 16, CA125), insulin-like growth factor-binding protein 3 (IGFBP3), midkine (MDK), vitamin k-dependent proteins (PROS1), and secretory leukoprotease inhibitor (SLPI) for which prior associations with ovarian cancer have been made. Numerous additional candidates have been identified that require further testing to confirm their relevance to ovarian cancer. Agrin (AGRN), cartilage oligomeric matrix protein (COMP), nidogen-2 (NID2), prostaglandin-h2 d-isomerase (PTGDS), and SDC4 (sydecan-4), among others, have few if any association with ovarian cancer in the literature (6).
A substantial enrichment of CDH1 (epithelial-cadherin) both on the cell surface and in culture medium was observed, suggesting potential utility of CDH1 as a blood biomarker (17). Numerous other soluble fragments of membrane proteins were found in the extracellular compartment, as in the case of mesothelin (MSLN), which has been proposed as a potential ovarian cancer biomarker (18). Additional proteins from the cadherin family of cell surface adhesion molecules were enriched in conditioned media, such as neural cadherin (CDH2), calsyntenin-1 (CLSTN1), alcadein β (CLSTN3), desmoglein-1 (DSG1), and desmoglein-2 (DSG2). Based on pathway analysis annotations, most of these membrane proteins are related to processes of cell adhesion and cell movement. Additionally, these proteins exhibit cadherin-type domains, suggestive of their shedding through the same mechanisms as CDH1. These shed proteins may have potential utility as blood-based biomarkers. Assays for the shed forms of these proteins has the potential to yield a means for diagnosing ovarian cancer. On the other hand, reduced representation of shed proteins on the cell surface and their occurrence in circulation diminishes their utility as imaging or therapeutic targets compared with stable, nonshed proteins. The list of proteins with no evidence of shedding thus detected exclusively in the cell surface that have potential as imaging or therapeutic targets included integrins α (ITGA6 and ITGAV), cell surface glycoprotein muc18, a protein associated with metastasis of melanoma and prostate cancer (19, 20), and several other receptors and adhesion molecules (6).
Conclusion
Challenges associated with the complexity of biological-fluid and tissue proteomes are gradually being overcome through advances in proteomic technologies. In-depth proteomic profiling of key compartments of ovarian cancer cells has yielded a rich data set of proteins that are secreted, shed, or remain attached to the cell surface. Several of these shed proteins represented cell adhesion molecules that play important roles in tumor adhesion and tumor metastasis. Deciphering the repertoire of secreted and shed proteins and of cell surface proteins that are resistant to shedding will yield biomarkers for diagnostics and targets for imaging and therapeutics.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–9. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 2.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract. 2008;5:588–99. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 3.Gortzak-Uzan L, Ignatchenko A, Evangelou AI, et al. A proteome resource of ovarian cancer ascites: integrated proteomic and bioinformatic analyses to identify putative biomarkers. J Proteome Res. 2008;7:339–51. doi: 10.1021/pr0703223. [DOI] [PubMed] [Google Scholar]
- 4.Kislinger T, Cox B, Kannan A, et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–86. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 5.Sodek KL, Evangelou AI, Ignatchenko A, et al. Identification of pathways associated with invasive behavior by ovarian cancer cells using multidimensional protein identification technology (MudPIT). Mol Biosyst. 2008;4:762–73. doi: 10.1039/b717542f. [DOI] [PubMed] [Google Scholar]
- 6.Faca VM, Ventura AP, Fitzgibbon MP, et al. Proteomic analysis of ovarian cancer cells reveals dynamic processes of protein secretion and shedding of extra-cellular domains. PLoS ONE. 2008;3:e2425. doi: 10.1371/journal.pone.0002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H, Sadygov RG, Yates JR., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Analyt Chem. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 8.Hamilton TC, Young RC, McKoy WM, et al. Characterization of a human ovarian carcinoma cell line (NIH:OVCAR-3) with androgen and estrogen receptors. Cancer Res. 1983;43:5379–89. [PubMed] [Google Scholar]
- 9.Karlan BY, Jones J, Slamon DJ, Lagasse LD. Glucocorticoids stabilize HER-2/neu messenger RNA in human epithelial ovarian carcinoma cells. Gynecol Oncol. 1994;53:70–7. doi: 10.1006/gyno.1994.1090. [DOI] [PubMed] [Google Scholar]
- 10.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 11.Naora H, Montell DJ. Ovarian cancer metastasis: integrating insights from disparate model organisms. Nat Rev. 2005;5:355–66. doi: 10.1038/nrc1611. [DOI] [PubMed] [Google Scholar]
- 12.Rosso O, Piazza T, Bongarzone I, et al. The ALCAM shedding by the metalloprotease ADAM17/TACE is involved in motility of ovarian carcinoma cells. Mol Cancer Res. 2007;5:1246–53. doi: 10.1158/1541-7786.MCR-07-0060. [DOI] [PubMed] [Google Scholar]
- 13.Symowicz J, Adley BP, Gleason KJ, et al. Engagement of collagen-binding integrins promotes matrix metal-loproteinase-9-dependent E-cadherin ectodomain shedding in ovarian carcinoma cells. Cancer Res. 2007;67:2030–9. doi: 10.1158/0008-5472.CAN-06-2808. [DOI] [PubMed] [Google Scholar]
- 14.Sawada K, Mitra AK, Radjabi AR, et al. Loss of E-cadherin promotes ovarian cancer metastasis via a 5-integrin, which is a therapeutic target. Cancer Res. 2008;68:2329–39. doi: 10.1158/0008-5472.CAN-07-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed N, Thompson EW, Quinn MA. Epithelialmesenchymal interconversions in normal ovarian surface epithelium and ovarian carcinomas: an exception to the norm. J Cell Physiol. 2007;213:581–8. doi: 10.1002/jcp.21240. [DOI] [PubMed] [Google Scholar]
- 16.Lutz AM, Willmann JK, Cochran FV, Ray P, Gambhir SS. Cancer screening: a mathematical model relating secreted blood biomarker levels to tumor sizes. PLoS Med. 2008;5:e170. doi: 10.1371/journal.pmed.0050170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Wever O, Derycke L, Hendrix A, et al. Soluble cadherins as cancer biomarkers. Clin Exp Metastasis. 2007;24:685–97. doi: 10.1007/s10585-007-9104-8. [DOI] [PubMed] [Google Scholar]
- 18.Scholler N, Fu N, Yang Y, et al. Soluble member(s) of the mesothelin/megakaryocyte potentiating factor family are detectable in sera from patients with ovarian carcinoma. Proc Natl Acad Sci U S A. 1999;96:11531–6. doi: 10.1073/pnas.96.20.11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nyormoi O, Bar-Eli M. Transcriptional regulation of metastasis-related genes in human melanoma. Clin Exp Metastasis. 2003;20:251–63. doi: 10.1023/a:1022991302172. [DOI] [PubMed] [Google Scholar]
- 20.Wu GJ, Fu P, Chiang CF, Huss WJ, Greenberg NM, Wu MW. Increased expression of MUC18 correlates with the metastatic progression of mouse prostate adenocarcinoma in the TRAMP model. J Urol. 2005;173:1778–83. doi: 10.1097/01.ju.0000154643.30048.2c. [DOI] [PubMed] [Google Scholar]