Abstract
A simple and straightforward technique for coating microplate wells with molecularly imprinted polymer nanoparticles (nanoMIPs) to develop ELISA type assays is presented here for the first time. NanoMIPs were synthesized by a solid phase approach with immobilized vancomycin (template) and characterized using Biacore 3000, dynamic light scattering and electron microscopy. Immobilization, blocking and washing conditions were optimized in microplate format. The detection of vancomycin was achieved in competitive binding experiments with a HRP-vancomycin conjugate. The assay was capable of measuring vancomycin in buffer and in blood plasma within the range 0.001-70 nM with a detection limit of 0.0025 nM (2.5 pM). The sensitivity of the assay was three orders of magnitude better than a previously described ELISA based on antibodies. In these experiments nanoMIPs have shown high affinity and minimal interference from blood plasma components. Immobilized nanoMIPs were stored for 1 month at room temperature without any detrimental effects to their binding properties. The high affinity of nanoMIPs and the lack of a requirement for cold chain logistics make them an attractive alternative to traditional antibodies used in ELISA.
INTRODUCTION
Immunoassays are routinely used in the clinical, environmental, agricultural/food and forensic industries for the analysis of proteins, hormones, viruses, microorganisms, DNA sequences and drugs.1,2 The enzyme-linked immunosorbent assay (ELISA) is probably the most commonly used method. In this format competition between the free analyte and an enzyme-labeled conjugate for binding to immobilized antibodies is used for quantitative determination of the analyte. The enzyme label reveals how much displacement has occurred by a colorimetric reaction, amplified by multiple turnovers of the enzymatic reaction.3 Immunoassays are rapid, sensitive and selective to the analyte of interest and are generally cost effective for large sample loads. However, as with any technology there are disadvantages; for example, the stability of reagents, the need for refrigerated transport and storage, batch to batch (or clone to clone) variability and the high cost of producing antibodies are often cited as problems.
In this regard molecularly imprinted polymers (MIPs) have already been identified as stable mimics of receptors or enzymes, suitable for use as substitutes for natural receptor molecules in assays or sensors.4-6 Their inherent stability, low cost, short development time and ease of preparation offer several major advantages over antibodies. MIPs however, are perceived to have several shortcomings. Among these are a heterogeneous distribution of binding sites, which is responsible for high levels of non-specific binding and the complex procedures required for their immobilization at surfaces. In particular, the absence of a reproducible method for coating microplate wells with MIPs restricts their application in assays where this format is preferred. Recently several examples of the application of MIPs to microplate-based assays have been described.7-15 Only a few of these examples however actually involved the application of MIPs to enzyme-linked assays for quantitative detection of the template.7-10 In the first of these, the surfaces of microplate wells were modified with a homopolymer of 3-aminophenylboronic acid, which was imprinted with epinephrine. The MIP-coated microplate was used successfully in an enzyme-linked assay for the detection of epinephrine at micromolar concentrations. That there are so few examples of MIP-based microplate assays can be due to several reasons: firstly the MIPs used in these assays resemble polyclonal antibodies, giving rise to high levels of non-specific binding. Secondly, their manufacture relies on manual, labor-intensive methods of synthesis. Thirdly, the immobilization protocols are often complex, affecting the reproducibility of their synthesis and hence the potential for a high degree of variability between measurements. Lastly, the developed MIP-based assays were not generic and required substantial modification to the analytical procedures traditionally used in ELISA. With the aim of resolving some of these problems, we recently developed a method for the solid-phase synthesis of MIP nanoparticles with pseudo-monoclonal binding properties.16 The MIP nanoparticles synthesized in a computer-controlled reactor were soluble in water and in organic solvents, and had uniform binding sites and high affinity to a range of targets used as the template. The main advantage of materials prepared in this manner is the possibility to directly replace antibodies with MIPs in standard ELISA-like assays with minimal modification of the immobilization and assay protocol. To demonstrate this potential we selected vancomycin as the target analyte. Vancomycin is a glycopeptide antibiotic derived from Amycolatopsis orientalis that acts by inhibiting cell wall biosynthesis and altering the permeability of the bacterial cell membrane. It has been used for the treatment of various serious gram-positive infections such as methicillin-resistant Staphylococcus aureus. Vancomycin is a very powerful antibiotic, which in high doses can be toxic to the ears and kidneys; whilst at low doses can cause hypersensitivity reactions. Thus, accurate measurement of vancomycin concentration in blood is advisable for the control of its administration to patients. The recommended target concentrations in blood achievable by administering vancomycin are 12-18 mg L−1, which can be measured with a Beckman Coulter Synchron competitive turbidimetric immunoassay.17,18 This measurement can however be affected by particle aggregation occurring via non-specific mechanisms (interactions with paraproteins), which can lead to an underestimate of the concentration in plasma.18 More accurate measurements can be achieved using ELISA, since this does not depend on particle aggregation. There are however very few examples of ELISA for vancomycin19 which justifies the importance of the development of novel assays.
MATERIALS AND METHODS
Materials
Vancomycin, amoxicillin, gentamicin, bleomycin, acrylic acid (AA), N-isopropylacrylamide (NIPAm) N,N′-methylene-bis-acrylamide (BIS), N-tert-butylacrylamide (TBAm), ammonium persulfate (APS), tetramethylethylenediamine (TEMED), 3-aminopropyltrimethyloxysilane (APTMS), sodium hydroxide (NaOH), glutaraldehyde (GA), bovine serum albumin (BSA), horseradish peroxidase (HRP), 3,3′,5,5′-tetramethylbenzidine, used in the form of TMB liquid substrate system for ELISA (Sigma, UK, catalogue number T0440), Tween-20, sodium dodecyl sulfate (SDS), (2-[morpholino]ethanesulfonic acid) (MES), N-hydroxysuccinimide (NHS), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) and acetone were from Sigma-Aldrich, UK. Phosphate buffered saline (PBS) was prepared, as specified, from PBS buffer tablets (Sigma-Aldrich, UK) and consisted of phosphate buffer (0.01 M), potassium chloride (0.0027 M) and sodium chloride (0.137 M), pH 7.4. Where PBS with pH 7.2 was used, the pH was adjusted with the addition of HCl. Double-distilled ultrapure water (Millipore, UK) was used for the experiments. All chemicals and solvents were of analytical or HPLC grade and were used without further purification. Microplates used were Nunclon 96 microwell plates (Thermo Scientific, UK).
Synthesis of MIP nanoparticles (nanoMIPs) imprinted with vancomycin
Preparation of vancomycin-derivatized glass beads
The protocol for the immobilization of vancomycin on glass beads has already been described elsewhere.16-21 Briefly glass beads were activated by boiling them in NaOH for 10 min and washed with double-distilled water followed by acetone and then dried. The beads were then incubated overnight in a solution of APTMS in toluene, washed with acetone and subsequently incubated overnight at 4 °C in a solution of GA in PBS pH 7.2. The surface immobilization of vancomycin was performed by incubating the beads with a solution of the antibiotic (5 mg mL−1) in PBS, pH 7.2, overnight at 4 °C. This method yields ca. 0.26 ligand molecules nm−2 of glass bead surface.16 Finally, the glass beads were washed with double-distilled water, dried under vacuum and stored at 4 °C until used.
Automated synthesis of vancomycin MIP nanoparticles (nanoMIPs)
An upgraded automated reactor as compared to the one used previously,16 was utilized for the synthesis of the nanoparticles. The reactor, manufactured by HEL Ltd., (Borehamwood, UK), was designed such that both photochemically- and chemically-initiated polymerizations can be performed. All processes were performed under computer control, requiring minimal intervention from the operator. For the preparation of nanoMIPs specific for vancomycin, the polymerization mixture consisted of NIPAM (39 mg), BIS (2 mg), TBAm (33 mg dissolved in 2 mL of ethanol) and AA (2.23 g). The components were dissolved in double distilled water (100 ml), sonicated for 10 minutes and degassed by bubbling with nitrogen for 30 minutes. For the automated synthesis of nanoMIPs, the vancomycin functionalized glass beads, used as the solid-phase (60 g), were added to the reaction cylinder. An aliquot of the polymerization mixture (60 mL) was added directly into the cylinder using a syringe pump and then stirred briefly to homogenize the contents. Polymerization was initiated by the addition of a solution of APS (60 mg mL−1, 600 μL) and TEMED (18 μL) followed by stirring. The monomer mixture was allowed to polymerize at ambient temperature for 1.5 hours and during this time brief stirring (30 s at 600 RPM) was applied at the end of each 30 min period. After 90 min the polymerization mixture was drained from the reaction vessel using a combination of suction and nitrogen purging. Subsequently, water at ambient temperature (2 aliquots of 50 mL) was added to the cylinder, the contents stirred and the washings, containing unreacted monomers and other low affinity materials, removed and discarded. Next, a further 3 aliquots of water (50 mL each) were individually added to the cylinder and the temperature was raised to 60 °C. At 60 °C the non-covalent interactions between the template attached to the solid-phase and the high-affinity nanoMIPs were disrupted, thus allowing the pure fraction of particles, free of residual template and monomers, to be collected. This entire process took approximately 3.5 hours. The stages of this synthesis were pre-programed into the WinISO® software such that, once started, the synthesis could be left to run without the necessity for user supervision. The solution of MIP nanoparticles was concentrated to a final volume of 100 mL by ultrafiltration on a Millipore Amicon Ultra centrifugal filter unit (30 kDa MWCO), and used for development of the biomimetic ELISA.
Analysis of the size of MIP nanoparticles
The size of the nanoparticles was determined by dynamic light scattering (DLS) using a Zetasizer Nano (Nano-S) from Malvern Instruments Ltd (Malvern, UK) and from images obtained on a Philips CM20 Transmission Electron Microscope (TEM). Prior to DLS measurements the solution of nanoMIPs was subjected to sonication for 5 minutes and measurements were performed at 25 °C. Prior to TEM analysis the solution of nanoMIPs was sonicated for 2 minutes after which a drop of the nanoMIP solution was placed on a carbon-coated copper grid and dried in air.
Preparation of the HRP-vancomycin (HRP-V) conjugate
HRP (10 mg) was dissolved in 0.1 M MES buffer, pH 6 (1 mL), to which EDC (0.4 mg), followed by NHS (0.6 mg) were added. The reaction was allowed to proceed at room temperature for 15 min. At this point the buffer was removed by ultrafiltration on a Millipore Amicon Ultra centrifugal filter unit (30 kDa MWCO). Activated HRP was collected from the ultrafiltration unit and immediately incubated with vancomycin (10 mL, 1 mg mL−1) in PBS buffer at pH 7.4 for 2 hours. The HRP-vancomycin conjugate (HRP-V) was then washed to remove free vancomycin on a Millipore Amicon Ultra centrifugal filter unit (30 kDa MWCO). For this 10 washes with PBS (5 mL) were performed. After washing, the conjugate was dissolved in deionized water (2 mL), its concentration estimated by comparison with the enzymatic activity of the free enzyme and stored frozen at −18 °C until use. This was used as the stock solution in the procedures described below.
Immobilization of nanoMIPs onto the surface of microplate wells
Vancomycin-imprinted nanoparticles (40 μL, 0.056 mg mL−1) were dispensed into the wells of a 96-well microplate and the solvent (water) allowed to evaporate overnight at room temperature.
Optimization of assay conditions
Several parameters such as the composition of the blocking and washing buffers, quantity of nanoMIPs, time of incubation and concentration of HRP-V were optimized in order to find the best conditions for the competitive assay.
For the optimization of the blocking conditions, microplate wells with immobilized nanoMIPs were conditioned by washing with PBS (250 μL per well). Solutions containing a surfactant, either Tween 20 (0-1%) or SDS (0-0.1%) as well as BSA (0-3%) in PBS (300 μL) were dispensed into the wells and the plate incubated for 1 hour. After washing with PBS (3 × 250 μL), a solution of either HRP-V (100 μL, diluted 1:400 from the stock solution) or of HRP at the same concentration was dispensed into the wells, followed by incubation for 2 hours. This was followed by washing with PBS (250 μL) containing 0.05% Tween 20. The HRP substrate, TMB reagent (100 μL) was added to each of the test wells, followed by incubation for 10 min, after which time the enzymatic reaction was stopped by the addition of H2SO4 (0.5 M, 100 μL). The plate was then read by determining the absorbance of each well at 450 nm using a UV/Visible microplate reader (Dynex, UK). Success criteria were the lowest color development from HRP alone, whilst maintaining a high value for the conjugate, indicative of a suppression of non-specific binding of HRP to the wells without disrupting the bond between the HRP-V and the immobilized nanoMIPs. A solution of PBS containing BSA (0.1 %) and Tween 20 (1 %) was selected for use in the following experiments.
Optimization of the washing conditions was performed in the same manner as described above, varying the composition of the washing solutions applied after incubation of the test wells with HRP-V (or HRP). Success criteria were the same as for the blocking step. A solution of PBS containing BSA (0.1 %) and Tween 20 (1 %) was selected for use in the following experiments.
For the optimization of the quantity of nanoMIPs, microplates were prepared, as described above, by dispensing a standard volume of a solution of vancomycin-imprinted nanoMIPs (40 μL) of different concentrations (from the stock concentration, 0.056 mg mL−1 to a 10-fold dilution) into the test wells. Evaporation of the solvent and performance of the blank assay (noncompetitive, in absence of free vancomycin) were as described above, using the previously optimized blocking and washing conditions. The optimum quantity of nanoparticles was determined to be that which gave the greatest difference in color development between experiments conducted with HRP and HRP-V. The highest concentration (0.056 mg mL−1) was selected and used in subsequent experiments.
For the optimization of the concentration of the conjugate (HRP-V), the blank assay was performed under the previously optimized conditions using different concentrations of HRP-V or HRP by diluting stock solutions from 1:200- to 1:1600-fold. The optimum concentration of the conjugate was determined to be that which gave the greatest difference in color development between experiments conducted with HRP and HRP-V. Dilution of the stock solution 1 in 800 was judged to be optimum and this concentration was used in subsequent experiments.
For the optimization of the incubation time of the conjugate, the blank assay was repeated under the previously optimized conditions using different times (30 min, 1 h or 2 h) for the incubation of HRP-V or HRP with the nanoMIP-coated microplate wells before washing and color development. The minimum incubation time required to maximize color development in the case of the conjugate compared to HRP alone was determined to be 1 h. This incubation time was used in all subsequent experiments.
Competitive assay
Following optimization of the assay conditions, the final protocol was then tested in a competitive assay conducted as follow: Microplate wells were coated with nanoMIPs by dispensing undiluted stock solution (40 μL) into each well followed by evaporation overnight. Each well was conditioned by washing with PBS (2 × 250 μL) followed by blocking by incubation with PBS (300 μL) containing BSA (0.1 %) and Tween 20 (1 %) for 1 h. Wells were then washed with PBS (3 × 250 μL). To each well a solution of HRP-V (100 μL, 1:800 dilution from stock) was added, containing free vancomycin at a final concentration of between 0.001-70 nM. Plates were incubated in the dark at room temperature for 1 h. Wells were then washed with PBS (300 μL) containing BSA (0.1%) and Tween 20 (1%), followed by addition of the TMB reagent (100 μL). After 10 minutes incubation, the enzymatic reaction was stopped by the addition of H2SO4 (0.5 M, 100 μL). Color development was determined by measuring the absorbance of each well at 450 nm using a UV/Visible microplate reader (Dynex, UK).
Analysis of vancomycin in plasma
For use in the following experiments, plasma was extracted from porcine blood, sourced from a local butcher, by centrifugation at 2,500 rpm (1,201g) for 30 minutes to remove the cellular components. The clear supernatant was then collected and stored in the freezer at – 20 °C until use. In order to demonstrate the analysis of vancomycin in biological media, the porcine plasma was spiked (3-50 μM) with vancomycin at levels spanning the clinically relevant concentration range. Plasma samples were then diluted 100,000-fold to fall within the calibration range of the competitive assay. Vancomycin concentrations in plasma were determined by analysis using the competitive assay described above. The concentration of vancomycin in plasma was calculated using the absorbance values read for the samples and the equation of the calibration curve obtained with the standards.
Cross-reactivity of the nanoMIP-based assay
To assess the cross-reactivity of nanoMIP-based assay, the competitive assay was performed with three other commonly used antibiotics (gentamicin, bleomycin and amoxicillin) over the concentration range 0.01-70 nM.
Stability of the nanoMIP-coated microplates
To assess the stability of microplate wells coated with nanoMIPs, several microplates were prepared as described above and subjected to the following storage trials: 1 month at room temperature or one week at 40 ° C in a humid environment. In each case this was followed by periodic testing by competitive assay for vancomycin, as described above.
RESULTS AND DISCUSSION
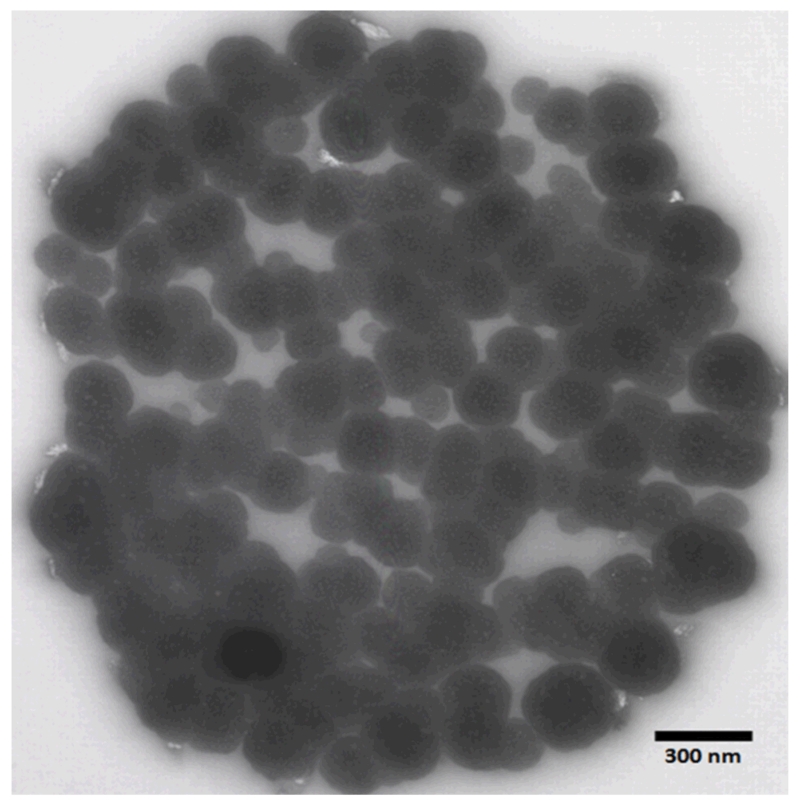
The synthesis of MIP nanoparticles (nanoMIPs) for vancomycin was performed using an automated solid-phase synthesis approach, as described earlier.16 Vancomycin was immobilized through its amino group by glutaraldehyde coupling to the surface of amine-derivatized glass beads which served as the template in nanoMIP synthesis. The polymer composition for the preparation of nanoMIPs was adapted from that published by Hoshino et al.22 The interaction between the imprinted polymer and the glycopeptide antibiotic is due to a combination of multiple weak electrostatic and hydrophobic interactions. The concentration of nanoparticles present in the stock solution (100 mL) was determined to be 0.056 mg mL−1, calculated by weighing a freeze-dried aliquot of the nanoparticle solution. The size of nanoparticles as measured by DLS was 170 ± 30 nm, and 200-300 nm by TEM (Figure 1).
Figure 1.
TEM image of nanoMIPs specific for vancomycin.
The apparent dissociation constant for the interaction between vancomycin-imprinted nanoMIPs and vancomycin immobilized on the surface of gold chips was determined by surface plasmon resonance using a Biacore 3000, as previously described,16 see Figure S1 in Supporting Information. The apparent dissociation constant was determined to be 0.48 nM.
In order to develop a quantitative assay analogous to ELISA, using MIP nanoparticles in place of antibodies, a simple immobilization procedure was required to be developed for the deposition of stable coatings of nanoMIPs on the surface of microplate wells. In ELISA, antibodies are frequently immobilized through physical adsorption to the walls of polystyrene microplates by hydrophobic binding. Previous MIP-based assays performed in microplates however, relied on in situ formation of the imprinted material through polymerization in the test wells.7-10 In our case we set out to show that MIP nanoparticles, previously prepared by solid-phase synthesis,16 could be used as convenient replacements for antibodies in an enzyme-linked competitive assay. It was found that stable coatings could be achieved by allowing a solution of nanoMIPs to evaporate to dryness within each of the microplate wells. The immobilized nanoMIPs were shown to remain attached to the microplate well surfaces (most likely due to physical adsorption), even after several washes with PBS. It can be estimated that each well, treated with 40 μL of the nanoMIP stock solution, would contain 2.2 μg of imprinted nanoparticles, 211 mg of nanoMIPs (which can be prepared in 1 week) would therefore be sufficient to coat up to one thousand 96-well microplates.
Having established that the nanoMIPs showed selective affinity for vancomycin and were capable of forming stable adsorbed coatings in microplates, the next component of the assay to be prepared was a conjugate between the analyte and an enzyme label. Horseradish peroxidase [EC 1.11.1.7] (HRP) was chosen, as it is commonly used in ELISA as a stable and efficient enzyme used for colorimetric detection. HRP was therefore coupled to vancomycin following activation with EDC-NHS to form the HRP-vancomycin conjugate (HRP-V). After conjugation the concentration of HRP-V was estimated to be 2.5 mg mL−1 by comparing its enzymatic activity with that of the free enzyme. A calibration curve of HRP-V and HRP for different dilutions of the substrate (TMB reagent) was also performed to ensure that the activity of the enzyme had not been compromised during conjugation. The results reported in Figure S2 show that there was no deterioration in the enzymatic activity of HRP-V, as the two calibration curves appeared to be nearly identical.
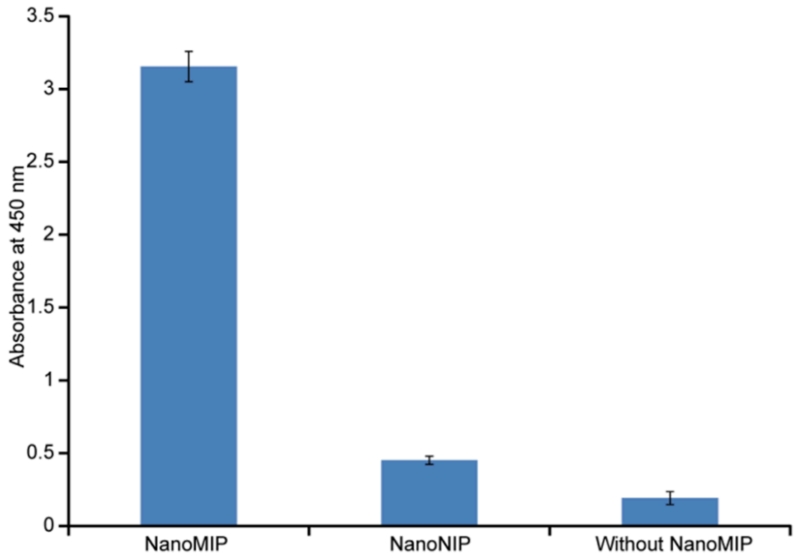
To establish that the immobilized nanoMIP layer retained affinity for vancomycin, coated wells were incubated with HRP-V, followed by washing and color development by reaction with TMB. Uncoated wells were similarly treated as a control. As an additional test of specificity, we also compared the binding of HRP-V to non-imprinted nanoparticles or nanoNIPs. In actuality these were nanoparticles imprinted with trypsin, synthesized under the same conditions, using the same composition as the vancomycin nanoMIPs, but using a trypsin-coated solid phase. The results, shown in Figure 2, show that much higher binding of HRP-V was seen in the case of the nanoMIPs than either with nanoNIPs or with bare wells.
Figure 2.
Testing the specificity of binding of the HRP-vancomycin conjugate to immobilized nanoMIPs. Binding of HRP-V to microplate wells coated with nanoMIPs, nanoNIPs or to bare wells, as revealed by color development with TMB reagent (100 μL ) for 10 min. Error bars represent ± 1 standard deviation for experiments performed in triplicate.
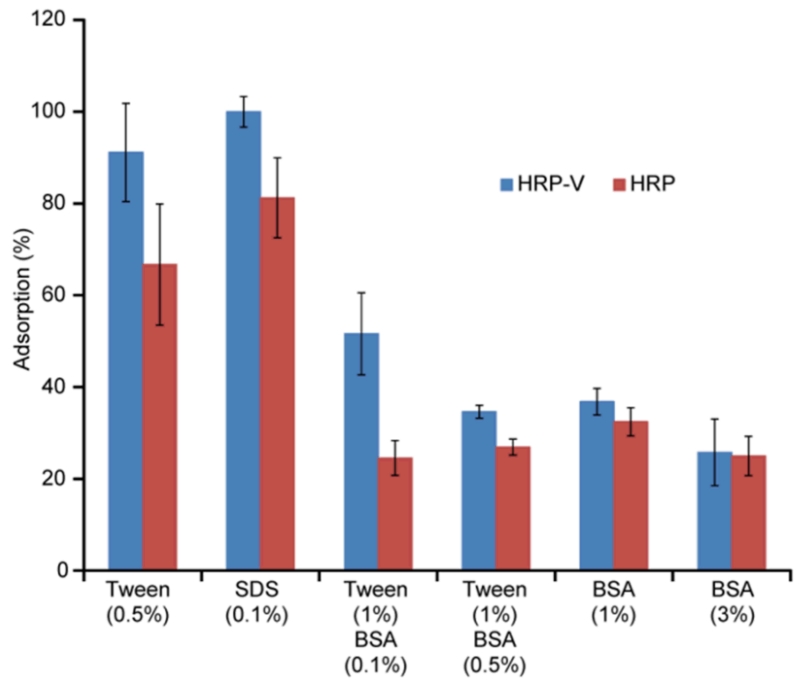
The blocking and washing protocols are very important for the development of a robust assay. Blocking refers to pre-treatment of the wells before addition of the HRP conjugate with the aim of reducing non-specific binding of the protein to the microplate well surfaces, without adversely affecting the specific interaction between the analyte portion of HRP-V (vancomycin in this case) and the MIPs. Washing is applied to the wells after the incubation period in order to remove non-bound conjugate, so that the color development accurately reflects the quantity of HRP-V remaining bound to the MIP layer following competition for free vancomycin. We tested PBS-based blocking buffers containing different amounts of albumin and either SDS or Tween 20 in our experiments, all of which are commonly used in antibody-based assays as components of the blocking solution. The effect of blocking buffer composition on binding of the HRP-V conjugate compared with adsorption of the free enzyme was analyzed in a series of blank assays (Figure 3). The quantity of adsorbed protein is presented as a percentage of the highest value obtained. The highest discrimination between HRP-V and HRP was shown in the case of PBS containing 1% Tween 20 and 0.1 % BSA.
Figure 3.
Optimization of blocking buffer. Aliquots (300 μL) of PBS containing different concentrations of a surfactant (Tween 20 or SDS) and BSA were incubated in microplate wells with immobilized nanoMIPs for 1 hour and the blank assay performed as explained in Material and Methods. Error bars represent ± 1 standard deviation for experiments performed in triplicate.
The optimization of washing buffer was performed in a similar way. The plate with immobilized nanoMIPs was incubated with HRP-V or HRP and washed 3 or 5 times with PBS containing different amounts of Tween 20 and BSA. The results, reported in Figure S3 in the supporting information, show that the optimum washing conditions were achieved by performing 3 washes with PBS containing 0.1% BSA and 1% Tween 20.
Having established conditions for minimizing non-specific interferences through optimization of blocking and washing buffers, the remaining parameters to be investigated were the optimum quantity of nanoMIPs in each microplate well and the concentration of the enzyme conjugate used during the competitive binding period. Several concentrations of nanoMIPs were obtained by dilution of the stock solution (0.056 mg/mL up to 10-fold dilution) to prepare coated microplate wells by evaporation of a 40 μL titre per well. The wells were subjected to the blank assay with either HRP-V or the free enzyme. The results, reported in the supporting information (Figure S4), show that wells coated with the undiluted stock solution produced the maximum difference in adsorption between the conjugate and enzyme. The immobilization of larger quantities of nanoparticles was also attempted by increasing the volume of stock solution added to each well, however this resulted in poor accuracy in the measurements, possibly due to partial “peeling” of thicker coatings from the microplate well surfaces. In the following experiments therefore, nanoparticles were used without further dilution, adding 40 μL per well.
A final optimization was then carried out to select the concentration of HRP-vancomycin conjugate (HRP-V) to be used in the assay. The stock solution of HRP-V and HRP were diluted from 1:200 to 1:1600 and incubated in the nanoMIP-coated wells for 2 hours. The results are shown in Table 1, comparing the absorbance values measured at 450 nm and their ratios for HRP-V and HRP after color development.
Table 1.
Optimization of the concentration of HRP-V in the blank assay: the absorbance values read at 450 nm for both HRP-V and HRP are reported, as well as the ratio between the two sets of data.
| Absorbance at 450 nm | |||
|---|---|---|---|
| Dilutions | HRP-V | HRP | Absorbance Ratio HRP-V/HRP |
| 1:200 | 1.804 ± 0.02 | 1.059 ± 0.18 | 1.70 |
| 1:400 | 1.670 ± 0.2 | 0.803 ± 0.10 | 2.08 |
| 1:800 | 0.878 ± 0.05 | 0.387 ±0.02 | 2.26 |
| 1:1600 | 0.572 ± 0.05 | 0.252 ± 0.01 | 2.27 |
Errors in the absorbance measurements represent ± 1 standard deviation and are for experiments performed in triplicate.
The table shows that the most dilute solutions (1:800 and 1:1600) produced the greatest differences between the binding of HRP-V and HRP to the nanoparticles. The 1:800 dilution was preferred however, as this allowed acceptable levels of color development to be achieved from between 10 to 30 minutes.
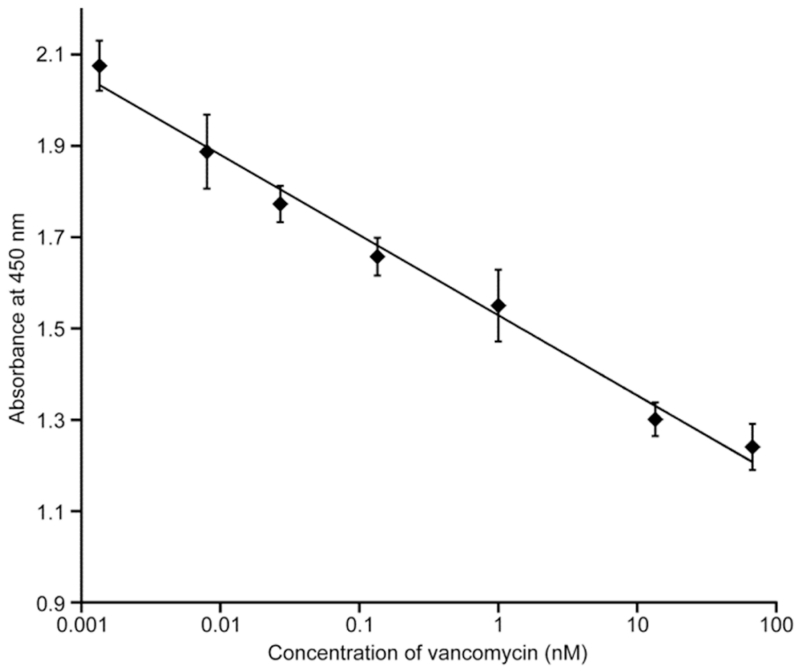
The optimized protocol was then applied to the performance of a competitive assay. Microplates, coated with vancomycin-imprinted nanoMIPs were tested in an enzyme-linked assay using competition between free vancomycin and the HRP-V conjugate. The concentration of free vancomycin was varied between 1 pM to 70 nM, added to the wells at the same time as the conjugate. The results, presented as the calibration curve shown in Figure 4, clearly indicate that competition for binding to the nanoMIP coating can be detected over 5 orders of magnitude, and is linearly proportional to the analyte (vancomycin) concentration when plotted on a logarithmic scale. As such it is suitable for use as a highly sensitive quantitative enzyme-linked assay for the target antibiotic.
Figure 4.
Calibration curve determined for the nanoMIP-based enzyme-linked competitive assay for vancomycin. Error bars represent ± 1 standard deviation and are for experiments performed in triplicate.
The assay showed linearity from 1 pM to 70 nM of vancomycin, with a limit of detection of 2.5 pM, calculated from the value of three times the standard deviation of the control (without free vancomycin). This is much lower than the detection limit of the immunoassay reported in the literature, which was only 0.1 μM.19 The sensitivity range demonstrated in this assay is in agreement with the value of the apparent dissociation constant for the interaction of the imprinted nanoparticles with vancomycin, as determined in experiments conducted on the Biacore (KD = 0.48 nM). The competitive assay started to show saturation at concentrations of vancomycin higher than 70 nM.
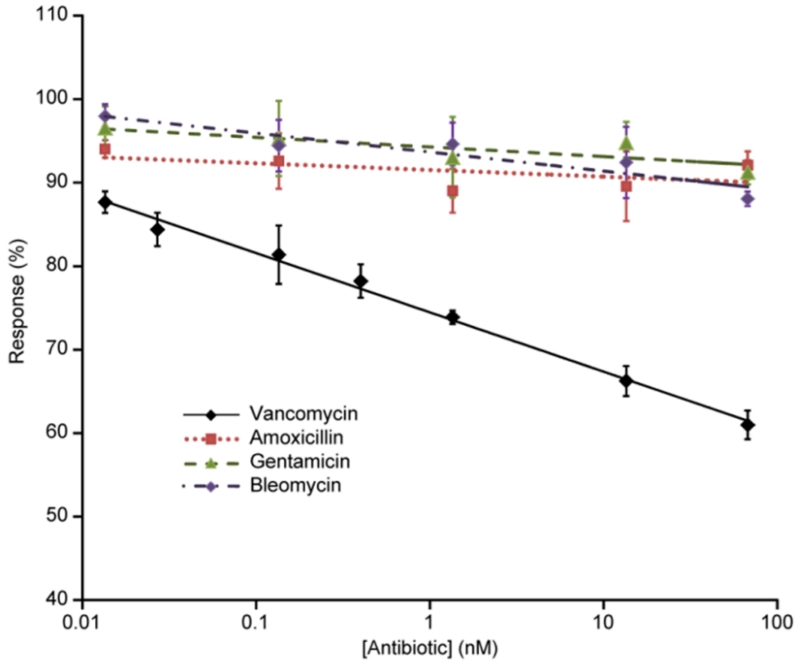
The cross-reactivity of the assay was measured by assessing the response to other commonly used antibiotics: amoxicillin, gentamicin and bleomycin and the results presented in Figure 5. In order to better compare the different assays, the responses are reported as a percentage, normalized to the response measured in the absence of free antibiotic. The results of this testing indicate that these analytes have much weaker binding to the MIP nanoparticles and compete poorly with the vancomycin conjugate.
Figure 5.
Calibration curves of the enzyme-linked nanoMIP-based competitive assay performed with vancomycin, amoxicillin, gentamicin and bleomycin. Error bars represent ± 1 standard deviation and are for experiments performed in triplicate.
These tests, together with the large difference in binding of the HRP-V conjugate to nanoMIPs and nanoNIPs (see Figure 2) clearly indicate the highly specific affinity of the imprinted nanoparticles for their template, which in this respect resembles the behavior of monoclonal antibodies.
Further experiments were performed in order to evaluate the stability of the nanoparticles and the coatings prepared from them. Binding of HRP-V to the MIP was evaluated in repeat measurements carried out during a period of one month. It was found that throughout this period the polymer retained its original activity and could still be used in an assay. As a test of the stability of coatings, nanoMIP-coated microplates were stored at 40 °C for 1 week to mimic unrefrigerated transportation and delivery to tropical countries (no cold chain). The results showed that even when subjected to these conditions, the plates could still be used in the vancomycin assay over the same concentration range (0.001-70 nM), with no deterioration in the detection limit.
Finally, to demonstrate the clinical relevance of the developed assay, porcine plasma was spiked with vancomycin at a number of concentrations spanning the clinically relevant range: 6-35 μM (10-50 mg/L) and subjected to the nanoMIP-based assay. The results demonstrated good correlation (mean recovery 98 ± 2.6 %) between the spike and concentrations of vancomycin measured in the assay (see Table 2). Clearly blocking of the nanoMIP-coated surfaces with protein and surfactant was sufficient to negate any impact that plasma components might have had on the analysis. The high sensitivity of the assay demanded extensive dilution of the serum samples which also helped to minimize potential interferences.
Table 2.
Testing of porcine plasma samples spiked with vancomycin.
| Spiked (μM) | Found (μM) | Recovery (%) |
|---|---|---|
| 3.0 | 2.9 ± 0.1 | 97 |
| 9.0 | 9.3 ± 0.3 | 103 |
| 15 | 14 ± 0.4 | 93 |
| 30 | 28 ± 0.4 | 93 |
| 50 | 51 ± 0.5 | 102 |
Errors in the absorbance measurements represent ± 1 standard deviation and are for experiments performed in triplicate.
CONCLUSIONS
This study demonstrates that molecularly imprinted nanoparticles (nanoMIPs), prepared by solid-phase synthesis,16 can be used in the development of a new, highly specific and sensitive, clinically relevant enzyme-linked assay for a currently prescribed antibiotic (vancomycin). The assay formulation was based on ELISA, with the exception that nanoMIPs were used as a direct substitute for antibodies. Coating microplate wells with the soluble MIP nanoparticles was achieved by simply allowing the solutions to evaporate to dryness in the wells, equivalent to physical adsorption of antibodies. The developed assay allowed the accurate determination of vancomycin over the concentration range 0.001-70 nM, with a limit of detection of 0.0025 nM (2.5 pM). The assay could be used to determine vancomycin in plasma at clinically relevant concentrations with a mean accuracy of 98% and very low cross-reactivity with three other antibiotics. In this respect the performance of nanoMIPs was comparable to high quality monoclonal antibodies. Furthermore, we have demonstrated that the nanoMIP-coated microplates can withstand exposure to high temperature for prolonged periods without affecting the sensitivity of the assay, suggesting that assays formulated in this manner do not require refrigeration during transportation and storage, which could have both social and economic benefits. While the assay presented here is for a specific analyte, vancomycin, the generic nature of nanoMIP preparation in solid-phase synthesis16,21,23,24 suggests that assays for many more analytes, including drugs, toxins, pesticides, pollutants, peptides, proteins, hormones, viruses and disease biomarkers, among others, can be created with relatively short development times.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank The Wellcome Trust (UK) for the granting of a Translational Award and the EU for the financial support through the project CHEBANA (FP7-PEOPLE-2010-ITN-204772).
Footnotes
Supporting Information. Figures showing the Biacore testing to estimate dissociation constant of nanoMIPs, the calibration curves of HRP-V and HRP for dilutions of TMB reagent and results of experiments carried out to optimize washing buffer and quantity of nanoparticles per well are supplied as Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Tremblay L, Van der Kraak G. Aquatic Toxicol. 1998;43:149–162. [Google Scholar]
- (2).Nakanishi K, Huang XF, Jiang H, Liu Y, Fang K, Huang DW, Choi SK, Katz E, Eldefrawi M. Bioorg. Med. Chem. 1997;5:1969–1988. doi: 10.1016/s0968-0896(97)00137-5. [DOI] [PubMed] [Google Scholar]
- (3).Viera A. Mol. Biotechnol. 1998;10:247–250. doi: 10.1007/BF02740845. [DOI] [PubMed] [Google Scholar]
- (4).Andersson LI, Müller R, Vlatakis G, Mosbach K. Proc. Natl. Acad. Sci. USA. 1995;92:4788–4792. doi: 10.1073/pnas.92.11.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Vlatakis G, Andersson LI, Müller R, Mosbach K. Nature. 1993;361:645–647. doi: 10.1038/361645a0. [DOI] [PubMed] [Google Scholar]
- (6).Piletsky SA, Piletskaya EV, Panasyuk TL, El’skaya AV, Levi R, Karube I, Wulff G. Macromolecules. 1998;31:2137–2140. [Google Scholar]
- (7).Piletsky SA, Piletska EV, Chen B, Karim K, Weston D, Barrett G, Lowe P, Turner APF. Anal. Chem. 2000;72:4381–4385. doi: 10.1021/ac0002184. [DOI] [PubMed] [Google Scholar]
- (8).Wang S, Xu ZX, Fang GZ, Zhang Y, Liu B, Zhu HP. J. Agric. Food Chem. 2009;57:4528–4534. doi: 10.1021/jf900505k. [DOI] [PubMed] [Google Scholar]
- (9).Zhao DY, Qiao XG, Xu ZX, Xu R, Yan ZH. J. Immunoassay Immunochem. 2013;34:16–29. doi: 10.1080/15321819.2012.668149. [DOI] [PubMed] [Google Scholar]
- (10).Tang YW, Fang GZ, Wang S, Sun JW, Qian K. J. AOAC Int. 2013;96:453–458. doi: 10.5740/jaoacint.10-387. [DOI] [PubMed] [Google Scholar]
- (11).Piletsky SA, Piletska EV, Bossi A, Karim K, Lowe P, Turner APF. Biosens. Bioelectron. 2001;16:701–707. doi: 10.1016/s0956-5663(01)00234-2. [DOI] [PubMed] [Google Scholar]
- (12).Surugiu I, Danielsson B, Ye L, Mosbach K, Haupt K. Anal. Chem. 2001;73:487–491. doi: 10.1021/ac0011540. [DOI] [PubMed] [Google Scholar]
- (13).Xu ZX, Gao HJ, Zhang LM, Chen XQ, Qiao XG. J. Food Sci. 2011;76:R69–R75. doi: 10.1111/j.1750-3841.2010.02020.x. [DOI] [PubMed] [Google Scholar]
- (14).Yonamine Y, Hoshino Y, Shea KJ. Biomacromol. 2012;13:2952–2957. doi: 10.1021/bm300986j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Lee MH, Thomas JL, Chen YC, Chin WT, Lin HY. Microchim. Acta. 2013 DOI: 10.1007/s00604-013-0995-6. [Google Scholar]
- (16).Poma A, Guerreiro A, Whitcombe MJ, Piletska E, Turner APF, Piletsky S. Adv. Funct. Mat. 2013;23:2821–2827. doi: 10.1002/adfm.201202397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Rodvold KA, Erdman SM, Pryka RD. In: Therapeutic drug monitoring. Schumacher GE, editor. Appleton and Lange; Norwalk, CT: 1995. pp. 587–632. [Google Scholar]
- (18).Simons SA, Molinelli AR, Sobhani K, Rainey PM, Hoofnaglea AN. Clin. Chem. 2009;55:578–580. doi: 10.1373/clinchem.2008.112946. [DOI] [PubMed] [Google Scholar]
- (19).Fujiwara K, Yoshizaki Y, Shin M, Miyazaki T, Saita T, Nagata S. Antimicrob Agents Chemother. 2012;56:5883–5891. doi: 10.1128/AAC.01267-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Piletsky SA, Guerreiro ARL, Whitcombe MJ. WO201106753 2011
- (21).Ambrosini S, Beyazit S, Haupt K, Tse Sum Bui B. Chem. Commun. 2013;49:6746–6748. doi: 10.1039/c3cc41701h. [DOI] [PubMed] [Google Scholar]
- (22).Hoshino Y, Kodama T, Okahata Y, Shea KJ. J. Am. Chem. Soc. 2008;130:15242. doi: 10.1021/ja8062875. [DOI] [PubMed] [Google Scholar]
- (23).Piletsky S, Piletska O, Guerriero A, Whitcombe M, Poma A. WO2013041861 2013
- (24).Moczko E, Poma A, Guerreiro A, Perez de Vargas Sansalvador I, Caygill S, Canfarotta F, Whitcombe MJ, Piletsky S. Nanoscale. 2013;5:3733–3741. doi: 10.1039/c3nr00354j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.