Abstract
Aristolochic acid (AA) is a potent dietary cytotoxin and carcinogen, and an established etiological agent underlying severe human nephropathies and associated upper urinary tract urothelial cancers, collectively designated aristolochic acid nephropathy (AAN). Its genome-wide mutational signature, marked by predominant A:T > T:A transversions occurring in the 5′-CpApG-3′ trinucleotide context and enriched on the nontranscribed gene strand, has been identified in human upper urinary tract urothelial carcinomas from East Asian patients and in experimental systems. Here we report a whole-exome sequencing screen performed on DNA from formalin-fixed, paraffin-embedded renal cell carcinomas (RCC) arising in chronic renal disease patients from a Balkan endemic nephropathy (EN) region. In the EN regions, the disease results from the consumption of bread made from wheat contaminated by seeds of Aristolochia clematitis, an AA-containing plant. In five of eight (62.5%) tested RCC tumor specimens, we observed the characteristic global mutational signature consistent with the mutagenic effects of AA. This signature was absent in the control RCC samples obtained from patients from a nonendemic, metropolitan region. By identifying a new tumor type associated with the AA-driven genome-wide mutagenic process in the context of renal disease, our results suggest new epidemiological and public health implications for the RCC incidence worldwide, particularly for the high-risk regions with unregulated use of AA-containing traditional herbal medicines.
Keywords: aristolochic acid, endemic nephropathy, renal cell carcinoma, whole-exome sequencing, mutational signature
Ingestion of nephrotoxic and carcinogenic aristolochic acid (AA) can lead to aristolochic acid nephropathy (AAN) marked by chronic tubulointerstitial nephropathy (CTN) and recurrent upper urinary tract urothelial cell carcinomas (UTUC), involving the renal pelvis and upper ureter. AAN has been designated a significant public health problem with millions of people world-wide being at risk.1–3 Endemic nephropathy (EN) is an environmental form of AAN affecting particular regions of several Balkan countries, and manifesting by increased rates of chronic kidney disease (CKD) and upper urinary tract urothelial carcinogenesis that have been causally linked to the intake of AA through consumption of home-made bread prepared from wheat grains contaminated with seeds of Aristolochia clematitis.4,5 Aristolactam-DNA adducts detected in renal cortex and/or A:T > T:A mutations in the 5′-CpApG-3′ context accumulating on the nontranscribed strand of the TP53 gene in CTN and UTUC were reported as biomarkers of AA exposure in this geographical region.4–6 Recent studies performed in Taiwanese patients with documented history of use of Aristolochia-containing traditional herbal medicine demonstrated that the A:T > T:A transversion, originally observed in the TP53 gene,7,8 is the predominant genome-wide mutation type in the UTUC.9,10 The detailed characteristics of this somatic alteration such as its predominance among other mutation types, gene strand orientation bias and sequence context are highly specific to the genotoxic effects of AA. AA is classified as Group 1 carcinogen by the World Health Organization-International Agency for Research on Cancer (WHO IARC) and its broader carcinogenic effects were demonstrated in animal models, by the induction of precancerous lesions and tumors in the forestomach, urinary tract and of fibrohistiocytic sarcomas at the AA injection site.11–13 A limited number of hepatocellular carcinoma (HCC) cases of East Asian origin studied for the etiological effects of hepatitis B virus manifested with the AA signature.10,14,15 The presence of the aristolactam-DNA adducts in the renal cortex has been reported previously in Taiwanese renal cancer patients7 and observed in rats in other target tissues including forestomach, liver, kidney, urinary tract,16 suggesting a wider tissue spectrum targeted by this highly potent mutagen. However, the association of AA with human malignancies other than UTUC and HCC remains largely unexplored.
In the last decade, a higher frequency of renal cell carcinomas (RCC) with distinct epidemiological and clinical features has been registered in the Croatian Centre for Endemic Nephropathy.17 We thus aimed to investigate a possible role of AA in the etiology of RCC among CKD patients from the EN regions and close vicinity, by analyzing the genome-wide mutation spectra in the tumor DNA.
Materials and Methods
Patient samples
Eight RCC patients from the farming villages were analyzed: five from an EN area previously associated with exposure to AA due to consumption of contaminated bread5,18 and three from villages close to the EN region with no EN cases reported in the past. In addition, two RCC cases from the metropolitan area of Croatia were analyzed as controls unlikely to have been exposed to AA. The clinical features of the patients are listed in Table 1. The study protocol included the patients’ informed consent and ethical approvals were obtained from the Ethical Boards of the School of Medicine, University of Zagreb, of the General Hospital in Slavonski Brod and from the IARC Ethics Committee. Of the eight EN RCC patients, we identified four (EN-01, EN-02, EN-04 and EN-05) who had been baking own bread, three of whom were farmers harvesting grain from locally grown wheat; one patient presented with CTN (EN-01), one with concurrent UTUC (EN-02) and one (EN-06) had been diagnosed with UTUC five years prior to the diagnosis of RCC (see Table 1).
Table 1.
Demographic and clinicopathological features of the studied RCC cases
| Case ID | Domicile | Gender | Age at surgery (years) |
RCC subtype |
ICD-O | Tumor grade |
Tumor stage |
Preoperative s-creatinine (µmol/L) |
CKD stage |
eGFR | Other kidney disease |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EN-01 | EN | M | 66 | Clear cell | 8310/3 | G4 | pT3NxMx | 710 | 5 | 7.0 | Yes1 |
| EN-02 | EN | M | 65 | Clear cell | 8310/3 | G2 | pT1N0Mx | 162 | 3 | 39.6 | UTUC2 |
| EN-03 | Near EN | M | 42 | Clear cell | 8310/3 | G2 | pT1N0Mx | 88 | 1 | 93.0 | No |
| EN-04 | EN | M | 78 | Clear cell | 8310/3 | G2 | pT1NxMx | 757 | 5 | 6.4 | No |
| EN-05 | Near EN | M | 55 | Clear cell | 8310/3 | G1 | pT1NxMx | 100 | 2 | 72.0 | Yes3 |
| EN-06 | EN | M | 76 | Unclassified | 8312/3 | G4 | pT3N2Mx | 364 | 4 | 15.1 | UTUC4 |
| EN-07 | Near EN | F | 64 | Chromophobe | 8317/3 | G3 | pT2NxMx | 75 | 2 | 73.0 | No |
| EN-08 | EN | F | 47 | Clear cell | 8310/3 | G2 | pT2N0Mx | 95 | 3 | 58.1 | No |
| Ctrl-01 | Non-EN | F | 56 | Clear cell | 8310/3 | G3 | pT2N0Mx | 62 | 1 | 91.8 | No |
| Ctrl-02 | Non-EN | M | 74 | Clear cell | 8310/3 | G2 | pT3aN0Mx | 193 | 3 | 31.5 | No |
Estimated glomerular filtration rate based on the Modification of Diet in Renal Disease (MDRD) equation and not measured by isotope dilution mass spectrometry (IDMS). The last column indicates the patient’s kidney disease other than RCC (UTUC: upper urinary tract urothelial carcinoma).
Chronic tubulointerstitial nephropathy (CTN).
Concurrent tumor.
Nephrolithiasis and RCC in the contralateral kidney.
UTUC diagnosed 5 years prior to RCC.
Abbreviations: EN: endemic region; near EN: close vicinity; F: female; M: male; ICD-O: the International Classification of Diseases for Oncology Code; CKD: chronic kidney disease; eGFR: estimated glomerular filtration rate.
DNA isolation
Hematoxylin-eosin preparations from the formalin-fixed paraffin- embedded (FFPE) tumor blocks were used to identify tumor tissue free of necrotic areas by digital scanning at 20× magnification (Leica SCN400 Scanner, Leica Biosystems). The tumor areas to be macro-dissected were measured using the ImageJ free software or SlidePath Gateway Client, Leica Biosystems. Ten µm sections prepared by Leica RM 2145 microtome (Leica Microsystems) were used to isolate genomic DNA (2–3 µg, yield 5–10 ng/mm2). Prior to genomic DNA isolation, slides were de-paraffinized for 5 min in 100% xylene, followed by 5 min in absolute ethanol, 5 min in 85% ethanol, 5 min in 75% ethanol and kept in milliQ water. DNA isolation was carried out using the QIAamp DNA FFPE Tissue kit (Qiagen). DNA yields and concentrations were measured using the Picogreen assay (Life Technologies) and Fluoroskan Ascent FL microplate fluorometer (Thermo Fisher Scientific). The purity was evaluated by the NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific). The integrity of genomic DNA was assessed by 0.8% agarose gel electrophoresis.
Library preparation and whole-exome sequencing (WES)
Two hundred and fifty ng of genomic DNA were sheared using the adaptive focused acoustics™ method (Covaris) to fragments of ~300 base-pair size on average, with water temperature of 4°C, one cycle at 175 Watt peak power, 10 duty factor and 200 cycles per burst. Resulting fragment size was assessed using the 2100 Bioanalyzer and the High Sensitivity DNA kit (Agilent Technologies). The sheared DNA went into library preparation using the KAPA LTP Library Preparation Kit (Kapa Biosystems). Briefly, the fragmented DNA was first subjected to end repair reaction followed by poly-A-tailing and adapter ligation. Excess adapters were removed by double solid-phase reversible immobilization clean-up using Agencourt AMPure XP beads (Beckman Coulter). Eight cycles of PCR were performed to amplify the libraries with correct adapter sequences on both ends. Next, exome capture was performed with pools of five libraries per hybridization (200 ng of each sample) using Nimblegen SeqCap EZ Exome v3.0 reagent. The exome-enriched populations were further amplified in a tencycle PCR amplification step. The post-enrichment libraries were pooled together to a final concentration of 6 pM in 420 µl. This volume was loaded on one lane of the rapid run mode flow cell for cluster generation on the HiSeq2500 (Illumina) and the samples were sequenced in paired-end 50 bp cycle run.
Sequencing data processing and analysis
Sequencing reads were aligned by Burrows-Wheeler Aligner, variants called by the Genome Analysis Toolkit (GATK, Broad Institute), annotated by ANNOVAR and filtered stringently to remove genetic variants observed in general population, using data from the public projects 1,000 genomes (1,000g, http://www.1000genomes.org/), Exome Sequencing Project (ESP, http://exome.gs.washington.edu/) and SNP database build 137 (dbSNP, http://www.ncbi.nlm.nih.gov/SNP/). Variants exhibiting a frequency higher than 0.1% in either 1,000g or ESP databases, annotated in dbSNP database and an in-house panel of germ line variants generated using data obtained from 560 cases from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) were removed, as were the variants in fragments mapping to repetitive sequences of the genome with a homology higher than 90%. The complete final list of 4,031 SBS variants in the tested as well as control RCC samples will be provided upon request. The raw sequencing data (fastQ and alignment (.bam) files) have been deposited in the Sequence Read Archive (SRA, access ID SRP049084) of the National Center for Biotechnology Information (NCBI) and can be obtained through the NCBI’s database for Genotypes and Phenotypes (dbGaP) authorized access system.
Determination of the AA mutational signature
The genome-wide AA signature in UTUC tumors had been defined previously based on varying criteria, e.g., ≥40 SBS per sample, increased A:T > T:A presence (20–80% of all SBS types) in the predominant 5′-CpApG-3′ context, and 1.25- to 2-fold bias for A:T > T:A accumulation on the nontranscribed strand.9,10 In the sequencing data presented here we first followed analogous criteria, considering ≥15% of A:T > T:A per tumor (10.5% is the maximum frequency seen in the COSMIC database upon excluding the HCC class containing a small number of potentially AA signature-positive tumors found by previous studies14,19) and the concurrent predominance of the 5′-CpApG-3′ sequence context. For strand bias analysis, we applied additional stringency by calculating its statistical significance using Pearson χ2 test (prop.test function available in the stats R package). For each sample the test calculated a p-value as the probability that the proportion of SBS in the nontranscribed strand is equal to 0.5, as expected by chance. As multiple conditions were assessed in parallel, a false discovery rate (FDR) correction was applied using the p.adjust function from the stats R package. The identified signature based on these criteria was visualized by customized R functions. To extract comprehensive gene signatures from all ten RCC samples and to validate the presence of the AA signature by an independent method, non-negative matrix factorization (NMF) was performed under optimized conditions using an R package.20 The input matrix contained one column per sample and the rows contained the frequency of the six possible mutation types within a two-flank sequence context (e.g., A > T in the C_G context et cetera). The context distribution was normalized to reflect the trinucleotide frequencies occurring in the portion of the human genome corresponding to the exome capture reagent coverage. The similarity between the NMF RCC signatures and published signatures from cancer and experimental settings was further evaluated by each signature represented as a vector in a 96-dimensional space. The tangent of the angle between each pair of vectors was taken as the distance metric. This distance was used to compute the grid of distance from each of the two RCC signatures to each of the 24 reference signatures and converted for graphical presentation to a similarity matrix by taking the negative logarithm of the distance.
Results
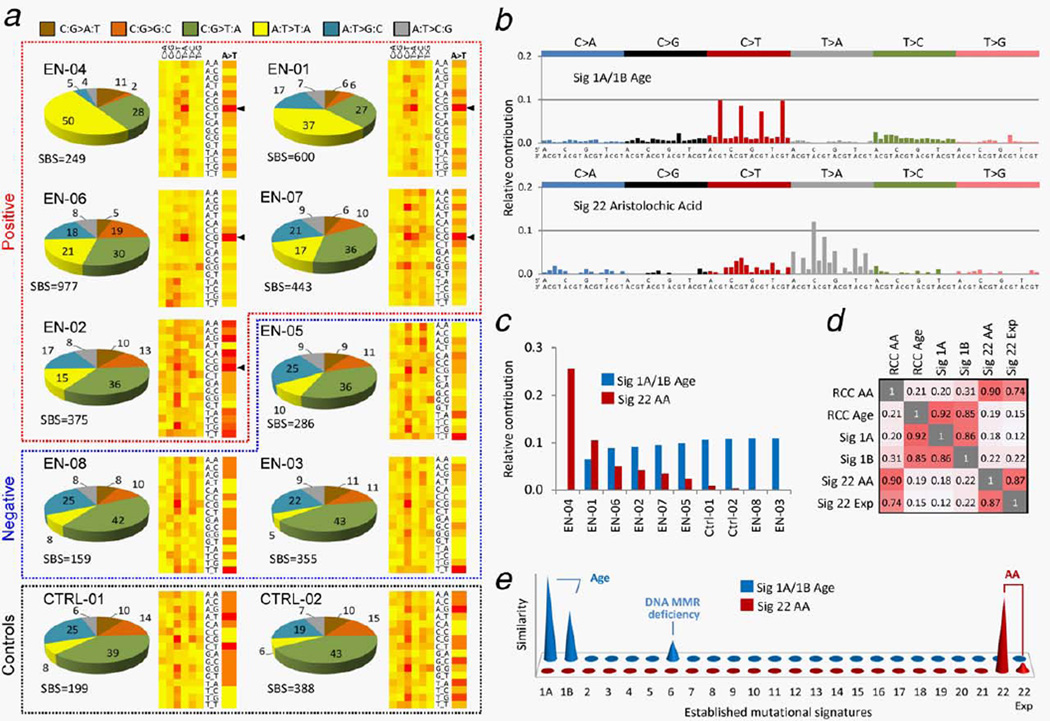
The WES analysis of DNA macrodissected from FFPE tumor specimens revealed the AA signature in five of eight (62.5%) RCC samples of patients from the EN regions. These findings were based on increased A:T > T:A transversion rates (1.2–3.5 SBS/Mbp, A:T > T:A frequencies of 17–50% and strand orientation bias ratios of 1.6–2.7) in four tested EN RCC samples (EN-01, EN-04, EN-06 and EN-07; Fig. 1a and Table 2). Interestingly, the EN-07 case was a resident of a village not considered an EN village. This observation is in line with recent evidence suggesting that ingestion of AA via contaminated bread was more widespread than previously thought, and cases of EN could be identified in villages and regions outside the established EN areas.5 One EN RCC case (EN-02) showed lower presence of A:T > T:A (15.2%, 0.9 per Mbp) but a significant strand orientation bias of 2.8, and also enrichment for the 5′-CpApG-3′ sequence context for the A:T > T:A transversions (Table 2). It was thus considered borderline positive considering the AA signature features previously described in Taiwanese UTUC patients9,10 and in experimental model systems.10,21 In contrast, the two non-EN RCC controls exhibited low A:T > T:A frequencies (Ctrl-01: 7.5% and Ctrl-02: 6.4%), as did the remaining two RCC cases from villages near to the EN area (EN-03 4.8%, EN-05 9.8%) and also one negative EN RCC case (EN-08, 8.2%) (see Table 2). In order to position these findings in a broader context, we analyzed 518 RCC samples unlikely to be exposed to AA, available from The Cancer Genome Atlas (TCGA). The 62.5% AA signature positivity rate in the samples of the EN region provenance appeared significant (p < 2.2e–16, using two-sample χ2 distribution test for equality of proportions with continuity correction) as the TCGA set contained only one sample meeting all the parameters of the AA signature (i.e., simultaneous occurrence of ≥50 SBS and ≥15% A > T, ≥20% 5′-CpApG-3′ context and >1.5 strand bias ratios with significant p values and FDR corrected q values, see Table 2), and it contained another six samples with elevated A:T > T:A and strand bias (with nonsignificant q values) but lacking the expected predominant 5′-CpApG-3′ sequence context for the A:T > T:A transition.
Figure 1.
Summary of WES results for the AA signature-positive RCC cases and RCC controls. (a) Pie charts show distribution of mutation types (color-code shown on top), as percentage of all single base substitution (SBS) counts in each sample. Groups of AA signature-positive, negative samples and controls are demarcated by dotted lines. Two-dimensional heat-maps indicate the relative distribution of 96 possibilities of six mutation type categories in a nonstranded view (columns, the labels C > A, C > G, etc. represent C:G > A:T, C:G > G:C, etc.) and their trinucleotide context (rows). A detailed view of the relative distribution of the A:T > T:A context only is shown as a single column to the right under the A > T header. The most frequent 5′-CpApG-3′ context (labeled C_G) for the A:T > T:A transversion in the positive samples is marked by black arrowheads. Heat-map color code: Red-to-yellow = high-to-low context frequency. (b) Comprehensive identification of the mutational signatures in all 10 studied samples, using NMF. X-axis: 96 trinucleotide contexts; Y-axis: normalized values of each column represent the contribution of each mutation type in a particular sequence context to each signature, expressed as the value of a given factor obtained from the NMF decomposition process. (c) Relative contribution of the mutational load identified in individual samples to the NMF signatures shown in (b), expressed as the given factor value obtained from the decomposition process. See Table 2 for contributions expressed as mutation counts. (d) Correlation matrix (1 equals 100% similarity) of RCC NMF signatures (RCC AA and RCC Age) with previously validated signatures 1A and 1B (age),22 with NMF-processed AA signatures in UTUC (Sig 22 AA)9 and in AA-exposed clonally immortalized mouse embryonic fibroblasts (Sig 22 Exp).21 (e) Graphical representation of the similarity distance of the RCC signatures (front-back axis) to 23 human cancer signatures (horizontal axis, 1A through 229,22) and to an AA signature established experimentally in cultured cells (22 Exp).21 Y-axis: similarity of signatures between the two systems, expressed as negative log(tan(angle)), see Materials and Methods. Negative values below the x–z plane correspond to angles >45° representing dissimilarity, and are thus suppressed.
Table 2.
Summary of the SBS mutation data and results of the AA signature analysis
| Case ID | Total SBS |
SBS per Mbp |
A > T SBS |
A > T per Mbp |
% A > T | % CAG context |
SB A > T | SB p value |
SB FDR q value |
Contribution to Sig 1A/1B (Age) |
Contribution to Sig 22 (AA) |
AA signature |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EN-01 | 600 | 9.4 | 224 | 3.5 | 37.3 | 23.0 | 1.6 (128/78) | 0.00064 | 0.010 | 228 (38%) | 372 (62%) | Yes |
| EN-02 | 375 | 5.9 | 57 | 0.9 | 15.2 | 11.4 | 2.8 (39/14) | 0.00098 | 0.013 | 256 (68%) | 119 (32%) | Yes1 |
| EN-03 | 355 | 5.5 | 17 | 0.3 | 4.8 | 7.0 | 2.5 (10/4) | 0.181 | 0.746 | 355 (100%) | 0 (0%) | – |
| EN-04 | 249 | 3.9 | 124 | 1.9 | 49.8 | 22.3 | 2.7 (82/30) | 0 | 0 | 0 (0%) | 249 (100%) | Yes |
| EN-05 | 286 | 4.5 | 28 | 0.4 | 9.8 | 8.2 | 1.3 (14/11) | 0.689 | 1 | 231 (81%) | 55 (19%) | – |
| EN-06 | 977 | 15.3 | 201 | 3.1 | 20.6 | 35.0 | 2.6 (140/53) | 0 | 0 | 623 (64%) | 354 (36%) | Yes |
| EN-07 | 443 | 6.9 | 76 | 1.2 | 17.2 | 23.7 | 1.8 (46/26) | 0.025 | 0.231 | 325 (73%) | 118 (27%) | Yes |
| EN-08 | 159 | 2.5 | 13 | 0.2 | 8.2 | 0.0 | 1.8 (7/4) | 0.546 | 1 | 159 (100%) | 0 (0%) | – |
| Ctrl-01 | 199 | 3.1 | 15 | 0.2 | 7.5 | 0.0 | 0.6 (5/8) | 0.579 | 1 | 185 (93%) | 14 (7%) | – |
| Ctrl-02 | 388 | 6.1 | 25 | 0.4 | 6.4 | 7.5 | 0.9 (12/13) | 1 | 1 | 376 (97%) | 12 (3%) | – |
A > T: A:T > T:A transversion; SB A > T: strand bias expressed as the ratio of A:T > T:A transversions on the nontranscribed versus transcribed strand (the number of respective transversions is shown in brackets). % CAG context: percentage of A:T > T:A transversions in the most frequent context reported for the AA signature. SB p value and false discovery rate (FDR) q value = measures of significance of the strand bias, see Materials and Methods. Contribution to Sig (Sig = non-negative matrix factorization-determined mutational signature) is shown as the number of SBS and the corresponding percentage (in parentheses) of the total SBS number per sample. AA signature—sample positivity for AA signature considering the simultaneous occurrence of ≥50 SBS and ≥15% A > T, ≥20% CAG context and its significance (SB p and/or q value) and mutation load contribution to Sig 22 (AA).
Lower percentage of A > T and CAG context but supported by NMF.
Abbreviation: SBS: single base substitutions.
To further strengthen the AA signature determination criteria and to identify additional mutational patterns reflecting effects of other etiological agents, we applied NMF, a mixed-pattern decomposition method used previously to successfully extract mutational signatures from human cancers and to pinpoint etiological factors.22 NMF established that samples EN-01, EN-02, EN-04, EN-06 and EN-07 were positive for the AA signature (NMF Signature 22 described previously in human urothelial and liver tumors23,24 and in immortalized clones arising from cultured, AA-treated primary embryonic fibroblasts21), and that all tumors except for EN-04 harbored high contents of C:G > T:A transitions within the 5′-XpCpG-3′ context (Figs. 1b and 1c). This observation is consistent with the age-specific signature 1A/1B22 reflecting the generally advanced patients’ age at the time of surgery (see Table 1). The NMF approach thus unequivocally grouped the borderline RCC sample EN-02 with the four AA signature-specific samples confirming the presence of the canonical AA signature in this tumor and providing additional information on sample-specific mutational load toward each signature (Fig. 1c and Table 2). The samples EN-03, EN-05, EN-08 and the two non-EN controls (Ctrl-01 and Ctrl-02) were negative for the AA signature but exhibited the signature of age (Figs. 1b and 1c). No additional markedly distinct signatures apart from the two shown in Figure 1b were found by the NMF approach when the number of expected signatures was increased. Next, we performed similarity analysis matching the two NMF signatures found in the tested RCC set (Fig. 1b) against the previously published data on known mutational signatures in human cancers22,23 including AA-associated UTUC tumors,9 as well as against the AA signature induced experimentally in cultured cells.21 While the EN RCC age signature matched the published the Signatures 1A, 1B, and to a lesser extent the Signature 6 (attributed to DNA mismatch repair deficiency),22 the RCC NMF AA signature matched the AA signatures previously identified both in the UTUC samples from Taiwanese patients and in cultured cells harboring an experimentally generated AA signature. The results are summarized graphically in Figures 1d and 1e.
Discussion
Our study identifies the specific AA mutational signature in a set of five RCC tumors from patients from a Balkan EN region and near vicinity, providing molecular evidence for AA-driven mutagenic process associated with carcinogenesis in the renal cortex and implicating AA in the etiology of this cancer type in the context of CKD. While the mutation load in the AA signature-positive EN RCC confirms the highly mutagenic effects of AA,9,10 it appears lower (4–15 SBS/Mbp) in our RCC set in comparison to the previously reported AA signature-positive UTUC (2–659 and 8–3510 SBS/Mbp). These differences warrant further epidemiological and laboratory investigations using larger patient sets, in order to establish whether they reflect varying AA exposure modes in each geographical region, the metabolizing rates in the distinct target cell types, the sampling procedures affecting the heterogeneity of tested tissues or other factors.
Given the unregulated global market with AA-containing herbs that might be putting millions of people at risk, particularly in Eastern Asia,3 our finding has potentially profound implications for increased awareness of additional cancer types associated with AAN. Robust designs of molecular epidemiology studies of AAN should now include RCC cases alongside patients with UTUC and HCC, to improve the worldwide surveillance of the disease. Furthermore, we document in a new region and a new cancer type that sensitive genome-wide screens coupled with sophisticated mutation pattern decomposition methods such as NMF, can identify AA as an environmental cancer risk factor in AAN and provide a robust evidence-base for diagnostic and preventive approaches toward eradication of this serious public health problem.3,25
What’s new?
Ingestion of aristolochic acid (AA) causes severe nephropathies and carcinomas of the upper urinary tract, and represents a significant public health problem with millions of people at risk worldwide. In this study of renal disease patients in an endemic region, the authors identified a previously unrecognized type of renal cell carcinoma that harbors the mutational signature of this potent carcinogen. Their findings suggest that the putative causal role of AA in renal cortex carcinogenesis should be broadly addressed in high-risk regions marked by inadvertent exposure to AA or widespread use of AA-containing herbal remedies.
Acknowledgements
We thank Drs. Želimir Stipančić and Tvrtko Hudolin for assistance with collection of clinical samples and data, Dr. Stephanie Villar and Ms. Christine Carreira for their assistance with archived tumor sample processing, and Dr. Magali Olivier for helpful comments on the sequencing data analysis. We thank the New York University Genome Technology Center at the New York University Laura and Isaac Perlmutter Cancer Center, for expert assistance with the Illumina HiSeq2500 sequencing experiments.
Grant sponsor: Croatian Science Foundation; Grant number: 04-38; Grant sponsor: NIH/NCI, partial support to the Genome Technology Center at the Laura and Isaac Perlmutter Cancer Center, New York University; Grant number: NIH/NCI P30CA016087; Other Support: IARC Regular Budget
Abbreviations
- AA
aristolochic acid
- AAN
aristolochic acid nephropathy
- bp
base-pair
- CKD
chronic kidney disease
- CTN
chronic tubulointerstitial nephropathy
- eGFR
estimated glomerular filtration rate
- EN
endemic nephropathy
- FFPE
formalin-fixed, paraffin-embedded
- HCC
hepatocellular carcinoma
- IARC
International Agency for Research on Cancer
- IDMS
isotope dilution mass spectrometry
- Mbp
megabase-pairs
- MDRD
Modification of Diet in Renal Disease
- NMF
Non-negative Matrix Factorization
- PCR
polymerase chain reaction
- RCC
renal cell carcinoma
- SBS
single base substitution
- TCGA
The Cancer Genome Atlas
- UTUC
upper urinary tract urothelial cell carcinomas
- WES
whole-exome sequencing
- WHO
World Health Organization
References
- 1.Debelle FD, Vanherweghem JL, Nortier JL. Aristolochic acid nephropathy: a worldwide problem. Kidney Int. 2008;74:158–169. doi: 10.1038/ki.2008.129. [DOI] [PubMed] [Google Scholar]
- 2.Gokmen MR, Cosyns JP, Arlt VM, et al. The epidemiology, diagnosis, and management of aristolochic acid nephropathy: a narrative review. Ann Intern Med. 2013;158:469–477. doi: 10.7326/0003-4819-158-6-201303190-00006. [DOI] [PubMed] [Google Scholar]
- 3.Grollman AP. Aristolochic acid nephropathy: Harbinger of a global iatrogenic disease. Environ Mol Mutagen. 2013;54:1–7. doi: 10.1002/em.21756. [DOI] [PubMed] [Google Scholar]
- 4.Grollman AP, Shibutani S, Moriya M, et al. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci USA. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jelakovic B, Karanovic S, Vukovic-Lela I, et al. Aristolactam-DNA adducts are a biomarker of environmental exposure to aristolochic acid. Kidney Int. 2012;81:559–567. doi: 10.1038/ki.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moriya M, Slade N, Brdar B, et al. TP53 Mutational signature for aristolochic acid: an environmental carcinogen. Int J Cancer. 2011;129:1532–1536. doi: 10.1002/ijc.26077. [DOI] [PubMed] [Google Scholar]
- 7.Chen CH, Dickman KG, Moriya M, et al. Aristolochic acid-associated urothelial cancer in Taiwan. Proc Natl Acad Sci USA. 2012;109:8241–8246. doi: 10.1073/pnas.1119920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollstein M, Moriya M, Grollman AP, et al. Analysis of TP53 mutation spectra reveals the fingerprint of the potent environmental carcinogen, aristolochic acid. Mutat Res. 2013;753:41–49. doi: 10.1016/j.mrrev.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoang ML, Chen CH, Sidorenko VS, et al. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci Transl Med. 2013;5:197ra02. doi: 10.1126/scitranslmed.3006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poon SL, Pang ST, McPherson JR, et al. Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci Transl Med. 2013;5:197ra01. doi: 10.1126/scitranslmed.3006086. [DOI] [PubMed] [Google Scholar]
- 11.Cosyns JP, Goebbels RM, Liberton V, et al. Chinese herbs nephropathy-associated slimming regimen induces tumours in the forestomach but no interstitial nephropathy in rats. Arch Toxicol. 1998;72:738–743. doi: 10.1007/s002040050568. [DOI] [PubMed] [Google Scholar]
- 12.Cosyns JP, Dehoux JP, Guiot Y, et al. Chronic aristolochic acid toxicity in rabbits: a model of Chinese herbs nephropathy? Kidney Int. 2001;59:2164–2173. doi: 10.1046/j.1523-1755.2001.00731.x. [DOI] [PubMed] [Google Scholar]
- 13.Debelle FD, Nortier JL, De Prez EG, et al. Aristolochic acids induce chronic renal failure with interstitial fibrosis in salt-depleted rats. J Am Soc Nephrol: JASN. 2002;13:431–436. doi: 10.1681/ASN.V132431. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, Deng Q, Wang Q, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–1121. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 15.Sung WK, Zheng H, Li S, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 16.Schmeiser HH, Schoepe KB, Wiessler M. DNA adduct formation of aristolochic acid I and II in vitro and in vivo. Carcinogenesis. 1988;9:297–303. doi: 10.1093/carcin/9.2.297. [DOI] [PubMed] [Google Scholar]
- 17.Belicza M, Demirovic A, Tomic K, et al. Comparison of occurrence of upper urinary tract carcinomas in the region with endemic villages and nonendemic nephropathy region in Croatia. Coll Antropol. 2008;32:1203–1207. [PubMed] [Google Scholar]
- 18.Hranjec T, Kovac A, Kos J, et al. Endemic nephropathy: the case for chronic poisoning by aristolochia. Croat Med J. 2005;46:116–125. [PubMed] [Google Scholar]
- 19.Kan Z, Zheng H, Liu X, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422–1433. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olivier M, Weninger A, Ardin M, et al. Modelling mutational landscapes of human cancers in vitro. Sci Rep. 2014;4:4482. doi: 10.1038/srep04482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poon SL, McPherson JR, Tan P, et al. Mutation signatures of carcinogen exposure: genome-wide detection and new opportunities for cancer prevention. Genome Med. 2014;6:24. doi: 10.1186/gm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gokmen MR, Lord GM. Aristolochic acid nephropathy. BMJ. 2012;344:e4000. doi: 10.1136/bmj.e4000. [DOI] [PubMed] [Google Scholar]